Abstract
Myelin sheath thickness is precisely regulated and essential for rapid propagation of action potentials along myelinated axons. In the peripheral nervous system, extrinsic signals from the axonal protein neuregulin 1 (NRG1) type III regulate Schwann cell fate and myelination. Here we ask if modulating NRG1 type III levels in neurons would restore myelination in a model of congenital hypomyelinating neuropathy (CHN). Using a mouse model of CHN, we improved the myelination defects by early overexpression of NRG1 type III. Surprisingly, the improvement was independent from the upregulation of Egr2 or essential myelin genes. Rather, we observed the activation of MAPK/ERK and other myelin genes such as peripheral myelin protein 2 and oligodendrocyte myelin glycoprotein. We also confirmed that the permanent activation of MAPK/ERK in Schwann cells has detrimental effects on myelination. Our findings demonstrate that the modulation of axon-to-glial NRG1 type III signaling has beneficial effects and improves myelination defects during development in a model of CHN.
Introduction
The neuregulin 1 (NRG1) family of ligands and cognate ErbB tyrosine kinase receptors (heterodimeric ErbB2/B3) regulate much of Schwann cell development (1–8). NRG1 function depends on isoform and amount of expression (9–11). In particular, the level of expression of the axonal membrane–bound NRG1 type III is a potent instructive signal that determines whether a Schwann cell will form myelin, as well as its thickness (6,7). A series of mouse genetics experiments have elucidated the complex molecular network downstream of NRG1/ErbB2/ErbB3 that regulates myelination during development and remyelination (11–13). Intrinsic Schwann cell signals downstream of NRG1 include PI3K/AKT, MAPK/ERK and calcineurin (CaN)/NFATc4, which are required to precisely regulate myelin gene transcription and protein synthesis required for myelin formation (14–20).
Expressed almost exclusively by myelin-forming Schwann cells, myelin protein zero (MPZ/P0) is the major protein in myelin of the peripheral nervous system (PNS) and is necessary for normal myelin structure and function (21). In addition to structural interactions with the membrane, the P0 cytoplasmic tail is thought to contain motifs that assemble signal complexes to stimulate myelin gene expression and therefore modulate myelin thickness (21,22). A human nonsense mutation in the Mpz gene generates a premature stop codon in the P0 protein at glutamine 215 (P0Q215X), which truncates the P0 cytoplasmic tail. P0Q215X mutation is associated with a severe congenital hypomyelinating neuropathy (CHN) in patients, characterized by weakness, hypotonia and areflexia from birth and with a severely reduced nerve conduction velocity (NCV) (23,24). The molecular mechanisms leading to hypomyelination in CHN are not well understood (25).
In this study, we used a mouse model of CHN with the targeted mutation of Mpz encoding the P0Q215X mutant protein. Mice heterozygous for Q215X (MpzQ215X/+) show a mild CHN neuropathy, less severe than the human pathology due to reduced expression of the MpzQ215X allele (25). Despite these difference in severity, MpzQ215X/+ mice are a genetic authentic model of the human neuropathy and recapitulate a mild CHN human nerve pathology, with diminished motor performance, and hypomyelination (25). We previously showed that P0Q215X does not elicit an unfolded protein response, which is a key mechanism for other pathogenic MPZ mutations (26–29), but is instead aberrantly trafficked to non-myelin plasma membranes. Genetic experiments suggest that the Q215X phenotype results from a combination of loss and gain of function (25). Here we find that the neuregulin axis is intact in Q215X nerves, but myelin gene expression is reduced. Although overexpression of the Mpz gene does not impact on the Q215X phenotype (25), we reasoned that augmented activity of Nrg1 type III might compensate for hypomyelination in CHN by increasing myelin gene expression and myelin thickness (7). To test this, we crossed Q215X mice with a transgenic mouse overexpressing HA-Nrg1 type III in neurons (30). We observed an improvement in myelin thickness and NCV in Nrg1 type III:Q215X mice. Transcriptional and biochemical analyses identified MAPK/ERK as the likely signaling pathway responsible for rescuing Q215X defects in Schwann cells.
Results
Myelin protein levels are reduced in Q215X mice
We showed that MpzQ215X/+ mice present a hypomyelination in sciatic nerves (Fig. 1A and B), caused by a technical loss of function and a dose-dependent gain of abnormal function (25). Thus, although the Q215X mutant allele caused a mild phenotype, the Q215X model displayed features of a human CHN with non-progressive hypomyelination throughout development (25).
Figure 1.
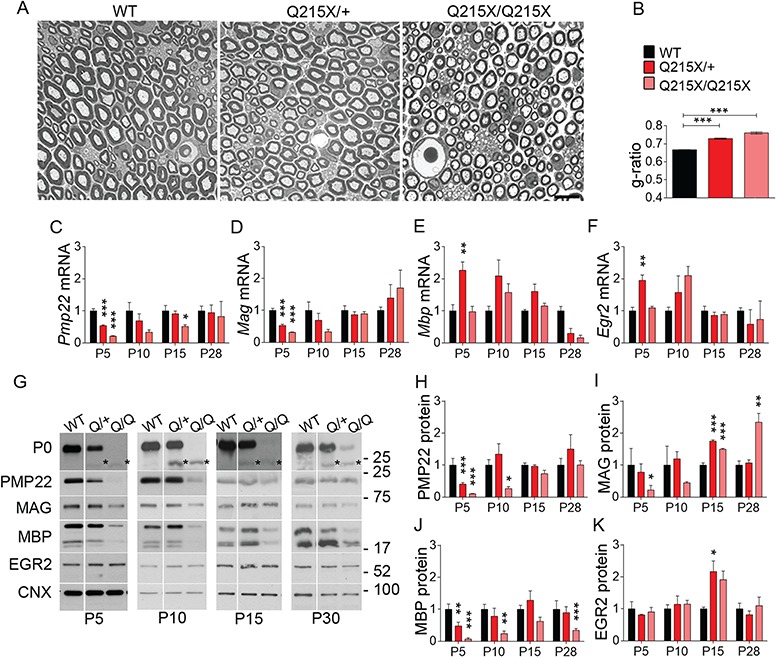
The Q215X mutation in P0 causes hypomyelination and alters myelin genes expression. (A) Semithin cross sections stained with toluidine blue from WT, P0Q215X heterozygote (Q215X/+) and P0Q215X homozygote (Q215X/Q215X) sciatic nerves at 1 month. Q215X/+ and Q215X/Q215X fibers are hypomyelinated as compared to WT. Scale bars, 10 μm. (B) g-ratio was increased in Q215X mutants from 0.66 ± 0.0005 (WT) to 0.72 ± 0.003 (Q215X/+) and 0.76 ± 0.005 (Q215X/Q215X). (C–F) mRNA relative quantification by real time PCR for Pmp22, Mag, Mbp and Erg2 in WT, Q215X/+ and Q215X/Q215X sciatic nerves at P5, P10, P15 and P30. (G–K) Western analysis and densitometries from WT, Q215X (Q/+) and Q215X/Q215X (Q/Q) sciatic nerves during development at P5, P10, P15 and P30 days. Blots were probed for P0, PMP22, MAG, MBP and EGR2. The Q215X allele causes a reduction of P0 stability (25). Calnexin was used as a loading control. Analyses were performed using three mice per genotype. Western blots were cropped. Error bars indicate s.e.m. Statistical analyses were performed using one-way ANOVA with Bonferroni’s multiple comparison. *P < 0.05, **P < 0.01, ***P < 0.001.
To determine if the hypomyelination in MpzQ215X/+ Schwann cells is caused by a misregulation of myelin genes, we quantified RNA and protein levels of the major myelin genes in Q215X mutants during development. Both Q215X heterozygote and homozygote sciatic nerves showed a reduction of Pmp22 and Mag mRNA at early time points P5 and/or P10, when myelination is initiating (Fig. 1C–F). In addition, Q215X heterozygote and homozygote sciatic nerves had reduced levels of PMP22, MAG and MBP at P5 (Fig. 1G–J). We analyzed the mRNA and protein levels for Egr2, a key transcriptional regulator of myelin genes, but did not find any dysregulation, suggesting that the underexpression of myelin genes in Q215X is independent of Egr2 (Fig. 1F and K). These results suggest that the Q215X allele affects myelin protein expression and is well suited for the study of therapeutical approaches aiming at rescuing hypomyelination.
Overexpression of NRG1 type III improves myelin thickness in Q215X mice
NRG1 type III expressed on axons determines whether Schwann cells will form myelin, and its thickness, through the activation of the master myelin gene transcription factor Egr2 (6,18). Overexpressing NRG1 type III isoform in sensory and motor neurons significantly increases myelin thickness in the PNS (7,30). Q215X mutants present a hypomyelination with decreased myelin gene expression, likely due to a loss of transcriptional signals. Thus, we expect that introducing exogenous expression of hemagglutinin-tagged NRG1 (HA-NRG1) type III in a hypomyelinating context (Q215X) could activate transcription of myelin genes and improve myelin thickness. We bred Q215X mice with the HA-NRG1 type III hemizygous transgenic mouse model (Nrg1t3) that overexpresses HA-NRG1 type III under the control of the neuronal Thy1.2 promoter (31,32).
Morphological and ultrastructure analyses of Nrg1t3;Q215X sciatic nerves were performed at 1 month of age and compared to Q215X heterozygote (Q215X) littermates. First, we confirmed by morphologic measurements that the newly generated Q215X and Nrg1t3 mice present hypomyelination and hypermyelination, with respectively 0.72 ± 0.01 and 0.53 ± 0.02 as g-ratio, as compared to 0.67 ± 0.005 in wild-type (WT) littermates (Fig. 2A–C). Second, we observed that double mutant animals present a recovery of myelin thickness when compared to Q215X and return to a myelin thickness close to WT animals with a g-ratio of 0.62 ± 0.01 (Fig. 2A–C).
Figure 2.
