Abstract
Transmission electron microscopy is the technique of choice to visualize the spatial relationships between nanoconstructs and cells, and especially to monitor the uptake process of nanomaterials. It is therefore crucial that the cell surface be preserved in its integrity, to obtain reliable ultrastructural evidence: the plasmalemma represents the biological barrier the nanomaterials have to cross, and the mode of membrane-nanoconstruct interaction is responsible for the intracellular fate of the nanomaterials. In this paper, we describe a simple and inexpensive method to process cell monolayers for ultrastructural morphology and immunocytochemistry, ensuring consistent preservation of the cell surface and of the occurring interactions with nanoparticles of different chemical composition.
Key words: Cell surface, nanoparticle uptake, endocytosis, ultrastructure, immunocytochemistry
Introduction
During the last decades, nanotechnology has seen an enormous progress, with the development of a plethora of nanomaterials designed for multiple applications in materials science, electronics, food industry and biomedicine. The analysis of the interaction of nanomaterials on living organisms, on humans in particular, has consequently become the matter of numerous studies aimed at exploring either their possible adverse effects as pollutants or their useful applications as therapeutic and diagnostic tools. In both cases, the first investigation step is generally performed using in vitro cultured cells that ensure simple and controlled experimental conditions to dynamically track the nanoparticulates inside the cells and to assess the occurrence of cell stress, damage or death. It is known that the rate of nanomaterial uptake is related to the physicochemical characteristics of the nanoconstruct itself but also depends on the target cell features. On the one hand, the shape, size and surface chemical properties determine the ability of the nanoconstruct to interact with the cell membrane, the mechanisms and efficiency of its internalization, and the intracellular pathway;1-3 on the other hand, the cell type, organ origin, size, shape, proliferation rate and cell membrane composition play a major role in conditioning the nanomaterials’ interactions.4,5
Transmission electron microscopy (TEM) is the technique of choice to visualize the spatial relationships between nanoconstructs and cells6,7 thanks to its high resolution and direct visualization of nanomaterials in the intracellular milieu, although histochemical methods are sometimes needed to make low-density nanoparticles unequivocally recognizable.8,9 In particular, TEM allows to monitor the uptake process of nanomaterials, by revealing the fine morphological modifications of the cell membranes when in contact with the nanoconstructs, the internalization modalities, the nanoconstructs’ interactions with (and their possible damaging action on) the cell organelles, their intracellular degradation/ accumulation and their possible extrusion from the cell.
In this view, it is crucial that the cell structure be preserved in its integrity, to obtain reliable ultrastructural evidence. This is especially true for the cell surface: the plasmalemma actually represents the biological barrier the nanomaterials have finally to cross; moreover, the mode of membrane- nanoconstruct interaction is responsible for the intracellular fate of the nanomaterials, and impacts on cell metabolism.
In the tissues in vivo, the cells establish molecular contacts either with other cells or with the extracellular matrix, that are essential for their structural organization and function; also, most of the cultured cell systems used for investigating the effects of nanomaterials on living cells actually grow adhering to a solid substrate and this growth mode may influence cell shape, intercellular contacts and intracellular organization. Thus, often the interaction of the nanoconstructs cannot uniformly take place over the entire cell surface, and it is mandatory that sample handling for TEM examination is carefully performed to maintain as much as possible the original cell organization and plasma membrane morphology, to avoid misleading artifacts.
In this paper, we describe a simple and inexpensive method to process cell monolayers for ultrastructural morphology and immunocytochemistry, ensuring consistent preservation of the cell surface and of the occurring interactions with nanoparticles of different chemical composition.
Materials and Methods
Different adhering cells were selected for the present study, that have already been used by our research group in previous investigations: 3T3-L1 mouse preadipocytes, 10 C2C12 immortalized mouse myoblasts,11 rat B50 neuronal cells,8,9,12 HeLa human cervical adenocarcinoma cells,13-15 human primary adipose-derived adult stem cells isolated from liposuction samples,10 human primary myoblasts isolated from skeletal muscle biopsies.16 The cells were grown in 75 cm2 plastic flasks (Sarstedt, Nümbrecht, Germany) in appropriate media as detailed in the above mentioned articles, and maintained at 37°C in a 5% CO2 humidified atmosphere.
