Abstract
CRISPR-Cas systems function as adaptive immune systems by acquiring nucleotide sequences called spacers that mediate sequence-specific defence against competitors. Uniquely, the phage ICP1 encodes a Type I-F CRISPR-Cas system that is deployed to target and overcome PLE, a mobile genetic element with anti-phage activity in Vibrio cholerae. Here, we exploit the arms race between ICP1 and PLE to examine spacer acquisition and interference under laboratory conditions to reconcile findings from wild populations. Natural ICP1 isolates encode multiple spacers directed against PLE, but we find that single spacers do not interfere equally with PLE mobilization. High-throughput sequencing to assay spacer acquisition reveals that ICP1 can also acquire spacers that target the V. cholerae chromosome. We find that targeting the V. cholerae chromosome proximal to PLE is sufficient to block PLE and is dependent on Cas2-3 helicase activity. We propose a model in which indirect chromosomal spacers are able to circumvent PLE by Cas2-3-mediated processive degradation of the V. cholerae chromosome before PLE mobilization. Generally, laboratory-acquired spacers are much more diverse than the subset of spacers maintained by ICP1 in nature, showing how evolutionary pressures can constrain CRISPR-Cas targeting in ways that are often not appreciated through in vitro analyses.
This article is part of a discussion meeting issue ‘The ecology and evolution of prokaryotic CRISPR-Cas adaptive immune systems’.
Keywords: CRISPR, phage, mobile genetic elements, cholera
1. Introduction
Phages often vastly outnumber their bacterial hosts in a variety of environments [1]. As such, bacteria have evolved numerous mechanisms for phage defence, including adaptive immunity via clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins [2,3]. CRISPR-Cas systems are composed of a CRISPR array—a series of ‘spacers’ of foreign sequence alternating with repeats that are transcribed into CRISPR RNAs (crRNAs)—and cas genes. Together with crRNAs, Cas proteins defend against foreign nucleic acids, such as the genome of an infecting phage, through a three-step process: adaptation, crRNA and cas gene expression, and interference. During adaptation, a foreign DNA fragment is incorporated into the CRISPR array to provide a molecular memory of the challenges that the host cell has faced. This CRISPR array is expressed and processed into individual crRNAs, which complex with Cas proteins and survey the cell for complementary invading nucleotides. Upon finding a complementary sequence, termed a protospacer, a Cas nuclease is recruited to the site to mediate interference by cleaving the substrate, ultimately leading to the destruction of the invader [3,4]. Across CRISPR-Cas containing bacteria and archaea, Class 1 Type I CRISPR-Cas systems employing a Cas3 enzyme for DNA unwinding and degradation [5] are the most prevalent [6].
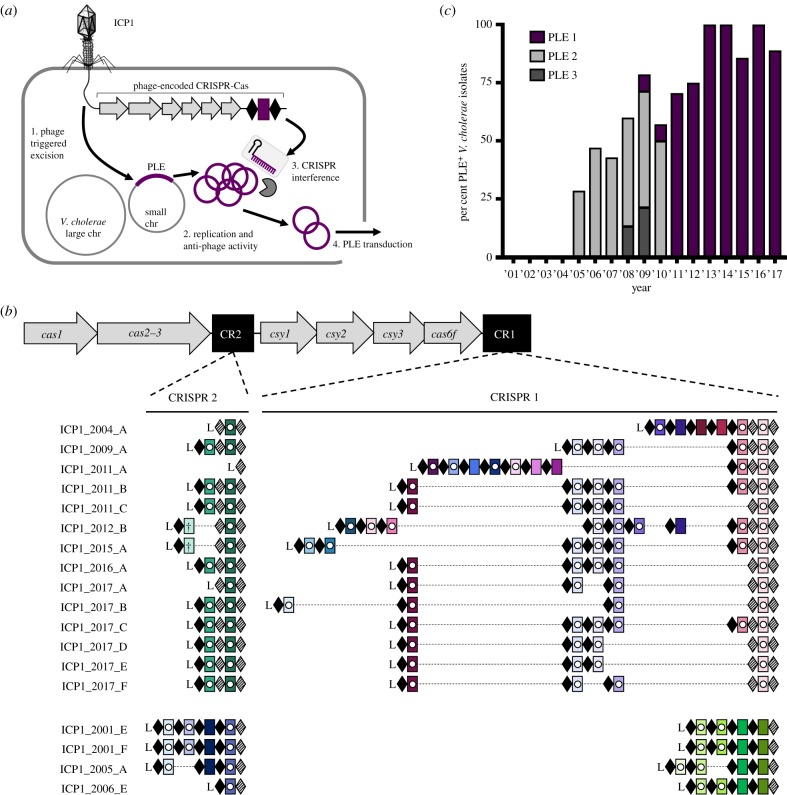
CRISPR-Cas systems do not discriminate between horizontally acquired traits based on fitness gain or loss. Hence, CRISPR-Cas systems are equally capable of halting harmful invading phage DNA as they are halting beneficial mobile genetic elements, including those encoding antibiotic resistance and pathogenicity genes [7–9]. As such, some pathogens only have alternative anti-phage defence systems [10]. For example, the currently circulating biotype of epidemic Vibrio cholerae, the causative agent of the diarrheal disease cholera, does not rely on CRISPR-Cas for phage defence [11]. Instead, V. cholerae evolved to use phage inducible chromosomal island-like elements (PLEs) to defend against the prevalent lytic phage, ICP1 [12]. PLEs are mobile genetic elements that reside integrated in the small chromosome of V. cholerae [12]. During ICP1 infection of PLE(+) V. cholerae, PLE excises from the host chromosome, replicates to high copy and is horizontally transduced to naive neighbouring cells, all the while inhibiting phage replication through unknown mechanisms (figure 1a).
