Abstract
Sexual selection is a powerful agent of evolution, driving microevolutionary changes in the genome and macroevolutionary rates of lineage diversification. The mechanisms by which sexual selection might influence macroevolution remain poorly understood. For example, sexual selection might drive positive selection for key adaptations that facilitate diversification. Furthermore, sexual selection might be a general driver of molecular evolutionary rate. We lay out some of the potential mechanisms that create a link between sexual selection and diversification, based on causal effects on other life-history traits such as body mass and the rate of molecular evolution. Birds are ideally suited for testing the importance of these relationships because of their diverse reproductive systems and the multiple evolutionary radiations that have produced their astounding modern diversity. We show that sexual selection (measured as the degree of polygyny) interacts with the rate of molecular evolution and with body mass to predict species richness at the genus level. A high degree of polygyny and rapid molecular evolution are positively associated with the net rate of diversification, with the two factors being especially important for explaining diversification in large-bodied taxa. Our findings further suggest that mutation rates underpin some of the macroevolutionary effects of sexual selection. We synthesize the existing theory on sexual selection as a force for diversity and propose avenues for exploring this association using genome data.
Keywords: sexual selection, speciation rate, life-history traits, mutation rate, substitution rate, body mass
1. Introduction
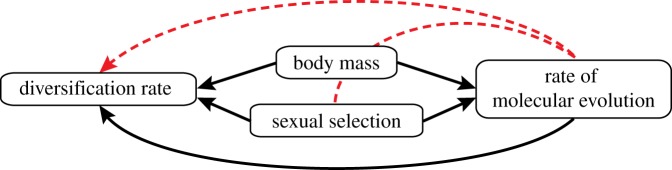
Sexual selection is often invoked as a major driver of speciation or extinction [1], and several comparative analyses have concluded that it elevates the net rate of diversification (speciation minus extinction) [2–4] (but see [5–7]). However, there is limited evidence about the mechanisms underlying this association, partly because the relationship between sexual selection and diversification is often explored in isolation from other well-known evolutionary consequences of sexual selection. For example, the reported association between sexual selection and diversification could have arisen because other traits that evolve in response to sexual selection directly affect rates of speciation and/or extinction. Two such traits that have been repeatedly linked to speciation and extinction rates are the rate of molecular evolution and adult body size [8–10] (figure 1).
Figure 1.
Hypothesized causal relationships between sexual selection, the rate of molecular evolution and the net rate of diversification. The red arrow shows the proposed hypothesis that sexual selection acts through other species traits, including those associated with body mass and the rate of molecular evolution, to drive higher rates of speciation or lower rates of extinction.
There are several ways in which sexual selection could directly influence speciation and extinction. First, female mate choice can drive the evolution of ecologically relevant traits that are genetically correlated with preferred male traits [11,12], increasing positive selection in those parts of the genome that facilitate local adaptation and lead to ecological divergence between populations [11,13–16]. Second, reduced gene flow between populations can arise from the rapid coevolution of male–female sexual traits along different arbitrary trajectories, caused by either mate choice (e.g. Fisherian runaway [11]) or sexual conflict (‘arms race’ between sexually antagonistic traits [15,17,18]). Third, sexual selection can elevate the risk of extinction by reducing the environmental ‘fit’ of individuals. This might be due to sexual conflict, a trade-off in trait expression between natural and sexual selection [19,20] or a reduction in the effective population size because only some individuals of the competitive sex reproduce [21].
Sexual selection acts on mutations that increase reproductive competitiveness, seductiveness, choosiness and mating resistance. This should increase the rate of evolution in regions of the genome encoding for these functional traits. Furthermore, sexual selection can accelerate the purging of deleterious mutations that affect resource acquisition and assimilation (i.e. condition). This is because these mutations are likely to have detrimental effects on the multitude of ecologically relevant traits that affect the expression of condition-dependent, sexually selected traits [22,23]. This purging reduces genomic changes that are harmful to population persistence, accelerates adaptive evolution [24], increases population fitness [25] and can even lower the costs of sex [23] (but see [26]). Sexual selection might also indirectly affect the type and number of mutations that become fixed if it lowers the effective population size, which leads to stronger genetic drift and a reduced efficacy of selection for beneficial mutations (but see [22]).
Another mechanism by which sexual selection can influence diversification is to favour a higher mutation rate. Directional female mate choice for exaggerated male traits is expected to reduce additive genetic variation, thereby decreasing the benefits of choice. However, this reduced genetic diversity is not observed in nature (the ‘lek paradox’ [27]). One explanation for this is an increased input of novel genetic variation through a higher mutation rate [28]. Theory suggests that ‘mutator’ alleles can spread if nonlinear benefits of increased trait expression (i.e. larger traits disproportionately increase fitness) select for greater phenotypic variability in sexual ornaments [9,28], but this idea is highly controversial and has little empirical support. There is, however, evidence that sperm competition, which increases the rate of mitosis in testes and is prevalent in most species under strong sexual selection, leads to greater accumulation of copy errors in the germline [9]. Sexual selection could therefore be linked to diversification rates if an elevated mutation rate accelerates the appearance of genetic incompatibilities between populations [29].