Various nanoconstructs suitable for therapeutic or diagnostic purposes were considered: liposomes,11,14,16 polymeric nanoparticles,8,9,11-14,16 mesoporous silica nanoparticles,11,14,16 and paramagnetic nanoparticles.10,15 To investigate the nanomaterial- cell interactions, the cells were seeded on glass coverslips of appropriate diameter in 6- or 12-multiwell microplates (Sarstedt), and exposed to the nanoconstructs one day post-seeding: to do this, the culture medium was replaced with a fresh one containing the nanoconstructs at biocompatible concentrations (see the specific articles8,10,11,14-16), and the cells were incubated for increasing time lengths (from 1 h to 14 days) to investigate the whole process of interaction, internalization, intracellular fate and degradation/extrusion of the nanoconstructs. At the end of each incubation time, the cells were fixed and processed as described below. To fix the cells while adhering to the substrate, the medium was removed and the fixative solution was gently poured into the well with a pipette, paying attention not to put the solution directly onto the glass coverslips in order to avoid cell detachment. For conventional ultrastructural morphology, the cell monolayers were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer saline (PBS), pH 7.4, for 2 h at 4°C. After washes in PBS, the cells were post-fixed with 1% OsO4 and 1.5% potassium ferrocyanide for 1 h at room temperature. The cells were then dehydrated in graded acetone, and impregnated with Epon 812 resin (Electron Microscopy Sciences, Hatfield, PA, USA). For ultrastructural immunocytochemistry, cell monolayers were fixed with 4% paraformaldehyde, in 0.1 M PBS, pH 7.4, for 2 h at 4°C. After washes in PBS, free aldehydes were blocked with 0.5 M NH4Cl in PBS for 30 min at 4°C. The cells were then dehydrated in graded ethanol, and impregnated with LRWhite resin (Electron Microscopy Sciences).
In both procedures, all passages consisted in removing and replacing the reagent in the well by pipettes, always paying attention to avoid cell detachment. As a final step, after resin impregnation, the coverslips were taken with thin tweezer, the resin excess was adsorbed with filter paper (Figure 1a), and the coverslips placed onto an aluminium foil (Figure 1b) (adsorbing the resin excess is essential to make the detachment of the bloc from the glass easy, at the end of the procedure). Then, gelatin capsules were filled with the resin (Figure 1c) and turned upside-down onto the coverslips with the aid of tweezers, carefully avoiding the formation of air bubbles (Figure 1d). To induce resin polymerization, the coverslips with the gelatin capsules were put either in the oven at 60°C (this is suitable for both epoxy and acrylic resins) or under UV light (suitable for acrylic resins only). After polymerization, the resin blocks were detached from the coverslips by dipping into liquid nitrogen for a few seconds (Figure 1 e-g): by such a procedure, the cell monolayers become exposed at the flat surface of the resin blocks (the cell surface originally attached to the coverslips facing-up) (Figure 1h). The suitable area was selected by observing the cell monolayer under a stereo microscope, the blocks were properly trimmed, and ultrathin sections were cut with an UltraCut E ultramicrotome (Reichert-Jung, Leica Microsystems, Wetzlar, Germany). As an example of immunohistochemical application, we used ultrathin sections from paraformaldehydefixed, LRWhite-embedded B50 cells that had previously been treated with chitosan nanoparticles loaded with the hypometabolising drug [D-Ala2, D-Leu5] enkephalin1:7 the sections were incubated with an anti-Leu5-enkephalin monoclonal antibody (Medicorp, Cranford, NJ, USA) then revealed by a specific gold-conjugated secondary antibody (Jackson Immuno- Research, Ely, UK) (for details see18).
Figure 1.