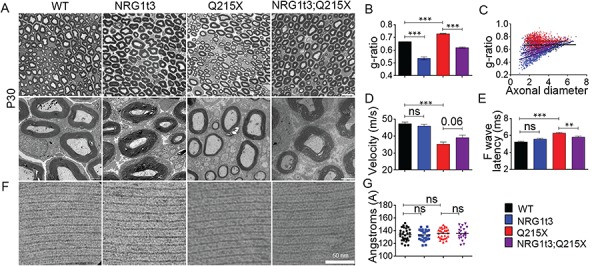
Neuronal overexpression of NRG1 type III improves myelination in P0Q215X. (A) Semithin cross sections stained with toluidine blue (top panel) and electron micrographs (bottom panel) from WT, Nrg1t3 hemizygote (Nrg1t3), P0Q215X heterozygote (Q215X) and Nrg1t3 hemizygote and P0Q215X heterozygote (Nrg1t3;Q215X) sciatic nerves at 1 month. NRG1t3 and Nrg1t3;Q215X fibers are hypermyelinated as compared to WT and Q215X. Scale bars, 10 μm (top panel) and 2 μm (bottom panel). (B) g-ratio was increased in Q215X (0.72 ± 0.003) and decreased in Nrg1t3 (0.53 ± 0.02), indicating hypomyelination and hypermyelination, respectively. (C) g-ratio as a function of axonal diameter showed preferential hypermyelination of small fibers in Nrg1t3 and Nrg1t3;Q215X as compared to WT and Q215X. (D) Neurophysiological studies showed a significant reduction of NCV in Q215X mice when compared to WT (47.28 ± 0.97 m/s). A recovery of NCVs can be observed in Nrg1t3;Q215X (39 ± 1.5 m/s) as compared to Q215X (35 ± 1.28 m/s) (P = 0.06). (E) F-wave latencies were increased in Q215X (6.54 ± 0.11 m/s) when compared to WT (5.32 ± 0.08 m/s). A significant recovery of F-wave latencies is observed in Nrg1t3;Q215X (6 ± 0.12 m/s), suggesting a proximal myelination improvement. (F–G) Myelin compaction is not altered in single or double mutant as indicated in electron micrographs (F) and periodicity quantification (G). Scale bar, 50 nm. Analyses were performed using three mice per genotype. Error bars indicate s.e.m. Statistical analyses from three independent experiments using one-way ANOVA with Bonferroni’s multiple comparison test. **P < 0.01, ***P < 0.001.
As previously described with transgenic expression of HA-NGR1 type III (7,30), the increase in myelin thickness was predominantly observed in small caliber axons (Fig. 2C). We also observed that Nrg1t3 animals present increased internodal length, as well as more myelinated small caliber fibers from 1 to 2 μm (Supplementary Material, Fig. 1).
As expected, in Q215X animals, the hypomyelination observed in 1-month-old animals correlated with a reduction of NCVs (Fig. 2D). In the Nrg1t3, mice sciatic nerve axons were well myelinated, and there was no alteration of NCV and F-wave latency (Fig. 2D and E). The increase of myelin thickness in Nrg1t3;Q215X led to an improvement of NCVs (Fig. 2D). However, the NCVs do not return to the WT level, likely due to the pronounced effect of Nrg1 type III on small caliber fibers that contributes in a minor way to electrophysiological measurements of NCV. The F-wave latencies were also improved in Nrg1t3;Q215X, suggesting an increase in proximal myelination (Fig. 2E).
P0 is essential for proper myelin compaction (33–36). To exclude that the changes in myelin thickness were due to a defect in compaction, we analyzed the periodicity of myelin sheath, but no difference between mutants and WT was found (Fig. 2F and G).
Q215X mutant displayed early postnatal altered motor performance (25). Therefore, we assessed the effect of HA-NRG1 type III on motor performance using the grid-walking test at P15. Unlike the first transgenic mouse model generated that developed mild tremor (7), no visible tremor phenotype was described in the Nrg1t3 transgenic mouse (30). However, the grid-walking test at P15 revealed motor coordination deficits (more ‘foot faults’ were recorded as compared to WT) in Nrg1t3 mice, possibly due to an effect of NRG1 in the central nervous system (37–42). This interfered with the behavioral investigations to assess a possible motor rescue in Nrg1t3;Q215X mice (Supplementary Material, Fig. 2).
Expression of NRG1 type III is uncoupled from major myelin gene transcription and myelin protein expression
We further assessed the molecular mechanism responsible for the increase in myelin thickness and the functional improvements we observed. We first confirmed that the active form of NRG1 type III was expressed in Nrg1t3 and Nrg1t3;Q215X adult mice in spinal cord lysates (Fig. 3A). Despite this, the level and activation of NRG1 type III receptors in Schwann cells, tyrosine kinase receptors ErbB2 and ErbB3 were unchanged in the presence of transgenic expression of Nrg1t3 at P30 (Fig. 3B). Next, we quantified the level of major compact (P0, PMP22, MBP) and non-compact myelin (MAG) proteins in 1-month-old nerves. Despite exogenous expression of the NRG1 type III active form, no significant increase in the major myelin proteins was detected (Fig. 3C–D, Supplementary Material, Fig 3). To further confirm that the increase in myelin thickness in Nrg1t3 animals was independent from an increase in myelin gene expression, we measured P0 protein levels in the myelin sheath using immunoelectron microscopy. We observed a decrease of P0 density in the myelin sheath of Nrg1t3 mutants (Fig. 3E), corresponding to an increase in myelin area without any increase in P0 protein levels. Thus, P0 proteins are more diluted in the myelin of Nrg1t3 mutants. This quantification supports our biochemical results of unchanged level in P0 protein despite the hypermyelination (Fig. 3C–D).
Figure 3.
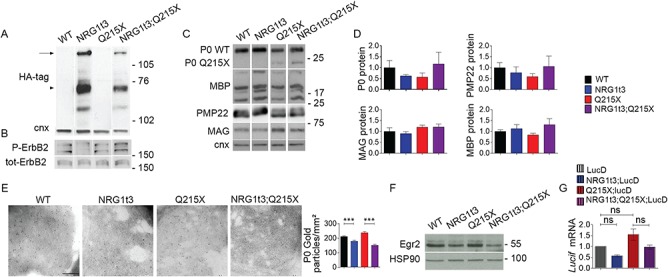
Expression of HA-NRG1 type III does not affect myelin gene expression. (A) Western blot analyses from WT, Q215X, Nrg1t3;Q215X and Nrg1t3 spinal cord at P30. Blots were probed for HA tag. HA is tagged to the NRG1 type III transgene. Calnexin was used as a loading control. We detected exogenous neuregulin precursor protein level and its cleavage products. The upper band corresponds to the full-length NRG1 type III inactive isoform (135 kDa, arrow); the 75-kDa band corresponds to the active form (arrow head) of NRG1 type III produced by BACE1-cleavage, as described in the literature (30,48). (B, C, F) Western blot and (D) densitometries analyses from WT, Q215X, Nrg1t3;Q215X and Nrg1t3 sciatic nerves at P30. Blots were probed for P-ErbB2, ErbB2 (B), P0, MBP, PMP22, MAG (C) and EGR2 (F). Calnexin and HSP90 were used as loading controls. Protein levels remain steady in all genotypes. (E) Immunogold labeling of P0 protein from cross sections of WT, Nrg1t3, Q215X and Nrg1t3;Q215X sciatic nerves at 1 month. Quantification of gold particles associated with P0 from cross sections of sciatic nerve at P30. Scale bar, 200 nm. (G) Relative quantification of luciferase reporter gene mRNA (Lucif) from Luc D hemizygotes (Luc D), Q215X;LucD, Nrg1t3;Luc D and Nrg1t3;Q215X;LucD sciatic nerves at P15. Luc D mice harbor a combinatorial EGR2/SOX10 binding site upstream of basal Hsp68 promoter. No significant differences were found at P15. Analyses were performed using three mice per genotype. Error bars indicate s.e.m. Statistical analyses were performed using one-way ANOVA with Bonferroni’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001.
EGR2 is the key regulator of myelination, myelin protein and cholesterol/lipid biosynthesis gene expression (43–46). Consistent with unchanged myelin gene transcription in hypermyelinated sciatic nerves of Nrg1t3 mice, Egr2 expression was not altered in Nrg1t3 mice at 1 month of age (Fig. 3F, Supplementary Material, Fig. 3). We also crossed our mice with mice expressing the luciferase reporter gene mRNA (Lucif) under the control of the Hsp68 promoter fused to a PMP22 enhancer with EGR2/SOX10 binding sites [Luc D hemizygotes (Luc D) mice] (47). Quantification of the relative expression of Luc D in Q215X;LucD, Nrg1t3;Luc D and Nrg1t3;Q215X;LucD sciatic nerves at P15 indicated that EGR2 transcriptional activity was not affected (Fig. 3G). These results suggest that exogenous HA-NRG1 type III promotes an increase in myelin thickness independent of the most common myelin genes and the modulation of the master Egr2 transcription factor.
The signaling pathways downstream of NRG1 type III are activated in Nrg1t3 mutant mice
We next sought to determine if increased NRG1 type III signaling in Nrg1t3 mice elicits activation of known signaling pathways downstream of ErbB2/3 that are necessary for Schwann cell development, differentiation and myelination, PI3K/AKT, CaN/NFATc4 and MAPK/ERK (14–18). These three signaling pathways are thought to trigger Egr2-dependent gene activation and, subsequently, myelination downstream of NRG1 type III.
The activity of AKT was increased in both Nrg1t3 and Nrg1t3;Q215X 1-month-old sciatic nerves (Fig. 4A) that correlate with previous in vitro and in vivo observation (6,48,49). In addition, we found hyperactivation of ERK1/2 pathway in both Nrg1t3 mutant mice (Fig. 4B), in agreement with recent findings that sustained and augmented MAPK/ERK signaling increases myelination (50,51).
Figure 4.
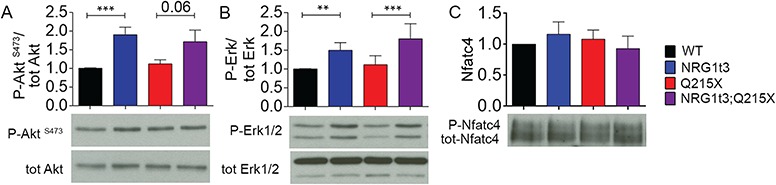
Expression of HA-NRG1 type III triggers the activation of ERK and AKT. Densitometries and western blot analyses of (A) P-AKT, AKT, (B) P-ERK, ERK, (C) P-NFATc4 and NFATc4 were performed on WT, Nrg1t3, Q215X and Nrg1t3;Q215X sciatic nerves lysates at 1 month. Increased activation of AKT and ERK was observed in NRG1t3 and Nrg1t3;Q215X. NFATc4 activation level was unchanged. Western blots were cropped. Analyses were performed using three mice per genotype. Error bars indicate s.e.m. Statistical analyses were performed using one-way ANOVA with Bonferroni’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001.
The total amount and the active form of NFATc4 were unchanged (Fig. 4C). Altogether, these results suggest that NRG1 type III overexpression induces the activation of both PI3K/AKT and MAPK/ERK, independently of the transcription factor EGR2.