Sexual selection therefore has several opposing effects on genome evolution that might each generate predictable, but opposing, macroevolutionary patterns [22]. In the light of the processes already mentioned, one possible long-term effect of sexual selection is an accelerated mutation rate, which facilitates adaptation because of greater production of beneficial mutations. Species with a history of stronger sexual selection might then be more likely to persist due to faster molecular evolution in regions of the genome that are under natural selection. Alternatively, these regions might show reduced molecular evolution if the predominant effect of sexual selection is not to elevate the mutation rate but simply to eliminate slightly deleterious mutations. Or it might be the case that sexual selection has a negligible effect on fixation of either beneficial or detrimental mutations and that solely by elevating the mutation rate [28] it drives genetic incompatibilities associated with promoting speciation.
There has been a limited success in disentangling how molecular evolution, sexual selection and other life-history traits affect species diversification. Although numerous traits have been proposed as drivers of both species diversification and molecular evolution, body size can be singled out as a key variable that affects most life-history traits and demographic parameters, including generation time, lifespan and population size. These, in turn, are all factors that are believed to affect rates of molecular evolution [30]. Indeed, there is evidence that smaller body size is positively associated with species richness in many taxa [31]. To date, however, there have been few attempts to determine if, and how, sexual selection, rates of molecular evolution and key life-history traits interact to affect species diversification.
Here, we test whether body size, sexual selection and molecular evolution interact to predict variation in rates of species diversification among bird genera. We collated data on diversification rates, body mass and sexual selection using a measure of the degree of polygyny as a proxy for sexual selection. We estimated non-synonymous (dN) and synonymous (dS) rates of molecular evolution, and their ratio (dN/dS, ω), using both nuclear and mitochondrial sequence data (n = 173 and 354 genera, respectively). Then, we tested whether the degree of polygyny (i.e. sexual selection) is associated with net diversification rates while taking into account any interactive effects of the rate of molecular evolution or body mass. We predicted that avian genera with a high degree of polygyny, small body size and higher rates of molecular evolution would have the highest rates of diversification. We tested for the effects of molecular rates that are associated with selection (dN and ω) or the mutation rate (dN and dS), or both.
2. Material and methods
(a). Data collection
We compiled information about the degree of polygyny and body mass in 954 species of birds in 558 genera, using existing data compendia [32,33] (electronic supplementary material). We used data about the degree of polygyny (i.e. variance in mating success of males) in which species were ranked from 1 to 5 [32,34]: 1 = polyandry; 2 = monogamy (less than 5% polygyny); 3 = mostly monogamy, but occasional polygyny (5–15% polygyny); 4 = mostly polygyny (greater than 15% polygyny); and 5 = lekking or promiscuous. This ordinal variable indicates the potential for sexual selection due to male–male competition for mates. For the statistical analyses, however, we used the polygyny score as a continuous variable which was the average value for each avian genus examined (see below). We used an estimate of the complete avian phylogeny and estimated the net diversification rate of every bird species using the reciprocal of the equal splits measure of diversity (strongly correlated with other metrics of net diversification rates; see electronic supplementary material, figure S1) [35]. We then identified all of the avian genera for which we had data for polygyny, body mass and nucleotide sequences of four mitochondrial (n = 354) and four nuclear genes (n = 173; GenBank accessions sourced from Jetz et al. [36]). The data on the degree of polygyny and body mass were averaged across the species within each genus.
To estimate rates of molecular evolution, we aligned the nucleotide sequences across genera (electronic supplementary material, table S9) and estimated dN and dS substitutions per lineage using the software HyPhy v2.2 [37]. We used the MG94 codon evolution model [38], paired with a general time-reversible model of nucleotide substitution selected automatically using the Akaike information criterion. The terminal branch lengths inferred in this procedure represent genus-average estimates of dN and dS. In order to avoid making misleading estimates of relative molecular evolutionary rates, we removed any genera for which terminal branch length was zero (electronic supplementary material). Estimates of dN and dS on terminal branches are confounded by the elapsed time over which taxa have been able to accumulate nucleotide changes [39], so we estimated relative rates of dN and dS evolution for each genus using a model that relaxes the molecular-clock assumption using penalized likelihood [40] (electronic supplementary material). Throughout the text, we differentiate molecular rates that are associated with selection (dN and ω) and with the mutation rate (dN and dS). To clarify, dS estimates the rate of silent substitutions that do not affect amino acid coding. It is mainly influenced by the mutation rate. By contrast, dN is an estimation of nucleotide changes that affect amino acid coding and is influenced by both the mutation rate and selection. In consequence, ω is expected to be influenced by selection but not by the mutation rate [8].