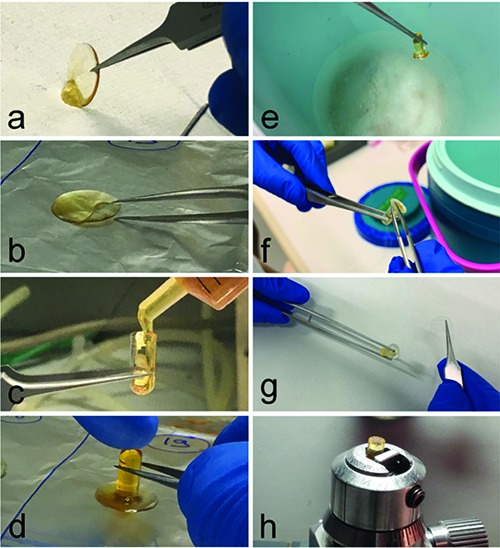
Sample handling after resin impregnation. a) The resin excess is adsorbed with filter paper. b) The glass coverslip is placed on a holder covered with aluminium foil. c) The gelatin capsule is filled with resin. d). The capsule is positioned upside-down onto the coverslip. e) After polymerization, the sample is dipped in liquid nitrogen and (f, g) the coverslip is detached from the resin bloc. h) The sample is placed in a specimen holder, ready to be trimmed and sectioned.
Ultrathin sections were observed either unstained or after weak staining with a 2.5% uranyl acetate aqueous solution. Observations were made in a Philips Morgagni TEM (FEI Company Italia Srl, Milan, Italy), operating at 80 kV and equipped with a Megaview II camera for digital image acquisition.
Results and Discussion
The described procedure is a simple, cheap and versatile method based on the use of standard multiwell plates and glass coverslips to process for TEM cell samples treated with nanomaterials. In the literature, various methods to process cell monolayers for investigations at TEM are available: several epoxy resins have been tested for embedding cell monolayers grown on plastic supports;19 cell monolayers grown on glass coverslips have been flat-embedded in a hydrophilic resin to allow cytochemical and immunocytochemical studies;20 cell monolayers grown on gridded glass supports have been used to correlate light and electron microscopy.21,22 The technique we have described in the present paper is especially suitable to study the spatial relationships between the cell surface and the nanoconstructs; in fact, cells are fixed and embedded as they were on the growing substrate, without being submitted to scraping and pelleting. The result is a cell monolayer placed at the surface of the resin bloc, ready to be trimmed and cut at the microtome.
Observations at TEM of the various samples considered in this study (Figure 2) demonstrated that our method is suitable for different nanoconstructs and different cell types, giving reliable results for both morphological and immunocytochemical analyses. The cell surface was always well preserved and, depending on the cell type and the nanoconstruct used, microvilli (Figure 2 a,b,d), endocytic invaginations (Figure 2 a,e,f) and phagocytic finger-like protrusions were observed (Figure 2c). The nanoconstructs were found in close proximity to the cell surface (Figure 2 c,e), or adhering to the plasmalemma (Figure 2 a,b,d,f) or enclosed in endocytic/phagocytic invaginations (Figure 2 a,c,e), as either single units or clustered particles. When cells grown in flasks are mechanically or enzymatically detached from the substrate and pelleted by centrifugation, the plasmalemma inevitably undergoes morphological modifications; moreover, under these processing conditions, only the nanomaterials firmly bound to the cell surface (or inside the cell) are preserved. On the contrary, by our procedure, the original spatial relationships are maintained, and also the nanoparticulates that lie close to the plasmalemma during the earliest phases of their interaction with the cell can be kept in place.
Figure 2.
