Abstract
The aims of the present study were to evaluate the presence of Salmonella in five fermented sausage processing plants and their products during the production process, and to trace the possible sources of contamination. A total of 270 samples were collected: mixture of ground pork meat and fat, products at the end of acidification, sausages at the end of ripening and, during production stages, surfaces in contact with meat and surfaces not in contact with meat. For samples of ground meat, product at the end of acidification and sausages at the end of ripening, the pH and water activity (aw), were determined. All the samples were tested for the presence of Salmonella. Thirtytwo Salmonella isolates were obtained, subjected to serotyping and PFGE. The sausages at the end of ripening pH and aw mean values were 5.39±0.24 and 0.91±0.03, respectively. Salmonella was detected in three processing plants with an overall prevalence of 16.7% in food samples and 5.8% in environmental samples. Salmonella prevalence was 24% in ground meat and products at the end of acidification and was also detected in a sample of sausage at the end of ripening (2%). In environmental samples, Salmonella was detected in 6.6% of surfaces in contact with meat and 5% of surfaces not in contact with meat. Five serotypes were identified among 32 isolates: S. Derby (37.5%), S. Typhimurium and S. Rissen (both 25%), S. Give and monophasic S. Typhimurium (both 6.25%). Six different pulsotypes were obtained with PFGE. The serotypes and the PFGE pattern of the strains were specific for each facility with no overlapping between different processing plants. The same observation can be pointed out considering different sampling days for the same processing plants, thus presumably indicating the raw material (ground pork meat and fat) as the source of contamination. The detection of Salmonella in a sample of sausage at the end of ripening highlights the ability of the pathogen to survive during manufacturing process.
Key words: Salmonella, meat products, pork, pulsotype
Introduction
Salmonella is a zoonotic pathogen present in the intestinal tract of a wide range of domestic and wild animals which may be recovered in a variety of foodstuffs, of both animal and plant origin, as consequence of either direct or indirect contamination. In 2016, 94.530 confirmed cases of human salmonellosis were reported in the European Union (EFSA and ECDC, 2017). Although a statistically significant decreasing trend of salmonellosis has been observed between 2008 and 2016, in recent years (2012-2016) the trend has stabilized and the number of reported cases and EU notification has slightly increased. Among the different food categories, a high proportion of Salmonellapositive units were reported for meat products intended to be eaten raw posing a serious risk for human health (EFSA and ECDC, 2017). In 2014, pork meat and products thereof were responsible for 9.3% of foodborne outbreaks caused by Salmonella, representing the type of meat most frequently associated with salmonellosis in humans (EFSA and ECDC, 2015). In Southern Europe countries pigs account for 43.6% of all salmonellosis cases (Pires et al., 2011).
Ready-to-eat (RTE) pork meat products have been linked to several Salmonella outbreaks: salami and sausages in Italy (Pontello et al., 1998; Luzzi et al., 2007; Scavia et al., 2013; Scaltriti et al., 2015; Andreoli et al., 2017) and north-European countries (Emberland et al., 2006; Hjertqvist et al., 2006; Kuhn et al., 2011), chorizo in Spain (Hernandez-Arricibita et al., 2015) and Denmark (Nygard et al., 2007). In 2015, an outbreak of monophasic and biphasic S. Typhimurium and S. Derby associated with the consumption of dried pork sausage was reported in Spain (Arnedo-Pena et al., 2016).
The raw materials used for the production of fermented meat products may be contaminated by Salmonella at the slaughterhouse (Pearce et al., 2004; Piras et al., 2011; Gomes-Neves et al., 2012; Bonardi et al., 2013; Fois et al., 2017), during the production process (Delhalle et al., 2009; Manios et al., 2015; D’Ostuni et al., 2016) and the post-processing stages (Barbuti and Parolari, 2002; Ferreira et al., 2009).
Traditional ripened Sardinian sausage is a typical Mediterranean-style, semi-dry, naturally fermented (without use of starter cultures) pork meat product. It is the main meat product of the meat supply chain in Sardinia (Italy), included in the national list of traditional food products (Ministerial Decree 8 September 1999, n.350; Italian Republic, 1999). Ground pork meat and pork fat are the main ingredients. The safety of fermented sausages is ensured by the combined effect of different factors, known as “hurdles” (Leistner, 2000). In fact, for the manufacturing of dry fermented sausages, carbohydrates, starter cultures and curing salts (NaCl, nitrates and nitrities) are added. The fermentation of the carbohydrates by lactic acid starter cultures leads to pH decrease to values of ca. 5.3, and the drying step decreases the water activity (aw) to values of ca. 0.920 (Christieans et al., 2018). The use of sodium chloride (NaCl) is essential, other than to solubilize proteins and emulsify fat, to control spoilage and pathogenic bacteria (Bonardi et al., 2017). Also nitrates and nitrites can be added, at maximum concentrations of 150 mg/kg each (Reg. EC 1129/2011). These additives are used due to their important role on color and flavor development and their antimicrobial activity against pathogenic bacteria such as Clostridium botulinum and Enterobacteriaceae (Hospital et al., 2014). Nevertheless, in case of initial contamination of the raw materials with high levels of pathogenic bacteria and/or insufficient control of the antimicrobial factors, the safety of these products can be compromised (Ducic et al., 2016). Survival of Salmonella in fermented meat products depends upon dry-curing and physicochemical conditions created by the hurdles. Water activity decrease is a key factor for Salmonella inactivation, but its effect depends also on contemporary pH decrease, as well as salt and nitrite concentration (Messier et al., 1989).