(b). Statistical analyses
We used phylogenetic generalized least squares (PGLS) to examine the relationships between the degree of polygyny (calculated as the mean of the available species values in each genus), diversification rate, relative rate of molecular evolution and body mass (see electronic supplementary material for information on the calculation of scores). We used the avian tree containing the sampled genera to correct for phylogenetic non-independence. In addition, we investigated how phylogenetic uncertainty affected our results. Phylogenetic regression models were replicated with each of 100 randomly selected trees from a published posterior distribution of avian relationships [36]. To linearize relationships, we log-transformed the rates of diversification, rates of molecular evolution and body mass (all continuous variables).
In PGLS, we assumed a lambda model of trait evolution [41], which provided a better fit than three other commonly used models (Brownian motion, Kappa and an Ornstein–Uhlenbeck process). The parameters of the PGLS regression models were optimized using maximum likelihood in the nlme R package [42], and model residuals were inspected for normality. Regression models that included two-way and three-way interactions were re-run with interactions excluded. If removal did not reduce the model fit (as determined by a likelihood-ratio test), we interpreted parameter estimates from the reduced model. Otherwise, we interpreted the estimates of retained interaction terms (electronic supplementary material, tables S1–S5).
We ran PGLS separately for mitochondrial and nuclear genes to address the two main questions described below. The links between these questions are illustrated in figure 1. We then explored the directionality of these relationships using phylogenetic path analysis [43]. We based our four hypotheses of path models (electronic supplementary material, figure S2) on a previous study of the association between life history, molecular evolution rate and species richness in angiosperms [44].
(i). Do the degree of polygyny and body mass predict the rate of molecular evolution?
We treated the degree of polygyny, body mass and their two-way interaction as fixed factors, and tested whether they explain variation in dN, dS or ω. When testing for the influence of the degree of polygyny and body mass on dN, we also repeated the model with dS as a covariate. This equates to exploring the effect of the degree of polygyny and body mass on dN independent of the influence of the mutation rate (estimated as dS).
(ii). Do the rate of molecular evolution, degree of polygyny and body size interact to predict the rate of diversification?
Molecular evolution, the degree of polygyny and body mass are likely to be related to each other and might have interactive effects on net rates of diversification (figure 1). We therefore ran models that tested whether the molecular rates dS, dN or ω, in conjunction with the degree of polygyny, body mass and all two-way and three-way interactions, explain variation in the net rate of diversification.
3. Results
(a). Do the degree of polygyny and body mass predict the rate of molecular evolution?
A higher rate of synonymous mutation dS for mitochondrial genes was associated with a higher mean score for polygyny (estimate ± s.e. = 0.097 ± 0.036, p = 0.008), but not with body mass or the interaction between body mass and the degree of polygyny (electronic supplementary material, table S1). For nuclear genes, there was no effect of body mass, degree of polygyny or their interaction on dS.
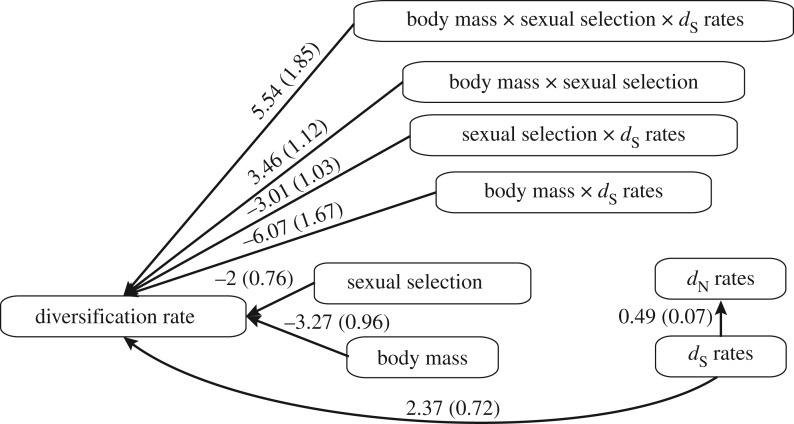
For the rate of non-synonymous mutation dN for mitochondrial genes, the PGLS model showed no effect of body mass, degree of polygyny or their interaction (electronic supplementary material, table S2). By contrast, dN for nuclear genes was positively associated with the degree of polygyny (estimate ± s.e. = 0.215 ± 0.099, p = 0.031; electronic supplementary material, table S2), but not with body mass nor with an interaction between the degree of polygyny and body mass. When we then ran models that included dS as a covariate, it was the main predictor of dN (mitochondrial: estimate ± s.e. = 0.442 ± 0.041, p < 0.001; nuclear: estimate ± s.e. = 0.685 ± 0.094, p < 0.001; electronic supplementary material, figure S3); the degree of polygyny, body mass and their interaction failed to explain any additional variation. The use of path analysis supported our conclusion that dS is the main driver of dN in nuclear genes (figure 2), but not in mitochondrial genes. There was no evidence that ω for either mitochondrial or nuclear genes is associated with the degree of polygyny, body mass or their interaction (electronic supplementary material, table S4).