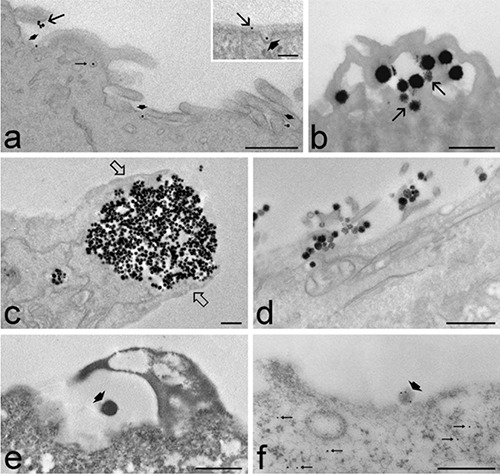
Transmission electron micrographs of cell samples processed for ultrastructural morphology (a-e) or immunocytochemistry (f ). a) Human primary adipose-derived adult stem cells treated with superparamagnetic iron oxide nanoparticles; some nanoparticles adhere to the cell surface (arrows), others occur inside plasmalemma invaginations (arrowheads), and others inside endocytic vacuoles (thin arrow); note the good preservation of microvilli at the cell surface. b) HeLa cells treated with liposomes; liposomes enter the cell by fusion with the plasmalemma, and undergo disaggregation in the outer cytoplasmic region (arrows). c) HeLa cells treated with mesoporous silica nanoparticles; a large cluster of nanoparticles is almost entirely enclosed by finger-like cytoplasmic protrusions (open arrows). d) C2C12 myotubes treated with mesoporous silica nanoparticles; numerous nanoparticles adhere to the cell surface showing many microvilli. e) Human primary myoblasts treated with poly(lactide-co-glycolide) nanoparticles; the nanoparticle (arrowhead) adhering to the substrate is partially enclosed by a cytoplasmic protrusion. f ) B50 neuronal cell treated with chitosan nanoparticles loaded with [D-Ala2, D-Leu5] enkephalin; immunogold labelling with anti-Leu5-enkephalin antibody; the signal is present inside the nanoparticle adhering to the cell surface (arrowhead) as well as in the cytoplasm (thin arrows). Scale bars: 500 nm; inset :100 nm.
This method can be applied also to samples embedded in acrylic resins, to perform cytochemical8,15 and pre- or post-embedding immunocytochemical procedures on thin sections (Figure 2f),12 although the handling of these samples is more challenging: in fact, the higher fluidity of the acrylic resins makes the upside-down positioning of the resin-filled capsules tricky, while the polymerization time may be much longer when performed under UV light at low temperature.
A weak staining of the ultrathin sections (or even the staining omission) may help to detect poorly electron dense nanoconstructs in the intracellular milieu; this has been especially useful to visualize remnants of disaggregating liposomes (Figure 2b)14 or to unequivocally detect at electron microscopy the presence of fluorescently-labelled nanoparticulates after procedures of diaminobenzidine photo-oxidation.8,13,15
Embedding cell monolayers is advantageous for several reasons. First of all, the integrity of the cell-to-growing support and cell-to-cell contacts are preserved: thus, in the ultrastructural study of cell surface it is possible to distinguish the plasmalemmal areas interacting with the substrate from the upper cell surface (in direct contact with the nanoparticulate-containing medium) and the lateral parts (where structural and functional contacts may occur between neighboring cells). In addition, the preparation of samples made of monolayers requires relatively lower amounts of cells (5-10 x 103 cells per cover glass) in comparison to conventional cell pellets (2-3 x 105 cells): this is a negligible factor when using established cell lines, but it becomes crucial when working with precious primary cell cultures, especially from unique biopsy material. A possible limiting factor of our method is related to the thinness of the cell monolayer. It is actually challenging to use the same resin block for both semithin and ultrathin sectioning: this result can however be obtained by alternatingly cutting semithin sections of about 0.5 μm and ultrathin sections of 60-70 nm. In addition, the number of sections obtainable from a cell monolayer is definitely much lower than from a cell pellet; however, the sample size may be enlarged by increasing the number of cover glasses from the cell culture batch and by placing more than one gelatin capsule on the same cover glass. To this aim, small gelatin capsules (about 5 mm in diameter) are especially suitable; in our experience, small capsules are also more easily detachable from the glass than larger ones, after dipping in liquid nitrogen. However, it must be underlined that this is true when epoxy resins are used, whereas capsules of larger size (about 8 mm in diameter) are necessary for acrylic resins since polymerization along the capsule border is impaired by oxygen23 and this makes the inner part only of the block surface suitable for sectioning.
