Abstract
The mammalian guanine deaminase (GDA), called cypin, is important for proper neural development, by regulating dendritic arborization through modulation of microtubule (MT) dynamics. Additionally, cypin can promote MT assembly in vitro. However, it has never been tested whether cypin (or other GDA orthologs) binds to MTs or modulates MT dynamics. Here, we address these questions and characterize Xenopus laevis GDA (Gda) for the first time during embryonic development. We find that exogenously-expressed human cypin and Gda both display a cytosolic distribution in primary embryonic cells. Furthermore, while expression of human cypin can promote MT polymerization, Xenopus Gda has no effect. Additionally, we find that the tubulin-binding CRMP homology domain is only partially conserved between cypin and Gda. This likely explains the divergence in function, as we discovered that the cypin region containing the CRMP homology and PDZ-binding domain is necessary for regulating MT dynamics. Finally, we observed that gda is strongly expressed in the kidneys during late embryonic development, although it does not appear to be critical for kidney development. Together, these results suggest that GDA has diverged in function between mammals and amphibians, and that mammalian GDA plays an indirect role in regulating MT dynamics.
Keywords: cypin, GDA, microtubule dynamics, development
Introduction
The proper establishment of neuronal morphology is essential for the correct communication between neurons, and the cytoskeleton plays a key role in determining neuronal morphology during development. In particular, microtubules (MTs) orchestrate formation of neuronal polarity and ensure proper axon and dendrite morphology. MT dynamics are widely regulated by proteins that bind to the plus-end of the MT, called plus-end tracking proteins (+TIPs), in addition to other types of MT-binding proteins (Bearce et al., 2015; Cammarata et al., 2016; Lasser et al., 2018). While many studies have begun to investigate MT plus-end dynamics in the developing nervous system, it is clear that much more information is needed to reveal a comprehensive picture for how MT dynamics regulate neural development.
One important MT regulator in the developing nervous system is the mammalian guanine deaminase (GDA), called cypin (cytosolic PSD-95 interactor) (Akum et al., 2004). Cypin has been linked to autism spectrum disorders and has been described as a risk biomarker for these disorders (Braunschweig et al., 2013). In addition, cypin levels increase in rat striatal neurons in response to nigro-striatal degeneration, which models the degeneration of neurons in Parkinson’s disease (Fuller et al., 2014). Multiple neurodevelopmental disorders are associated with defects in axon guidance and dendritogenesis. Furthermore, it has been shown that cypin plays a key role in establishing neuronal morphology by antagonizing PSD-95 function and increasing dendrite branching (Firestein et al., 1999; Riefler et al., 2003; Akum et al., 2004). Cypin contains a domain with homology to the collapsin response mediator protein (CRMP), which binds tubulin and has a suggested role in regulating MT assembly in the growth cone (Schmidt and Strittmatter, 2007). In addition, cypin can bind tubulin heterodimers and regulate MT assembly in vitro (Akum et al., 2004). Moreover, the binding partner of cypin, PSD-95, binds to the known +TIP, EB3, in neurons and hence regulates MT dynamics (Sweet et al., 2011). However, while cypin has previously been proposed to regulate MT dynamics, it has not yet been determined whether cypin can actually localize to MT plus-ends or alter MT dynamics in living cells.
Multiple +TIPs have been described and characterized using Xenopus laevis (Lee et al., 2004; Lowery et al., 2013; Marx et al., 2013; Nwagbara et al., 2014; Lucaj et al., 2015; Rutherford et al., 2016; Erdogan et al., 2017). Xenopus embryos can be easily manipulated, and their primary embryonic neural cells are facile to obtain, culture, and image, as they display large growth cones (10 microns or more), which are useful for imaging cytoskeletal dynamics (Erdogan et al., 2016; Slater et al., 2017). Live cell imaging is particularly important, as some +TIPs only bind to growing MTs and, thus, their localization dynamics cannot be visualized using immunohistochemistry of fixed cells (Nwagbara et al., 2014).
In this work, we investigated whether human GDA (cypin) and/or Xenopus laevis GDA (Gda) can localize to MT plus ends, perhaps functioning as a +TIP, and tested whether it has a role in regulating MT dynamics. Additionally, we examined gda expression in Xenopus laevis during embryonic development, as its expression and function had not been well characterized. We visualized exogenously-expressed cypin and Gda for the first time in living primary embryonic neural growth cones using fluorescent confocal microscopy and found that neither of them co-localize with MT plus-ends, suggesting that GDA does not function as a +TIP. Despite the lack of MT binding, we found that expression of cypin, but not Gda, promotes MT assembly in neuronal growth cones. Moreover, we found that the CRMP homology domain, which has been described as important for binding tubulin and regulating MT assembly, is not fully conserved in Xenopus. By generating a chimera containing the N-terminal region from Gda plus the cypin domain containing the CRMP homology and PDZ-binding domain, we found that this cypin region is necessary for regulating MT dynamics. Finally, using whole-mount in situ hybridization, we observed that gda is highly expressed in the kidneys and may be important for proper kidney function. Our results suggest that the function of GDA to regulate MT dynamics is not conserved in Xenopus.
Results
The CRMP homology domain is not conserved between human and Xenopus GDA
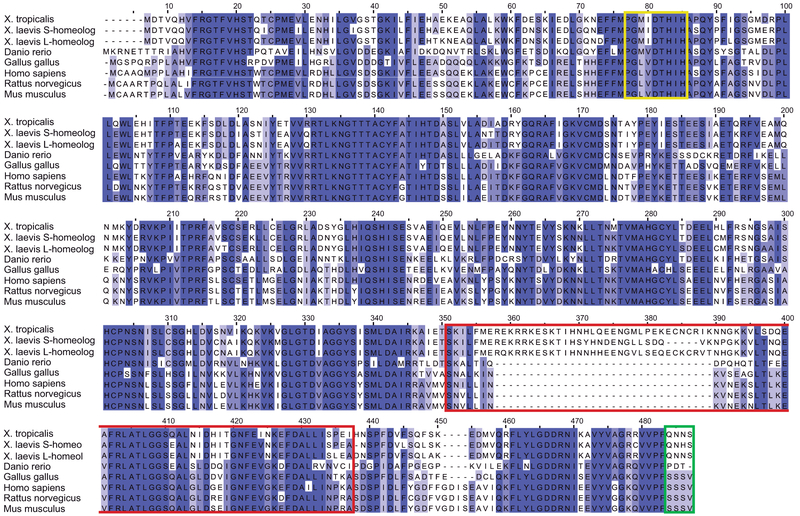
While a previous phylogeny study reported that the guanine deaminase (GDA) DNA sequence has been conserved from prokaryotes to higher eukaryotes (Fernandez et al., 2009), this early study did not examine the entire amino acid sequence nor did it look specifically at the CRMP domain sequence. X. laevis, X. tropicalis, Danio rerio, Gallus gallus, Homo sapiens, Mus musculus and Rattus norvegicus form part of the same related clusters of GDA sequences (Fernandez et al., 2009). Here, we compared the GDA protein sequences of all the aforementioned species. X. laevis is an allotetraploid organism and has two GDA homeologs, S and L; thus, we considered both homeologs for protein sequence comparison. We found the following sequence homologies of each organism with human cypin: X. laevis GDA S-homeolog 60%; X. laevis GDA L-homeolog 61%; X. tropicalis 61%; zebrafish 59%; chicken 72%; rat 91%; and mouse 91% (Fig. 1).