In Sardinian sausages mean values of pH between 5.3-5.5 and aw between 0.90-0.92 at the end of ripening indicate a correct acidification and drying processes that allow obtaining safe and stable products (Mazzette et al., 1998; Greco et al., 2005; Mangia et al., 2007). To our knowledge, there are no published data on the occurrence of Salmonella in Sardinian sausages. Therefore, the objective of the present study was to evaluate the presence of Salmonella in five fermented sausage processing plants and their products.
Materials and Methods
Features of the processing plants
The survey was conducted in five sausage processing plants (S1, S2, S3, S4 e S5) located in Sardinia (Italy) during one year. The processing plants included in the survey were representative of the regional fermented sausage production characterized by similar sausage manufacturing process (Figure 1). All the processing plants processed raw materials (fresh meat) supplied by Italian regions (Northern Italy) other than Sardinia. S1, S2 and S4 had an average production of 300-500 tons meat/year, while S3 and S5 of 50-100 tons meat/year. S5 declared to use microbial starter, consisting of lactic bacteria or staphylococci coagulase negative (MI100, Briostarter CAP, dried SH101 Texel, SA301 Milan). All plants used natural bowel (mutton or beef). The ripening step had an average duration of 16.2 days (range 9-20). Table 1 shows some of the features of the processing plants included in the study.
Figure 1.
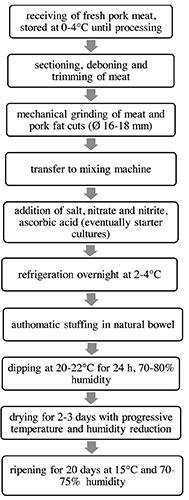
Process flow diagram of Sardinian sausage.
Table 1.
Characteristics of the processing plants included in the study.
| Production tons meat/year | Origin of the raw meat | Characteristics of the raw meat | Use of starter culture | Ripening duration (days) | |
|---|---|---|---|---|---|
| S1 | 300-500 | National | Fresh | No | 20 |
| S2 | 300-500 | National | Fresh | No | 18 |
| S3 | 50-100 | National | Fresh | No | 15 |
| S4 | 300-500 | National | Fresh | No | 15 |
| S5 | 50-100 | National | Fresh | Yes | 18 |
Samples collection
Each plant was visited twice within one month, collecting a total of 270 samples. Overall, 150 raw materials, semi-finished and finished products were collected and analyzed: n. 50 mixture of raw ground pork meat and fat (RM), n. 50 products at the end of acidification (PA, two days after stuffing) and n.50 sausages at the end of ripening (SR, n. days after stuffing variable according to the processing plant). Moreover, n. 120 environmental samples were collected by means of hydrated-sponges pre-moistened with 10 ml Buffered Peptone Water Broth (BPW, 3M Health Care, Milan, Italy): a) 60 samples from surfaces in contact with meat (SCM): work tables, trolleys, hooks, meat grinder machine, mixer, stuffing machine; b) 60 samples from surfaces not in contact with meat (SNCM): drains and a pool of walls+floor of ground meat store room, drying and ripening room. Environmental sampling was performed during the production stages. All samples were transferred to the laboratory under aseptic and refrigerated conditions in portable insulated cold-boxes. Samples were kept at 4°C and analyzed within 24h.
Chemical-physical analysis
For samples of RM, PA and SR, the pH and water activity (aw), were determined. The potentiometric measurement of pH was carried out with 10 g of sample in 1:1 ratio with sterile distilled water using a pH meter GLP 22 (Crison Instruments SA, Barcelona, Spain) directly. Water activity (aw) was determined at 25°C, using an Aqualab CX3 (Decagon, Pullman, Washington, USA).
Salmonella detection, serotyping and phage typing
All the samples were tested for the presence of Salmonella according to EN ISO 6579-1:2017. Salmonella suspect colonies were tested for biochemical properties with API® 20E micro-substrate system (bioMerieux, Marcy l’Etoile, France). A single confirmed Salmonella isolate from each positive sample was serotyped according to Kauffmann-Le Minor scheme by slide agglutination with O and H antigen specific sera (Staten Serum Institute, Copenhagen, Denmark). Definitive identification of S. enterica ser. 4,[5],12:i:- was obtained by a previously described PCR protocol (Barco et al., 2011). Isolates identified as S. Typhimurium and the monophasic variant of S. Typhimurium were phagetyped according to standard methods (Anderson et al., 1977) and following the interpretative guidelines defined for S. Typhimurium by the international federation for enteric phage typing (IFEPT, International Federation for Enteric Phage Typing, Colindale, London, United Kingdom).