Figure 1.
ICP1 uses CRISPR-Cas to overcome epidemic V. cholerae PLE. (a) Lytic phage ICP1 infects V. cholerae triggering PLE excision. PLE replicates and exerts anti-phage activity, ultimately leading to PLE transduction. Concurrently, ICP1-encoded CRISPR-Cas is expressed to interfere with PLE activity. (b) The architecture of the ICP1 CRISPR-Cas system and comparison of spacer composition between phage isolates. For each CRISPR locus, the repeat (28 bp) and spacer (32 bp) content is detailed as black diamonds and coloured rectangles, respectively. Repeats that match the repeat consensus [13] are shown by solid diamonds, and degenerate repeats are indicated by hatched black diamonds. An AT-rich leader sequence (L) precedes each CRISPR locus. Identical spacers shared between isolates are shown as rectangles with identical colours. Spacers containing a white circle target PLE, and spacers containing a cross target the V. cholerae large chromosome. (c) Percentage of V. cholerae isolates harbouring PLE recovered from epidemic sampling at the International Centre for Diarrhoeal Disease Research, Bangladesh over time (n = 230 strains analysed).
In order to overcome the anti-phage activity encoded by V. cholerae PLE, some ICP1 isolates use a Type I-F CRISPR-Cas system that directly targets PLE (figure 1a), making the CRISPR-Cas system essential for the phage to form plaques on PLE(+) V. cholerae [13]. Type I-F systems are composed of three Csy proteins that make up the Csy complex along with Cas6f, a protein involved in crRNA processing [14]. This complex interacts with the processed crRNA to search DNA for a complementary protospacer with an appropriate self versus non-self discrimination sequence, known as the protospacer adjacent motif (PAM) [15]. Upon finding a match with an appropriate PAM, the trans-acting Cas2-3 fusion protein is recruited to degrade the target DNA. In addition to endonuclease activity, Cas2-3 has a helicase domain that unwinds DNA as the protein translocates away from the target DNA, allowing for continued processive degradation of adjacent DNA in vitro [16,17]. Recently, sequence analysis identified phages that are predicted to encode CRISPR arrays and/or cas genes [18,19]; however, ICP1 is the only phage shown to encode a fully functional CRISPR-Cas system [12,13].
As is true when CRISPR-Cas is harnessed by a prokaryotic host for genome defence, the ICP1-encoded CRISPR-Cas system is tasked with targeting and degrading a hostile mobile genetic element. However, there are additional challenges associated with a phage encoding and relying on CRISPR-Cas for its own survival. The ICP1 infection cycle occurs over a 20 min period, and current data suggest that ICP1 synthesizes its CRISPR-Cas machinery de novo upon infection of V. cholerae [13]. PLE is induced to excise within minutes of infection through interactions with an early phage-encoded gene product [20]. Thus, in order to overcome PLE, CRISPR synthesis and interference must outpace a rapidly replicating target.