Figure 1. The CRMP homology domain is not conserved between human and Xenopus GDA.
Sequence alignment of X. laevis homeologs; X.tropicalis; Danio rerio; Gallus gallus; Homo sapiens; Rattus norvegicus; and Mus musculus GDA. The GDA N-terminal, including the Zn2+ binding domain (yellow square), is well conserved among all species, while the CRMP homology (red square) and the PDZ-binding domains (green square) are less conserved.
In addition, while all GDAs were previously described to have well-conserved zinc-binding domains, the CRMP homology and PDZ-binding domains are not completely conserved. Gallus gallus, Homo sapiens, Mus musculus and Rattus norvegicus all have a PDZ-binding domain, which is absent from X. laevis, X. tropicalis and Danio rerio (Fig. 1). Moreover, even though the CRMP homology domain was present in all the organisms, it is not well conserved. While chicken, rat, and mouse have strong conservation with human (82%, 91%, 91%, respectively), the CRMP homology domains in X. laevis GDA S and L, X. tropicalis, and zebrafish are not as similar (60%, 53%, 53%, 58% conservation with human GDA, respectively) (Fig. 1). Notably, neither the PDZ-binding domain nor the CRMP domains are conserved between amniotes and amphibians/fish.
GDA has a cytosolic distribution and does not accumulate at MT plus-ends
In order to explore the possibility that GDA may function as a +TIP and thereby bind to the plus-ends of MTs to regulate MT dynamics (Bearce et al., 2015), we first examined whether human (cypin) and/or X.laevis (Gda) GDA localize to MT plus-ends or not. We co-injected mRNAs coding for GFP-tagged N- or C-terminus cypin and Gda along with mKate2-MACF43, a known marker of growing MTs that localizes to the MT plus-ends (Honnappa et al., 2009), into X. laevis embryos at the 2-cell stage. We prepared spinal cord explants from stages 20–24 Xenopus embryos, and we used live imaging to visualize the exogenously-expressed GDAs. We found that neither cypin nor Gda localized to the MT plus-end in neuronal growth cones (Fig. 2A), nor in neural-derived mesenchymal cells (Fig. 2B), as GDAs did not co-localize with mKate2-MACF43. Rather, in all cases, GDA was diffused throughout the cytosol and did not appear to localize to MTs at all, consistent with what has been previously described for cypin localization (Firestein et al., 1999; Akum et al., 2004; Chen and Firestein, 2007; Sweet et al., 2011). These data suggest that neither human nor Xenopus GDA accumulate on MTs plus-ends, and thus, GDA does not act as a +TIP in Xenopus embryonic cells.
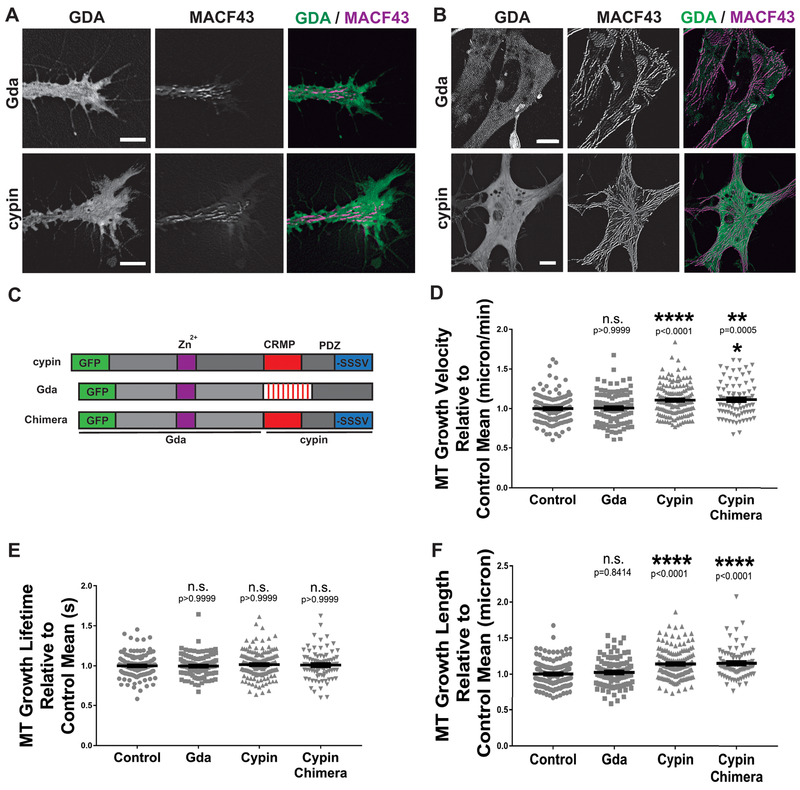
Figure 2. GDAs do not localize to the MT plus-end and only cypin promotes MT polymerization.
A-B. Maximum intensity montage of 31 frames from a one-minute time-lapse image series, of Gda and cypin (green), and mKate2-MACF43 tracks (magenta), in cultured neuronal growth cones (A) and neural-derived embryonic mesenchymal cells (B), obtained from Xenopus embryos at stage 20–24. Scale bars 8 μm (A) and 5 μm (B). C. Schematic representation of the GFP-tagged constructs used, showing the critical domains. D-F. Quantification of mean values of MT growth velocity (D), growth lifetime (E) and growth length (F) in neuronal growth cones upon GFP, Gda, cypin and cypin chimera over-expression. (See Supplementary Movie 1 for a representative movie of mKate2-MACF43.) Bars on dot plots show mean and SEM. ** P < 0.01; *** P < 0.001; **** P < 0.0001; n.s., not significant. A total of 138 control; 107 Gda; 154 cypin and 83 cypin chimera growth cones, from three independent experiments, were analyzed.
Exogenous expression of cypin, but not Gda, promotes MT polymerization in living cells
Considering that some proteins regulate MT dynamics through indirect mechanisms, for example, by binding tubulin heterodimers or other proteins that bind to MTs (Andersen, 2000; Akhmanova and Steinmetz, 2008), we sought to determine whether over-expression of cypin or Gda (Fig. 2C) could affect MT dynamics indirectly in living cells. We measured MT behavior in cultured primary embryonic neuronal growth cones obtained from Xenopus stages 20–24, by quantifying the MT growth track velocity, lifetime and length of mKate2-MACF43 comets after GDA over-expression (Supplementary Movie 1). We observed that axonal growth cones over-expressing cypin showed an 11% increase in MT growth track velocity and a 17% increase in MT growth track length compared to those of control cells, while no change was observed in mKate2-MACF43 comet lifetime (Fig. 2D–F). Similar results were observed for both N- and C-terminal tagged GFP-cypin (data not shown). However, we observed no changes in MT dynamics in cells over-expressing Gda compared to control cells (Fig. 2D–F). These data show that cypin over-expression increases MT polymerization rate in living Xenopus embryonic cells, and that human and Xenopus GDAs are different in their ability to modulate MT dynamics.