Pulsed field gel electrophoresis
Pulsed Field Gel Electrophoresis PFGE typing was performed according SalmGene protocol by Peters et al. (2003) with XbaI (Invitrogen, Carlsbad, CA, USA) restriction of DNA. The comparison of PFGE patterns was made using the BioNumerics software v7.1 (Applied Maths, Saint-Martens-Platen, Belgium). Cluster analysis was performed using the Dice similarity coefficient, with 0.5% optimization and 1.5% tolerance, and the unweighted pair group method with arithmetic mean (UPRMA).
Results
Chemical-physical analysis
The pH values (mean±s.d.) of RM, PA and SR were 5.79±0.16, 5.57±0.28 and 5.39±0.24, respectively. The aw values (mean±s.d.) were 0.97±0.02, 0.95±0.01 and 0.91±0.03 in RM, PA and SR, respectively.
Table 2 shows the physico-chemical parameters of sausages at the end of ripening and ripening duration (days) in the different facilities.
Table 2.
Distribution of Salmonella serotypes and pulsotypes in samples of ground pork meat and fat, product at the end of acidification, sausages at the end of ripening and environmental samples of the processing plants. For samples of sausages at the end of ripening physico-chemical parameters and ripening duration (days) are shown.
| Plant | Sampling day | GM | PA | SR | SCM | SNCM | |||
|---|---|---|---|---|---|---|---|---|---|
| Salmonella serotype/pulsotype | Salmonella serotype/pulsotype | Salmonella serotype/pulsotype | pH mean (range) | aw mean (range) | Ripening duration, days | Salmonella serotype/pulsotype | Salmonella serotype/pulsotype | ||
| S1 | 1 | Typhimurium /Typh1 | Typhimurium /Typh1 | - | 5.46 (5.38-5.56) | 0.895 (0.890-0.893) | 20 | Typhimurium/Typh1 | - |
| 2 | Rissen/Riss1 | Rissen /Riss1 | - | 5.44 (5.38-5.51) | 0.902 (0.899-0.909) | 19 | - | Rissen/Riss1 | |
| S2 | 1 | - | - | - | 5.70 (5.49-6.20) | 0.901 (0.897-0.900) | 20 | - | - |
| 2 | - | - | - | 5.32 (5.10-5.43) | 0.909 (0.901-0.919) | 14 | - | - | |
| S3 | 1 | - | - | - | 5.44 (5.39-5.47) | 0.902 (0.900-0.905) | 18 | - | - |
| 2 | Give/Give1 | Derby/Der1 | Derby/Der1 | 5.39 (5.10-5.49) | 0.922 (0.914-0.941) | 14 | Derby/Der1 | Derby/Der1 | |
| S4 | 1 | 4,[5],12:i:—/Monof1 | 4,[5],12:i:—/Monof1 | - | 5.25 (5.05-5.82) | 0.945 (0.939-0.950) | 9 | - | - |
| 2 | Derby/Der2 | Derby/Der2 | - | 4.94 (4.91-4.96) | 0.945 (0.938-0.950) | 13 | - | - | |
| S5 | 1 | - | - | - | 5.63 (5.50-5.81) | 0.860 (0.818-0.889 | 21 | - | - |
| 2 | - | - | - | 5.39 (5.29-5.53) | 0.918 (0.901-0.927) | 14 | - | - | |
RM: mixture of ground pork meat and fat; PA: product at the end of acidification; SR: sausages at the end of ripening; SC: surfaces in contact with meat; SNC: surfaces not in contact with meat.
Salmonella prevalence
Salmonella was detected in three out of five processing plants (S1, S3 and S4), with an overall prevalence of 16.7% (25/150) in semi-finished and finished samples and 5.8% (7/120) in environmental samples (Table 3). Different processing plants showed different prevalence: S1 showed the highest prevalence (16/54; 29.6%), followed by S3 (10/54; 18.5%) and S4 (6/54; 11.1%). Between food samples, RM and PA, showed the highest prevalence (both 12/50; 24%) followed by SR (1/50; 2%). Among environmental samples, prevalence of 6.6% (4/60) and 5% (3/60) were detected, respectively in SCM and SNCM. At S1, Salmonella was detected from RM (60%; 6/10) and PA (60%; 6/10) during both sampling days. As regard to environmental samples, at S1 Salmonella was detected during the first sampling day from SCM (2/5; 40%: trolley and work table), and during the second sampling day from SNCM (2/6; 33%: drain and pool floor + walls of drying room). At S3, Salmonella was detected only during the second sampling day, from RM (20%; 2/10), PA (40%; 4/10) and also from a sample of SR (10%; 1/10). Moreover, during the same sampling day, Salmonella was detected from SCM (2/5; 40%: work table and meat grinder machine) and SNCM (20%; 1/6: floor + walls of ripening room). Finally, at S4 Salmonella was detected during both sampling days, from samples of RM, (40%; 4/10) and PA (20%; 2/10).
Table 3.
