Abstract
Objective
In previous studies of associations of variants in the genes UCP2, UCP3, PPARG2, CART, GRL, MC4R, MKKS, SHP, GHRL, and MCHR1 with obesity, we have used a case-control approach with cases defined by a threshold for BMI. In the present study, we assess the association of seven abdominal, peripheral, and overall obesity phenotypes, which were analyzed quantitatively, and thirteen candidate gene polymorphisms in these ten genes in the same cohort.
Methods
Obese Caucasian men (n = 234, BMI ≧31.0 kg/m2) and a randomly sampled non-obese group (n = 323), originally identified at the draft board examinations, were re-examined at median ages of 47.0 or 49.0 years by anthropometry and DEXA scanning. Obesity phenotypes included BMI, fat body mass index, waist circumference, waist for given BMI, intra-abdominal adipose tissue, hip circumference and lower body fat mass (%). Using logistic regression models, we estimated the odds for defined genotypes (dominant or recessive genetic transmission) in relation to z-scores of the phenotypes.
Results
The minor (rare) allele for SHP 512G>C (rs6659176) was associated with increased hip circumference. The minor allele for UCP2 Ins45bp was associated with increased BMI, increased abdominal obesity, and increased hip circumference. The minor allele for UCP2 –866G>A (rs6593669) was associated with borderline increased fat body mass index. The minor allele for MCHR1 100213G>A (rs133072) was associated with reduced abdominal obesity. None of the other genotype-phenotype combinations showed appreciable associations.
Conclusion
If replicated in independent studies with focus on the specific phenotypes, our explorative studies suggest significant associations between some candidate gene polymorphisms and distinct obesity phenotypes, predicting beneficial and detrimental effects, depending on compartments for body fat accumulation.
Key Words: Obesity, Body composition, Body fat distribution, Genetics, Case-cohort study
Introduction
Genetic susceptibility to common diseases may include common variants in many genes with relatively small effects [1, 2]. Several studies have tried to elucidate the mechanism behind genetic predisposition to common obesity, which is polygenic and probably heterogeneous [2, 3]. Although, mutations in specific genes explain a few rare cases of monogenic forms of human obesity, no specific gene variants responsible for the more common forms of obesity have consistently shown association so far, except for the robust association between the FTO gene and BMI that was recently discovered and replicated in genome-wide association studies [4, 5]. Previous association studies on single nucleotide polymorphisms (SNPs) and obesity have mainly addressed obesity in case-control settings with cases defined by a threshold in BMI [3], which is also the case in our cohort. We have reported on a panel of polymorphisms in relation to obesity defined by a BMI threshold [6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19], and except for one polymorphism in MC4R (Tyr35Ter) [14], none of the SNPs showed any association with the obesity phenotype under study.
However, obesity examinations should not be limited to BMI, and not to a case versus control comparison, neglecting the quantitative feature of the phenotype. We suggest that the lack of significant associations between gene variants and BMI is in part related to the fact that it is a mixed phenotype and that the quantitative feature is not exploited. It is possible to identify specific and distinct obesity phenotypes. Thus, several proxy markers of abdominal obesity are available such as waist circumference, waist-to-hip ratio, waist circumference for given BMI and sagittal abdominal diameter. In previous studies of obesity, all of these markers have proven to be more qualified markers for the adverse health effects of obesity than BMI [20, 21, 22, 23]. Moreover, the peripheral obesity has been associated with protective effects for several adverse health outcomes [24, 25, 26]. A recent study showed that, when mutually adjusted, waist and hip circumferences show opposite associations with all-cause mortality [27]. We hypothesize that the different prognostic effects of body fat accumulation markers imply differences in etiology, including genetic predisposition. This is supported by analysis of obesity phenotypes in twin studies, which show a genetic correlation of less than one that indicates only partly overlap between the genes determining these phenotypes [28, 29]. In some of our previous studies, we did include analyses of waist and/or waist-to-hip ratios in relation to some gene variants, but the quantitative feature throughout the population range was not used in the analyses. A deeper phenotypic exploration of gene polymorphisms is warranted. Thus, this paper aims to systematically examine and compare the association between seven specific abdominal, peripheral and overall obesity phenotypes, analyzed as quantitative traits throughout a broad range of obesity, with a panel of putative obesity-related candidate-gene variants that have been previously analyzed in relation to BMI in a case-control setting in order to elucidate effects of the gene variants on different aspects of body fat distribution.
Patients and Methods
Study Population
The study population consisted of Danish men originally identified from the mandatory draft board examinations of approximately 360,000 men at a median age of 19 years in the metropolitan area of Copenhagen and surrounding counties from year 1943–1977. Two groups were selected from this study population: one group of all obese men with a BMI >31 kg/m2 (n = 1,930), corresponding to 35% overweight according to a national standard scale in use when the obese sample was identified, and a randomly selected control group (n = 3,601). The non-obese group was selected as 1% random sample of all men at draft board examination in the same region and time period. The prevalence of obesity was thus as defined 1,930/360,000 = 0.54%, which means that all obese were above the 99th percentile of BMI in this population.
All men in the obese cohort and half of the randomly selected group, who were alive, under 65 years of age at the initiation of the survey, and still residing within the same region, were invited to participate in a thorough re-examination in 1998–2000. In total 234 of the obese and 323 of the controls participated and underwent anthropometry and examination of body composition. The case-cohort study design implies that the cohort of the randomly selected group of 323 non-obese participants is representative of approximately 64,600 young Danish men (control group of 323 subjects times 200, since they were originally a 1% random sample of which half were invited to the follow-up study). The obese group consisted only of subjects who at the draft board examination were above the 99th percentile in this segment of the study population.