Figure 2.
Diagram of the significant paths exploring the association between a proxy of sexual selection (degree of polygyny), body mass, rates of nuclear molecular evolution and diversification rate (n = 173 genera). Values show the estimated correlation coefficients with standard errors in parentheses. dN = non-synonymous substitution rate; dS = synonymous substitution rate.
(b). Do the rate of molecular evolution, degree of polygyny and body mass interact to predict the rate of diversification?
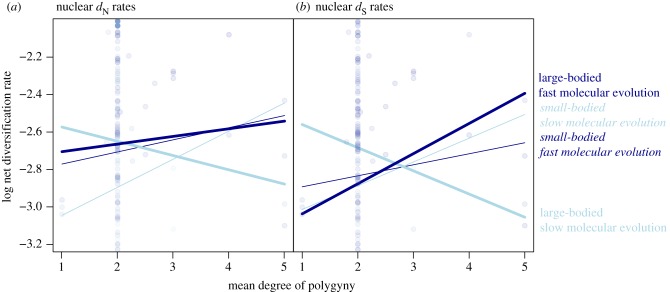
The net rate of diversification was explained by a three-way interaction between the degree of polygyny, body mass and dN for both mitochondrial and nuclear genes (mitochondrial interaction estimate ± s.e. = 0.044 ± 0.022, p = 0.039; nuclear interaction estimate ± s.e. = 0.05 ± 0.022, p = 0.024). The same interaction was present for nuclear dS (interaction estimate ± s.e. = 0.129 ± 0.043, p = 0.003; figure 3), but was absent for mitochondrial dS, and for either mitochondrial or nuclear estimates of ω. The significant three-way interactions for mitochondrial and nuclear dN and nuclear dS all showed a positive association between the degree of polygyny and the diversification rate in small-bodied taxa that was unaffected by the rate of molecular evolution (electronic supplementary material, table S5). In large-bodied taxa, there was a positive association between the degree of polygyny and diversification rates, but only when the rate of molecular evolution was high.
Figure 3.
Degree of polygyny (as a proxy for the potential for sexual selection due to variance in male mating success) interacts with body mass and the rate of molecular evolution in the nuclear genome (n = 173 genera) to drive net diversification rates in bird genera (all continuous variables). The association between the degree of polygyny (x-axis) and net diversification rate (y-axis) is always positive in small-bodied genera (thin line, first quartile of body mass). In large-bodied genera (thick line, third quartile of body mass), this association is mediated by rates of molecular evolution: only those with high rates of molecular evolution (dark blue) showed a positive association between the degree of polygyny and net diversification rate. For a better representation of the interaction, the y-axis is truncated to a range between −2.0 and −3.2 log units of net diversification rate. The complete figure is available in the electronic supplementary material, figure S3.
In models where the three-way interaction was not significant and was removed, the diversification rate was associated with an interaction between the degree of polygyny and body mass (electronic supplementary material, table S5). The single exception occurred for the nuclear ω, which was positively associated with diversification rate (estimate ± s.e. = 0.1 ± 0.04, p = 0.014). We found that the results of replicate phylogenetic regression analyses on a sample of 100 trees were consistent with those using the published summary tree estimate (electronic supplementary material, figure S4). One exception was the analyses of the three-way interaction for mitochondrial dN, which led to p-values above 0.05 in 24% of posterior trees.
Phylogenetic path analyses supported our finding of a three-way interaction between the degree of polygyny, body mass and dS (but not dN) for nuclear genes (figure 2), but not for mitochondrial genes. In sum, our data are broadly consistent with the hypothesis that the effect of the degree of polygyny on diversification is mediated by mutation rates (dN and dS) and body mass (or life-history traits that scale with body mass).
4. Discussion
Our data provide evidence that high degrees of polygyny are associated with an increased rate of net diversification across bird genera with small average body size. This finding of an association between sexual selection and net diversification is consistent with theoretical expectations [15], and with the results of previous studies (e.g. [2–4]). More interestingly, we found that molecular evolutionary rates influenced by the mutation rate (dN and dS) interact with the degree of polygyny and body mass to explain some of the variation in diversification rates in large-bodied taxa. The degree of polygyny we used for our analysis is related to the social mating system of species and therefore represents a good proxy for the strength of sexual selection [34]. However, it should be noted that the degree of polygyny is correlated with other species traits, such as the degree of parental care (see electronic supplementary material).