MAPK/ERK is enhanced by the exogenous expression of axonal NRG1 type III
While myelin thickness was increased with NRG1 type III transgene expression, EGR2 was unaltered. Therefore, we asked whether we could identify the transcriptomic signature of one of the canonical pathways downstream of NRG1 type III that could explain the increase in myelination in sciatic nerves of Nrg1t3 and Nrg1t3;Q215X mice.
We conducted a transcriptomic analysis of WT, Q215X, Nrg1t3 and Nrg1t3;Q215X sciatic nerves at 28 days of age, using mRNA next-generation sequencing. We looked for available gene libraries of signaling pathways downstream ErbB2/3/NRG1 type III, such as EGR2, NFATc4 and MAPK/ERK. We then systematically calculated the Pearson’s correlation coefficient (r) as measure of linear correlation on fold-change expression between our subset of gene-regulated downstream of NRG1 type III and subset of gene known to be regulated downstream of NRG1 type III–related pathways.
We first used a list of gene-regulated downstream of a hypomorphic Egr2 allele (Egr2Lo/Lo) expressed in mouse sciatic nerve at 1 month old (46), which mimics an Egr2 loss of function. Thus, a negative correlation between Nrg1t3 and Egr2Lo/Lo mice transcriptome should reveal the activation of EGR2 downstream of ErbB2/3/NGR1 type III signaling pathway. The absence of correlation, indicated by a coefficient r = 0.27 between Erg2Lo/Lo and Nrg1t3 and r = 0.19 between Erg2Lo/Lo and Nrg1t3;Q215X (Fig. 5A), confirmed that the hypermyelination mediated by HA-NRG1 type III was uncoupled from EGR2-regulated genes.
Figure 5.
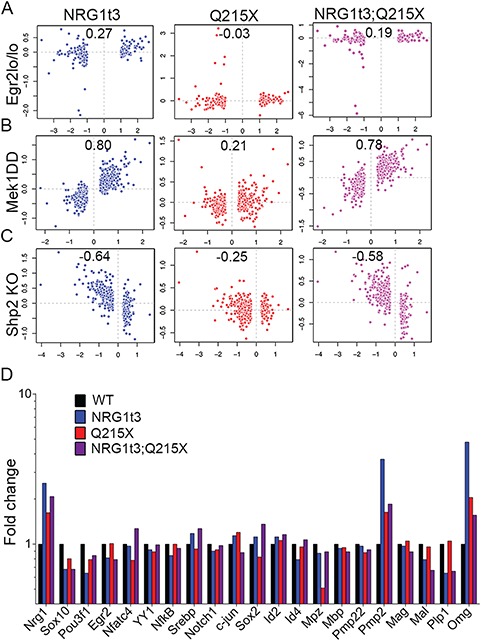
Transcriptomic signature in Nrg1t3 sciatic nerves correlates with the signature of activation of the MAPK/ERK pathway. (A–C) Pearson’s correlation between RNA deep sequencing from 1-month-old sciatic nerve of NRG1t3 (left panel in blue), Q215X (middle panel in red) and Nrg1t3;Q215X (right panel in purple) and targets downstream of EGR2 and ERK. (A) Pearson’s correlation analysis with combined ChIP-Sequencing and arrays data sets obtained from mice homozygous for an Egr2 hypomorphic allele (Egr2lo/lo). Pairwise correlation analysis showed no correlation between the exogenous expression of Nrg1t3 and EGR2 downstream regulated genes. (B) Pearson’s correlation analysis with arrays data sets obtained from conditional Schwann-specific constitutive active form of MEK1 (Mek1DD;Egr2Cre), which phosphorylates ERK. Pairwise correlation showed a high correlation between the exogenous expression of NRG1 type III and MEK1 downstream–regulated genes (panel B blue and purple). (C) Pearson’s correlation analysis with arrays data set generated from Schwann cell conditional knockout for Shp2 (Egr2Cre), which dephosphorylates ERK. Pairwise correlation showed an anticorrelation between exogenous expression of HA-NRG1 type III and Shp2 loss of function (panel C blue and purple). (D) Fold change of transcripts for NRG1, transcription factors regulating myelination and myelin genes obtained from mRNA deep sequencing in Nrg1t3, Q215X and Nrg1t3;Q215X in 1 month mice sciatic nerve. The quantification showed an overall unchanged level of the major regulators and components of the myelin, apart from Pmp2 (a fatty acid binding protein, Fabp8) and Omg.
Second, although NFATc4 is thought to regulate Sox10 and Egr2 expression (18), no comprehensive analysis of NFATc4 target genes in Schwann cells has been published. Given that NFATc4 is a nuclear partner of SOX10 (18) and that SOX10 is a transcriptional activator of myelination, we used a hypergeometric-based test and data mining to first assess whether the sets of regulated genes from RNA sequence analysis of experimental nerves were enriched for genes identified by ChIP-Seq analysis of enhancers bound by SOX10. Previous studies had localized peripheral nerve enhancers through ChIP-seq of histone H3K27 acetylation, and Sox10-bound enhancers were identified (45,52,53). Sox10-bound enhancers were analyzed for the presence of NFAT motifs and annotated to the nearest expressed gene in peripheral nerve. By dividing these putative SOX10 target genes into two categories—either with or without NFATc4 motifs within 50 kb up or downstream of the gene—we inferred putative NFATc4 regulation. This analysis showed that genes regulated in nerves from either Nrg1t3 versus WT, or NRg1t3;Q215X versus Q215X, were enriched specifically for SOX10 target genes with NFATc4 motifs (Supplementary Material, Table 1). Surprisingly, those genes were significantly downregulated (Supplementary Material, Table 1), indicating that exogenous expression of HA-NRG1 type III activates less, or may even repress, NFATc4/SOX10 target genes.
Finally, to assess if genes known to be regulated downstream of the MAPK/ERK pathway correlate with those regulated downstream of ErbB2/3/NRG1 type III, we used an expression profile obtained from 1-month-old transgenic mice expressing a Schwann cell–specific constitutive active form of the kinase MEK1 (Mek1DD) that promotes the sustained activation of ERK1/2 (51,54). Pearson’s correlation analysis between the two gene lists showed a striking correlation, with a coefficient r = 0.8 between Mek1DD and Nrg1t3 and r = 0.78 between Mek1DD and Nrg1t3;Q215X (Fig. 5B). No correlation was found between Q215X and Mek1DD (Fig. 5B), indicating no contribution of the P0 mutant protein itself to the regulation of MEK1-ERK pathway.
We further confirmed the activation of ERK-related genes downstream of NRG1 type III, as we found a negative correlation between our subsets and the phosphatase Shp2 KO gene subset, an upstream regulator of ERK1/2 phosphorylation (15) (Fig. 5C).
Our transcriptomic results showed that among the known canonical pathways downstream of ErbB2/3/NRG1 type III, the activation of MAPK/ERK is highly correlated to an increased level of NRG1 type III. Among the genes downstream the MAPK/ERK activation pathway, we found the upregulation of fatty acid basic protein Pmp2 (encoded by Fabp8) and of myelin oligodendrocytes glycoprotein [encoded by oligodendrocyte myelin glycoprotein (Omg)] (Fig. 5D). Furthermore, we found that none of the major myelin genes, such as Mpz, Pmp22, Mag and Mbp, as well as Egr2, were found deregulated in Nrg1t3 transgenic mice (Fig. 5D). To summarize, our analyses revealed that the gene molecular signature downstream of NRG1 type III transgenic expression is very similar to the constitutive activation of MAPK/ERK pathway. Altogether, these results suggest that axonal HA-NRG1 type III expression stimulates myelin growth in an ERK-dependent manner and activates myelin genes such as peripheral myelin protein 2 (Pmp2) and oligodendrocyte myelin glycoprotein (Omg) (55,56).
MAPK/ERK constitutive activation in Schwann cells increases myelination in a model of congenital hypomyelination
Our data indicated that the ERK pathway is triggered downstream of the exogenous expression of HA-NRG1 type III. Therefore, we assessed whether the Schwann cell–specific activation of the MAPK/ERK pathway could also improve myelin functionality in the Q215X neuropathic context. The expression of a constitutively activated form of MEK1 was restricted to Schwann cells by breeding transgenic mice hemizygous for Mek1DD (50) with P0-Cre mice (Mek1DD;P0Cre). Morphological and ultrastructure analyses from cross sections of sciatic nerves of 1-month-old animals showed a strong increase in myelin thickness in Mek1DD;P0Cre;Q215X sciatic nerves, with a g-ratio of 0.39 ± 0.002 in Mek1DD;P0-Cre (Fig. 6A and B), which is even lower than Nrg1t3;Q215X animals (Fig. 2B). Similarly to Nrg1t3 sciatic nerves, we observed that small caliber axons were predominantly hypermyelinated with Mek1DD;P0Cre and present numerous small caliber myelinated axons (Fig. 6C; Supplementary Material, Fig. 4). Also, no defect in myelin compaction was observed in Mek1DD;P0Cre mutants at 1 month (Fig. 6F–G).
Figure 6.
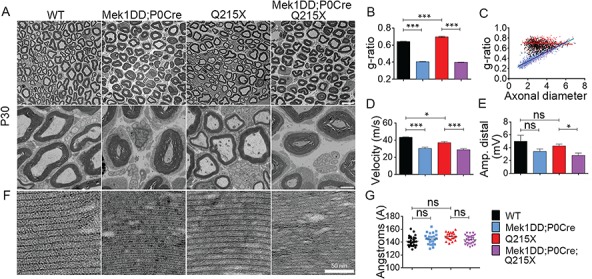
Constitutive activation of MAPK/ERK increases peripheral myelination in normal and Q215X genetic context, but it is functionally detrimental. (A) Semithin cross sections stained with toluidine blue (top panel) and electron micrographs (bottom panel) from WT, Q215X, Mek1DD hemizygote and P0-Cre hemizygote (Mek1DD;P0Cre) and Mek1DD hemizygote, P0-Cre hemizygote and Q215X (Mek1DD;P0Cre;Q215X) sciatic nerves at 1 month. Mek1DD;P0Cre and Mek1DD;P0Cre;Q215X fibers are hypermyelinated, and Q215X is hypomyelinated. Scale bars, 10 μm (top panel) and 2 μm (bottom panel). (B) g-ratio was increased in Q215X (0.70 ± 0.008) and decreased in Mek1DD;P0Cre (0.40 ± 0.002) and Mek1DD;P0Cre;Q215X (0.39 ± 0.002) compared to WT (0.64 ± 0.002), indicating hypomyelination and strong hypermyelination, respectively. (C) g-ratio as a function of axonal diameter showed preferential hypermyelination of small fibers in Mek1DD;P0Cre and Mek1DD;P0Cre;Q215X. (D, E) Neurophysiological studies showed significantly reduced NCV in all mutants. The amplitude of compound muscle action potentials was decreased in Mek1DD;P0Cre;Q215X, suggesting axonal loss. (F, G) Myelin compaction (F) and periodicity (G) were not altered in Mek1DD;P0Cre and Mek1DD;P0Cre;Q215X sciatic nerves. Scale bar, 50 nm. Analyses were performed using three mice per genotype. Error bars indicate s.e.m. Statistical analyses were performed using one-way ANOVA with Bonferroni’s multiple comparison test. *P < 0.05, **P < 0.01, ***P < 0.001.