Prevalence of Salmonella in mixture of ground pork meat and fat, product at the end of acidification, sausages at the end of ripening and environments of sausages processing plants.
| Plant | RM | PA | SR | SCM | SNCM | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive/total | % | Positive/total | % | Positive/total | % | Positive/total | % | Positive/total | % | |
| S1 | 6/10 | 60 | 6/10 | 60 | 0/10 | 0 | 2/12 | 16.7 | 2/12 | 16.7 |
| S2 | 0/10 | 0 | 0/10 | 0 | 0/10 | 0 | 0/12 | 0 | 0/12 | 0 |
| S3 | 2/10 | 20 | 4/10 | 40 | 1/10 | 10 | 2/12 | 16.7 | 1/12 | 8.3 |
| S4 | 4/10 | 40 | 2/10 | 20 | 0/10 | 0 | 0/12 | 0 | 0/12 | 0 |
| S5 | 0/10 | 0 | 0/10 | 0 | 0/10 | 0 | 0/12 | 0 | 0/12 | 0 |
| Total | 12/50 | 24 | 12/50 | 24 | 1/50 | 2 | 4/60 | 6.6 | 3/60 | 5 |
RM: mixture of ground pork meat and fat; PA: product at the end of acidification; SR: sausages at the end of ripening; SC: surfaces in contact with meat; SNC: surfaces not in contact with meat.
Salmonella serotyping and phage typing
Five different serotypes were identified among 32 S. enterica isolates (Table 4). S. enterica ser. Derby was the most prevalent (37.5%; 12/32), followed by S. Typhimurium and S. Rissen (both 25%; 8/32). Finally, S. Give and monophasic variant of S. Typhimurium (antigenic formula 4, [5], 12:i-) accounted both for two isolates (6.25%).
Table 4.
Prevalence of Salmonella serotypes in samples of ground pork meat and fat, product at the end of acidification, sausages at the end of ripening and environmental samples.
| Salmonella serovars | RM | PA | SR | SCM | SNCM | Total (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S3 | S4 | S1 | S3 | S4 | S1 | S3 | S4 | S1 | S3 | S4 | S1 | S3 | S4 | ||
| Derby | 3 | 4 | 1 | 1 | 2 | 1 | 12 (37.4) | |||||||||
| Typhimurium DT 193 | 3 | 3 | 2 | 8(25) | ||||||||||||
| Rissen | 3 | 3 | 2 | 8(25) | ||||||||||||
| Give | 2 | 2 (6.3) | ||||||||||||||
| 4,[5], 12:i:– (untypable) | 1 | 1 | 2 (6.3) | |||||||||||||
| Total | 6 | 2 | 4 | 6 | 4 | 2 | 1 | 2 | 2 | 2 | 1 | 32(100) | ||||
RM: mixture of ground pork meat and fat; PA: product at the end of acidification; SR: sausages at the end of ripening; SC: surfaces in contact with meat; SNC: surfaces not in contact with meat.
Phage typing of S. Typhimurium isolates resulted into DT193, while S. Typhimurium monophasic variant were untypable.
In samples of RM, the following serotypes were identified: S. Derby (n.3), S. Typhimurium (n.3), S. Rissen (n.3), S. Give (n.2) and S. Typhimurium monophasic variant (n.1). In PA S. Derby (n.5), S. Typhimurium (n.3), S. Rissen (n.3) and S. Typhimurium monophasic variant (n.1) were identified. In SR, only S. Derby was identified. As regard to environmental samples, in SCM S. Derby (n.2) and S. Typhimurium (n.2) were detected and the same serotypes were identified in SNCM (n.1 for S. Derby and n.2 for S. Typhimurium).
Pulsed field gel electrophoresis
Six different pulsotypes were identified. Among S. Derby isolates, it was possible to identify two different pulsotypes (Der1 and Der2). Among the other serotypes, only one pulsotype was detected.
Table 2 shows the distribution of Salmonella serotypes and pulsotypes in food and environmental samples.
Discussion
This study showed the overall prevalence of Salmonella enterica in different samples of Sardinian fermented sausages and processing plants environment.
Our results on Salmonella prevalence in samples of raw ground pork meat and fat (24%) were higher than those reported by other authors that observed a prevalence between 4.3% (Delhalle et al., 2009) and 14% (Bonardi et al., 2017). Moreover, this prevalence was higher than those detected in pig carcasses at slaughterhouse where prevalence between 7 and 16% were detected (Pearce et al., 2004; Vieira-Pinto et al., 2005; De Busser et al., 2011), and fresh pig meat where mean prevalence of 3-5% were observed (Zhao et al., 2001; Busani et al., 2005; Hansen et al., 2010).
S. Derby was the most prevalent serotype (37.4%). This result was expected as this serotype is well known to be adapted to pigs (Matiasovic et al., 2014). In fact, it is one of the most common in slaughter pigs in Italy (Bonardi et al., 2003 and 2016; Piras et al., 2011 and 2014; Fois et al., 2017; Pesciaroli et al., 2017) and other European countries (Nollet et al., 2004; Hauser et al., 2011; Gomes-Neves et al., 2012; Kerouanton et al., 2013; Sanchez- Rodriguez et al., 2018). Moreover, S. Derby is between the top five most commonly reported serovars in human salmonellosis acquired in EU during 2016 (EFSA and ECDC, 2017) and it has also been recently associated to outbreaks linked to pork meat products (Arnedo-Pena et al., 2016)
S. Typhimurium has been identified in samples of ground meat, sausages at the end of acidification and environmental samples (SCM). Also this serotype, which is the second most reported in human salmonellosis, has been increasingly described both in slaughter pigs (Arguello, et al., 2013; Van Damme et al., 2018; Morales-Partera et al., 2018) and in cases of human salmonellosis attributed to pork meat and meat products (Luzzi et al., 2007; Andreoli et al., 2017).