Additionally, considering the differential regulation of MT dynamics by cypin and Gda, and the striking differences between the cypin and Gda CRMP homology and PDZ-binding domain, we decided to evaluate if this cypin region is responsible for the effect on MT dynamics. We generated a chimera GDA containing the N-terminal half from Gda and the CRMP homology plus PDZ-binding domain from cypin (Fig. 2C) and examined the effect of the chimera GDA on MT dynamics. We found that the chimera GDA was able to increase MT growth track velocity and MT growth track length compared to those of control, similar to cypin (Fig. 2D–F). Our results suggest that the cypin region containing both CRMP homology and PDZ-binding domains plays an important role in promoting MT polymerization.
Xenopus GDA is strongly expressed in the embryonic kidney and thus may play a role in kidney function
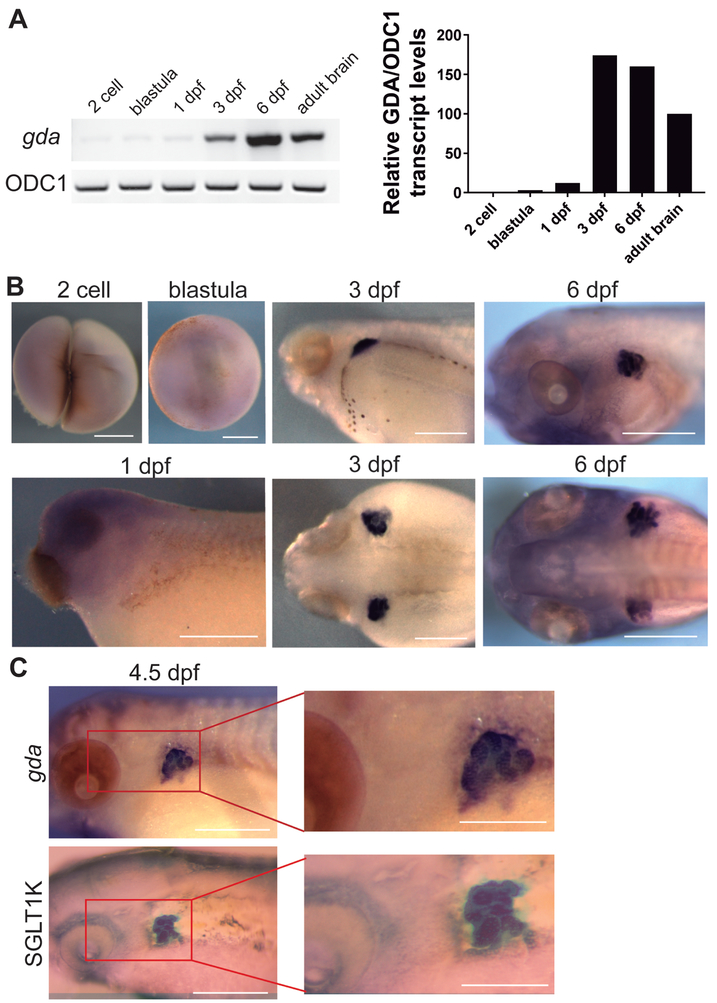
Given that our results suggested that Xenopus GDA did not function as a promoter of MT polymerization, we wondered if it was still expressed in the developing brain, like cypin. In order to characterize gda expression during Xenopus embryonic development, we used reverse transcription (RT) PCR and whole-mount in situ hybridization at different stages of development. First, we performed RT-PCR using cDNA from embryos at various stages including 2 cell, blastula, neurula, 3 dpf and 6 dpf, as well as cDNA from the adult brain. We found that strong gda expression was not apparent until 3–6 dpf and persisted, although to a lesser extent, through adulthood within the brain (Fig. 3A). Whole-mount in situ hybridization showed no staining in early stages including 2-cell, blastula, and 1 dpf, confirming the RT-PCR results (Fig. 3B). However, specific accumulation of gda expression in two bilateral regions behind the embryonic eye became apparent at 3 dpf and remained at a high level to 6 dpf (Fig. 3B). Following the stages of Xenopus laevis development (Nieuwkoop and Faber, 1994), and considering the localization and the tubular structures (Fig. 3B), we identified these areas to be the developing kidneys. Thus, we used the proximal tubule kidney marker, xSGLT1K (Zhou and Vize, 2004) to perform in situ hybridization in 4.5 dpf embryos. Our results showed that both gda and xSGLT1K are expressed in the same location (Fig. 3C), confirming the expression of gda in the kidneys.
Figure 3. Xenopus GDA is expressed in the developing kidney.
A. RT-PCR showing gda expression in the different developmental stages: 2 cell, blastula, neurula, 3 dpf, 6 dpf, and adult. ODC1 was used as an internal RT-PCR reference. B. Whole-mount in situ hybridization using an antisense probe specific to gda in different developmental stages. Scale bar: 0.5 mm C. Whole-mount in situ hybridization using an antisense probe specific to GDA or the kidney marker xSGLT-1K in 4.5 dpf embryos. Both gda and xSGLT-1K can be seen in a similar embryonic location. Scale bar: 0.5 mm and 0.25 mm for magnifications.