Anthropometric Measurements and Body Composition at the Follow-Up Examination
Height was measured without shoes, to the nearest 0.5 cm, with the participants standing against a wall-mounted stadiometer. Body weight was measured on an electronic scale (Lindell 8000, Kristianstad, Sweden) to the nearest 0.05 kg with the subjects standing in their underwear and without shoes. BMI (kg/m2) at re-examination was calculated as weight (kg) divided by height (m) squared. Waist circumference was measured to the nearest 0.5 cm with the subjects standing, using a nonexpendable linen tape measure, according to WHO recommendations [30]. The measurement of waist was repeated three times, and the mean was calculated. Fat mass (FM; kg) of the entire skeleton and truncal region was evaluated by total body scanning employing dual-energy X-ray absorptiometry (DEXA; Lunar DXA-IQ DEXA, Madison, WI, USA). All scans were performed in slow mode and analyzed using Lunar smart scan version 4.6c with the slowest scan mode [31].
Definition and Measurement of Obesity Phenotypes
Fat mass in percent of total tissue mass in the particular region was calculated [32] and converted to a fat body mass index (FBMI; kg/m2), which was calculated as total body fat mass (kg), derived from the DEXA scan, divided by height (m) squared.
Waist and BMI measure different aspects of body fatness but it has also been suggested that BMI and waist independently contribute to the prediction of abdominal fat [33, 34]. As others, we believe that waist conditioned by BMI represent an obesity phenotype separate from waist or BMI [34, 35]. Calculating the residuals from a linear regression model with waist as the dependent and BMI as the independent variable assessed the variation in waist for given BMI. Intra-abdominal adipose tissue (IAAT, cm2) was calculated from DEXA scans and anthropometry using the equation [36]: y = –208.2+ 4.62 (sagittal diameter, cm) + 0.75 (age, years) + 1.73 (waist, cm) + 0.78 (trunk fat, %).
Hip circumference and lower body fat mass (%) (LBFM (%)) were included as indicators of peripheral obesity phenotypes. LBFM (%) is derived from DEXA scans and defined as the fat mass (%) distributed at the gluteofemoral region, including buttocks, thighs and lower legs [32, 37].
Altogether, seven obesity phenotypes were analyzed in the present study: BMI, FBMI, waist circumference, waist for given BMI, IAAT, LBFM (%), and hip circumference.
Selection and Genotyping of Variants
The selection of gene variants in our previous publications [6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19] was based on their putative contribution to obesity phenotypes [2, 3], with known or assumed association to hypothalamic regulation of appetite, efficiency of energy expenditure, regulation of adipocyte differentiation and function, and lipid and glucose metabolism (table 1).
Table 1.
Genotype distribution and allelic frequency of gene variants in the ADIGEN study
| Gene name | Gene | Rs | Variant | Genotype Controlsa (n = 323) |
Obesea (n = 234) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Wt | He | Ho | Wt | He | Ho | ||||
| Efficiency of energy expenditure | |||||||||
| Uncoupling protein 2 [7] | UCP2 | None | Ins45bp | 166 | 127 | 21 | 104 | 94 | 24 |
| 6593669 | -866G>A | 114 | 131 | 49 | 88 | 96 | 41 | ||
| Uncoupling protein 3 [8] | UCP3 | 1900849 | -55C>T | 159 | 132 | 19 | 129 | 78 | 13 |
| Regulation of adipocyte differentiation and function | |||||||||
| Peroxisome proliferation-activated receptor-gamma 2 [12, 15] | PPARG2 | 1801282 | Pro12Ala | 224 | 83 | 5 | 158 | 59 | 6 |
| Hypothalamic regulation of appetite | |||||||||
| Cocaine and amphetamine related transcript [9] | CART | 586860 | +21del | 270 | 36 | 3 | 191 | 33 | 0 |
| 41271753 | +39A>G | 257 | 53 | 4 | 180 | 43 | 1 | ||
| Ghrelin [13] | GHRL | 696217 | Leu72Met | 271 | 44 | 1 | 199 | 30 | 0 |
| 4684677 | Gln90Leu | 256 | 47 | 3 | 177 | 38 | 5 | ||
| Melanocortin 4 receptor [14] | MC4R | 13447324 | Tyr35Ter | 256 | 0 | 0 | 222 | 3 | 0 |
| Melanin-concentrating hormone receptor 1 [16] | MCHR1 | 133072 | 100213G>A | 123 | 149 | 46 | 96 | 107 | 28 |
| McKusick-Kaufman syndrome gene [6] | MKKS | 1547 | Arg517Cys | 249 | 59 | 4 | 175 | 42 | 5 |
| Regulation of lipid and glucose metabolism | |||||||||
| Small heterodimer partner [11] | SHP | 6659176 | 512G>C | 240 | 44 | 1 | 182 | 40 | 2 |
| Glucocorticoid receptor gene [10] | GRL | 6195 | Asn363Ser | 278 | 24 | 2 | 195 | 21 | 0 |
Wt = wild type, He = heterozygotes, Ho = homozygotes.
For some variants the number of control and/or obese individuals may be lower than the reported total number for both groups.