ICP1 and V. cholerae are consistently co-isolated from patient stool samples in regions where cholera is endemic, such as Bangladesh [12,21,22]. Five genetically distinct PLE variants in V. cholerae have appeared in temporally discrete waves across cholera epidemics [12]. Previous analysis revealed that ICP1-encoded CRISPR-Cas can adapt and acquire new spacers against PLE under laboratory conditions [13], however the rules governing spacer acquisition and targeting efficacy for this system are not known. Further, recent comparative genomics of 18 ICP1 isolates collected from Bangladesh between 2001 and 2012 found that 50% carry CRISPR-Cas [23], however the contemporary state of circulating ICP1 and V. cholerae PLE in the region are not known.
Here, we provide an up-to-date understanding of the genomic variants of ICP1 and PLE circulating in Bangladesh. We find that natural ICP1 isolates encode multiple anti-PLE spacers and experimentally validate that increased PLE targeting by ICP1 is required to fully abolish PLE mobilization. Significantly, using a high-throughput spacer acquisition assay and experimental validation, we show that non-canonical PAMs and indirect protospacers in the V. cholerae small chromosome can unexpectedly provide protection against PLE. Our results support a model in which ICP1-encoded CRISPR-Cas that is directed against the V. cholerae small chromosome is in a race to reach PLE before it excises from the chromosome to exert its anti-phage activity. Taken together, our study highlights the differences between interference competent spacers under laboratory conditions and those that are selected for in nature to provide mechanistic insight into the evolutionary pressures governing the interactions between epidemic V. cholerae and its longstanding battle with the predatory phage ICP1.
2. Methods
(a). Strains, growth conditions and genomic analysis
Phage, bacterial strains and plasmids used in this study are listed in electronic supplementary material, tables S1–S3. Bacteria were routinely grown at 37°C on lysogeny broth (LB) agar or in LB broth with aeration. Media was supplemented with ampicillin (50 µg ml−1), kanamycin (75 µg ml−1), spectinomycin (100 µg ml−1) and/or streptomycin (100 µg ml−1) when appropriate. Phage susceptibility was determined by standard soft agar overlays as described [11] and phage plaque spot plates were performed as described previously [20]. Images are representative of at least two independent assays. Cholera stool samples collected and stored at the ICDDR,B between 2015 and 2017 were probed for the presence of phage by standard soft agar overlays, and V. cholerae isolates were recovered by plating on Thiosulfate Citrate Bile Salts Sucrose selective media (Difco). ICP1-specific primers [13,22] and PLE-specific primers (electronic supplementary material, table S4) were used for preliminary screening of isolates from stool samples. The presence of CRISPR-Cas in ICP1 and PLE in V. cholerae was validated by whole-genome sequencing. Genomic libraries were generated using NEBNext Ultra II DNA Library preparation kit for Illumina (New England Biolabs), according to the manufacturer's recommended protocols. Paired-end sequencing (2 × 150 bp) was performed on an Illumina HiSeq4000 (University of California, Berkeley QB3 Core Facility). Sequencing assembly/mapping and detection of CRISPR was performed as described [23]. The genome of the V. cholerae clinical isolate KS393 was sequenced on Illumina HiSeq4000, PacBio Sequel and Oxford Nanopore MinION sequencers (University of California, Berkeley QB3 Core Facility). Assembly of KS393 sequences was performed using the canu assembler v1.6 [24] to combine the PacBio and Oxford Nanopore reads into genomic scaffolds for the large and small chromosomes using default settings and an expected genome size of 4033460 bp. This generated two scaffolds of the expected sizes for each chromosome which were then polished with the Illumina paired-end sequences using Pilon v1.22 [25] with the ‘fix all’ command to generate a high-quality genomic assembly in a fasta format of both chromosomes (electronic supplementary material, File S1). The sequencing data for strain KS393 have been deposited in the Sequence Read Archive (SRA) database under accession codes SRR7826356, SRR7826357 and SRR7826358.