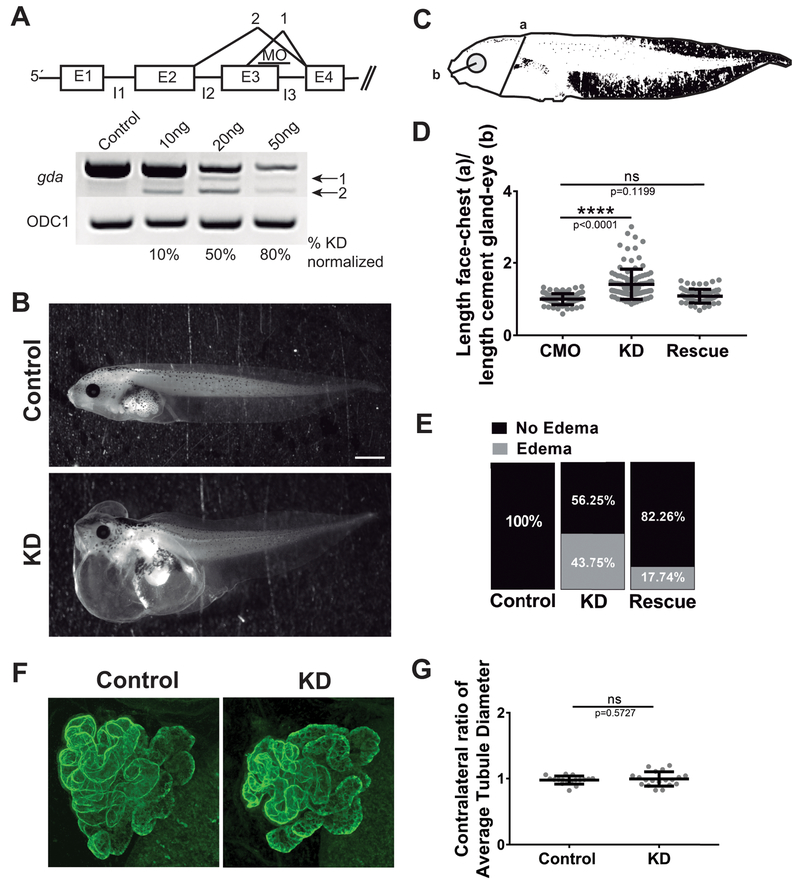
We were also interested in determining a possible function of Gda in Xenopus. Since our over-expression analysis suggested that Gda does not play a role in modulating MT dynamics, and our expression analysis showed that gda is not present in most embryonic cells during early neural development, we reasoned that Gda cannot be playing a role to promote MT polymerization in the early nervous system. Given its strong expression in the kidneys beginning at 3 dpf, we wondered if Gda might play a role in kidney development or function. However, pronephros development in Xenopus begins at stage 12.5, and kidneys become initially functional by stage 38, which corresponds to 2 dpf (Vize et al., 1995; Wessely and Tran, 2011). Thus, it did not seem likely that Gda was required for initial kidney development. Still, we wondered whether Gda might be important for kidney function. It has been demonstrated that an altered osmoregulatory function of the kidney leads to edema formation due to water retention (Wessely and Tran, 2011; Krneta-Stankic et al., 2017) and evaluation of edema has been widely used to assess kidney function (Getwan and Lienkamp, 2017; Krneta-Stankic et al., 2017). Thus, we used an antisense oligonucleotide to knock down (KD) gda by 50% (Fig. 4A), and at 4 dpf, we analyzed edema formation in the chest as a proxy to study kidney function (Fig 4B–C). We observed that gda KD induced edema in 44% of tadpoles, while no edema was observed in the controls (Fig. 4B–E). This phenotype was rescued by adding back gda mRNA to the KD condition (Fig. 4E, D), attributing these effects specifically to Gda. However, as cardiac and other developmental anomalies can also lead to edema, we also performed targeted injections at the 4-cell stage to knock down gda function in a more restricted cell lineage. With these targeted injections, we did not observe significant edema (not shown), suggesting that the edema formation in the gda knockdown embryos was not necessarily due to defects in kidney function alone. We also examined gross morphology of kidney development by immunostaining for anti-kidney antibodies, 4A6 and 3G8, at 4 dpf and observed no apparent differences with gda knockdown compared to controls (Fig. 4F–G). Thus, these data suggest that Gda does not appear to be involved in kidney development, but it may still play a role in later kidney function in Xenopus.
Figure 4. Xenopus GDA knockdown results in edema formation but kidney morphology appears to be normal.
A. Schematic representation of the MO effect on gda RNA splicing (above) RT-PCR showing gda knockdown (KD) when injecting increasing concentrations of MO (below). B. Representative images at 4 dpf control and Gda KD tadpoles. Scale bar: 1 mm. C. Schematic representation of a tadpole showing the measurements for edema identification. D. Quantification of the ratio: length face-to-chest (a) over length of cement gland-to-eye (b). E. Quantification of the percentage of tadpoles showing edema. F. Representative images of immunofluorescence using the kidney marker 3G8 in addition to 4A6 at 4 dpf control and gda KD tadpoles. F. Quantification of the ratio: average tubule diameter of the gda MO-injected side over that of the uninjected side of the same tadpole. Bars on dot plots show mean and SEM. ns: not significant. **** P < 0.0001.
Discussion
In this study, we first compared the GDA amino acid sequence from different species, which were related phylogenetically. We determined that the CRMP homology domain is not present in amphibians or fish. Additionally, we visualized fluorescently-tagged cypin and Gda using live imaging for the first time and determined that GDA protein is cytosolic but is not enriched at the MT plus-end or the MT lattice in Xenopus embryonic cells. We also showed that over-expression of cypin influenced MT dynamics in embryonic cells, and that this effect of GDA is not conserved among all vertebrates, since the overexpression of Gda has no effect in Xenopus cells. In addition, we showed that the CRMP homology domain and the PDZ-binding domain of cypin are responsible for the effect on MT dynamics, as we could recapitulate the increased MT polymerization by expressing a cypin chimera containing the N-terminal region of Gda combined with the cypin CRMP homology and PDZ-binding domains. Finally, we characterized the spatiotemporal expression of gda throughout Xenopus embryonic development using RT-PCR and whole-mount in situ hybridization. We show that the highest expression of gda occurs at 3 dpf and that it is strongly expressed in the kidneys, where it may play a role in kidney function. Thus, our work provides, for the first time, a characterization of gda expression during embryonic development along with a comparison between human and Xenopus GDA, revealing divergence in sequence and function.
The CRMP homology domain is not conserved between human and Xenopus GDA
A GDA evolutionary study had determined that the Rattus norvegicus, Xenopus laevis and Homo sapiens GDA proteins are part of the same sequence cluster, meaning that their sequences are evolutionarily similar (Fernandez et al., 2009). While the GDA enzymatic activity of guanine deamination into xanthine and ammonia has been conserved across many species (Fernandez et al., 2009), few studies have examined GDA function during embryonic development. One of the most prominent studies of GDA function has examined the role of the mammalian ortholog, cypin, during neuronal development (Akum et al., 2004; Chen et al., 2005; Charych et al., 2006; Chen and Firestein, 2007). Importantly, GDAs contain a PDZ-binding domain that is important for interaction and clustering with other proteins (Sheng and Sala, 2001). The cypin PDZ-binding domain is necessary for binding to PSD-95 protein (Firestein et al., 1999), which has been described to increase dendritic branching (Firestein et al., 1999) and indirectly affect MT dynamics (Sweet et al., 2011). Interestingly, while the enzymatic active site and divalent cation binding domain are conserved among these species, the PDZ-binding domain is not present in either of the Xenopus homeologs (Fernandez et al., 2009).
Additionally, GDAs also have a CRMP homology domain that is known to bind tubulin heterodimers (Firestein et al., 1999; Fukata et al., 2002). The CRMP homology domain is necessary for inducing MT polymerization in vitro (Akum et al., 2004). Our work is the first to evaluate the sequence homology of the CRMP homology domain between cypin and Gda, showing that the CRMP homology domain is not fully conserved. These differences in the PDZ-binding and CRMP homology domains between cypin and Gda likely explain the divergence in function between the proteins as it relates to their ability to regulate MT dynamics.