For the present study, we restricted the analyses of the previously published gene variants to those where the allele frequency gave us a fair chance to reveal a statistical association with the various phenotypes depending on the mode of transmission. Thus, when the minor allele is rare, the distinction between additive and dominant transmission and identification of recessive transmission is not possible. Therefore additive, and dominant effects have only been analyzed if the number of heterozygous subjects was 20 or above. Likewise, the recessive effects were only analyzed if 20 or more homozygous subjects were present. In accordance with these criteria, we did not analyze the POMC, LEPR and ADRB2 gene variants [17, 18, 19]. We have also investigated a number of variants in the MC4R gene [14], including the V103I polymorphism, but none of these met the selection criteria. However, in our previous study we discovered a MC4R variant Tyr35ter (rs13447324), which was strongly associated with obesity, and the only one we have found to be associated with obesity in all our studies. Thus, in spite of the very low allele frequency, according to our inclusion criteria for analysis, we did include it in the present study. Thus, in total thirteen polymorphisms in ten genes were analyzed (table 1).
The 3‘UTR Ins45bp polymorphism in UCP2 consisted of deletion/ deletion (DD), insertion/deletion (ID), and insertion/insertion (II) genotypes. However, all genotypes were categorized and have been presented as wildtype (Wt), heterozygote (He), and homozygote (Ho). PPARG2 Pro12Ala (rs1801282) is in strong linkage disequilibrium with our other published PPARG2 variants (1431C>T, –792A>G, –882T>C, –2604T>C, –2953A>G). We therefore have chosen to only include Pro12Ala in our study. Table 1 presents all variants analyzed in this study. Genotyping was done as described previously [6, 7, 8, 9, 10, 11, 12, 13, 14, 16].
Statistical Methods
All obesity phenotypes had individual units, e.g. kg/m2 and cm, but a direct comparison of the association between different obesity phenotypes and the genotype needs similar units. Therefore, the measures of obesity phenotypes were converted to z-scores, which indicate the deviations from the population mean values in SD units. The z-scores were calculated using the mean and SD values of the control group and applied to the entire study population. This conversion into z-scores enables us to directly compare the strength of the association between the genotype and the various obesity phenotypes.
For the present study the two groups of obese and controls have been analyzed together. Owing to the sampling design with massive enrichment of the right tail of the BMI distribution, the data cannot be analyzed with BMI or BMI-associated outcomes as response variables in common regression models, which assumes normal distribution of the residual variation of the response variables. However, to take advantage of the greater statistical power and much wider coverage of the phenotypes by keeping the obese and non-obese groups in the same analyses, we reversed the analyses of the associations in order to examine the probability of finding particular genotypes for a given level of the phenotypes. Using the logistic regression modeling, this can be done without distributional assumptions about the phenotypes. Hence, this model was used to estimate the odds ratios (OR) of the genotype (response variable) in relation to the phenotypes (covariates) in the combined case and control groups. The OR for being carrier of a given genotype according to BMI z-score should be interpreted as an increment in odds for being carrier of a given genotype per unit increase in BMI z-score.
To determine the transmission mode of the gene, we compared the fit of three models, assuming an additive, a dominant, or a recessive model. This was done by likelihood ratio test comparing each model with the multinomial logistic model unspecified with regard to genetic transmission. We used proc logistic in SAS (version 9.1; SAS Institute Inc, Cary, NC, USA), which can analyze both the multinomial logistic model (the unspecified model), the ordinal model were the SNP is scored 0, 1, 2 (co-dominant or additive model) and the binary logistic model (recessive and dominant). In a dominant model, the genotype is determined when at least one copy of the minor allele ‘a’ is present (AA versus Aa and aa). In a recessive model, homozygosity of the minor allele ‘a’ is required for the expression of the phenotype (AA and Aa versus aa). On basis of the tests, we chose the simplest transmission mode most compatible with our data. The tests for the additive and dominant model fitted data equally well. Therefore, a dominant transmission mode was chosen for all genes except two; for the UCP2 and MKKS polymorphisms the recessive model fitted data best. All analyses were adjusted for age as a continuous variable.
The significance level was set to 0.05 (two-sided), and no correction for multiple testing was applied, but the consequences hereof for the interpretation of the outcome of the analyses have been addressed in the Discussion (see below).
Ethics
We certify that all applicable institutional and governmental regulations concerning the ethical use of human volunteers were followed during this research. The Danish Data Protection Agency and the regional Ethical Committee approved the study to be in accordance with the Helsinki Declaration II. All participants signed a written consent before participating.
Results
Descriptive Analyses
In Table 1 the allele distribution of the variants is given as two wild type alleles (major), one wild type variant allele, and two variant alleles (minor). No significant departures from Hardy Weinberg equilibrium were observed for the gene polymorphisms. The genotype distribution showed no differences between obese subjects, defined by the original BMI threshold in the sampling from the draft board population, and control subjects for any of the examined polymorphisms. In our previous publications significant differences were observed between obese and control subjects for MC4R Tyr35Ter (p = 0.004) and MKKS Arg517Cys (rs1547) (p = 0.048), but these results however were based on a larger segment of the original cohort of obese and controls examined on an earlier occasion with less detailed phenotyping of obesity [6, 14].
Table 2 shows that mean age and the different anthropometric measurements including body composition, but not height, differed between the control and obese groups.
Association Analyses
The OR from the logistic regression analyses for MC4R Tyr35Ter are given in table 3. They showed no significant tendencies to associations with all obesity phenotypes except for waist for given BMI.