NCVs were significantly reduced in Mek1DD;P0Cre mutants (Fig. 6D), and the distal amplitude of Mek1DD;P0Cre;Q215X was reduced, suggesting an axonal loss (Fig. 6E). Overall, MAPK/ERK pathway activation seems to have contrasting effects depending on its mode of activation. Beneficial effect on myelination was seen downstream of HA-NRG1 type III transgenic expression, while the direct constitutive activation of MAPK/ERK pathway showed increased myelin thickness with detrimental effect on myelin functionality.
HA-Nrg1 type III expression promotes changes in myelin composition
Similar to the constitutive activation of MEK1 (Mek1DD;P0Cre or Mek1DD;Egr2Cre) (51), the transgenic expression of HA-NRG1 type III increased myelin thickness independent of the major myelin gene transcription factor EGR2 and of myelin protein synthesis (Fig. 5D and 7A). However, among the genes correlated to MAPK/ERK activation pathway, we found increased levels of Pmp2 transcript and protein (Fig. 7A; Supplementary Material, Fig. 5), a myelin protein with functions in lipid homeostasis in myelinated Schwann cells (55). As in several previous studies (55,57–59), we observed that cellular expression of PMP2 in WT cross sections of sciatic nerve was found in compact myelin but also had cytoplasmic expression with a mosaic distribution between fibers (Supplementary Material, Fig. 5B). In the presence of HA-NRG1 type III transgenic expression, PMP2 level was clearly increased while maintaining its mosaic distribution in sciatic nerve cross section at 1 month (Supplementary Material, Fig. 5B). The specific increase in PMP2 suggests that NRG1 type III overexpression may determine a change in myelin protein composition.
Figure 7.
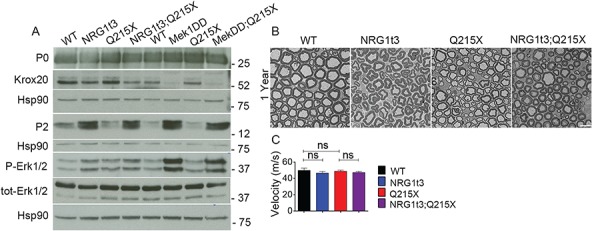
Expression of HA-NRG1 type III transgene increases PMP2 protein level. (A) Western blot analysis from WT, Nrg1t3, Q215X, Nrg1t3;Q215X, Mek1DD;P0Cre and Mek1DD;P0Cre;Q215X sciatic nerves at 1 month. Expression of HA-NRG1t3 transgene in sciatic nerves or constitutive activation of the MAPK/ERK pathway in Schwann cells leads to an increase in ERK phosphorylation, a decrease in EGR2 protein levels and an increase in PMP2 protein levels. These effects are enhanced in Mek1DD;P0Cre mutants as compared to HA-NRG1t3 mutants. Western blots were cropped. (B) Semithin cross sections stained with toluidine blue (top panel) and electron micrographs (bottom panel) from WT, Nrg1t3 hemizygote (Nrg1t3), P0Q215X heterozygote (Q215X) and Nrg1t3 hemizygote and P0Q215X heterozygote (Nrg1t3;Q215X) sciatic nerves at 1 year.
In conclusion, both elevated NRG1 type III expression and MEK1 activation increase ERK phosphorylation but have different outcomes on myelin structure. The activation of MAPK/ERK pathway results in hypermyelination associated to NCV deficits (Fig. 6D), and its prolonged activation results in the formation of aberrant myelin structure, i.e. misfolded myelin structures increasing with age, onion bulbs and compressed axons (51,60). On the contrary, sustained HA-NRG1 type III transgenic expression for 1 year did not alter nerve physiology, and we did not observe any abnormal myelin structure (Fig. 2A and 7B), as previously reported (7,30).
Discussion
In this study, we explored a suitable treatment for a severe early-onset Charcot–Marie–Tooth disease subtype, CHN. We developed and characterized a novel mouse model of CHN, associated with a truncation of P0 cytoplasmic tail (P0Q215X) (25). We showed that animals harboring the Q215X mutation present decreased expression of major myelin genes and proteins and displayed the hypomyelinating features of a human CHN.
In the PNS, extrinsic signals from axonal NRG1 type III regulate Schwann cell fate and myelination (6). Thus, the modulation of NRG1 signaling is considered as a possible approach for the restoration of myelination defects in myelin diseases (12,61). NRG1 type III promotes myelination by stimulating several signaling pathways, including PI3K/AKT, CaN/NFATc4 and MAPK/ERK, which are thought to converge on the activation of EGR2 (Fig. 8). These pathways are tightly regulated. PI3K/AKT or MAPK/ERK pathways are both necessary for myelin protein expression (20,62,63), and imbalances in PI3K/AKT and MAPK/ERK pathways have been reported in two different demyelinating neuropathies, Charcot–Marie–Tooth type 1A (12,64) and type 1X (65). Conversely, sustained activation of either PI3K/AKT or MAPK/ERK pathways leads to hypermyelination in part through the enhancement of protein synthesis (51,60,62).
Figure 8.
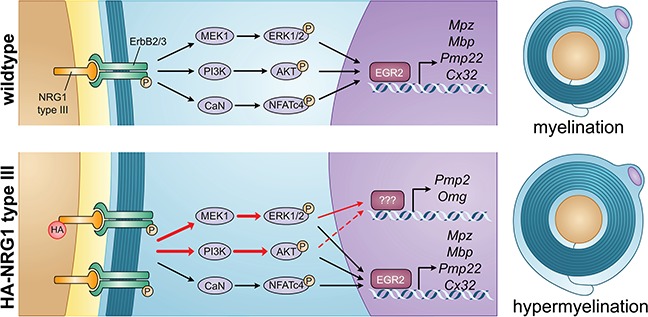
Regulation of myelination by NRG1 type III. In WT animals, NRG1 type III/ ErbB2/3 promotes myelination by stimulating several signaling pathways, including PI3K/AKT, MAPK/ERK and CaN/NFATc4, which are thought to converge on the activation of EGR2. In Nrg1t3 animals, the exogenous expression of NRG1 type III leads to the activation of both PI3K/AKT and MAPK/ERK pathways (red arrows) and the increased transcription of Pmp2 and Omg.
Here, we show that in two different mouse models of hypomyelinated diseases i.e. CHN (this paper) and CMT1B (see accompanying paper by Scapin et al.), no initial alteration in PI3K/AKT and MAPK/ERK levels or activation was observed. Therefore, we expected that expression of exogenous NRG1 type III could improve functional outcomes in these pathologies. Indeed, we obtained the first evidences that in a CHN animal model, overexpression of NRG1 type III leads to increased activation of both PI3K/AKT and MAPK/ERK pathways, triggered the expression of genes restricted to the MAPK/ERK pathway and increases myelin thickness. We then showed that the sustained activation of MEK1-ERK in Schwann cell promotes an even stronger increase in myelin thickness but accompanied by an alteration in NCV in both normal (as suggested in previous studies (51,60)) and pathologic contexts, providing further evidences for the contrasting effect of ERK phosphorylation depending on its level of expression (60). Finally, while NRG1 type III overexpression and MEK1-ERK–sustained activation increased myelin thickness, both were uncoupled from increased expression of EGR2 or major myelin genes and proteins.
Through a comprehensive transcriptomic analysis of signaling downstream of NRG1 type III in the context of neuropathy, we documented that in NRG1 type III overexpressing nerves, genes regulated by the MAPK/ERK pathway were upregulated and may contribute to increased myelin thickness in Schwann cells. Among these genes we identified Pmp2, encoding for the PMP2, and Omg, encoding for the OMG. Pmp2 is expressed in myelinating cells (66), but only some myelinated fibers appear to maintain PMP2 (55,59). PMP2 seems to have a role in lipid homeostasis of myelinating Schwann cells, with recent evidences that PMP2 binds fatty acids (55,67). This is particularly interesting as NRG1 was also identified upstream of MAF, a regulator of cholesterol biosynthesis (68), and in mice overexpressing NRG1 type III, cholesterol along with saturated and unsaturated fatty acids are increased (see accompanying paper Scapin et al.). On the other hand, there are conflicting reports on the expression of OMG in Schwann cells (69–71), and the function of OMG is unknown. Total knockout mice revealed that both PMP2 and OMG are dispensable to the formation or maintenance of myelin structure in the PNS and central nervous system, respectively (55,71). However, two recent studies in humans revealed that de novo dominant mutations in PMP2 were identified in families with type 1 Charcot–Marie–Tooth demyelinated disease (72,73). Thus, it has been suggested that mutations in PMP2 result to a probable toxic gain of function in myelinating Schwann cells causing demyelination (72,73). Here, the hyperactivation of the MAPK/ERK pathway in Nrg1t3 animals appears to stimulate specifically the transcription of these less-studied myelin proteins. The understanding of PMP2 and OMG function in Nrg1t3 animals could help us to understand further the mechanisms that drive hypermyelination.
MAPK/ERK activation has been shown to have beneficial promyelinating function during normal development (14,15,74) and to promote remyelination after injury (63). Paradoxically, the activation of MAPK/ERK in Schwann cells has been also associated to detrimental effects on Schwann cell survival, development, myelination, remyelination (16,17,49,63) and axonal regeneration (75). The discrepancies between these studies could be explained in part by the context, the timing and the level of MAPK/ERK activation. A limited activation of MAPK/ERK, as in Nrg1t3 mice, was not associated to myelin structural or functional alterations, in accordance with previous histological examination in Nrg1t3 sciatic nerve (7,30). However, we and others showed that a sustained activation of MEK1 is associated to aberrant myelin structures and affects axon and myelin integrity (51,60). Interestingly, we noticed that a strong decrease of EGR2 protein level specifically correlated with the sustained activation of MEK1 in Schwann cells, suggesting a dose effect of ERK phosphorylation levels on the regulation of EGR2 expression.