The primary finding of our study is that the association between net diversification rates and the degree of polygyny is mediated by body mass and the rate of molecular evolution. We found that the interaction between the degree of polygyny and body mass explains net diversification rates in phylogenetic regression models (electronic supplementary material, table S5). However, an association between polygyny and net diversification rate was only general in small-bodied genera. Among large-bodied genera, it is only those with high rates of molecular evolution that show a positive association between the degree of polygyny and net diversification rate (electronic supplementary material, table S5). One explanation for this pattern is that the benefits of sexual selection in facilitating local adaptation are greater in smaller-bodied genera. Smaller species tend to have larger effective population sizes, which are expected to increase the likelihood that beneficial mutations reach fixation [45]. Alternatively, larger-bodied taxa might be more reliant than smaller-bodied taxa on mechanisms that increase their genetic variability: larger-bodied organisms generally have greater long-term susceptibility to extinction in the face of environmental change [46,47]. This greater susceptibility might be partly due to their lower rate of molecular evolution (per unit time). Slower speciation or increased extinction in some larger-bodied taxa might therefore be caused by the combination of smaller population size and lower reproductive potential [48], and reduced genetic diversity due to sexual selection [27] unless compensated for by a higher mutation rate [28].
Strikingly, we found that the association between net diversification rates and the interaction between the degree of polygyny, body mass and molecular evolution was mediated by the mutation rate (estimated as dN and dS) rather than by the strength of selection (estimated as dN and ω). One explanation for the lack of a mediating effect of molecular evolutionary rates attributed to selection is that their impact might only be detectable in specific genes, such as those coding for sexual traits [49]. Future explorations of molecular evolutionary rates in those regions of the genome will provide additional insights into the relative impact of sexual selection across different genomic regions (e.g. faster evolution of seminal-protein genes [50]).
Nonetheless, our results are broadly consistent with our hypothesis that an elevated mutation rate in taxa with strong sexual selection can accelerate reproductive isolation. Despite natural selection across metazoans to reduce the mutation rate [51], sexual selection due to female mate choice has controversially been suggested to adaptively increase the mutation rate to elevate phenotypic variability in traits with nonlinear fitness returns [9,28]. This might manifest as sexual selection enhancing the spread of mutator alleles that hitch-hike with beneficial mutations [28] that increase the probability of adaptation and the resilience of populations to environmental change. It should be noted, however, that the degree of polygyny was only significantly associated with the estimated mutation rate (dS) in mitochondrial genes, and not in nuclear genes (question (i); electronic supplementary material, table S1). Consequently, we cannot reject the hypothesis that the association seen in mitochondrial genes is driven by another process.
Finally, we found that the direction of the observed associations in several of our models was similar for both nuclear and mitochondrial genes, lending some robustness to our findings. Sexual selection is expected to have a stronger effect in nuclear genes coding for traits involved in male–male competition and mate choice. Similarly, mitochondrial genomes are subject to a wide range of selective constraints that might influence estimates of dN and dS. One noteworthy example is the preference for codons ending in A and T in many protein-coding regions [52], which can decrease the power to estimate proxies of mutation rate accurately. This is also the case with other influences on molecular rate estimates that might overshadow the estimate of interest, such as substitutional saturation. These considerations can be examined further when genome-scale data become available.
Some processes involved in sexual selection might play an important role in driving the evolution of mitochondrial genes. For example, mitochondria with high rates of energy production are essential for male–male competition but can be detrimental for females, leading to sexually antagonistic selection and rapid mitochondrial evolution [53]. Furthermore, mitochondrial genomes can play a key role in the process of speciation. In species with high mitochondrial mutation rates, allopatric populations can develop mito-nuclear incompatibilities that will tend to accelerate reproductive isolation [54]. Our results suggest that this process is potentially quicker in species with sexual selection, perhaps because sex differences in the optimal expression of mitochondrial genes lead to an evolutionary arms race. In future work, it will be useful to disentangle the effects of nuclear and mitochondrial evolution on the link between sexual selection and diversification.
5. Conclusion
Our study provides evidence that sexual selection and molecular evolution play a joint role in driving rates of diversification in birds. Through its impact on rates of molecular evolution, sexual selection staves off extinction, facilitates the appearance of key innovations and generates reproductive incompatibilities that drive speciation [55,56]. Future research using genome-scale data across many species will help to determine the impact of sexual selection on molecular evolution on a macroevolutionary scale [49]. Molecular data at the genome scale across bird genera will be available within the next few years and will help to connect these micro- and macroevolutionary processes.