GDA does not localize to the MT plus end
Our results show that exogenous expression of GFP-tagged cypin and Gda present a cytosolic distribution in living cells and are not enriched at MT plus-ends, where the majority of the proteins that regulate MT plus-end dynamics are localized. Nevertheless, our observations are consistent with previous immunocytochemistry studies showing that cypin has a cytosolic localization in rat primary hippocampal neurons (Firestein et al., 1999; Akum et al., 2004; Chen and Firestein, 2007; Sweet et al., 2011). Additionally, while GDA is obtained primarily in the soluble fractions after subcellular fractionation, it is also present in other intracellular compartments, where it exists in isoenzymic forms (Kumar et al., 1965; Kumar et al., 1967). Considering this evidence, even though GDA localization is not enriched on MTs, we cannot discard a possible GDA activity near MT lattice or plus ends, which could still allow cypin to have an indirect effect on MT dynamics.
Overexpression of cypin, but not Gda, affects MT dynamics in vivo
Using live imaging, we showed that cypin over-expression increases the MT growth velocity and MT track length in neuronal growth cones. These results are in accordance with previous studies showing that rat cypin promotes MT assembly and polymerization using in vitro assays (Akum et al., 2004). Additionally, it is well known that cypin increases dendritic arbor length (Akum et al., 2004) and that it interacts with PSD-95 (Firestein et al., 1999). Cypin interaction with PSD-95 leads to decreased dendrite branching, potentially caused by the disruption of the MT cytoskeleton (Charych et al., 2006). Interestingly, PSD-95 was found to bind and reduce the MT binding of the known +TIP EB3, as well as reduce EB3 comet lifetime and velocity (Sweet et al., 2011). Thus, given that cypin has been shown to oppose PSD-95 function, and that PSD-95 reduces MT polymerization, it is possible that cypin expression in our study perturbs the ability of PSD-95 to inhibit MT polymerization.
However, we observed that expression of mammalian cypin, but not Xenopus Gda, promotes MT polymerization. This result is consistent with the fact that the CRMP homology domain, which has been shown to bind tubulin and regulate MT assembly (Schmidt and Strittmatter, 2007), is not fully conserved between cypin and Gda. Additionally, the cypin PDZ-binding domain that has been described to bind PSD-95 and is responsible for regulating MT dynamics (Sweet et al., 2011) is absent from Gda. Finally, we found that by adding the cypin CRMP homology and PDZ-binding domains to the N terminal region of Xenopus Gda, we observed MT dynamics regulation similar to that of cypin. This result further supports the importance of the CRMP and PDZ-binding domain in regulating MT dynamics.
Xenopus GDA is strongly expressed in the embryonic kidney
We found that gda is highly expressed at 3 dpf, which corresponds to stages 41–44, a relatively late time point in Xenopus embryonic development. By this point, significant dendrite branching and elongation in Xenopus retinal ganglion cells has already occurred (stages 32–34) and the specialization into various dendritic subtypes is in process (stages 34–46) (McFarlane and Lom, 2012). While we cannot overlook the possibility of Gda being involved in this second phase of dendrite branching and differentiation, this hypothesis is unlikely due to the fact that whole-mount in situ hybridization showed minimal gda expression in the brain or neural tube in Xenopus. While the lack of gda staining in the neural tube is unsurprising given that cypin is absent from the mammalian spinal cord (Firestein et al., 1999), the lack of gda expression in the brain is striking, as cypin is highly expressed in the developing brain in mammals. This suggests that cypin may have evolved to serve a different function in mammals than that observed in Xenopus.
In the present work, we demonstrated by in situ hybridization that gda is expressed primarily in the kidney. This expression pattern is more restricted than that observed for cypin, which is expressed in the human nervous system, placenta, liver, and kidney, and absent from the heart, lung, muscle, and pancreas (Firestein et al., 1999). Moreover, cypin protein is expressed in rat kidney, liver, lung, brain, and spleen (Firestein et al., 1999). Thus, rat cypin expression pattern contrasts with that of human cypin, which is not expressed in the lung, suggesting that even within mammals, cypin has a slightly different expression pattern.
To date, most studies relating to GDA function have focused on its role in maintenance of homeostasis of triphosphate nucleotides, its role as a nitrogen source, its signal transduction pathway, and, lately, on its role in neuronal development. Although serum GDA function is often altered in kidney diseases (Caraway, 1966; Prodanov and Astrug, 1971; Kebejian and al-Khalidi, 1973; Kuzmits et al., 1980), whether GDA functions during kidney development or maintenance had not been addressed. Xenopus begin developing a primitive kidney called pronephroi by stage 12.5, which becomes fully functional by stage 38, approximately 2 days post-fertilization. Later in development, the pronephroi is replaced by a more complex kidney structure, the mesonephroi (Vize et al., 1995; Wessely and Tran, 2011). In the present work, we showed that the peak gda expression is at 3 dpf, which is temporally coincident with a fully functional primitive kidney. Thus, a Gda effect on kidney functioning or maturation to mesonephroi might be expected. Moreover, knockdown of Xenopus GDAs resulted in edema formation, which has been shown to be indicative of kidney malfunction (Wessely and Tran, 2011; Krneta-Stankic et al., 2017). Nevertheless, at the Xenopus developmental time point that we performed our studies, the pronephroi has still not matured to mesonephroi, thus, it remains to be determined if Gda has an effect on kidney maturation to mesonephroi, or if the effect is restricted to kidney function. Additionally, given that edema also results from cardiac and other developmental issues, it is also possible that other organ systems are affected by reduced Gda function.
Conclusion
The present study shows that human cypin and Xenopus Gda have different protein sequences and functions. In addition, our MT dynamics analysis suggests that the role of GDA in regulating MT dynamics is not conserved among species. Furthermore, gda is expressed with high specificity in the kidneys during Xenopus development. Thus, our study suggests a novel function for Gda and warrants further study of its potential role in kidney maturation and function.
Experimental Procedures
Culture of Xenopus embryonic explants
Eggs obtained from female X. laevis frogs (NASCO, Fort Atkinson, WI) were fertilized in vitro, dejellied, and cultured following standard methods (Sive et al., 2010). Embryos were staged according to Nieuwkoop and Faber (1994), and embryonic explants were dissected at stage 22–24 and cultured as described (Lowery et al., 2012). All experiments were approved by the Boston College Institutional Animal Care and Use Committee and performed according to national regulatory standards.