It is possible that in Nrg1t3 nerves, other molecular mechanisms downstream of NRG1 type III limit MAPK/ERK activation over time, in contrast to the constitutively activated MAPK/ERK pathway. For example, we found a decreased activation of genes with NFATc4 motifs, while the total amount and the active (nuclear) form of NFATc4 were unchanged. In addition, we saw an upregulation of the PI3K/AKT pathway, which might be important to balance MAPK/ERK signaling, as suggested previously (12). Also, possible upstream mechanisms such as laminin and laminin receptors can limit NRG1 type III effect on myelination (76–79).
Independently of the mechanism, we show that exogenous expression of HA-NRG1 type III has no detrimental effects on peripheral myelination and improved myelin thickness and NCV in both CHN and CMT1B (see accompanying paper by Scapin et al.). Thus, our observations suggest that controlled activation of NRG1 type III could represent a favorable therapeutic approach in several human peripheral neuropathies associated with hypomyelination. During Schwann cell myelination, NRG1 type III activity is tightly regulated by complex and stepwise proteolytic cleavages by BACE1, TACE, GPR44 and I-CLips (48,80–83). It is therefore possible to regulate the activity of specific proteases to favor the cleavage products of NRG1 type III beneficial for myelination. By this principle, inhibition of TACE improved myelination in a hypermyelinating model CMT4B, showing that modulation of NRG1 type III signaling in both directions can be beneficial in different CMT neuropathies (61). Interestingly, the pathway-involved downstream of NRG1-III appears to be different: AKT in the CMT4B study and ERK in our study. This difference may be due to the fact that the CMT4B1 model presents focal hypermyelination (outfoldings) that may be controlled by local AKT levels [see also (84)], while the thickness of myelin itself is more likely to be controlled by ERK [see also (50)]. However, the modulation of a single protease in vivo can be challenging because inhibition of these proteases in regions other than peripheral nerves could have unwanted corollary effects; for example, TACE inhibition might promote amyloid production in the brain, and sustained activation of MAPK/ERK in non-myelinating Schwann cells promotes corneal neurofibromas (85). Finally, NRG1 type III is expressed in neurons of the central nervous system, and exogenous expression of HA-NRG1 type III induces hypermyelination in the corpus callosum (19,86). The consequences of this increase in myelination in the brain have not been fully investigated, but several studies have linked the increase of NRG1 type III to schizophrenia, possibly by disrupting cortical neuronal synaptic plasticity and altering synaptic transmission (37–39). Thus, a better understanding of the function of NRG1 type III and its proteases in adult Schwann cells is required to manipulate NRG1 signaling, specifically in the PNS.
In conclusion, this study shows encouraging results for the application of NRG1 type III modulations in improving myelination in the context of peripheral hypomyelinated neuropathies.
Materials and Methods
Animal model
All animal experimentations were performed in strict accord with Institutional Animal Care and Use Committee and approved protocols of San Raffaele Institute and University at Buffalo (UB) (n. 363 and UB1188M, respectively). P0Q215X knock-in mice (hereafter called Q215X) contain a targeted mutation in Mpz exon 5 (25). NRG1 type III hemagglutinin-tagged transgenic mice (hereafter called Nrg1t3), expressing Nrg1 type III under the control of neuronal promoter Thy1.2, were previously described (30). The R26StopFLMek1DD mice (hereafter called Mek1DD), expressing conditionally an activated form of MEK1, were previously described (87). Mek1DD mice were crossbred with P0-Cre transgenic mice that express Cre recombinase starting at E13.5 (88,89). We used a reporter mouse model containing a basal Hsp68 promoter upstream of the luciferase reporter gene activated by the +11 kb human PMP22 intronic enhancer containing a combinatorial Egr2/Sox10 binding site, transgenic luciferase line D (hereafter called Luc D) (47).
Q215X and P0-Cre mice were in FVB/N congenic background. Nrg1t3 and Mek1DD mice were in C57BL/6 background. F1 mice were respectively generated by crossbreeding Q215X mice with the transgenic Nrg1t3 mice and Q215X with Mek1DD in C57BL/6xFVB/N mixed background. Luciferase mice were in mixed background C57BL/6xFVB/N/CBA. Genotyping of mutant P0-Cre, Nrg1t3 and Mek1DD mice was performed by polymerase chain reaction (PCR) on tail genomic DNA, as described previously (7,47,87,88). The genotype of Q215X animals was determined by PCR analysis with the Mpz forward primer 5′-CAACCTCTCTTGCCACAGTG-3′ and the Mpz reverse primer 5′-GCTAACCGCTATTTCTTATCC-3′ that amplified 400 and 280 nucleotide fragments for Q215X and WT allele, respectively. PCR conditions were 94°C for 1 min, 56°C for 1 min and 72°C for 1 min (29 cycles), followed by 10 min extension at 72°C, in a standard PCR reaction mix.
No animals were excluded from the study. Animals were housed in cages of no more than five animals in 12 h light/dark cycles. Mutant and control littermates from either sex were sacrificed at the indicated ages, and sciatic nerves were dissected.
Functional analyses
Grid-walking test was used to measure the frequency of step errors on rungs of an elevated grid. Littermate mice were allowed to walk on a mesh grid for five trials of 5 min each, with the first three trials considered as the training period. Hindpaw placement errors (foot faults) defined as passage of the hindpaw through an opening in the grid were determined as a proportion of total hindpaw steps on horizontal grid mesh (1.2 × 1.2 cm openings). Sciatic nerve motor conduction velocity and amplitude were obtained as previously described (90,91).
Morphology
Semithin section and electron microscopic analyses of sciatic nerves were performed as previously described (92). For g-ratio (axon diameter/fiber diameter) and axonal distribution, four semithin images per sciatic nerve were acquired with the 100× objective. Three to four animals per age per genotype were analyzed. Axon and fiber diameters were quantified using the Leica QWin software (Leica Microsystem, Buffalo Grove, IL). Data were analyzed using GraphPad Prism 6.01. Periodicity was measured at 20000× magnification as previously described (26). Immunoelectron microscopy was performed as in Quattrini et al. (92). Primary antibody was chicken anti-P0 (Aves Labs, Tigard, OR). Gold-conjugated secondary antibodies were 10 nm for P0 (British Biocell International, Cardiff, UK). For teased fibers, sciatic nerves were fixed in 2% glutaraldehyde overnight at 4°C, washed three times in phosphate buffer (79 mm Na2HPO4, 21 mm NaH2PO4, pH 7.4). Sciatic nerves were stained in 1% osmium, washed four times in phosphate buffer and incubated at 55°C for 12 h in 30% glycerol, followed by 12 h in 60% glycerol and 12 h in 100% glycerol.
Western analysis
Sciatic nerves were dissected, frozen in liquid nitrogen, pulverized and resuspended in lysis buffer [150 mm NaCl, 25 mm HEPES, 0.3% CHAPS, pH 7.4, 1 mm Na3VO4, 1 mm NaF and 1:100 Protease Inhibitor Cocktail (Roche Diagnostic, Florham Park, NJ)]. Protein lysates were centrifuged at 13 000 g for 20 min at 4°C. Supernatant protein concentrations were determined by bicinchoninic acid assay protein assay (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions. Equal amounts of homogenates were diluted 3:1 in 4× Laemmli (250 mm Tris-HCl, pH 6.8, 8% sodium dodecyl sulfate, 8% β-Mercaptoethanol, 40% Glycerol, 0.02% Bromophenol Blue), denatured 5 min at 100°C, resolved on SDS-polyacrylamide gel and electroblotted onto PVDF membrane. Blots were then blocked with 5% bovine serum albumin in 1× Phosphate-buffered saline (PBS), 0.05% Tween-20 and incubated overnight with the following appropriate antibodies: Cell Signaling Technology, Danvers, MA: anti-AKT (9272) 1/1000, anti-p-AKT Ser473 (9271) 1/1000, anti-p-AKT Thr308 (5106) 1/1000, anti-ERK 1/2 (9102) 1/1000, anti-p-ERK 1/2 p44/42 (9101) 1/1000, anti-NFATc4 (2183) 1/1000; Aves Labs, Tigard, OR: anti-P0 (PZO) 1/5000; Covance, San Diego, CA: anti-MBP (SMI94) 1/1000, anti-TUJ1 (MMS-435P) 1/5000; Sigma, St. Louis, MO: anti-GAPDH (G9545) 1/2000, anti-Calnexin (C4731) 1/3000; Santa Cruz: anti-PMP2 (sc-374058), anti-HSP90 (sc-7947) 1/1000; Abcam, Cambridge, MA: anti-PMP22 (ab15506) 1/1000, anti-ErbB2 (ab2428) 1/200, anti p-ErbB2 Y1248 (ab47755) 1/1000. Invitrogen, Waltham, MA: anti-MAG (34-6200) 1/1000; Roche Diagnostic, Florham Park, NJ: anti-HA (11867423001) 1/200. anti-EGR2 1/200 was gifted by Dr Meijer of the Centre for Neuroregeneration, Edinburgh. Membranes were then rinsed in 1× PBS and incubated for 1 h with secondary antibodies. Blots were developed using ECL, ECL plus (GE Healthcare, Chicago, IL) or Odyssey CLx infrared imaging system (Li-Cor, Lincoln, NE). Western blots were quantified using Image J software (http://imagej.nih.gov/ij). Each experiment was performed at least two times on nerves from at least three animals.
Immunofluorescence and immunohistochemistry
For immunohistochemistry, sciatic nerves were fixed with 4% paraformaldehyde and embedded in Optimal Cutting Temperature compound. Cross sections were permeabilized with cold MetOH. Sections were rinsed in 1× PBS, blocked for 1 h in 20% fetal bovine serum, 2% BSA and 0.1% Triton X-100 in 1× PBS. The following primary antibodies were incubated overnight: Proteintech, Chicago, IL anti-PMP2 (12717-AP) 1/500 and Aves Labs, Tigard, OR anti-P0 (PZO) 1/1000. Sections were rinsed in 1× PBS, incubated 1 h with Jackson, Bar Harbor, ME TRITC-conjugated (711-025-152) 1/200 and with Jackson, Bar Harbor, ME DyLight 488 1/1000, stained with DAPI and mounted with Vectashield (Vector Laboratories, Burlingame, CA). Images were acquired with a Zeiss ApoTome.