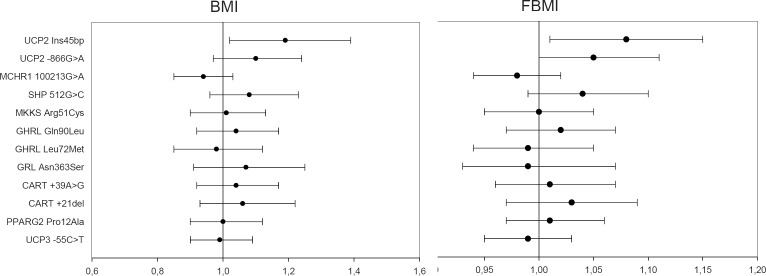
In figure 1 plots for OR (95% confidence intervals (95% CI)) of the minor alleles of all variants show that UCP2 Ins45bp was significantly associated with an increase of BMI (OR = 1.19 (1.02–1.39); p = 0.03) and of FBMI (OR = 1.08 (1.01–1.15); p = 0.04). UCP2 –866G>A was also associated with a borderline increase of FBMI (OR = 1.05 (1.00–1.11); p = 0.06). As an example, the OR of 1.19 for being homozygous for the risk allele according to BMI z-score should be interpreted as an increment in odds of 19% for being homozygous for the risk allele per unit increase in BMI z-score.
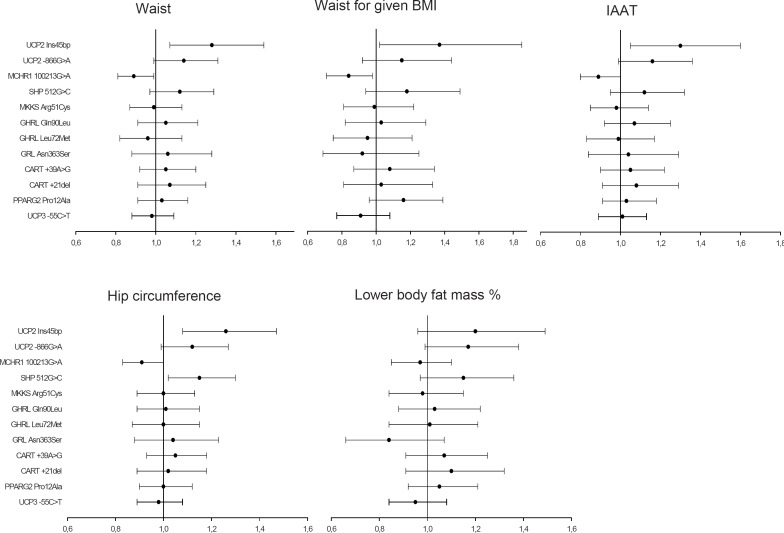
Figure 2 shows the associations with the various phenotypic measures of abdominal obesity, and only the following revealed notable associations: UCP2 Ins45bp was associated with an increase of waist and IAAT (range: OR = 1.28–1.30 (1.05–1.60); p = 0.01–0.02), with an increase of waist for given BMI (OR = 1.37 (1.02–1.85); p = 0.04). MCHR1 100213G>A was associated with a decreased waist and IAAT (range OR = 0.89–0.90 (0.80–1.00); p = 0.03–0.05) and with a decreased waist for given BMI (range: OR = 0.84 (0.71–0.99); p = 0.03).
Figure 2 also showed associations with peripheral obesity assessed by hip circumference and LBFM (%). UCP2 Ins45bp and SHP 512G>C were associated with an increase of hip circumference (OR = 1.26 (1.08–1.47); p = 0.003 and OR = 1.15 (1.02–1.30); p = 0.02, respectively), whereas MCHR1 100213G>A was associated with a reduced hip circumference (OR = 0.91 (0.83–1.00); p = 0.05). There were no other notable associations with hip circumference, and none were observed for LBFM (%).
Thus, the results suggest that, particularly the minor allele of UCP2 Ins45bp under a recessive transmission mode is positively associated with overall, abdominal and peripheral obesity. Furthermore, the results from the dominant genetic models suggest that the minor allele of SHP predicts an increase of hip circumference, and that MCHR1 predicts a decrease of waist for given BMI and of IAAT.
Discussion
The present study aimed at examining and comparing the association between seven specific obesity phenotypes and a panel of thirteen candidate gene polymorphisms. The results confirmed our hypothesis that BMI may not always be the appropriate obesity phenotype and that the obesity phenotypes so far examined in relation to potential obesity candidate genes have been too unspecific [3]. Only one out of ten significant associations was seen between BMI and a candidate gene polymorphism. Various specific obesity phenotypes, except for LBFM (%), were significantly associated with UCP2 Ins45bp, UCP2 –866G>A, SHP 512G>C and MCHR1 100213G>A when assuming dominant or recessive genetic models as appropriate.
Strengths of this study include its epidemiological case-cohort design and the minimization of confounding due to the homogenous study population of Danish Caucasians. Furthermore, the availability of several anthropometric measures in the same individuals have given the unique opportunity of analyzing and comparing inexpensively and easily measurable obesity phenotypes (BMI, waist and hip circumference) especially suitable for larger cohort studies and the more precise, but also more costly and time-consuming measurements of obesity (FBMI, IAAT and LBFM (%)) in relation to a broad panel of obesity-related candidate gene variants. However, in the present study the predictive value of the phenotypes did not deviate according to whether the phenotypes were inexpensive or costly. Hence, the main obesity phenotypes used for the present study, i.e. BMI, waist and hip circumference, are applicable with minimum efforts both in clinical and public health practice and for the health-concerned individual.
The study population consisted of men only, because they were recruited from the draft board examination, which is a mandatory for the Danish population of young men only. We acknowledge that there might be gender-related differences in the genotype-phenotype associations. Attendance of the follow-up examination may be prone to selection bias, since it is plausible that the most healthy volunteers participated and this may question whether the observed allele distributions represent the actual allele distributions in the original study population and hence in the general population of men. An apparent limitation of the study is its relative small sample size for a genetic epidemiological examination, but it should be emphasized that the sampling design implied that the randomly selected group is representative of 64,600 men examined at the draft board examination, and that the obese group therefore can be assumed to be among the most obese within this population as originally screened, except for the non-attendance of such obese. Furthermore, our results in fact show narrow confidence intervals, which means that we have a high power to predict that the true OR of the associations are within the observed 95 CI. We find it unlikely that the attrition of the samples from the original identification to the follow-up examination is related to the particular associations between genotypes and phenotypes.