Guanine deaminase constructs and sequence alignment
The X. laevis GDA L homeolog (GDA) sequence was designed based on the GDA annotated sequence from the laevis genome, version 9.1 (www.xenbase.org) using the transcript model of chr1L:128,874,669..128,903,678 (+ strand), which was predicted to contain the X. laevis GDA genomic sequence. The GDA sequence was acquired from Biomatik (Biomatik, Atlanta, GA, USA) in pBluescript, and then subcloned into the GFP-pCS2+ vector to add a GFP tag to the N-terminus of GDA. The human cypin sequence, based on NCBI reference sequence NM_004293.4, was obtained from Origen (Origen, Rockville, MD, USA) and subcloned into a GFP-pCS2+ to generate GFP-cypin and cypin-GFP. The cypin-chimera was obtained by removing the GDA region, beginning with the CRMP domain, and replacing it with the human cypin CRMP homology and PDZ-binding domain. The NCBI Reference Sequences for the GDA protein sequence comparison were the following: Xenopus laevis S homeolog (XP_018099342.1); Xenopus laevis L homeolog (NP_001083074.1); Homo sapiens (NP_001229434.1); Xenopus tropicalis (XP_004910825.1); Danio rerio (NP_001018510.1); Rattus norvegicus (NP_113964.2); Mus musculus (NP_034396.1); Gallus gallus (XP_424835.3). The GDA protein sequences were compared using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/; RRID: SCR_001591).
RNA and morpholino
Capped mRNA was transcribed in vitro using the SP6 mMessage mMachine Kit (Thermo Fisher Scientific, Waltham, MA, USA). RNA was purified with LiCl precipitation and re-suspended in nuclease-free water. Constructs coding for GFP, GFP-GDA, GFP-cypin and cypin-GFP, cypin-chimera, and mKate2-MACF43 (a gift from Hoogenraad Lab sub-cloned into pCS2+) were used as templates for in vitro transcription. Embryos were injected at the two-to-four-cell stage in each dorsal blastomere (in 0.1× MMR containing 5% Ficoll) with total mRNA amount per embryo of 1000 pg of cypin, gda and 200 pg of mKate2-MACF43.
A morpholino (MO) was designed specific to the Xenopus GDA L homeolog with the sequence 5’ TCCCCAAACCAAAGTCCTTACCACA 3’ and targeted to the exon 3 – intron 3 splice junction (Fig. 4C) (Gene Tools, LLC, Philomath OR, USA). A standard control MO (5’-cctcttacctcagttacaatttata-3’) (Gene Tools, LLC, Philomath OR, USA) was used as control MO. The GDA MO was injected at concentrations of 10, 20, and 50 ng per embryo in order to assess knock down of GDA (Fig. 4C). The GDA transcript levels were measured by reverse-transcription PCR, and the PCR product was sent for sequencing to ensure the introduction of a stop codon. A 50% KD was obtained when injecting 20 ng of the MO (Fig. 4A), which was used for the rest of the experiments in order to generate a partial KD phenotype.
Reverse-transcription PCR
Whole RNA was extracted from embryos at various stages and adult Xenopus brain using Trizol reagent (Invitrogen, LifeTechnologies) and reverse transcribed using SuperScript IV reverse transcriptase (Invitrogen, LifeTechnologies). PCR was then performed with HotStarTaq Plus DNA Polymerase (Qiagen, Hilden, Germany) and primers specific to Xenopus gda: forward 5’ CCCTTGTGCTGGCCGATATTAC 3’ and reverse 5’ GTAGCAGCCATGAGCCATCAC 3’ PCR was performed on the same samples of cDNA, also using primers specific to the housekeeping gene ODC1: forward 5’GCCATTGTGAAGACTCTCTCCATTC 3’ and reverse 5’TTCGGGTGATTCCTTGCCAC3’ as a RT-PCR control.
Microscopy
Live images of cultured cells were obtained with a CSU-X1M 5000 spinning-disk confocal (Yokogawa, Tokyo, Japan) on a Zeiss Axio Observer inverted motorized microscope with a Zeiss 63× Plan Apo 1.4 numerical aperture lens (Zeiss, Thornwood, NY). Images were acquired with an ORCA R2 charge-coupled device camera (Hamamatsu, Hamamatsu, Japan) controlled with Zen software. For time-lapse, images were collected every 2 sec for 1 min. For two-color images, the red and green channels were imaged sequentially. The images were deconvolved using the “Iterative Deconvolve 3D” plugin within ImageJ.
Lateral images of embryos at 4 days post-fertilization (dpf), for quantification of edema formation, in addition to in situ hybridization images, were obtained with a SteREO Discovery.V8 microscope using a Zeiss 1X objective for edema formation experiments and 5X or 8X objectives for in situ hybridization, and Axiocam 512 color camera (Zeiss, Thornwood, NY).
Images of immunostained fluorescent kidneys at 4 dpf were obtained with a Leica TCS SP5 scanning confocal inverted microscope (Leica, Buffalo Grove, IL) using a 10X objective. Lateral images of embryos were obtained in a BABB solution on a MatTek glass bottom dish.
plusTipTracker software analysis
MT dynamics were analyzed from mKate2-MACF43 movies from embryonic mesenchymal cells using plusTipTracker, as previously described (Applegate et al., 2011; Lowery et al., 2013; Stout et al., 2014). The same parameters were used for all movies: maximum gap length, eight frames; minimum track length, three frames; search radius range, 5–12 pixels; maximum forward angle, 50°; maximum backward angle, 10°; maximum shrinkage factor, 0.8; fluctuation radius, 2.5 pixels; and time interval 2 sec. Only cells with a minimum number of 10 MT tracks in a 1-min time lapse were included for analysis. All raw data of automated tracking of mKate2-MACF43 comets were normalized to the same-day control means. Dot plots were made using GraphPad Prism (GraphPad Software, La Jolla, CA). To determine statistical differences, unpaired two-tailed t tests were used for comparing two conditions, after confirming normalized data distribution (GraphPad). A total of 83–154 growth cones were analyzed for each condition, from three independent experiments.
Whole-mount in situ hybridization
Embryos at various developmental stages from two-cell to 6 dpf were fixed overnight at 4°C in a solution of 4% paraformaldehyde in phosphate-buffered saline (PBS), gradually dehydrated using ascending concentrations of methanol in PBS, and stored in methanol at −20°C before in situ hybridization, which was performed as previously described (Sive et al., 2007). Embryos were treated with proteinase K for: 1–2 min in the case of 1 dpf embryos and 3–5 min for 3–6 dpf embryos, and then bleached under a fluorescent light in 1.8× saline–sodium citrate, 1.5% H2O2, and 5% (vol/vol) formamide for 45 min before prehybridization. Probes for both GDA and the proximal tubule kidney marker, xSGLT1K (sodium glucose transporter), were used at a concentration of 0.5 μg/ml and hybridized overnight. Sglt1K construct pCMV-SPORT6, SalI/T7 was a gift from the Wessely Lab (Cleveland Clinic, Lerner Research Institute, Cleveland, OH, USA). The full-length antisense digoxigenin-labeled hybridization RNA probes were transcribed in vitro using the T7 MAXIscript kit (Thermo Fisher Scientific, Waltham, MA, USA). The probes were purified using ammonium acetate precipitation and resuspended in nuclease-free water.