Serotyping and PFGE allowed to presumably indicate the raw materials (pork meat and fat) as the primary source of contamination of Sardinian sausages. In fact, the serovar and the PFGE patterns of the strains were specific for each facility with no exchange between different abattoirs. The same observation can be pointed out considering different sampling days for the same slaughterhouse. This result seems to demonstrate the effective application of cleaning and disinfection procedures that do not allow to the pathogen to become part of the resident flora in the investigated processing plants.
The evolution of pH and aw detected in our survey during the process was similar to that described for other Italian typical fermented meat products (Bonardi et al., 2017; Montanari et al., 2018). Aw mean values between 0.89 and 0.91 in Sardinian semidry fermented sausage are usually detected after 20 days of ripening (Mazzette et al., 1998; Meloni et al., 2014). In the same conditions, pH at the end of ripening is between 5.03 and 5.28 (Greco et al., 2005; Mangia et al., 2007). These conditions, along with NaCl concentration, should allow to obtain shelf-stable and safe meat products.
It is known that Salmonella is able to growth within a broad range of pH (4.1-9) and with minimum aw value of 0.95 (Cox et al., 2000). Messier et al. (1989) showed that in fermented meat products with NaCl % between 2 and 3.3 and pH < 4.9 Salmonella survival is inhibited with aw between 0.93 and 0.96.
We isolated Salmonella in a sample of sausage at the end of ripening that had been dried for 13 days and showed a pH of 4.92 and an aw value of 0.950, thus sufficient to allow Salmonella survival and growth. This can be the consequence of an empirical application of the hurdle’s technology in the manufacturing of traditional products like Sardinian sausages that, most often when consumers demand increases (e.g. during Christmas or Easter time), leads to a shortage of the processing steps, particularly of the ripening, not allowing to obtain a correct drying of the product.
Conclusions
Reduction of Salmonella contamination of raw materials used for sausage production should be seen as a multicomponent action along the pork production chain, including lowering of infection prevalence in swine population and adoption of proper hygiene procedures during slaughter and meat processing. The presence of Salmonella in the ripened Sardinian sausages confirms that the hurdles of microbial growth (mainly the aw decrease) should be properly applied during the ripening in order to prevent the survival of Salmonella that can contribute to the consumer exposure.
Funding Statement
Funding: none.
References
- Anderson ES, Ward LR, De Saxe MJ, De Sa JD. 1977. Bacteriophage-typing designations of Salmonella Typhimurium. J Hyg (London) 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoli G, Merla C, Valle CD, Corpus F, Morganti M, D’incau M, Colmegna S, Marone P, Fabbi M, Barco L, Carra E, 2017. Foodborne salmonellosis in Italy: characterization of Salmonella enterica serovar Typhimurium and monophasic variant 4,[5],12:i-isolated from salami and human patients. J Food Protect 14:632-9. [DOI] [PubMed] [Google Scholar]
- Arguello H, Carvajal A, Naharro G, Arcos M, Rodicio MR, Martin MC, Rubio P, 2013. Sero and genotyping of Salmonella in slaughter pigs, from farm to cutting plant, with a focus on the slaughter process. Int J Food Microbiol 161:44-52. [DOI] [PubMed] [Google Scholar]
- Arnedo-Pena A, Sabater-Vidal S, Herrera- León S, Bellido-Blasco JB, Silvestre- Silvestre E, Meseguer-Ferrer N, Yague-Muñoz A, Gil-Fortuño M, Romeu-García A, Moreno-Muñoz R, 2016. An outbreak of monophasic and biphasic Salmonella Typhimurium, and Salmonella Derby associated with the consumption of dried pork sausage in Castellon (Spain). Enferm Infec Micr Cl 34:544-50. [DOI] [PubMed] [Google Scholar]
- Barbuti S, Parolari G, 2002. Validation of manufacturing process to control pathogenic bacteria in typical dry fermented product. Meat Sci 62:323-9. [DOI] [PubMed] [Google Scholar]
- Barco L, Lettini AA, Ramon E, Longo A, Saccardin C, Dalla Pozza MC, Ricci A, 2011. A rapid and sensitive method to identify and differentiate Salmonella enterica serotype Typhimurium and Salmonella enterica serotype 4,[5],12:i:— by combining traditional serotyping and multiplex polymerase chain reaction. Foodborne Pathog Dis 8:741-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi S, Brindani F, Pizzin G, Lucidi L, D’Incau M, Liebana E, Morabito S, 2003. Detection of Salmonella spp., Yersinia enterocolitica and verocytotoxin- producing Escherichia coli O157 in pigs at slaughter in Italy. Int J Food Microbiol, 85:101-10. [DOI] [PubMed] [Google Scholar]