Vibrio cholerae mutants were constructed by natural transformation as described [26]. Mutations in ICP1 were generated using CRISPR-Cas mediated genome engineering with the V. cholerae classical biotype Type I-E system as described [11] (electronic supplementary material, tables S3 and S4). Engineered phage ± Cas1 D244A with spacer 9 were validated by plaquing on a permissive PLE 1 host and determining the frequency of phage with a newly acquired spacer by calculating the efficiency of plaquing on the permissive PLE 1 host to a PLE 1 host with the protospacer deleted (PLE 1ΔPS9). The limit of detection is met when the phage is unable to form a plaque on the restrictive host at the highest concentration while still being able to productively infect a permissive host, with at least 6 orders of magnitude tested. Examination of PLE replication and transduction during phage infection was as reported previously [12].
(b). High-throughput spacer acquisition, data processing and analyses
Three independent experiments were performed as follows: a 50 ml culture of PLE 1 V. cholerae was grown to OD600 = 0.3 and infected at a multiplicity of infection of 1 with ICP1_2011_A ΔS9, which harbours spacer 8 at the leading edge of the CRISPR 1 array that targets PLE 1 and allows for phage replication [13]. Infected cells were incubated for 90 min at 37°C with aeration, at which point lysis was observed. The lysate was treated with chloroform and centrifuged to remove bacterial debris. Phage were precipitated with 10% (w/v) polyethylene glycol (PEG) 8000 at 4°C overnight. Phage pellets were collected by centrifugation at 4°C and the passaging was repeated as above. After three passages, the resulting pools were plated on a host possessing silent mutations in protospacer 8 (PLE 1PS8*) that inhibits spacer 8-mediated CRISPR interference, which enabled the selection of phage with expanded arrays that allow plaque formation. Phage DNA libraries were generated by homopolymer tail-mediated PCR (HTM-PCR) as previously described [27]. As ICP1_2011_A possesses only a single functional CRISPR array (figure 1b), the expanded phage CRISPR 1 array was amplified from genomic DNA libraries by PCR using custom barcoded primers (electronic supplementary material, table S4) to sequence the leader proximal spacer. 50 bp single-end sequencing was performed on an Illumina HiSeq 2500 (Tufts University Core Facility) using a custom sequencing primer. The resulting reads as fastq files were mapped to the large and small chromosome of V. cholerae strain KS393 using Bowtie v1.2.2 [28] with a seed_length of 31 and allowing for 0 max_total_mismatches which ensured that spacer to protospacer matches were 100% identical. These mappings were performed in two parallel ways: first, to obtain all possible spacer mapping locations regardless of the number of identical protospacer targets (i.e. translucent spacers in figure 3b) and second, restricting max_alignments to 1 which only mapped spacers with exactly one unique mapping location across both chromosomes. With a custom Python script (https://git.io/fNVqZ) we extracted the PAM sequences and GG PAM slippage locations from the restricted unique mappings. We also used this script to generate spacer mapping location graphs for both set of mappings using Biopython's GenomeDiagram module [29]. The amplicon sequencing data have been deposited in the SRA database under accession codes SRR7827053, SRR7827054 and SRR7827055.
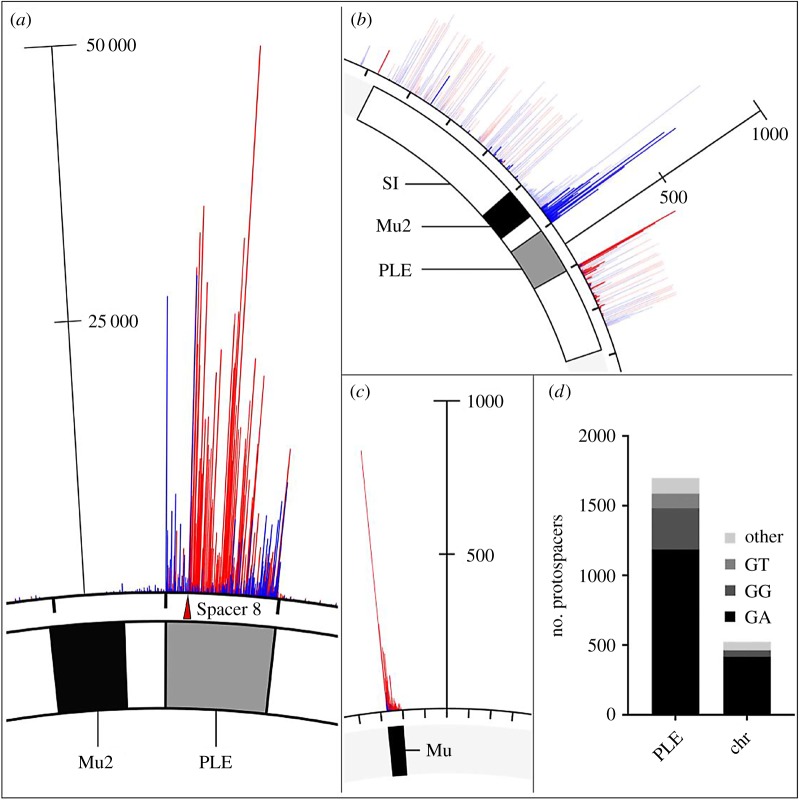
Figure 3.