In conclusion, the cell sample processing method we have described has an excellent potential in disclosing the interactions between nanomaterials and cell membrane at a high-resolution level, preserving the spatial relationships as close as possible to the ones the cells had under the experimental conditions.
This method may thus represent an added value in ultrastructural research to investigate the nano-bio interface, whose detailed knowledge is crucial to assess the safety of the increasingly used nanomaterials in electronic, environmental, and industrial applications,24,25 as well as to design efficient strategies in the biomedical field for nanoconstruct targeting26,27 or the crossing of biological barriers.28-30
References
- 1.Sahay G, Alakhova DY, Kabanov AV. Endocytosis of nanomedicines. J Control Release 2010;145:182-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akinc A, Battaglia G. Exploiting endocytosis for nanomedicines. Cold Spring Harb Perspect Biol 2013;5:a016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang S, Gao H, Bao G. Physical principles of nanoparticle cellular endocytosis. ACS Nano 2015;9:8655-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yalçın S, Özlüer Ö, Gündüz U. Nanoparticle-based drug delivery in cancer: the role of cell membrane structures. Ther Deliv 2016;7:773-81. [DOI] [PubMed] [Google Scholar]
- 5.Mahmoudi M. Debugging nano-bio interfaces: systematic strategies to accelerate clinical translation of nanotechnologies. Trends Biotechnol 2018;36:755-69. [DOI] [PubMed] [Google Scholar]
- 6.Malatesta M. Transmission electron microscopy for nanomedicine: novel applications for long-established techniques. Eur J Histochem 2016;60:2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reifarth M, Hoeppener S, Schubert US. Uptake and intracellular fate of engineered nanoparticles in mammalian cells: Capabilities and limitations of transmission electron microscopy-polymer- based nanoparticles. Adv Mater 2018;30. doi:10.1002/adma.20170 3704 [DOI] [PubMed] [Google Scholar]
- 8.Malatesta M, Giagnacovo M, Costanzo M, Conti B, Genta I, Dorati R, et al. Diaminobenzidine photoconversion is a suitable tool for tracking the intracellular location of fluorescently labelled nanoparticles at transmission electron microscopy. Eur J Histochem 2012; 56:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malatesta M, Pellicciari C, Cisterna B, Costanzo M, Galimberti V, Biggiogera M, et al. Tracing nanoparticles and photosensitizing molecules at transmission electron microscopy by diaminobenzidine photo-oxidation. Micron 2014;59: 44-51. [DOI] [PubMed] [Google Scholar]
- 10.Marinozzi MR, Pandolfi L, Malatesta M, Colombo M, Collico V, Lievens PM, et al. Innovative approach to safely induce controlled lipolysis by superparamagnetic iron oxide nanoparticlesmediated hyperthermic treatment. Int J Biochem Cell Biol 2017;93:62-73. [DOI] [PubMed] [Google Scholar]
- 11.Costanzo M, Vurro F, Cisterna B, Boschi F, Marengo A, Montanari E, et al. Uptake and intracellular fate of biocompatible nanocarriers in cycling and noncycling cells. Nanomedicine (Lond) 2019;14:301-16. [DOI] [PubMed] [Google Scholar]
- 12.Malatesta M, Galimberti V, Cisterna B, Costanzo M, Biggiogera M, Zancanaro C. Chitosan nanoparticles are efficient carriers for delivering biodegradable drugs to neuronal cells. Histochem Cell Biol 2014;141:551-8. [DOI] [PubMed] [Google Scholar]