Quantification of the edema formation
Embryos were injected with 20 ng of the control MO, Xenopus GDA MO or Xenopus GDA MO plus 1000 pg GDA mRNA, for rescue experiments. From lateral images of 4 dpf injected tadpoles, the formation of edema was analyzed by measuring the length from the top of the face to the bottom of the chest (face-chest), and as the size of the head can vary from tadpole to tadpole, the length from the middle of the cement gland to the middle of the eye (cement gland-eye) was used as an internal reference (Fig. 4C). The ratios of the former over the latter measurement were compared among the different conditions, to normalize the data for differences in tadpole size. The presence of edema in the embryo was defined as when the above ratio was 2×greater than standard deviation of ratios from the control MO injected embryos. Dot plots were produced using GraphPad Prism (GraphPad Software, La Jolla, CA).
Whole-mount immunostaining
Embryos were injected at the 4-cell stage in the left ventral blastomere to target the developing kidney on the left side of the embryo, leaving the right side as an uninjected internal control. Embryos were injected with 20 ng of control MO with 300 pg of GFP mRNA or 20 ng of Xenopus GDA MO with 300 pg of GFP mRNA. At 4 dpf, the embryos were fixed overnight at 4° C in a solution of 4% paraformaldehyde in phosphate-buffered saline (PBS). After 24 hours, the 4% paraformaldehyde solution was removed and embryos were dehydrated in multiple methanol washes, and subsequently rehydrated in a methanol/PBS series. The eyes and surrounding cartilage were removed from each embryo for ease of mounting when imaging. Embryos were bleached under a fluorescent lamp in a 1.8X saline-sodium citrate, 1.5% H2O2, and 5% (vol/vol) formamide solution for 30 minutes. After incubating the embryos in blocking solution (3% bovine serum albumin, 1% Triton X-100 in PBS) for two hours, embryos were incubated overnight at 4° in 2% 4A6 and 2% 3G8 monoclonal antibodies (European Xenopus Resource Center) in blocking solution. After multiple rinses, embryos were incubated with 0.1% anti-mouse Alexa 488 overnight at 4° C. Embryos were then cleared overnight in BABB solution (33% benzyl alcohol, 66% benzyl benzoate).
Quantification of pronephric tubule diameter
The embryonic kidneys were immunostained using both 3G8 and 4A6 antibodies, specific markers of pronephric tubules and nephrostomes, respectively, in order to obtain a full picture of the kidney. The diameter of each tubule at its widest point was measured at 4 layers through the immunostained embryonic kidney: 35%, 45%, 55%, and 65% through the sample. The average of the widest diameter was obtained for each sample, then a ratio was calculated between the average tubule diameter of the injected and uninjected sides. The Xenopus GDA knockdown condition was compared with the control MO condition, and the results were presented with a dot plot produced with GraphPad Prism (GraphPad Software, La Jolla, CA).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism (GraphPad Software Inc). The D’Agostino and Pearson normality test was used to determine if the data was distributed normally. For MT dynamics analyses, the unpaired Student’s t-test was performed when comparing two conditions. To assess statistical significance when quantifying the edema formation, the Mann Whitney test was used, as GDA KD population did not pass the normality test. At least 3 independent experiments were carried out for each condition. The alpha value was set at 0.05 for all statistical tests, and the P values are represented as follows: *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001. Values are expressed as mean ± standard error of the mean.
Supplementary Material
Control growth cone expressing mKate2-MACF43 to depict the growing microtubule plus-ends, in a 1 minute movie, with a 2 second time interval between frames. After acquisition of the movies, fluorescent comets were tracked using automated tracking software (see Experimental Procedures for details).
Acknowledgements
We thank members of the Lowery Lab for helpful discussions, and Laurie Hayrapetian, Claire Stauffer, Torrey Mandigo and Samantha Dyckman, for early technical help on this project. We also thank Nancy McGilloway and Todd Gaines for excellent Xenopus husbandry. We thank Bret Judson and the Boston College Imaging Core for infrastructure and support. This material is based upon work supported by the National Science Foundation under Grant No. 1626072. In addition, we thank the National Xenopus Resource RRID:SCR_013731 and Xenbase RRID:SCR_003280 for their support.
This work was supported by, the National Institutes of Health to LAL (NIH R01 MH109651–01, NIH R00 MH095768 and NIH/NIDCR R03 DE025824–01), postdoctoral fellowship from CONICYT to PGS, Boston College Undergraduate Thesis Supplementary Grant to JTB, and McNair Scholars Grant to OY.
Abbreviations
- cypin
cytosolic PSD-95 interactor
- GDA
guanine deaminase
- MT
microtubule
- F-actin
actin filament
- MO
morpholino
- +TIPs
plus-end-tracking proteins
- CRMP
collapsin response mediator protein
- KD
knock down
- dpf
days post fertilization
References
- Akhmanova A, Steinmetz MO. 2008. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 9:309–322. [DOI] [PubMed] [Google Scholar]
- Akum BF, Chen M, Gunderson SI, Riefler GM, Scerri-Hansen MM, Firestein BL. 2004. Cypin regulates dendrite patterning in hippocampal neurons by promoting microtubule assembly. Nat Neurosci 7:145–152. [DOI] [PubMed] [Google Scholar]
- Andersen SS. 2000. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol 10:261–267. [DOI] [PubMed] [Google Scholar]
- Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. 2011. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol 176:168–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearce EA, Erdogan B, Lowery LA. 2015. TIPsy tour guides: how microtubule plus-end tracking proteins (+TIPs) facilitate axon guidance. Front Cell Neurosci 9:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig D, Krakowiak P, Duncanson P, Boyce R, Hansen RL, Ashwood P, Hertz-Picciotto I, Pessah IN, Van de Water J. 2013. Autism-specific maternal autoantibodies recognize critical proteins in developing brain. Transl Psychiatry 3:e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarata GM, Bearce EA, Lowery LA. 2016. Cytoskeletal social networking in the growth cone: How +TIPs mediate microtubule-actin cross-linking to drive axon outgrowth and guidance. Cytoskeleton (Hoboken) 73:461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraway WT. 1966. Colorimetric determination of serum guanase activity. Clinical chemistry 12:187–193. [PubMed] [Google Scholar]