High-throughput interference-driven spacer acquisition mapping. (a) The locations of the ICP1 CRISPR leader-proximal spacer on the V. cholerae small chromosome. The location of the interference-efficient spacer (S8) is indicated with the red triangle. (b) Spacer locations on the V. cholerae small chromosome (PLE mappings not shown for clarity). Uniquely mapped spacers are shown in solid blue or red, while translucent bars show mapping of spacers to all possible locations. (c) Spacer locations on the V. cholerae large chromosome. For panels (a–c), spacers on the plus and minus strand are indicated in red and blue, respectively. The scale bar measures the number of mapped spacers, and the tick marks around the chromosome are in 18 kb intervals. The white box represents the superintegron (SI), the black box is the mu-like region and the grey box is PLE 1. (d) Proportion of unique protospacers with a GA or other dinucleotide PAM sequence in PLE or in the small chromosome (chr).
3. Results
(a). ICP1-encoded CRISPR-Cas is fixed in the natural phage population
We set out to compare ICP1 and PLE from contemporary cholera patient stool samples to previously identified isolates from the International Centre for Diarrhoeal Disease Research, in Dhaka, Bangladesh (ICDDR,B) [13,21]. We isolated eight new ICP1 isolates from cholera patient stool samples collected between 2015 and 2017 and found that all isolates harbour CRISPR-Cas. Thus, it appears that ICP1 isolates lacking CRISPR have not been identified in Bangladesh since 2006 [23]. Analysis of the CRISPR arrays indicates a strong selection for spacers specifically targeting PLE (figure 1b; electronic supplementary material, table S5), supporting the function of the ICP1-encoded CRISPR-Cas system as a counter-attack against the anti-phage island PLE [13]. To evaluate if the fixation of CRISPR in ICP1 is necessitated by co-circulating PLE in epidemic V. cholerae, we determined the prevalence of PLE over the same near two-decade long period in Dhaka. Combined with previous analyses [12,21], we observed an increase in the prevalence of PLE(+) V. cholerae in epidemic sampling over time (figure 1c). Of note is the high prevalence of PLE 1 V. cholerae over the past 6 years, indicating that this variant of the anti-phage island is currently dominating the epidemic landscape in Dhaka. Despite the relatively long period over which PLE 1 has been dominant in Dhaka, and consistent with previous results [12,21], whole-genome sequencing of eight PLE 1 V. cholerae isolates showed that PLE 1 is 100% identical at the nucleotide level in all strains.