To what extent is speciation due to the selection that creates incompatibilities between populations, as opposed to being a by-product of the stochastic fixation of mutations? Strikingly, the results of several studies, including ours, point towards the latter [29]. In addition to the impact of sexual selection on diversification rates being mediated by other life-history traits (e.g. body size in our study), it might also vary across environments (e.g. covary with altitude [57] or ecosystem productivity [36]). Our analyses provide a theoretical and methodological framework to examine the mechanisms by which sexual selection can drive genomic evolution and species diversity patterns that have shaped the tree of life.
Supplementary Material
Acknowledgements
We thank Megan Head for comments, and Kristal Cain and Jordi Figuerola for advice on the extraction of data from published sources.
Data accessibility
The data used in this paper are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.p39d12d [58].
Authors' contributions
M.I.-C., M.D.J., S.Y.W.H. and D.A.D. conceived and designed the project; M.I.-C. and D.A.D. collected and analysed the data; all authors contributed to the final version of the manuscript.
Competing interests
We have no competing interests.
Funding
D.A.D., S.Y.W.H. and M.D.J. were supported by the Australian Research Council.
References
- 1.Kraaijeveld K, Kraaijeveld-Smit FJL, Maan ME. 2011. Sexual selection and speciation: the comparative evidence revisited. Biol. Rev. 86, 367–377. ( 10.1111/j.1469-185X.2010.00150.x) [DOI] [PubMed] [Google Scholar]
- 2.Stuart-Fox D, Owens IPF. 2003. Species richness in agamid lizards: chance, body size, sexual selection or ecology? J. Evol. Biol. 16, 659–669. ( 10.1046/j.1420-9101.2003.00573.x) [DOI] [PubMed] [Google Scholar]
- 3.Arnqvist G, Edvardsson M, Friberg U, Nilsson T. 2000. Sexual conflict promotes speciation in insects. Proc. Natl Acad. Sci. USA 97, 10 460–10 464. ( 10.1073/pnas.97.19.10460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janicke T, Ritchie MG, Morrow EH, Marie-Orleach L. 2018. Sexual selection predicts species richness across the animal kingdom. Proc. R. Soc. B 285, 20180173 ( 10.1098/rspb.2018.0173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gage MJG, Parker GA, Nylin S, Wiklund C. 2002. Sexual selection and speciation in mammals, butterflies and spiders. Proc. R. Soc. B 269, 2309–2316. ( 10.1098/rspb.2002.2154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Rabosky DL. 2014. Sexual selection and diversification: reexamining the correlation between dichromatism and speciation rate in birds. Am. Nat. 184, E101–E114. ( 10.1086/678054) [DOI] [PubMed] [Google Scholar]
- 7.Morrow E, Pitcher T, Arnqvist G. 2003. No evidence that sexual selection is an ‘engine of speciation’ in birds. Ecol. Lett. 6, 228–234. ( 10.1046/j.1461-0248.2003.00418.x) [DOI] [Google Scholar]
- 8.Lanfear R, Ho SYW, Love D, Bromham L. 2010. Mutation rate is linked to diversification in birds. Proc. Natl Acad. Sci. USA 107, 20 423–20 428. ( 10.1073/pnas.1007888107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Møller A, Cuervo J. 2003. Sexual selection, germline mutation rate and sperm competition. BMC Evol. Biol. 3, 6 ( 10.1186/1471-2148-3-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petrie M, Doums C, Møller AP. 1998. The degree of extra-pair paternity increases with genetic variability. Proc. Natl Acad. Sci. USA 95, 9390–9395. ( 10.1073/pnas.95.16.9390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie MG. 2007. Sexual selection and speciation. Annu. Rev. Ecol. Evol. Syst. 38, 79–102. ( 10.1146/annurev.ecolsys.38.091206.095733) [DOI] [Google Scholar]
- 12.Turelli M, Barton NH, Coyne JA. 2001. Theory and speciation. Trends Ecol. Evol. 16, 330–343. ( 10.1016/S0169-5347(01)02177-2) [DOI] [PubMed] [Google Scholar]
- 13.Boughman JW. 2001. Divergent sexual selection enhances reproductive isolation in sticklebacks. Nature 411, 944–948. ( 10.1038/35082064) [DOI] [PubMed] [Google Scholar]
- 14.Safran RJ, Scordato ESC, Symes LB, Rodríguez RL, Mendelson TC. 2013. Contributions of natural and sexual selection to the evolution of premating reproductive isolation: a research agenda. Trends Ecol. Evol. 28, 643–650. ( 10.1016/j.tree.2013.08.004) [DOI] [PubMed] [Google Scholar]
- 15.Parker GA, Partridge L. 1998. Sexual conflict and speciation. Phil. Trans. R. Soc. Lond. B 353, 261–274. ( 10.1098/rstb.1998.0208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veen T, Otto SP. 2015. Liking the good guys: amplifying local adaptation via the evolution of condition-dependent mate choice. J. Evol. Biol. 28, 1804–1815. ( 10.1111/jeb.12696) [DOI] [PubMed] [Google Scholar]