RNA extraction and quantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted from sciatic nerves using TRIzol® (Life Technologies, Carlsbad, CA). Several pools of nerves of two to six nerves were used, depending on the animal age. For RT-QPCR analysis, first strand cDNA was synthesized from 1 μg total RNA using polythymine and random primers with Superscript III RNAse H reverse transcriptase (Invitrogen, Waltham, MA). The expression of selected mRNAs was determined by quantitative real-time PCR using 20 ng of cDNA. Samples were processed in triplicate, and reactions without target cDNA were used as negative control for each reaction. PCRs were performed on 96-well plates using the Power SYBR Green PCR Master Mix (Applied Biosystems, Waltham, MA), Taqman probe assays or FastStart Universal Probe Master (Roche Diagnostic, Florham Park, NJ) following the manufacturer’s conditions. Data were analyzed using the threshold cycle (Ct) and 2(-ΔΔCt) method. Actb, Hsp90, Gapdh or Pgk1 genes were used as reference in normalizing the data. The Taqman probes used are the following: Actb (Mm00607939-S1), Gapdh (Mm99999915-g1), Pgk1 (Mm00435617), Pmp2 (Mm03015237_m1), Mpz (Mm00485139-m1), Mbp (Mm01262035), Mag (Mm00487541-m1), Egr2 (Mm00456650-m1) and Pmp22 (Mm01333393_m1).
RNAseq analysis
Sciatic nerves at P30 were dissected, frozen in liquid nitrogen and pulverized. The total RNA was prepared from pools of two nerves with TRIzol (Roche Diagnostic, Florham Park, NJ), then purified with RNeasy column (Qiagen, Germantown, MD). Samples were quantified using Ribogreen Assay (Invitrogen, Waltham, MA), and the quality of samples was checked using Agilent Bioanalyzer 2100 RNA nano 6000 chip (Agilent, Santa Clara, CA). Illumina TruSeq RNA sample preparation kit (Illumina, San Diego, CA) was used to prepare cDNA libraries from RNA samples. Samples were poly A selected to isolate mRNA, the mRNA was cleaved into fragments and the first strand reverse transcribed to cDNA using SuperScript II reverse Transcriptase (Invitrogen, Waltham, MA) and random primers, followed by second strand cDNA synthesis using Second Strand Master Mix supplied with the kit. After the end repair, the addition of a single ‘A’ base and ligation with adapters, the products were enriched and purified with PCR to create the final cDNA library as per the manufacturer’s protocol. cDNA libraries were quantified using Picogreen Assay (Invitrogen, Waltham, MA) and Library Quantification kit (Kapa Biosystems). Agilent Bioanalyzer 2100 DNA 7500 chip was used to confirm the quality and size of the cDNA libraries. The cDNA libraries were then normalized, pooled and paired-end sequenced (100 standard cycles) using the Illumina HiSeq2500, following the manufacturer’s instructions at the UB Genomics and Bioinformatics Core Facility (Buffalo, NY). Reads were aligned to the mouse reference genome (mm10) with the transcript annotation (The University of California, Santa Cruz (UCSC) Genome Browser mm10 annotation downloaded from TopHat website) using TopHat (v2.0.9). Quantification for the gene expression and differential expression analysis were done using the Cufflinks software (v2.2.0). Data are available at http://www.ncbi.nlm.nih.gov/geo/ with accession number GSE101808.
Experimental design
Experiments were not randomized, but data collection and analysis were performed blindly to the conditions of the experiments. Researchers blinded to the genotype performed behavioral analyses, NCVs and morphometric analyses. This study was not preregistered. Most experiments were conducted with three animals per age and per genotype. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally employed in the field.
Statistical analyses
The data obtained are presented in mean ± s.e.m. One-way analysis of variance (ANOVA) with Bonferroni’s comparison test was used for statistical analysis of the differences among multiples groups. Values of P < 0.05 were considered to represent a significant difference.
Each Pearson’s correlation coefficient (r) was calculated from sets of genes selected with a fold change cutoff of ≥1.2. Correlation plots show the linear relationship between gene sets obtained from P30 sciatic nerves.
A hypergeometric-based test (Fisher’s exact test) interrogated for enrichment of regulated genes from RNA sequencing from nerves of various genotypes, in subsets of regulated genes selected from Sox10 ChIP-Seq (45,52) with or without NFATc4 motif enrichment. Motifs (MA0152.1 Jaspar) were called using the Homer annotate Peaks program with option -m (93). The rationale for using the hypergeometric test was that if gene-activated downstream of Nrg1 type III had any biological or functional association with the CaN/NFATc4 pathway, a higher number of genes would be found regulated in nerve and present in ChIP-Seq libraries than expected by chance.
Supplementary Material
Acknowledgements
We thank Edward Hurley for semithin sections and electron microscopic preparations.
Conflict of Interest statement. None declared.
Funding
Telethon Grant (GPP10007 to L.W., M.L.F and C.T.); National Institute of Neurological Disorders and Stroke (R01NS096104 to L.W.); Charcot-Marie-Tooth Association (CMT1B-003 to L.W.); Peripheral Nerve Society Fellowship Grant for research in neuropathy 2012–2013 (to S.B.).
Author contributions
S.B. and L.W. designed the research and analyzed and interpreted the data. S.B., F.O. and G.S. performed the majority of the research. J.W. performed RNAseq analysis. C.L. and J.S. performed motif enrichment analysis. N.S. performed electrophysiology analysis. C.S., N.R., C.W., C.T., Y.P. and M.D. provided technical assistance. A.I., R.B., M.S. and K.N. contributed analytical tools. S.B., Y.P. and L.W. wrote the manuscript. M.L.F. and L.W. provided funding and scientific direction. J.S., R.B., M.S., P.F., Y.P., M.D. and M.L.F. critically reviewed the manuscript.
References
- 1. Meyer D. and Birchmeier C. (1995) Multiple essential functions of neuregulin in development. Nature, 378, 386–390. [DOI] [PubMed] [Google Scholar]
- 2. Riethmacher D., Sonnenberg-Riethmacher E., Brinkmann V., Yamaai T., Lewin G.R. and Birchmeier C. (1997) Severe neuropathies in mice with targeted mutations in the ErbB3 receptor. Nature, 389, 725–730. [DOI] [PubMed] [Google Scholar]
- 3. Lin W., Sanchez H.B., Deerinck T., Morris J.K., Ellisman M. and Lee K.F. (2000) Aberrant development of motor axons and neuromuscular synapses in erbB2-deficient mice. Proc. Natl. Acad. Sci. U. S. A., 97, 1299–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garratt A.N., Voiculescu O., Topilko P., Charnay P. and Birchmeier C. (2000) A dual role of erbB2 in myelination and in expansion of the schwann cell precursor pool. J. Cell Biol., 148, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birchmeier C. and Nave K.A. (2008) Neuregulin-1, a key axonal signal that drives Schwann cell growth and differentiation. Glia, 56, 1491–1497. [DOI] [PubMed] [Google Scholar]
- 6. Taveggia C., Zanazzi G., Petrylak A., Yano H., Rosenbluth J., Einheber S., Xu X., Esper R.M., Loeb J.A., Shrager P. et al. (2005) Neuregulin-1 type III determines the ensheathment fate of axons. Neuron, 47, 681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Michailov G.V., Sereda M.W., Brinkmann B.G., Fischer T.M., Haug B., Birchmeier C., Role L., Lai C., Schwab M.H. and Nave K.A. (2004) Axonal neuregulin-1 regulates myelin sheath thickness. Science, 304, 700–703. [DOI] [PubMed] [Google Scholar]
- 8. Nave K.A. and Salzer J.L. (2006) Axonal regulation of myelination by neuregulin 1. Curr. Opin. Neurobiol., 16, 492–500. [DOI] [PubMed] [Google Scholar]
- 9. Gomez-Sanchez J.A., Lopez de Armentia M., Lujan R., Kessaris N., Richardson W.D. and Cabedo H. (2009) Sustained axon-glial signaling induces Schwann cell hyperproliferation, Remak bundle myelination, and tumorigenesis. J. Neurosci., 29, 11304–11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fricker F.R., Lago N., Balarajah S., Tsantoulas C., Tanna S., Zhu N., Fageiry S.K., Jenkins M., Garratt A.N., Birchmeier C. et al. (2011) Axonally derived neuregulin-1 is required for remyelination and regeneration after nerve injury in adulthood. J. Neurosci., 31, 3225–3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stassart R.M., Fledrich R., Velanac V., Brinkmann B.G., Schwab M.H., Meijer D., Sereda M.W. and Nave K.A. (2013) A role for Schwann cell-derived neuregulin-1 in remyelination. Nat. Neurosci., 16, 48–54. [DOI] [PubMed] [Google Scholar]
- 12. Fledrich R., Stassart R.M., Klink A., Rasch L.M., Prukop T., Haag L., Czesnik D., Kungl T., Abdelaal T.A., Keric N. et al. (2014) Soluble neuregulin-1 modulates disease pathogenesis in rodent models of Charcot–Marie–Tooth disease 1A. Nat. Med., 20, 1055–1061. [DOI] [PubMed] [Google Scholar]
- 13. Pereira J.A., Lebrun-Julien F. and Suter U. (2012) Molecular mechanisms regulating myelination in the peripheral nervous system. Trends Neurosci., 35, 123–134. [DOI] [PubMed] [Google Scholar]