- Bonardi S, Bassi L, Brindani F, D’Incau M, Barco L, Carra E, Pongolini S, 2013. Prevalence, characterization and antimicrobial susceptibility of Salmonella enterica and Yersinia enterocolitica in pigs at slaughter in Italy. Int J Food Microbiol 163:248-57. [DOI] [PubMed] [Google Scholar]
- Bonardi S, Bruini I, Bolzoni L, Cozzolino P, Pierantoni M, Brindani F, Bellotti P, Renzi M, Pongolini S, 2017. Assessment of Salmonella survival in dry-cured Italian salami. Int J Food Microbiol 262:99-106. [DOI] [PubMed] [Google Scholar]
- Busani L, Cigliano A, Taioli E, Caligiuri V, Chiavacci L, Di Bella C, 2005. Prevalence of Salmonella enterica and Listeria monocytogenes contamination in foods of animal origin in Italy. J Food Protect 68:729-33. [DOI] [PubMed] [Google Scholar]
- Christieans S, Picgirard L, Parafita E, Lebert A, Gregori T, 2018. Impact of reducing nitrate/nitrite levels on the behavior of Salmonella Typhimurium and Listeria monocytogenes in French dry fermented sausages. Meat Sci 137:160-7. [DOI] [PubMed] [Google Scholar]
- Cox J. Salmonella. Robinson KR, Batt CA, Pradip DP. Encyclopedia of food microbiology, vol III, Ed. Academic Press, 2000. [Google Scholar]
- De Busser EV, Maes D, Houf K, Dewulf J, Imberechts H, Bertrand S, De Zutter L, 2011. Detection and characterization of Salmonella in lairage, on pig carcasses and intestines in 5 slaughterhouses. Int J Food Microbiol 145:279-86. [DOI] [PubMed] [Google Scholar]
- Delhalle L, Saegerman C, Farnir F, Korsak N, Maes D, Messens W, 2009. Salmonella surveillance and control at post-harvest in the Belgian pork meat chain. J Food Protect 26:265-71. [DOI] [PubMed] [Google Scholar]
- D’Ostuni V, Tristezza M, De Giorgi MG, Rampino P, Grieco F, Perrotta C, 2016. Occurrence of Listeria monocytogenes and Salmonella spp. in meat processed products from industrial plants in Southern Italy. Food Control 62:104-9. [Google Scholar]
- Ducic M, Klisara N, Markov S, Blagojevic B, Vidakovic A, Buncic S. 2016. The fate and pasteurization-based inactivation of Escherichia coli O157 Salmonella Typhimurium and Listeria monocytogenes in dry fermented sausages. Food Control 59:400-6. [Google Scholar]
- Emberland KE, Nygård K, Heier BT, Aavitsland P, Lassen J, Stavnes TL, Gondrosen B, 2006. Outbreak of Salmonella Kedougou in Norway associated with salami, April–June 2006. Eurosurveillance 11:E060706.3. [DOI] [PubMed] [Google Scholar]
- European Commission (2011). Commission regulation (EU) n° 1129/2011 of 11 November 2011 amending Annex II to Regulation (EC) n° 1333/2008 of the European Parliament and of the Council by establishing a Union list of food additives. Official Journal of the European Union, L295, 1-177. [Google Scholar]
- EFSA (European Food Safety Agency) and ECDC (European Centre for Disease Prevention and Control), 2016. The European Union summary report on trends of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J 4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control), 2017. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J 5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira V, Barbosa J, Silva J, Gibbs P, Hogg T, Teixeira P, 2009. Microbiological profile of Salpicão de Vinhais and Chouriça de Vinhais from raw materials to final products: Traditional dry sausages produced in the north of Portugal. Innov Food Sci Emerg 10:279-83. [DOI] [PubMed] [Google Scholar]
- Fois F, Piras F, Torpdahl M, Mazza R, Consolati SG, Spanu C, Scarano C, De Santis EPL, 2017. Occurrence, characterization, and antimicrobial susceptibility of Salmonella enterica in slaughtered pigs in Sardinia. J Food Sci 82:969-76. [DOI] [PubMed] [Google Scholar]
- Gomes-Neves E, Antunes P, Tavares A, Themudo P, Cardoso M, Gartner F, Costa JM, Peixe L, 2012. Salmonella cross-contamination in swine abattoirs in Portugal: carcasses, meat, and meat handlers. Int J Food Microbiol 157:82-7. [DOI] [PubMed] [Google Scholar]
- Greco M, Mazzette R, De Santis EPL, Corona A, Cosseddu AM, 2005. Evolution and identification of lactic acid bacteria isolated during the ripening of Sardinian sausages. Meat Sci 69:733-9. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Christensen BB, Aabo S, 2010. Salmonella in pork cuttings in supermarkets and butchers’ shops in Denmark in 2002 and 2006. Zoonoses Public Hlth 57:23-9. [DOI] [PubMed] [Google Scholar]