We acknowledge that this study is not based upon HapMap-derived tagging SNPs of the selected candidate gene loci wherefore much stronger associations between gene variants and the specified estimates of body fat accumulation cannot be excluded. Moreover, our findings are not corrected for multiple comparisons and are by nature exploratory. Using the logistic regression ((2 × 13 (additive and dominant) + 4 recessive) × 7 phenotypes) 210 analyses were conducted. The number of significant statistical tests was slightly below what would be expected under repetitive testing of the same null hypothesis (10 significant tests out of 210 tests = 4.8%) and hence, implies that the significant genotype-phenotype associations must be interpreted cautiously and can only be used as tentative leads for further studies to elucidate the possible role of the analyzed panel of obesity-related gene variants.
We acknowledge that the accuracy to estimate body fat and fat distribution may vary between the seven specific obesity measurements in the present study. For example waist and hip circumference are useful measures of fat distribution, whereas their ability to measure total body fat is low. On the other hand, BMI may be used as a measure of total body fat, but it is obviously confounded by the variation in the other body compartments, and by definition it cannot measure fat distribution [3, 38]. All our anthropometric variables were objectively measured, and thus the bias related to self-reported measures is eliminated. The body composition measures were derived from DEXA scans which have a high capability of measuring both total body fat and some aspects of fat distribution [38]. Although IAAT may be a better measure of the abdominal adipose tissue than waist circumference, several validation studies have demonstrated that waist circumference is the most accurate proxy marker for the abdominal adipose tissue [38]. In addition, a recent review concluded that the protocol for measuring waist circumference has no substantial influence on the association between waist circumference and cardiovascular disease, diabetes and all-cause mortality [39]. We believe that the reliability and validity of the examined obesity phenotypes is high enough to justify the phenotypic differentiation of approach in genotype-phenotype studies.
By conditioning waist circumference for BMI, we obtained a measure that is independent of total body fatness. We examined whether waist circumference for given BMI better explained the genotypic predisposition, as an alternative phenotype of obesity, compared with waist, but it appeared that the results overall were not different from those obtained by using waist circumference alone. Whereas waist circumference reflects the abdominal fat, hip circumference reflects the composition of muscle mass, bone mass, and fat mass in the gluteofemoral area [38, 40]. In our study we observed different associations for waist and hip circumference.
In the present study an increased overall and abdominal obesity was seen for both UCP2 variants. In the previous publication of these polymorphisms we did not find any association with BMI [6, 7]. Our results support the studies suggesting an association between UCP2 and obesity [41, 42]. An increased hip circumference was associated with UCP2 Ins45bp and the SHP 512G allele, suggesting a beneficial effect on that aspect of body fat distribution.
We observed decreased levels of abdominal and peripheral obesity in the presence of the minor allele for MCHR1 100213G>A. In a previous publication divergent effects of the polymorphism were shown in a German, in a French, in an American and in our Danish population. The minor allele for MCHR1 was positively associated with obesity in the German cohort, but this finding was not confirmed in the other cohorts [16]. However, in the present study analyzing MCHR1 100213G>A with several abdominal obesity phenotypes shows protective effects of the minor alleles against abdominal obesity. However, we are unable to say whether this is a net beneficial effect because this polymorphism is also associated with reduced peripheral obesity and possibly reduced lean body mass [43].
We have systematically analyzed each gene variant in our cohort to distinct obesity phenotypes that cover the overall, abdominal and peripheral obesity in order to elucidate the potential impact of body fat distribution. These results imply that specific obesity phenotypes are needed when associations between gene polymorphisms and the obesity phenotype are explored. In the present study, we found that i) the minor allele of SHP 512G>C was associated with increased hip circumference, ii) the minor allele for MCHR1 was associated with decreased abdominal obesity, and iii) the minor alleles for UCP2 ins45bp and UCP2 –866G>A were associated with increased abdominal, peripheral and overall obesity. These findings will need to be replicated before they can be interpreted as possibly causal, but we suggest that the same approach should be applied to the other relevant obesity genes, such as other variants of the MC4R gene [2, 3] and the INSIG2 gene [44], and gene polymorphisms emerging from various genome-wide association studies, such as the recently explored FTO gene [4].
Conflicts of Interest
None.
Fig. 1.
Plots for odds ratio and 95% confidence intervals of all variants in relation to overall obesity sorted by recessive (UCP2 and MKKS variants) and dominant effects. Error bars represent 95% confidence intervals.
Fig. 2.
Plots for odds ratio and 95% confidence intervals of all variants in relation to abdominal and peripheral obesity sorted by recessive (UCP2 and MKKS variants) and dominant effects.
Table 2.
Distribution of age, anthropometric measures and body composition at the follow-up examination. Values are given as median and range.
| Anthropometric measure | Controls (95% CI) (n = 323) | Obese (95% CI) (n = 234) |
|---|---|---|
| Height, cm* | 178.5 (162.0–197.0) | 179.0 (156.0–196.0) |
| Weight, kg | 81.3 (55.3–134.3) | 111.8 (66.4–201.5) |
| Age, years | 49.0 (39.0–65.0) | 47.0 (39.0–64.0) |
| Overall obesity | ||
| BMI, kg/m2 | 25.7 (17.9–42.9) | 35.3 (23.2–56.4) |
| FBMI, kg/m2 | 6.0 (1.1–15.9) | 12.0 (3.8–24.8) |
| Abdominal obesity | ||
| Waist circumference, cm | 93.0 (69.0–127.0) | 117.0 (88.0–164.0) |
| IAAT, cm2 | 104.8 (23.0–226.7) | 182.8 (89.7–308.7) |
| Peripheral obesity | ||
| Hip circumference, cm | 97.0 (74.0–126.0) | 112.0 (92.0–155.0) |
| LBFM (%) | 22.0 (5.1–41.2) | 33.1 (15.4–47.8) |
FBMI = fat body mass index; IAAT = intra-abdominal adipose tissue; LBFM (%) = lower body fat mass (%).