- 14. Newbern J.M., Li X., Shoemaker S.E., Zhou J., Zhong J., Wu Y., Bonder D., Hollenback S., Coppola G., Geschwind D.H. et al. (2011) Specific functions for ERK/MAPK signaling during PNS development. Neuron, 69, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grossmann K.S., Wende H., Paul F.E., Cheret C., Garratt A.N., Zurborg S., Feinberg K., Besser D., Schulz H., Peles E. et al. (2009) The tyrosine phosphatase Shp2 (PTPN11) directs Neuregulin-1/ErbB signaling throughout Schwann cell development. Proc. Natl. Acad. Sci. U. S. A., 106, 16704–16709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maurel P. and Salzer J.L. (2000) Axonal regulation of Schwann cell proliferation and survival and the initial events of myelination requires PI 3-kinase activity. J. Neurosci., 20, 4635–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ogata T., Iijima S., Hoshikawa S., Miura T., Yamamoto S., Oda H., Nakamura K. and Tanaka S. (2004) Opposing extracellular signal-regulated kinase and Akt pathways control Schwann cell myelination. J. Neurosci., 24, 6724–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kao S.C., Wu H., Xie J., Chang C.P., Ranish J.A., Graef I.A. and Crabtree G.R. (2009) Calcineurin/NFAT signaling is required for neuregulin-regulated Schwann cell differentiation. Science, 323, 651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brinkmann B.G., Agarwal A., Sereda M.W., Garratt A.N., Muller T., Wende H., Stassart R.M., Nawaz S., Humml C., Velanac V. et al. (2008) Neuregulin-1/ErbB signaling serves distinct functions in myelination of the peripheral and central nervous system. Neuron, 59, 581–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He Y., Kim J.Y., Dupree J., Tewari A., Melendez-Vasquez C., Svaren J. and Casaccia P. (2010) Yy1 as a molecular link between neuregulin and transcriptional modulation of peripheral myelination. Nat. Neurosci., 13, 1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giese K.P., Martini R., Lemke G., Soriano P. and Schachner M. (1992) Mouse P0 gene disruption leads to hypomyelination, abnormal expression of recognition molecules, and degeneration of myelin and axons. Cell, 71, 565–576. [DOI] [PubMed] [Google Scholar]
- 22. Xu W., Manichella D., Jiang H., Vallat J.M., Lilien J., Baron P., Scarlato G., Kamholz J. and Shy M.E. (2000) Absence of P0 leads to the dysregulation of myelin gene expression and myelin morphogenesis. J. Neurosci. Res., 60, 714–724. [DOI] [PubMed] [Google Scholar]
- 23. Warner L.E., Hilz M.J., Appel S.H., Killian J.M., Kolodry E.H., Karpati G., Carpenter S., Watters G.V., Wheeler C., Witt D. et al. (1996) Clinical phenotypes of different MPZ (P0) mutations may include Charcot–Marie–Tooth type 1B, Dejerine–Sottas, and congenital hypomyelination. Neuron, 17, 451–460. [DOI] [PubMed] [Google Scholar]
- 24. Mandich P., Mancardi G.L., Varese A., Soriani S., Di Maria E., Bellone E., Bado M., Gross L., Windebank A.J., Ajmar F. et al. (1999) Congenital hypomyelination due to myelin protein zero Q215X mutation. Ann. Neurol., 45, 676–678. [DOI] [PubMed] [Google Scholar]
- 25. Fratta P., Ornaghi F., Dati G., Zambroni D., Saveri P., Belin S., D'Adamo P., Shy M., Quattrini A., Laura Feltri M. et al. (2019) A nonsense mutation in Myelin Protein Zero causes congenital hypomyelination neuropathy through altered P0 membrane targeting and gain of abnormal function. Hum. Mol. Genet., 28, 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wrabetz L., D'Antonio M., Pennuto M., Dati G., Tinelli E., Fratta P., Previtali S., Imperiale D., Zielasek J., Toyka K. et al. (2006) Different intracellular pathomechanisms produce diverse Myelin Protein Zero neuropathies in transgenic mice. J. Neurosci., 26, 2358–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pennuto M., Tinelli E., Malaguti M., Del Carro U., D'Antonio M., Ron D., Quattrini A., Feltri M.L. and Wrabetz L. (2008) Ablation of the UPR–mediator CHOP restores motor function and reduces demyelination in Charcot–Marie–Tooth 1B mice. Neuron, 57, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. D'Antonio M., Musner N., Scapin C., Ungaro D., Del Carro U., Ron D., Feltri M.L. and Wrabetz L. (2013) Resetting translational homeostasis restores myelination in Charcot–Marie–Tooth disease type 1B mice. J. Exp. Med., 210, 821–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bai Y., Wu X., Brennan K.M., Wang D.S., D'Antonio M., Moran J., Svaren J. and Shy M.E. (2018) Myelin protein zero mutations and the unfolded protein response in Charcot–Marie–Tooth disease type 1B. Ann. Clin. Transl. Neurol., 5, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Velanac V., Unterbarnscheidt T., Hinrichs W., Gummert M.N., Fischer T.M., Rossner M.J., Trimarco A., Brivio V., Taveggia C., Willem M. et al. (2012) Bace1 processing of NRG1 type III produces a myelin-inducing signal but is not essential for the stimulation of myelination. Glia, 60, 203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Campsall K.D., Mazerolle C.J., De Repentingy Y., Kothary R. and Wallace V.A. (2002) Characterization of transgene expression and Cre recombinase activity in a panel of Thy-1 promoter-Cre transgenic mice. Dev. Dyn., 224, 135–143. [DOI] [PubMed] [Google Scholar]
- 32. Caroni P. (1997) Overexpression of growth-associated proteins in the neurons of adult transgenic mice. J. Neurosci. Methods, 71, 3–9. [DOI] [PubMed] [Google Scholar]
- 33. Kirschner D.A. and Ganser A.L. (1980) Compact myelin exists in the absence of basic protein in the shiverer mutant mouse. Nature, 283, 207–210. [DOI] [PubMed] [Google Scholar]
- 34. Shapiro L., Doyle J.P., Hensley P., Colman D.R. and Hendrickson W.A. (1996) Crystal structure of the extracellular domain from P0, the major structural protein of peripheral nerve myelin. Neuron, 17, 435–449. [DOI] [PubMed] [Google Scholar]
- 35. Lemke G. (1988) Unwrapping the genes of myelin. Neuron, 1, 535–543. [DOI] [PubMed] [Google Scholar]
- 36. Ding Y. and Brunden K.R. (1994) The cytoplasmic domain of myelin glycoprotein P0 interacts with negatively charged phospholipid bilayers. J. Biol. Chem., 269, 10764–10770. [PubMed] [Google Scholar]
- 37. Agarwal A., Zhang M., Trembak-Duff I., Unterbarnscheidt T., Radyushkin K., Dibaj P., Martins de Souza D., Boretius S., Brzozka M.M., Steffens H. et al. (2014) Dysregulated expression of neuregulin-1 by cortical pyramidal neurons disrupts synaptic plasticity. Cell Rep., 8, 1130–1145. [DOI] [PubMed] [Google Scholar]
- 38. Hahn C.G., Wang H.Y., Cho D.S., Talbot K., Gur R.E., Berrettini W.H., Bakshi K., Kamins J., Borgmann-Winter K.E., Siegel S.J. et al. (2006) Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat. Med., 12, 824–828. [DOI] [PubMed] [Google Scholar]
- 39. Weickert C.S., Tiwari Y., Schofield P.R., Mowry B.J. and Fullerton J.M. (2012) Schizophrenia-associated HapICE haplotype is associated with increased NRG1 type III expression and high nucleotide diversity. Transl. Psychiatry, 2, e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee Y.I., Li Y., Mikesh M., Smith I., Nave K.A., Schwab M.H. and Thompson W.J. (2016) Neuregulin1 displayed on motor axons regulates terminal Schwann cell-mediated synapse elimination at developing neuromuscular junctions. Proc. Natl. Acad. Sci. U. S. A., 113, E479–E487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo X., He W., Hu X. and Yan R. (2014) Reversible overexpression of bace1-cleaved neuregulin-1 N-terminal fragment induces schizophrenia-like phenotypes in mice. Biol. Psychiatry, 76, 120–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Walther S. and Strik W. (2012) Motor symptoms and schizophrenia. Neuropsychobiology, 66, 77–92. [DOI] [PubMed] [Google Scholar]
- 43. Leblanc S.E., Srinivasan R., Ferri C., Mager G.M., Gillian-Daniel A.L., Wrabetz L. and Svaren J. (2005) Regulation of cholesterol/lipid biosynthetic genes by Egr2/Krox20 during peripheral nerve myelination. J. Neurochem., 93, 737–748. [DOI] [PubMed] [Google Scholar]
- 44. Topilko P., Schneider-Maunoury S., Levi G., Baron-Van Evercooren A., Chennoufi A.B., Seitanidou T., Babinet C. and Charnay P. (1994) Krox-20 controls myelination in the peripheral nervous system. Nature, 371, 796–799. [DOI] [PubMed] [Google Scholar]
- 45. Srinivasan R., Sun G., Keles S., Jones E.A., Jang S.W., Krueger C., Moran J.J. and Svaren J. (2012) Genome-wide analysis of EGR2/SOX10 binding in myelinating peripheral nerve. Nucleic Acids Res., 40, 6449–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Le N., Nagarajan R., Wang J.Y., Araki T., Schmidt R.E. and Milbrandt J. (2005) Analysis of congenital hypomyelinating Egr2Lo/Lo nerves identifies Sox2 as an inhibitor of Schwann cell differentiation and myelination. Proc. Natl. Acad. Sci. U. S. A., 102, 2596–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jones E.A., Lopez-Anido C., Srinivasan R., Krueger C., Chang L.W., Nagarajan R. and Svaren J. (2011) Regulation of the PMP22 gene through an intronic enhancer. J. Neurosci., 31, 4242–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. La Marca R., Cerri F., Horiuchi K., Bachi A., Feltri M.L., Wrabetz L., Blobel C.P., Quattrini A., Salzer J.L. and Taveggia C. (2011) TACE (ADAM17) inhibits Schwann cell myelination. Nat. Neurosci., 14, 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Syed N., Reddy K., Yang D.P., Taveggia C., Salzer J.L., Maurel P. and Kim H.A. (2010) Soluble neuregulin-1 has bifunctional, concentration-dependent effects on Schwann cell myelination. J. Neurosci., 30, 6122–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ishii A., Furusho M. and Bansal R. (2013) Sustained activation of ERK1/2 MAPK in oligodendrocytes and schwann cells enhances myelin growth and stimulates oligodendrocyte progenitor expansion. J. Neurosci., 33, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sheean M.E., McShane E., Cheret C., Walcher J., Muller T., Wulf-Goldenberg A., Hoelper S., Garratt A.N., Kruger M., Rajewsky K. et al. (2014) Activation of MAPK overrides the termination of myelin growth and replaces Nrg1/ErbB3 signals during Schwann cell development and myelination. Genes Dev., 28, 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lopez-Anido C., Sun G., Koenning M., Srinivasan R., Hung H.A., Emery B., Keles S. and Svaren J. (2015) Differential Sox10 genomic occupancy in myelinating glia. Glia, 25, 3055–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hung H.A., Sun G., Keles S. and Svaren J. (2015) Dynamic regulation of Schwann cell enhancers after peripheral nerve injury. J. Biol. Chem., 290, 6937–6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ishii A., Furusho M., Dupree J.L. and Bansal R. (2014) Role of ERK1/2 MAPK signaling in the maintenance of myelin and axonal integrity in the adult CNS. J. Neurosci., 34, 16031–16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zenker J., Stettner M., Ruskamo S., Domenech-Estevez E., Baloui H., Medard J.J., Verheijen M.H., Brouwers J.F., Kursula P., Kieseier B.C. et al. (2014) A role of peripheral myelin protein 2 in lipid homeostasis of myelinating Schwann cells. Glia, 62, 1502–1512. [DOI] [PubMed] [Google Scholar]