- 17.Martin OY, Hosken DJ. 2003. The evolution of reproductive isolation through sexual conflict. Nature 423, 979–982. ( 10.1038/nature01752) [DOI] [PubMed] [Google Scholar]
- 18.Gavrilets S. 2014. Is sexual conflict an ‘engine of speciation’? Cold Spring Harb. Pers. Biol. 6, a017723 ( 10.1101/cshperspect.a017723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chenoweth SF, Appleton NC, Allen SL, Rundle HD. 2015. Genomic evidence that sexual selection impedes adaptation to a novel environment. Curr. Biol. 25, 1860–1866. ( 10.1016/j.cub.2015.05.034) [DOI] [PubMed] [Google Scholar]
- 20.Long TAF, Agrawal AF, Rowe L. 2012. The effect of sexual selection on offspring fitness depends on the nature of genetic variation. Curr. Biol. 22, 204–208. ( 10.1016/j.cub.2011.12.020) [DOI] [PubMed] [Google Scholar]
- 21.Nunney L. 1993. The influence of mating system and overlapping generations on effective population size. Evolution 47, 1329–1341. ( 10.1111/j.1558-5646.1993.tb02158.x) [DOI] [PubMed] [Google Scholar]
- 22.Whitlock MC. 2000. Fixation of new alleles and the extinction of small populations: drift load, beneficial alleles, and sexual selection. Evolution 54, 1855–1861. ( 10.1111/j.0014-3820.2000.tb01232.x) [DOI] [PubMed] [Google Scholar]
- 23.Agrawal AF. 2001. Sexual selection and the maintenance of sexual reproduction. Nature 411, 692–695. ( 10.1038/35079590) [DOI] [PubMed] [Google Scholar]
- 24.Lorch PD, Proulx S, Rowe L, Day T. 2003. Condition-dependent sexual selection can accelerate adaptation. Evol. Ecol. Res. 5, 867–881. [Google Scholar]
- 25.Whitlock MC, Agrawal AF. 2009. Purging the genome with sexual selection: reducing mutation load through selection in males. Evolution 63, 569–582. ( 10.1111/j.1558-5646.2008.00558.x) [DOI] [PubMed] [Google Scholar]
- 26.Arbuthnott D, Rundle HD. 2012. Sexual selection is ineffectual or inhibits the purging of deleterious mutations in Drosophila melanogaster. Evolution 66, 2127–2137. ( 10.1111/j.1558-5646.2012.01584.x) [DOI] [PubMed] [Google Scholar]
- 27.Kirckpatrick M, Ryan MJ. 1991. The evolution of mating preferences and the paradox of the lek. Nature 350, 33–38. ( 10.1038/350033a0) [DOI] [Google Scholar]
- 28.Petrie M, Roberts G. 2007. Sexual selection and the evolution of evolvability. Heredity 98, 198–205. ( 10.1038/sj.hdy.6800921) [DOI] [PubMed] [Google Scholar]
- 29.Hua X, Bromham L. 2017. Darwinism for the genomic age: connecting mutation to diversification. Front. Genet. 8, 1–18. ( 10.3389/fgene.2017.00012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bromham L. 2009. Why do species vary in their rate of molecular evolution? Biol. Lett. 5, 401–404. ( 10.1098/rsbl.2009.0136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillimore AB, Freckleton RP, Orme CDL, Owens IPF. 2006. Ecology predicts large-scale patterns of phylogenetic diversification in birds. Am. Nat. 168, 220–229. [DOI] [PubMed] [Google Scholar]
- 32.Lislevand T, Figuerola J, Székely T. 2007. Avian body sizes in relation to fecundity, mating system, display behavior, and resource sharing. Ecology 88, 1605 ( 10.1890/06-2054) [DOI] [Google Scholar]
- 33.Dunning JB., Jr (ed). 2007. CRC handbook of avian body masses, 2nd edn Boca Raton, FL: CRC Press. [Google Scholar]
- 34.Dunn PO, Whittingham LA, Pitcher TE. 2001. Mating systems, sperm competition, and the evolution of sexual dimorphism in birds. Evolution 55, 161–175. ( 10.1111/j.0014-3820.2001.tb01281.x) [DOI] [PubMed] [Google Scholar]
- 35.Redding DW, Mooers AO. 2006. Incorporating evolutionary measures into conservation prioritization. Conserv. Biol. 20, 1670–1678. ( 10.1111/j.1523-1739.2006.00555.x) [DOI] [PubMed] [Google Scholar]
- 36.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. 2012. The global diversity of birds in space and time. Nature 491, 444–448. ( 10.1038/nature11631) [DOI] [PubMed] [Google Scholar]
- 37.Kosakovsky PSL, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. ( 10.1093/bioinformatics/bti079) [DOI] [PubMed] [Google Scholar]
- 38.Muse SV, Gaut BS. 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 11, 715–724. [DOI] [PubMed] [Google Scholar]
- 39.Lanfear R, Welch JJ, Bromham L. 2010. Watching the clock: studying variation in rates of molecular evolution between species. Trends Ecol. Evol. 25, 495–503. ( 10.1016/j.tree.2010.06.007) [DOI] [PubMed] [Google Scholar]
- 40.Sanderson MJ. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19, 101–109. ( 10.1093/oxfordjournals.molbev.a003974) [DOI] [PubMed] [Google Scholar]
- 41.Freckleton RP, Harvey PH, Pagel M. 2002. Phylogenetic analysis and comparative data: a test and review of evidence. Am. Nat. 160, 712–726. ( 10.1086/343873) [DOI] [PubMed] [Google Scholar]
- 42.Pinheiro J, Bates D, DebRoy S, Sarkar D. 2013. nlme: linear and nonlinear mixed effects models. See https://cran.r-project.org/package=nlme.