(b). Multiple spacers increase ICP1 CRISPR-Cas mediated PLE interference
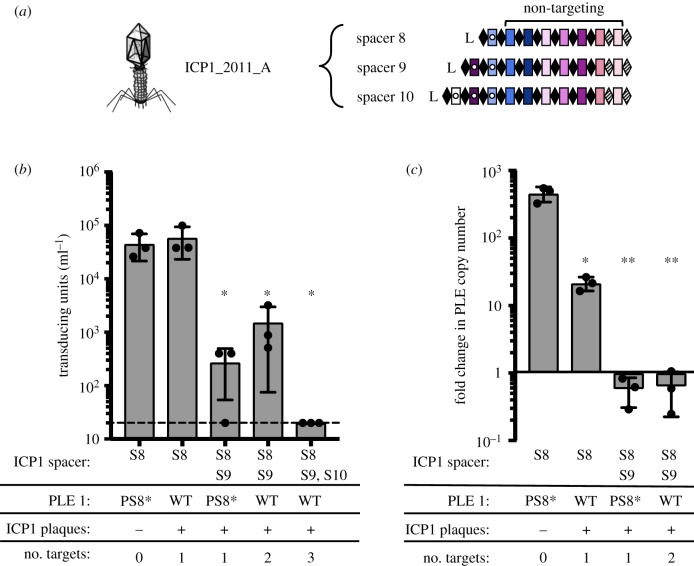
All of the natural phage we isolated encode multiple CRISPR spacers against PLE (figure 1b); however, previous work revealed that only one functional spacer is required for ICP1 to overcome PLE-mediated anti-phage activity as evaluated by plaque formation (electronic supplementary material, figure S1) [13]. Conversely, a single spacer against the PLE did not prevent transduction of PLE [12]. To investigate the consequences of varying spacer number and identity on PLE transduction and replication, we used co-isolated ICP1 and PLE 1 V. cholerae obtained from a cholera patient sample in 2011 [13]. This ICP1 isolate harbours two spacers (spacers 8 and 9) at the leading edge of the CRISPR 1 array that target PLE 1. We also used an isogenic phage with a spontaneous loss of spacer 9 [13], as well as one that acquired an additional 10th spacer targeting PLE in vitro (figure 2a). Despite the ability to overcome PLE and form plaques, spacer 8 targeting was not sufficient to decrease PLE transduction during ICP1 infection relative to an untargeted control (figure 2b). In comparison, two anti-PLE spacers decreased PLE transduction during ICP1 infection and three spacers completely abolished PLE transduction, showing that increased CRISPR targeting by ICP1 has a stronger anti-PLE effect. To evaluate potential differences between spacer 8 and spacer 9 on PLE targeting, we used PLE 1 with a protospacer mutation (PLE 1PS8*) that inhibits spacer 8-mediated PLE targeting [13]. Strikingly, just spacer 9 targeting PLE alone was able to decrease PLE transduction to the same level as when two spacers were targeting PLE (figure 2b).
Figure 2.
CRISPR can limit horizontal transmission of PLE. (a) ICP1_2011_A with anti-PLE 1 spacers S8, S9 and S10 (shown with internal white circles) tested in panels (b) and (c). (b) PLE transduction after infection with ICP1 with 0, 1, 2 or 3 spacers. The dashed line indicates the limit of detection for this assay. PS8* indicates silent mutations in protospacer 8 that abolish CRISPR interference [13]. A single spacer is necessary and sufficient to permit lytic growth of ICP1 on PLE 1 V. cholerae as seen by equal plaque formation (indicated by +, electronic supplementary material, figure S1). (c) PLE replication 20 min after infection with ICP1 with 0, 1 or 2 spacers as determined by qPCR. For panels (b,c), error bars indicate standard deviations of biological triplicates. Significance was determined by T-test, *p < 0.05, **p < 0.005.
We next analysed the copy number of PLE during infection with ICP1 encoding one or two targeting spacers to identify if the differences in reducing PLE transduction were due to differences in PLE copy number (figure 2c). In the absence of ICP1 CRISPR targeting, PLE replicates to high copy number, which facilitates horizontal transmission [12]. Targeting with only one spacer was sufficient to significantly decrease PLE replication, and in agreement with the transduction data, spacer 9 had a stronger inhibitory effect on PLE replication than spacer 8. Sequencing of the newly transduced PLEs showed no mutations in the protospacers. Thus, the transduced PLEs did not escape CRISPR targeting through mutation, but instead, the individual spacers possess different and incomplete abilities to fully block PLE mobilization. Altogether, these results demonstrate that not all spacers selected in nature equally interfere with PLE mobilization and that increasing the number of spacers provides enhanced capacity of ICP1 to interfere with PLE.