Comparison of the controls and obese by Student t-tests gave p values <0.0001, except for height (p = 0.6).
Table 3.
Odds ratio (OR) including 95% confidence intervals (CI) for MC4R Tyr35Ter in relation to overall, abdominal and peripheral obesity
| Obesity phenotype | OR (95% CI) | p value |
|---|---|---|
| Overall obesity | ||
| BMI, kg/m2 | 1.58 (0.94–2.65) | 0.08 |
| FBMI, kg/m2 | 1.16 (0.93–1.46) | 0.19 |
| Abdominal obesity | ||
| Waist, cm | 1.65 (0.86–3.17) | 0.14 |
| Waist for given BMI, cm | 0.67 (0.21–2.11) | 0.49 |
| IAAT, cm2 | 1.71 (0.78–3.72) | 0.18 |
| Peripheral obesity | ||
| Hip circumference, cm | 1.53 (0.95–2.47) | 0.08 |
| LBFM (%) | 1.27 (0.57–2.81) | 0.56 |
FBMI = fat body mass index. IAAT = intraabdominal adipose tissue; LBFM (%) = Lower body fat mass (%).
References
- 1.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 2.Hinney A, Hebebrand J. Polygenic obesity in humans. Obesity Facts. 2008:35–42. doi: 10.1159/000113935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clement KSørensen TIA. London Informa Healthcare. 2007. Obesity: Genomics and Postgenomics. [Google Scholar]
- 4.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, Harries LW, Barrett JC, Ellard S, Groves CJ, Knight B, Patch AM, Ness AR, Ebrahim S, Lawlor DA, Ring SM, Ben-Shlomo Y, Jarvelin MR, Sovio U, Bennett AJ, Melzer D, Ferrucci L, Loos RJ, Barroso I, Wareham NJ, Karpe F, Owen KR, Cardon LR, Walker M, Hitman GA, Palmer CN, Doney AS, Morris AD, Smith GD, Hattersley AT, McCarthy MI. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinney ANguyen TT, Scherag A, Friedel S, Bronner G, Müller TD, Grallert H, Illig T, Wichmann HE, Rief W, Schäfer H, Hebebrand J. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS ONE. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen KLEchwald SMLarsen LH, Hamid YH, Glumer C, Jorgensen T, Borch-Johnsen K, Andersen T, Sørensen TI, Hansen T, Pedersen O. Variation of the McKusick-Kaufman gene and studies of relationships with common forms of obesity. J Clin Endocrinol Metab. 2005;90:225–230. doi: 10.1210/jc.2004-0465. [DOI] [PubMed] [Google Scholar]
- 7.Dalgaard LTSørensen TI, Andersen T, Hansen T, Pedersen O. An untranslated insertion variant in the uncoupling protein 2 gene is not related to body mass index and changes in body weight during a 26-year follow-up in Danish Caucasian men. Diabetologia. 1999;42:1413–1416. doi: 10.1007/s001250051312. [DOI] [PubMed] [Google Scholar]
- 8.Dalgaard LTSørensen TI, Drivsholm T, Borch-Johnsen K, Andersen T, Hansen T, Pedersen O. A prevalent polymorphism in the promoter of the UCP3 gene and its relationship to body mass index and long term body weight change in the Danish population. J Clin Endocrinol Metab. 2001;86:1398–1402. doi: 10.1210/jcem.86.3.7301. [DOI] [PubMed] [Google Scholar]
- 9.Echwald SMSørensen TI, Andersen T, Hansen C, Tommerup N, Pedersen O. Sequence variants in the human cocaine and amphetamine-regulated transcript (CART) gene in subjects with early onset obesity. Obes Res. 1999;7:532–536. doi: 10.1002/j.1550-8528.1999.tb00710.x. [DOI] [PubMed] [Google Scholar]
- 10.Echwald SMSørensen TI, Andersen T, Pedersen O. The Asn363Ser variant of the glucocorticoid receptor gene is not associated with obesity or weight gain in Danish men. Int J Obes Relat Metab Disord. 2001;25:1563–1565. doi: 10.1038/sj.ijo.0801744. [DOI] [PubMed] [Google Scholar]
- 11.Echwald SMAndersen KL, Sørensen TI, Larsen LH, Andersen T, Tonooka N, Tomura H, Takeda J, Pedersen O. Mutation analysis of NR0B2 among 1545 Danish men identifies a novel c.278G>A (p.G93D) variant with reduced functional activity. Hum Mutat. 2004;24:381–387. doi: 10.1002/humu.20090. [DOI] [PubMed] [Google Scholar]
- 12.Ek J, Urhammer SA, Sørensen TI, Andersen T, Auwerx J, Pedersen O. Homozygosity of the Pro12Ala variant of the peroxisome proliferation-activated receptor-gamma2 (PPAR-gamma2): divergent modulating effects on body mass index in obese and lean Caucasian men. Diabetologia. 1999;42:892–895. doi: 10.1007/s001250051243. [DOI] [PubMed] [Google Scholar]