- 56. Mikol D.D., Gulcher J.R. and Stefansson K. (1990) The oligodendrocyte-myelin glycoprotein belongs to a distinct family of proteins and contains the HNK-1 carbohydrate. J. Cell Biol., 110, 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gillespie C.S., Trapp B.D., Colman D.R. and Brophy P.J. (1990) Distribution of myelin basic protein and P2 mRNAs in rabbit spinal cord oligodendrocytes. J. Neurochem., 54, 1556–1561. [DOI] [PubMed] [Google Scholar]
- 58. Kadlubowski M., Hughes R.A. and Gregson N.A. (1984) Spontaneous and experimental neuritis and the distribution of the myelin protein P2 in the nervous system. J. Neurochem., 42, 123–129. [DOI] [PubMed] [Google Scholar]
- 59. Trapp B.D., McIntyre L.J., Quarles R.H., Sternberger N.H. and Webster H.D. (1979) Immunocytochemical localization of rat peripheral nervous system myelin proteins: P2 protein is not a component of all peripheral nervous system myelin sheaths. Proc. Natl. Acad. Sci. U. S. A., 76, 3552–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ishii A., Furusho M., Dupree J.L. and Bansal R. (2016) Strength of ERK1/2 MAPK activation determines its effect on myelin and axonal integrity in the adult CNS. J. Neurosci., 36, 6471–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bolino A., Piguet F., Alberizzi V., Pellegatta M., Rivellini C., Guerrero-Valero M., Noseda R., Brombin C., Nonis A., D'Adamo P. et al. (2016) Niacin-mediated Tace activation ameliorates CMT neuropathies with focal hypermyelination. EMBO Mol. Med., 8, 1438–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Domenech-Estevez E., Baloui H., Meng X., Zhang Y., Deinhardt K., Dupree J.L., Einheber S., Chrast R. and Salzer J.L. (2016) Akt regulates axon wrapping and myelin sheath thickness in the PNS. J. Neurosci., 36, 4506–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Napoli I., Noon L.A., Ribeiro S., Kerai A.P., Parrinello S., Rosenberg L.H., Collins M.J., Harrisingh M.C., White I.J., Woodhoo A. et al. (2012) A central role for the ERK-signaling pathway in controlling Schwann cell plasticity and peripheral nerve regeneration in vivo. Neuron, 73, 729–742. [DOI] [PubMed] [Google Scholar]
- 64. Martini R. (2014) Neuregulin-1 alleviates Charcot–Marie–Tooth disease in rats. Nat. Med., 20, 984–985. [DOI] [PubMed] [Google Scholar]
- 65. Groh J., Klein I., Hollmann C., Wettmarshausen J., Klein D. and Martini R. (2015) CSF-1-activated macrophages are target-directed and essential mediators of Schwann cell dedifferentiation and dysfunction in Cx32-deficient mice. Glia, 63, 977–986. [DOI] [PubMed] [Google Scholar]
- 66. Patzig J., Jahn O., Tenzer S., Wichert S.P., Monasterio-Schrader P., Rosfa S., Kuharev J., Yan K., Bormuth I., Bremer J. et al. (2011) Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. J. Neurosci., 31, 16369–16386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Majava V., Polverini E., Mazzini A., Nanekar R., Knoll W., Peters J., Natali F., Baumgartel P., Kursula I. and Kursula P. (2010) Structural and functional characterization of human peripheral nervous system myelin protein P2. PLoS One, 5, e10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kim M., Wende H., Walcher J., Kuehnemund J., Cheret C., Kempa S., McShane E., Selbach M., Lewin G.R. and Birchmeier C. (2018) Maf links Neuregulin1 signaling to cholesterol synthesis in myelinating Schwann cells. Genes Dev., 32, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Huang J.K., Phillips G.R., Roth A.D., Pedraza L., Shan W., Belkaid W., Mi S., Fex-Svenningsen A., Florens L., Yates J.R. 3rd et al. (2005) Glial membranes at the node of Ranvier prevent neurite outgrowth. Science, 310, 1813–1817. [DOI] [PubMed] [Google Scholar]
- 70. Apostolski S., Sadiq S.A., Hays A., Corbo M., Suturkova-Milosevic L., Chaliff P., Stefansson K., LeBaron R.G., Ruoslahti E., Hays A.P. et al. (1994) Identification of Gal(beta 1-3)GalNAc bearing glycoproteins at the nodes of Ranvier in peripheral nerve. J. Neurosci. Res., 38, 134–141. [DOI] [PubMed] [Google Scholar]
- 71. Chang K.J., Susuki K., Dours-Zimmermann M.T., Zimmermann D.R. and Rasband M.N. (2010) Oligodendrocyte myelin glycoprotein does not influence node of ranvier structure or assembly. J. Neurosci., 30, 14476–14481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Motley W.W., Palaima P., Yum S.W., Gonzalez M.A., Tao F., Wanschitz J.V., Strickland A.V., Loscher W.N., De Vriendt E., Koppi S. et al. (2016) De novo PMP2 mutations in families with type 1 Charcot–Marie–Tooth disease. Brain, 139, 1649–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gonzaga-Jauregui C., Harel T., Gambin T., Kousi M., Griffin L.B., Francescatto L., Ozes B., Karaca E., Jhangiani S.N., Bainbridge M.N. et al. (2015) Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep., 12, 1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shin Y.K., Jang S.Y., Park S.Y., Park J.Y., Kim J.K., Kim J.P., Suh D.J., Lee H.J. and Park H.T. (2014) Grb2-associated binder-1 is required for neuregulin-1-induced peripheral nerve myelination. J. Neurosci., 34, 7657–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cervellini I., Galino J., Zhu N., Allen S., Birchmeier C. and Bennett D.L. (2018) Sustained MAPK/ERK activation in adult Schwann cells impairs nerve repair. J. Neurosci., 38, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ghidinelli M., Poitelon Y., Shin Y.K., Ameroso D., Williamson C., Ferri C., Pellegatta M., Espino K., Mogha A., Monk K. et al. (2017) Laminin 211 inhibits protein kinase A in Schwann cells to modulate neuregulin 1 type III-driven myelination. PLoS Biol., 15, e2001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mogha A., Harty B.L., Carlin D., Joseph J., Sanchez N.E., Suter U., Piao X., Cavalli V. and Monk K.R. (2016) Gpr126/Adgrg6 has Schwann cell autonomous and nonautonomous functions in peripheral nerve injury and repair. J. Neurosci., 36, 12351–12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Heller B.A., Ghidinelli M., Voelkl J., Einheber S., Smith R., Grund E., Morahan G., Chandler D., Kalaydjieva L., Giancotti F. et al. (2014) Functionally distinct PI 3-kinase pathways regulate myelination in the peripheral nervous system. J. Cell Biol., 204, 1219–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Monk K.R., Feltri M.L. and Taveggia C. (2015) New insights on Schwann cell development. Glia, 63, 1376–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hu X., Hicks C.W., He W., Wong P., Macklin W.B., Trapp B.D. and Yan R. (2006) Bace1 modulates myelination in the central and peripheral nervous system. Nat. Neurosci., 9, 1520–1525. [DOI] [PubMed] [Google Scholar]
- 81. Willem M., Garratt A.N., Novak B., Citron M., Kaufmann S., Rittger A., DeStrooper B., Saftig P., Birchmeier C. and Haass C. (2006) Control of peripheral nerve myelination by the beta-secretase BACE1. Science, 314, 664–666. [DOI] [PubMed] [Google Scholar]
- 82. Fleck D., Voss M., Brankatschk B., Giudici C., Hampel H., Schwenk B., Edbauer D., Fukumori A., Steiner H., Kremmer E. et al. (2016) Proteolytic processing of neuregulin 1 type III by three intramembrane-cleaving proteases. J. Biol. Chem., 291, 318–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Trimarco A., Forese M.G., Alfieri V., Lucente A., Brambilla P., Dina G., Pieragostino D., Sacchetta P., Urade Y., Boizet-Bonhoure B. et al. (2014) Prostaglandin D2 synthase/GPR44: a signaling axis in PNS myelination. Nat. Neurosci., 17, 1682–1692. [DOI] [PubMed] [Google Scholar]
- 84. Goebbels S., Oltrogge J.H., Wolfer S., Wieser G.L., Nientiedt T., Pieper A., Ruhwedel T., Groszer M., Sereda M.W. and Nave K.A. (2012) Genetic disruption of Pten in a novel mouse model of tomaculous neuropathy. EMBO Mol. Med., 4, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bargagna-Mohan P., Ishii A., Lei L., Sheehy D., Pandit S., Chan G., Bansal R. and Mohan R. (2017) Sustained activation of ERK1/2 MAPK in Schwann cells causes corneal neurofibroma. J. Neurosci. Res., 95, 1712–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Taveggia C., Thaker P., Petrylak A., Caporaso G.L., Toews A., Falls D.L., Einheber S. and Salzer J.L. (2008) Type III neuregulin-1 promotes oligodendrocyte myelination. Glia, 56, 284–293. [DOI] [PubMed] [Google Scholar]
- 87. Srinivasan L., Sasaki Y., Calado D.P., Zhang B., Paik J.H., DePinho R.A., Kutok J.L., Kearney J.F., Otipoby K.L. and Rajewsky K. (2009) PI3 kinase signals BCR-dependent mature B cell survival. Cell, 139, 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Feltri M.L., D'Antonio M., Previtali S., Fasolini M., Messing A. and Wrabetz L. (1999) P0-Cre transgenic mice for inactivation of adhesion molecules in Schwann cells. Ann. N. Y. Acad. Sci., 883, 116–123. [PubMed] [Google Scholar]
- 89. Feltri M.L., Graus Porta D., Previtali S.C., Nodari A., Migliavacca B., Cassetti A., Littlewood-Evans A., Reichardt L.F., Messing A., Quattrini A. et al. (2002) Conditional disruption of beta 1 integrin in Schwann cells impedes interactions with axons. J. Cell Biol., 156, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Poitelon Y., Lopez-Anido C., Catignas K., Berti C., Palmisano M., Williamson C., Ameroso D., Abiko K., Hwang Y., Gregorieff A. et al. (2016) YAP and TAZ control peripheral myelination and the expression of laminin receptors in Schwann cells. Nat. Neurosci., 19, 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Poitelon Y., Bogni S., Matafora V., Della-Flora Nunes G., Hurley E., Ghidinelli M., Katzenellenbogen B.S., Taveggia C., Silvestri N., Bachi A. et al. (2015) Spatial mapping of juxtacrine axo-glial interactions identifies novel molecules in peripheral myelination. Nat. Commun., 6, 8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Quattrini A., Previtali S., Feltri M.L., Canal N., Nemni R. and Wrabetz L. (1996) Beta 4 integrin and other Schwann cell markers in axonal neuropathy. Glia, 17, 294–306. [DOI] [PubMed] [Google Scholar]
- 93. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H. and Glass C.K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