- Hauser E, Hebner F, Tietze E, Helmuth R, Junker E, Prager R, Schroeter A, Rabsch W, Fruth A, Malorny B, 2011. Diversity of Salmonella enterica serovar Derby isolated from pig, pork and humans in Germany. Int J Food Microbiol 151:141-9. [DOI] [PubMed] [Google Scholar]
- Hernandez-Arricibita E, Santamaría Zuazua R, Ramos-López G, Herrera-León S, Karkamo-Zuñeda JA, Muniozguren-Zuazua N., 2016. Outbreak of Salmonella Typhimurium infections associated with consumption of chorizo in Bizkaia. Enferm Infec Micr Cl 34:577-8. [DOI] [PubMed] [Google Scholar]
- Hjertqvist M, Luzzi I, Löfdahl S, Olsson A, Rådal J, Andersson Y, 2006. Unusual phage pattern of Salmonella Typhimurium isolated from Swedish patients and Italian salami. Eurosurveillance 11:E060209.3. [DOI] [PubMed] [Google Scholar]
- Hospital XF, Hierro E, Fernández M, 2014. Effect of reducing nitrate and nitrite added to dry fermented sausages on the survival of Salmonella Typhimurium. Food Res Int 62:410-5. [Google Scholar]
- Italian Republic, 1999. Ministerial Decree 8 September 1999, n.350. [Regolamento recante norme per l’individuazione dei prodotti tradizionali di cui all’articolo 8, comma 1, del decreto legislativo 30 aprile1998, n. 173]. [Regulation in Italian]. In: Official Journal, 12 October 1999 n. 240. [Google Scholar]
- Kerouanton A, Rose V, Weill FX, Granier SA, Denis M, 2013. Genetic diversity and antimicrobial resistance profiles of Salmonella enterica serotype Derby isolated from pigs, pork, and humans in France. Foodborne Pathog Dis 10:977-84. [DOI] [PubMed] [Google Scholar]
- Kuhn K, Torpdahl M, Frank C, Sigsgaard K, Ethelberg S, 2011. An outbreak of Salmonella Typhimurium traced back to salami, Denmark, April to June 2010. Eurosurveillance 12:16. [PubMed] [Google Scholar]
- Luzzi I, Galetta P, Massari M, Rizzo C, Dionisi AM, Filetici E, 2007. An Easter outbreak of Salmonella Typhimurium DT 104A associated with traditional pork salami in Italy. Eurosurveillance 12:149-52. [DOI] [PubMed] [Google Scholar]
- Manios SG, Grivokostopoulos NC, Bikouli VC, Doultsos DA, Zilelidou EA, Gialitaki MA, 2015. A 3-year hygiene and safety monitoring of a meat processing plant which uses raw materials of global origin. Int J Food Microbiol 209:60-9. [DOI] [PubMed] [Google Scholar]
- Matiasovic J, Stepanova H, Kudlackova H, Havlickova H, Sisak F, Rychlik I, Chlebova K, Faldyna M, 2014. Immune response of pigs to Salmonella enterica serovar Derby and Typhimurium infections. Vet Microbiol 170:284-90. [DOI] [PubMed] [Google Scholar]
- Mangia NP, Murgia MA, Garau G, Deiana P, 2007. Microbiologia e valutazione igienico-sanitaria della salsiccia sarda. Ind Aliment Italy 46:533-6. [Google Scholar]
- Mazzette R, De Santis EPL, Greco M, Bean V, Pisanu S, 1998. Critical Control Points (CCPs) in processing ripened sausages. In Proceedings of the 52nd national congress of Italian Society of Veterinary Science (SISVet) (pp. 391– 392). Silvi Marina (TE), Italy. [Google Scholar]
- Meloni D, Consolati SG, Mazza R, Mureddu A, Fois F, Piras F, Mazzette R, 2014. Presence and molecular characterization of the major serovars of Listeria monocytogenes in ten Sardinian fermented sausage processing plants. Meat Sci 97:443-50. [DOI] [PubMed] [Google Scholar]
- Meloni D, Galluzzo P, Mureddu A, Piras F, Griffiths M, Mazzette R, 2009. Listeria monocytogenes in RTE foods marketed in Italy: Prevalence and automated EcoRI ribotyping of the isolates. Int J Food Microbiol 129:166-73. [DOI] [PubMed] [Google Scholar]
- Messier S, Smith HJ, Tittiger F, 1989. Survival of Salmonella typhimurium and Staphylococcus aureus in Genoa salami of varying salt concentration. Can J Vet Res 53:84-6. [PMC free article] [PubMed] [Google Scholar]
- Ministerial Decree (MD) (2000). Decreto Ministeriale n. 130, 18.07.2000. Elenco nazionale dei prodotti agroalimentari tradizionali. In Gazzetta Ufficiale 194, 21.08.2000. [Google Scholar]
- Montanari C, Gatto V, Torriani S, Barbieri F, Bargossi E, Lanciotti R, Grazia L, Magnani R, Tabanelli G, Gardini F, 2018. Effects of the diameter on physico- chemical, microbiological and volatile profile in dry fermented sausages produced with two different starter cultures. Food Biosc 22:9-18. [Google Scholar]