- 13.Larsen LHGjesing AP, Sørensen TI, Hamid YH, Echwald SM, Toubro S, Black E, Astrup A, Hansen T, Pedersen O. Mutation analysis of the pre-proghrelin gene: No association with obesity and type 2 diabetes. Clin Biochem. 2005;38:420–424. doi: 10.1016/j.clinbiochem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Larsen LHEchwald SM, Sørensen TI, Andersen T, Wulff BS, Pedersen O. Prevalence of mutations and functional analyses of melanocortin 4 receptor variants identified among 750 men with juvenile-onset obesity. J Clin Endocrinol Metab. 2005;90:219–224. doi: 10.1210/jc.2004-0497. [DOI] [PubMed] [Google Scholar]
- 15.Larsen TMLarsen LH, Torekov SK, Ek J, Black E, Toubro S, Astrup A, Sørensen TI, Hansen T, Pedersen O. Novel variants in the putative peroxisome proliferator-activated receptor γ promoter and relationships with obesity in men. Obes Res. 2005;13:953–958. doi: 10.1038/oby.2005.110. [DOI] [PubMed] [Google Scholar]
- 16.Wermter AKReichwald K, Buch T, Geller F, Platzer C, Huse K, Hess C, Remschmidt H, Gudermann T, Preibisch G, Siegfried W, Goldschmidt HP, Li WD, Price RA, Biebermann H, Krude H, Vollmert C, Wichmann HE, Illig T, Sørensen TI, Astrup A, Larsen LH, Pedersen O, Eberle D, Clement K, Blundell J, Wabitsch M, Schäfer H, Platzer M, Hinney A, Hebebrand J. Mutation analysis of the MCHR1 gene in human obesity. Eur J Endocrinol. 2005;152:851–862. doi: 10.1530/eje.1.01917. [DOI] [PubMed] [Google Scholar]
- 17.Echwald SMSørensen TI, Andersen T, Tybjaerg-Hansen A, Clausen JO, Pedersen O. Mutational analysis of the proopiomelanocortin gene in Caucasians with early onset obesity. Int J Obes Relat Metab Disord. 1999;23:293–298. doi: 10.1038/sj.ijo.0800814. [DOI] [PubMed] [Google Scholar]
- 18.Echwald SMSørensen TD, Sørensen TI, Tybjaerg-Hansen A, Andersen T, Chung WK, Leibel RL, Pedersen O. Amino acid variants in the human leptin receptor: lack of association to juvenile onset obesity. Biochem Biophys Res Commun. 1997;233:248–252. doi: 10.1006/bbrc.1997.6430. [DOI] [PubMed] [Google Scholar]
- 19.Echwald SMSørensen TI, Tybjaerg-Hansen A, Andersen T, Pedersen O. Gln27Glu variant of the human beta2-adrenoreceptor gene is not associated with early-onset obesity in Danish men. Diabetes. 1998;47:1657–1658. doi: 10.2337/diabetes.47.10.1657. [DOI] [PubMed] [Google Scholar]
- 20.Pouliot MCDespres JP, Nadeau A, Moorjani S, Prud’Homme D, Lupien PJ, Tremblay A, Bouchard C. Visceral obesity in men. Associations with glucose toleranceplasma insulin and lipoprotein levels. Diabetes. 1992;41:826–834. doi: 10.2337/diab.41.7.826. [DOI] [PubMed] [Google Scholar]
- 21.Ross R, Freeman J, Hudson R, Janssen I. Abdominal obesitymuscle composition and insulin resistance in premenopausal women. J Clin Endocrinol Metab. 2002;87:5044–5051. doi: 10.1210/jc.2002-020570. [DOI] [PubMed] [Google Scholar]
- 22.Bacha F, Saad R, Gungor N, Janosky J, Arslanian SA. obesity regional fat distribution and syndrome X in obese black versus white adolescents: race differential in diabetogenic and atherogenic risk factors. J Clin Endocrinol Metab. 2003;88:2534–2540. doi: 10.1210/jc.2002-021267. [DOI] [PubMed] [Google Scholar]
- 23.Despres JP, Moorjani S, Ferland M, Tremblay A, Lupien PJ, Nadeau A, Pinault S, Theriault G, Bouchard C, Adipose tissue distribution and plasma lipoprotein levels in obese women Importance of intra-abdominal fat. Arteriosclerosis. 1989;9:203–210. doi: 10.1161/01.atv.9.2.203. [DOI] [PubMed] [Google Scholar]
- 24.Lissner LBjorkelund C, Heitmann BL, Seidell JC, Bengtsson C. Larger hip circumference independently predicts health and longevity in a Swedish female cohort. Obesity Res. 2001;9:644–646. doi: 10.1038/oby.2001.85. [DOI] [PubMed] [Google Scholar]
- 25.Tanko LBBagger YZ, Alexandersen P, Larsen PJ, Christiansen C. Peripheral adiposity exhibits an independent dominant antiatherogenic effect in elderly women. Circulation. 2003;107:1626–1631. doi: 10.1161/01.CIR.0000057974.74060.68. [DOI] [PubMed] [Google Scholar]
- 26.Seeman EMelton LJ, III, O’Fallon WM, Riggs BL. Risk factors for spinal osteoporosis in men. Am J Med. 1983;75:977–983. doi: 10.1016/0002-9343(83)90878-1. [DOI] [PubMed] [Google Scholar]
- 27.Bigaard JFrederiksen K, Tjonneland A, Thomsen BL, Overvad K, Heitmann BL, Sørensen TI. Waist and hip circumferences and all-cause mortality: usefulness of the waist-to-hip ratio? Int J Obes Relat Metab Disord. 2004;28:741–747. doi: 10.1038/sj.ijo.0802635. [DOI] [PubMed] [Google Scholar]