- 13.Malatesta M, Grecchi S, Chiesa E, Cisterna B, Costanzo M, Zancanaro C. Internalized chitosan nanoparticles persist for long time in cultured cells. Eur J Histochem 2015;59:2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costanzo M, Carton F, Marengo A, Berlier G, Stella B, Arpicco S, et al. Fluorescence and electron microscopy to visualize the intracellular fate of nanoparticles for drug delivery. Eur J Histochem 2016;60(2):2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costanzo M, Scolaro L, Berlier G, Marengo A, Grecchi S, Zancanaro C, et al. Cell uptake and intracellular fate of phospholipidic manganese-based nanoparticles. Int J Pharm 2016;508:83-91. [DOI] [PubMed] [Google Scholar]
- 16.Guglielmi V, Carton F, Vattemi G, Arpicco S, Stella B, Berlier G, et al. Uptake and intracellular distribution of different types of nanoparticles in primary human myoblasts and myotubes. Int J Pharm 2019;560:347-56. [DOI] [PubMed] [Google Scholar]
- 17.Malatesta M, Luchetti F, Marcheggiani F, Fakan S, Gazzanelli G. Disassembly of nuclear bodies during arousal from hibernation: an in vitro study. Chromosoma 2001;110:471-7. [DOI] [PubMed] [Google Scholar]
- 18.Baldelli B, Vecchio L, Bottone MG, Muzzonigro G, Biggiogera M, Malatesta M. The effect of the enkephalin DADLE on transcription does not depend on opioid receptors. Histochem Cell Biol 2006;126:189-97. [DOI] [PubMed] [Google Scholar]
- 19.Murray AB, Schulze H, Blauw E. In situ embedding of cell monolayers cultured on plastic surfaces for electron microscopy. Biotech Histochem 1991;66:269-72. [DOI] [PubMed] [Google Scholar]
- 20.Steiner M, Schöfer C, Mosgoeller W. In situ flat embedding of monolayers and cell relocation in the acrylic resin LR white for comparative light and electron microscopy studies. Histochem J 1994;26:934-8. [PubMed] [Google Scholar]
- 21.Malatesta M, Fakan S, Fischer U. The Sm core domain mediates targeting of U1 snRNP to subnuclear compartments involved in transcription and splicing. Exp Cell Res 1999;249:189-98. [DOI] [PubMed] [Google Scholar]
- 22.Hanson HH, Reilly JE, Lee R, Janssen WG, Phillips GR. Streamlined embedding of cell monolayers on gridded glass-bottom imaging dishes for correlative light and electron microscopy. Microsc Microanal 2010;16:747-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayat MA. Basic techniques for transmission electron microscopy. London, Academic Press; 1986. [Google Scholar]
- 24.Erbis S, Ok Z, Isaacs JA, Benneyan JC, Kamarthi S. Review of research trends and methods in nano environmental, health, and safety risk analysis. Risk Anal 2016;36:1644-65. [DOI] [PubMed] [Google Scholar]
- 25.Higashisaka K, Nagano K, Yoshioka Y, Tsutsumi Y. Nano-safety research: examining the associations among the biological effects of nanoparticles and their physicochemical properties and kinetics. Biol Pharm Bull 2017;40:243-8. [DOI] [PubMed] [Google Scholar]
- 26.Swain S, Sahu PK, Beg S, Babu SM. Nanoparticles for cancer targeting: current and future directions. Curr Drug Deliv 2016;13:1290-302. [DOI] [PubMed] [Google Scholar]
- 27.Villaverde G, Baeza A. Targeting strategies for improving the efficacy of nanomedicine in oncology. Beilstein J Nanotechnol 2019;10:168-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beloqui A, des Rieux A, Préat V. Mechanisms of transport of polymeric and lipidic nanoparticles across the intestinal barrier. Adv Drug Deliv Rev 2016;106:242-55. [DOI] [PubMed] [Google Scholar]
- 29.Muoth C, Aengenheister L, Kucki M, Wick P, Buerki-Thurnherr T. Nanoparticle transport across the placental barrier: pushing the field forward! Nanomedicine (Lond) 2016;11: 941-57. [DOI] [PubMed] [Google Scholar]
- 30.Sharma G, Sharma AR, Lee SS, Bhattacharya M, Nam JS, Chakraborty C. Advances in nanocarriers enabled brain targeted drug delivery across blood brain barrier. Int J Pharm 2019; 559:360-72. [DOI] [PubMed] [Google Scholar]