- Morales-Partera AM, Cardoso-Toset F, Luque I, Astorga RJ, Maldonado A, Herrera-León S, Hernández M, Gómez-Laguna J, Tarradas C, 2018. Prevalence and diversity of Salmonella spp., Campylobacter spp., and Listeria monocytogenes in two free-range pig slaughterhouses. Food Control 92:208-15. [Google Scholar]
- Nollet N, Maes D, De Zutter L, Duchateau L, Houf K, Huysmans K, Imberechts H, Geers R, de Kruif A, Van Hoof J, 2004. Risk factors for the herd-level bacteriologic prevalence of Salmonella in Belgian slaughter pigs. Prev Vet Med 65:63-75. [DOI] [PubMed] [Google Scholar]
- Nygard K, Lindstedt BA, Wahl W, Jensvoll L, Kielso C, Molback K, 2007. Outbreak of Salmonella Typhimurium infection traced to imported cured sausage using MLVA-subtyping. Eurosurveillance 12:86-7. [DOI] [PubMed] [Google Scholar]
- Pearce RA, Bolton DJ, Sheridan JJ, McDowell DA, Blair IS, Harrington D, 2004. Studies to determine the critical control points in pork slaughter hazard analysis and critical control points system. Int J Food Microbiol 90:331-9. [DOI] [PubMed] [Google Scholar]
- Pesciaroli M, Cucco L, De Luca S, Massacci FR, Maresca C, Medici L, Paniccià M, Scoccia E, Staffolani M, Pezzotti G, Magistrali CF, 2017. Association between pigs with high caecal Salmonella loads and carcass contamination. Int J Food Microbiol 242:82-6. [DOI] [PubMed] [Google Scholar]
- Peters TM, Maguire C, Threlfall EJ, Fisher IS, Gill N, Gatto AJ, 2003. Salm-gene project, 2003. The Salm-gene project — a European collaboration for DNA fingerprinting. Eurosurveillance 8:46-50. [DOI] [PubMed] [Google Scholar]
- Piras F, Brown DJ, Meloni D, Mureddu A, Mazzette R, 2011. Investigation of Salmonella enterica in Sardinian slaughter pigs: prevalence, serotype, and genotype characterization. Int J Food Microbiol 151:201-9. [DOI] [PubMed] [Google Scholar]
- Piras F, Fois F, Mazza R, Putzolu M, Delogu ML, Lochi PG, Pani SP, Mazzette R, 2014. Salmonella prevalence and microbiological contamination of pig carcasses and slaughterhouse environment. Ital J Food Saf 3:210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires SM, de Knegt L, Hald T, 2011. Estimation of the relative contribution of different food and animal sources to human Salmonella infections in the European Union. Question No EFSAQ- 2010-00685. Published as an External Scientific Report on 28 July 2011. Available from: http//:www.efsa.europa.eu/en/supporting/pub/184e.htm. [Google Scholar]
- Pontello M, Sodano L, Nastasi A, Mammina C, Astuti M, Domenichini M, Gerosa E, Montagna A, 1998. A community-based outbreak of Salmonella enterica serotype typhimurium associated with salami consumption in Northern Italy. Epidemiol Infect 120: 209-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Rodríguez JA, Navas L, Vinuesa FM, Castells C, Martínez MA, Lopez A, Lindez B, Cabrera-Vique C, 2018. New insights on the risk factors associated with the presence of Salmonella on pig carcasses. Lessons from small slaughterhouses. Food Control 87:46-52. [Google Scholar]
- Scaltriti E, Sassera D, Comandatore F, Morganti M, Mandalari C, Gaiarsa S, Bandi C, Zehender G, Bolzoni L, Casadei G, Pongolini S, 2015. Differential single nucleotide polymorphism- based analysis of an outbreak caused by Salmonella enterica serovar Manhattan reveals epidemiological details missed by standard pulsed-field gel electrophoresis. J Clin Microbiol 53:1227-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavia G, Ciaravino G, Luzzi I, Lenglet A, Ricci A, Barco L, Pavan A, Zaffanella F, Dionisi AM, 2013. A multistate epidemic outbreak of Salmonella Goldcoast infection in humans, June 2009 to March 2010: the investigation in Italy. Eurosurveillance 18:204-24. [DOI] [PubMed] [Google Scholar]
- Van Damme I, Mattheus W, Bertrand S, De Zutter L, 2018. Quantification of hygiene indicators and Salmonella in the tonsils, oral cavity and rectal content samples of pigs during slaughter. Food Microbiol 71:120-8. [DOI] [PubMed] [Google Scholar]
- Vieira-Pinto M, Temudo P, Martins C, 2005. Occurrence of Salmonella in the ileum, ileocolic lymph nodes, tonsils, mandibular lymph nodes, and carcasses of pigs slaughtered for consumption. J Vet Med B 52:476-81. [DOI] [PubMed] [Google Scholar]
- Zhao C, Ge B, de Villena J, Sudler R, Yeh E, Zhao S, White DG, Wagner D, Meng J, 2001. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the Greater Washington, D.C., area. Appl Environ Micro 67:5431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