- 28.Kaprio JEriksson J, Lehtovirta M, Koskenvuo M, Tuomilehto J. Heritability of leptin levels and the shared genetic effects on body mass index and leptin in adult Finnish twins. Int J Obes Relat Metab Disord. 2001;25:132–137. doi: 10.1038/sj.ijo.0801526. [DOI] [PubMed] [Google Scholar]
- 29.Jordan JBrabant G, Brinsuk M, Tank J, Horn R, Luft FC, Busjahn A. Heritability of free and receptor-bound leptin in normal twins. Am J Physiol Regul Integr Comp Physiol. 2005;288:R1411–R1416. doi: 10.1152/ajpregu.00446.2004. [DOI] [PubMed] [Google Scholar]
- 30.World Health Organization: Physical status: The Use and Interpretation of Anthropometry Report of a WHO Expert Committee Geneva World Health Organization. 2003 [PubMed] [Google Scholar]
- 31.Black E, Petersen L, Kreutzer M, Toubro S, Sørensen TI, Pedersen O, Astrup A. Fat mass measured by DXA varies with scan velocity. Obes Res. 2002;10:69–77. doi: 10.1038/oby.2002.12. [DOI] [PubMed] [Google Scholar]
- 32.Buemann BSørensen TIA, Pedersen O, Black E, Holst C, Toubro S, Echwald S, Holst JJ, Rasmussen C, Astrup A. Lower-body fat mass as an independent marker of insulin sensitivity – the role of adiponectin. 2005;29:624–631. doi: 10.1038/sj.ijo.0802929. [DOI] [PubMed] [Google Scholar]
- 33.Janssen IHeymsfield SBAllison DB, Kotler DP, Ross R. Body mass index and waist circumference independently contribute to the prediction of nonabdominalabdominal subcutaneous and visceral fat. Am J Clin Nutr. 2002;75:683–688. doi: 10.1093/ajcn/75.4.683. [DOI] [PubMed] [Google Scholar]
- 34.Bigaard J, Tjonneland A, Thomsen BL, Overvad K, Heitmann BL, Sørensen TI. Waist circumferenceBMI, smoking and mortality in middle-aged men and women. Obes Res. 2003;11:895–903. doi: 10.1038/oby.2003.123. [DOI] [PubMed] [Google Scholar]
- 35.Janssen I, Katzmarzyk PT, Ross R. Waist circumference and not body mass index explains obesity-related health risk. Am J Clin Nutr. 2004;79:379–384. doi: 10.1093/ajcn/79.3.379. [DOI] [PubMed] [Google Scholar]
- 36.Treuth MSHunter GR, Kekes-Szabo T. Estimating intraabdominal adipose tissue in women by dual-energy X-ray absorptiometry. Am J Clin Nutr. 1995;62:527–532. doi: 10.1093/ajcn/62.3.527. [DOI] [PubMed] [Google Scholar]
- 37.Buemann B, Astrup A, Pedersen O, Black E, Holst C, Toubro S, Echwald S, Holst JJ, Rasmussen C, Sørensen TIA. Possible role of adiponectin and insulin sensitivity in mediating the favorable effects of lower body fat mass on blood lipids. Journal of Clin Endocrinol Metab. 2006;91:1698–1704. doi: 10.1210/jc.2005-1062. [DOI] [PubMed] [Google Scholar]
- 38.Snijder MB, van Dam RM, Visser M, Seidell JC. What aspects of body fat are particularly hazardous and how do we measure them? Int J Epidemiol. 2006;35:83–92. doi: 10.1093/ije/dyi253. [DOI] [PubMed] [Google Scholar]
- 39.Ross RBerentzen TBradshaw AJ, Janssen I, Kahn HS, Katzmarzyk PT, Kuk JL, Seidell JC, Snijder MB, Sørensen TI, Despres JP. Does the relationship between waist circumferencemorbidity and mortality depend on measurement protocol for waist circumference? Obes Rev. 2007 doi: 10.1111/j.1467-789X.2007.00411.x. DOI: 10.1111/j.1467-789X.2007.00411.x. [DOI] [PubMed] [Google Scholar]
- 40.Molarius A, Seidell JC. Selection of anthropometric indicators for classification of abdominal fatness-a critical review. Int J Obes Relat Metab Disord. 1998;22:719–727. doi: 10.1038/sj.ijo.0800660. [DOI] [PubMed] [Google Scholar]
- 41.Dalgaard LT, Pedersen O. Uncoupling proteins: functional characteristics and role in the pathogenesis of obesity and Type II diabetes. Diabetologia. 2001;44:946–965. doi: 10.1007/s001250100596. [DOI] [PubMed] [Google Scholar]
- 42.Buemann BSchierning B, Toubro S, Bibby BM, Sørensen TIA, Dalgaard L, Pedersen O, Astrup A. The association between the val/ala-55 polymorphism of the uncoupling protein 2 gene and exercise efficiency. Int J Obes Relat Metab Disord. 2001;25((4)):467–471. doi: 10.1038/sj.ijo.0801564. [DOI] [PubMed] [Google Scholar]
- 43.Kuk JL, Janiszewski PM, Ross R. Body mass index and hip and thigh circumferences are negatively associated with visceral adipose tissue after control for waist circumference. American Journal of Clinical Nutrition. 2007;85:1540–1544. doi: 10.1093/ajcn/85.6.1540. [DOI] [PubMed] [Google Scholar]
- 44.Herbert AGerry NP, McQueen MB, Heid IM, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu X, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]