- Charych EI, Akum BF, Goldberg JS, Jornsten RJ, Rongo C, Zheng JQ, Firestein BL. 2006. Activity-independent regulation of dendrite patterning by postsynaptic density protein PSD-95. J Neurosci 26:10164–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Firestein BL. 2007. RhoA regulates dendrite branching in hippocampal neurons by decreasing cypin protein levels. J Neurosci 27:8378–8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Lucas KG, Akum BF, Balasingam G, Stawicki TM, Provost JM, Riefler GM, Jornsten RJ, Firestein BL. 2005. A novel role for snapin in dendrite patterning: interaction with cypin. Mol Biol Cell 16:5103–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan B, Cammarata GM, Lee EJ, Pratt BC, Francl AF, Rutherford EL, Lowery LA. 2017. The microtubule plus-end-tracking protein TACC3 promotes persistent axon outgrowth and mediates responses to axon guidance signals during development. Neural Dev 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdogan B, Ebbert PT, Lowery LA. 2016. Using Xenopus laevis retinal and spinal neurons to study mechanisms of axon guidance in vivo and in vitro. Semin Cell Dev Biol 51:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez JR, Byrne B, Firestein BL. 2009. Phylogenetic analysis and molecular evolution of guanine deaminases: from guanine to dendrites. J Mol Evol 68:227–235. [DOI] [PubMed] [Google Scholar]
- Firestein BL, Firestein BL, Brenman JE, Aoki C, Sanchez-Perez AM, El-Husseini AE, Bredt DS. 1999. Cypin: a cytosolic regulator of PSD-95 postsynaptic targeting. Neuron 24:659–672. [DOI] [PubMed] [Google Scholar]
- Fukata Y, Itoh TJ, Kimura T, Menager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, Kaibuchi K. 2002. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol 4:583–591. [DOI] [PubMed] [Google Scholar]
- Fuller HR, Hurtado ML, Wishart TM, Gates MA. 2014. The rat striatum responds to nigro-striatal degeneration via the increased expression of proteins associated with growth and regeneration of neuronal circuitry. Proteome Sci 12:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getwan M, Lienkamp SS. 2017. Toolbox in a tadpole: Xenopus for kidney research. Cell Tissue Res 369:143–157. [DOI] [PubMed] [Google Scholar]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A. 2009. An EB1-binding motif acts as a microtubule tip localization signal. Cell 138:366–376. [DOI] [PubMed] [Google Scholar]
- Kebejian GY, al-Khalidi UA. 1973. Serum guanase in kidney diseases. Eur J Clin Invest 3:41–43. [DOI] [PubMed] [Google Scholar]
- Krneta-Stankic V, DeLay BD, Miller RK. 2017. Xenopus: leaping forward in kidney organogenesis. Pediatr Nephrol 32:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Josan V, Sanger KC, Tewari KK, Krishnan PS. 1967. Studies on guanine deaminase and its inhibitors in rat tissue. Biochem J 102:691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tewari KK, Krishnan PS. 1965. Guanine-Deaminase Activity in Rat Brain and Liver. Biochem J 95:797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmits R, Seyfried H, Wolf A, Muller MM. 1980. Evaluation of serum guanase in hepatic diseases. Enzyme 25:148–152. [DOI] [PubMed] [Google Scholar]
- Lasser M, Tiber J, Lowery LA. 2018. The Role of the Microtubule Cytoskeleton in Neurodevelopmental Disorders. Front Cell Neurosci 12:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Engel U, Rusch J, Scherrer S, Sheard K, Van Vactor D. 2004. The microtubule plus end tracking protein Orbit/MAST/CLASP acts downstream of the tyrosine kinase Abl in mediating axon guidance. Neuron 42:913–926. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Faris AE, Stout A, Van Vactor D. 2012. Neural Explant Cultures from Xenopus laevis. J Vis Exp:e4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery LA, Stout A, Faris AE, Ding L, Baird MA, Davidson MW, Danuser G, Van Vactor D. 2013. Growth cone-specific functions of XMAP215 in restricting microtubule dynamics and promoting axonal outgrowth. Neural Dev 8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucaj CM, Evans MF, Nwagbara BU, Ebbert PT, Baker CC, Volk JG, Francl AF, Ruvolo SP, Lowery LA. 2015. Xenopus TACC1 is a microtubule plus-end tracking protein that can regulate microtubule dynamics during embryonic development. Cytoskeleton (Hoboken) 72:225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A, Godinez WJ, Tsimashchuk V, Bankhead P, Rohr K, Engel U. 2013. Xenopus cytoplasmic linker-associated protein 1 (XCLASP1) promotes axon elongation and advance of pioneer microtubules. Mol Biol Cell 24:1544–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane S, Lom B. 2012. The Xenopus retinal ganglion cell as a model neuron to study the establishment of neuronal connectivity. Dev Neurobiol 72:520–536. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P, Faber J. 1994. Normal Table of Xenopus laevis (Daudin), 1994. New York: Garland; 3. [Google Scholar]
- Nwagbara BU, Faris AE, Bearce EA, Erdogan B, Ebbert PT, Evans MF, Rutherford EL, Enzenbacher TB, Lowery LA. 2014. TACC3 is a microtubule plus end-tracking protein that promotes axon elongation and also regulates microtubule plus end dynamics in multiple embryonic cell types. Mol Biol Cell 25:3350–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodanov K, Astrug A. 1971. Guanase activity in endemic Balkan nephropathy. Clin Chim Acta 35:445–448. [DOI] [PubMed] [Google Scholar]
- Riefler GM, Balasingam G, Lucas KG, Wang S, Hsu SC, Firestein BL. 2003. Exocyst complex subunit sec8 binds to postsynaptic density protein-95 (PSD-95): a novel interaction regulated by cypin (cytosolic PSD-95 interactor). Biochem J 373:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford EL, Carandang L, Ebbert PT, Mills AN, Bowers JT, Lowery LA. 2016. Xenopus TACC2 is a microtubule plus end-tracking protein that can promote microtubule polymerization during embryonic development. Mol Biol Cell 27:3013–3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EF, Strittmatter SM. 2007. The CRMP family of proteins and their role in Sema3A signaling. Adv Exp Med Biol 600:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Sala C. 2001. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci 24:1–29. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. 2007. Baskets for in situ hybridization and immunohistochemistry. CSH Protoc 2007:pdb prot4777. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. 2010. Microinjection of Xenopus embryos. Cold Spring Harb Protoc 2010:pdb ip81. [DOI] [PubMed] [Google Scholar]
- Slater PG, Hayrapetian L, Lowery LA. 2017. Xenopus laevis as a model system to study cytoskeletal dynamics during axon pathfinding. Genesis 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout A, D’Amico S, Enzenbacher T, Ebbert P, Lowery LA. 2014. Using plusTipTracker software to measure microtubule dynamics in Xenopus laevis growth cones. J Vis Exp:e52138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet ES, Previtera ML, Fernandez JR, Charych EI, Tseng CY, Kwon M, Starovoytov V, Zheng JQ, Firestein BL. 2011. PSD-95 alters microtubule dynamics via an association with EB3. J Neurosci 31:1038–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vize PD, Jones EA, Pfister R. 1995. Development of the Xenopus pronephric system. Dev Biol 171:531–540. [DOI] [PubMed] [Google Scholar]
- Wessely O, Tran U. 2011. Xenopus pronephros development--past, present, and future. Pediatr Nephrol 26:1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Vize PD. 2004. Proximo-distal specialization of epithelial transport processes within the Xenopus pronephric kidney tubules. Dev Biol 271:322–338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Control growth cone expressing mKate2-MACF43 to depict the growing microtubule plus-ends, in a 1 minute movie, with a 2 second time interval between frames. After acquisition of the movies, fluorescent comets were tracked using automated tracking software (see Experimental Procedures for details).