- 43.van der Bijl W. 2018. phylopath: easy phylogenetic path analysis in R. PeerJ 6, e4718 ( 10.7717/peerj.4718) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bromham L, Hua X, Lanfear R, Cowman PF. 2015. Exploring the relationships between mutation rates, life history, genome size, environment, and species richness in flowering plants. Am. Nat. 185, 507–524. ( 10.1086/680052) [DOI] [PubMed] [Google Scholar]
- 45.Ohta T. 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23, 263–286. ( 10.1146/annurev.es.23.110192.001403) [DOI] [Google Scholar]
- 46.Kozlowski JAN, Gawelczyk AT. 2002. Why are species' body size distributions usually skewed to the right? Proc. R. Soc. Lond. B 16, 419–432. [Google Scholar]
- 47.Clauset A, Erwin DH. 2008. The evolution and distribution of species body size. Science 321, 399–401. ( 10.1126/science.1157534) [DOI] [PubMed] [Google Scholar]
- 48.Cardillo M, Mace G, Jones K, Bielby J, Bininda-Emonds O, Sechrest W, Orme C, Purvis A. 2005. Multiple causes of high extinction risk in large mammal species. Science 309, 1239–1241. ( 10.1126/science.1116030) [DOI] [PubMed] [Google Scholar]
- 49.Malinsky M, et al. 2015. Genomic islands of speciation separate cichlid ectomorphs in an East African crater lake. Science 350, 1493–1498. ( 10.1126/science.aac9927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT. 2004. Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat. Genet. 36, 1326–1329. ( 10.1038/ng1471) [DOI] [PubMed] [Google Scholar]
- 51.Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. 2000. The evolution of mutation rates: separating causes from consequences. Bioessays 22, 1057–1066. () [DOI] [PubMed] [Google Scholar]
- 52.Broughton RE, Milam JE, Roe BA. 2001. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11, 1958–1967. ( 10.1101/gr.156801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallach M, Chandrasekaran C, Betrán E. 2010. Analyses of nuclearly encoded mitochondrial genes suggest gene duplication as a mechanism for resolving intralocus sexually antagonistic conflict in Drosophila. Genome Biol. Evol. 2, 835–850. ( 10.1093/gbe/evq069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee HY, Chou JY, Cheong L, Chang NH, Yang SY, Leu JY. 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135, 1065–1073. ( 10.1016/j.cell.2008.10.047) [DOI] [PubMed] [Google Scholar]
- 55.Salisbury CL, Seddon N, Cooney CR, Tobias JA. 2012. The latitudinal gradient in dispersal constraints: ecological specialisation drives diversification in tropical birds. Ecol. Lett. 15, 847–855. ( 10.1111/j.1461-0248.2012.01806.x) [DOI] [PubMed] [Google Scholar]
- 56.Maia R, Rubenstein DR, Shawkey MD. 2013. Key ornamental innovations facilitate diversification in an avian radiation. Proc. Natl Acad. Sci. USA 110, 10 687–10 692. ( 10.1073/pnas.1220784110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quintero I, Jetz W. 2018. Global elevational diversity and diversification of birds. Nature 555, 246–250. ( 10.1038/nature25794) [DOI] [PubMed] [Google Scholar]
- 58.Iglesias-Carrasco M, Jennions MD, Ho SYW, Duchêne DA. 2019. Data from: Sexual selection, body mass and molecular evolution interact to predict diversification in birds Dryad Digital Depository. ( 10.5061/dryad.p39d12d) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Iglesias-Carrasco M, Jennions MD, Ho SYW, Duchêne DA. 2019. Data from: Sexual selection, body mass and molecular evolution interact to predict diversification in birds Dryad Digital Depository. ( 10.5061/dryad.p39d12d) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The data used in this paper are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.p39d12d [58].