(c). Interference-driven spacer acquisition in ICP1 reveals indirect targets and non-canonical protospacer adjacent motifs
Since spacer composition variability in nature was lower than we expected (figure 1b), we next set out to experimentally sample the repertoire of spacers that ICP1 can acquire to overcome PLE. Low-throughput experiments previously demonstrated that ICP1 can acquire new spacers targeting the PLE under laboratory conditions without the need to overexpress cas genes [13]. To further analyse the natural process of interference-driven spacer acquisition in this system, we performed high-throughput sequencing of expanded CRISPR arrays of phage selected on PLE 1 V. cholerae. We infected PLE 1 V. cholerae with ICP1 containing spacer 8 (figure 2a), and the recovered lysate was probed for ICP1 progeny with newly acquired spacers that allowed for plaque formation on a PLE 1PS8* host. Illumina sequencing of the leader-proximal spacer in CRISPR 1 allowed us to sample over 106 acquired spacers in each replicate experiment (electronic supplementary material, figure S2 and table S6). In order to accurately map the spacers to the PLE 1 V. cholerae host, we performed complete whole-genome sequencing and assembly of the bacterial genome (electronic supplementary material, File S1). As was previously reported [12], we found that PLE 1 was integrated in a V. cholerae repeat, of which over 100 repeats intersperse the V. cholerae small chromosome in a gene-capture region, the superintegron [30]. In total, 96% of the acquired spacers mapped to PLE (electronic supplementary material, figure S3), while, interestingly, the other 4% mapped to V. cholerae chromosomes (electronic supplementary material, table S6).
Mapping of the spacers to the small chromosome showed a pattern of strand bias that reflected previous observations in primed acquisition experiments performed in other Type I-F systems [31], with a distribution of acquired spacers 5′ of the protospacer on the non-targeted strand and 3′ of the protospacer on the targeted strand (figure 3a). The distribution of spacers acquired 5′ of the protospacer on the non-targeted strand were split between the small chromosomal region proximal to the PLE 1 integration site (figure 3b, electronic supplementary material, figure S4), as well as the 3′ end of PLE. Acquired spacers mapping to the V. cholerae chromosome were not evenly distributed between the large and small chromosome, but instead approximately 90% of the chromosomal spacers mapped to the small chromosome (figure 3b; electronic supplementary material table S6). Spacers that mapped to the large chromosome were restricted to a mu-like region (figure 3c), which was duplicated in this strain and was also in the small chromosome proximal to PLE (figure 3b). Acquired spacers mapped uniformly throughout the superintegron, however, this is likely an artefact as the superintegron is highly repetitive. When considering spacers that map to a single site in the small chromosome, we observed an obvious bias for acquired spacers mapping closer to the PLE integration site (figure 3b; electronic supplementary material, figure S4).
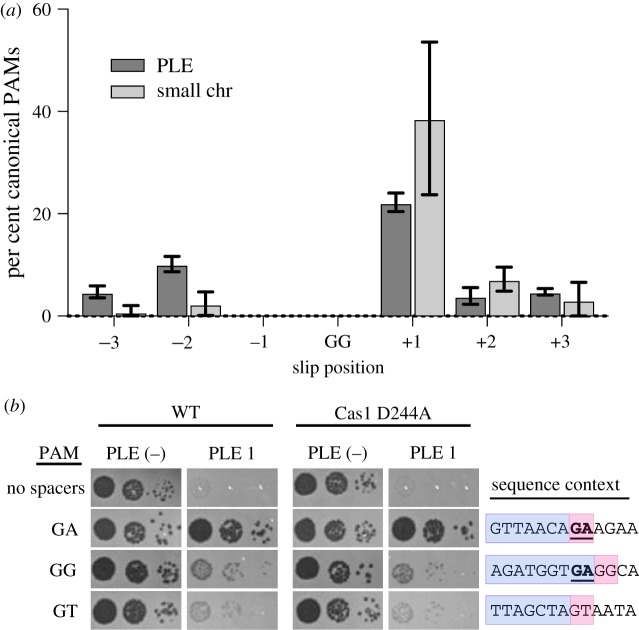
Consistent with CRISPR+ ICP1 isolates from nature (electronic supplementary material, figure S5), the majority (approx. 70%) of the spacers acquired experimentally targeted protospacers in PLE 1 that were flanked by a 3′ GA PAM (figure 3d). Variations from the canonical GA motif would be expected to abolish CRISPR interference. However, approximately 30% of protospacers in PLE had non-canonical PAMs, and of those, the majority were GG or GT. Previous CRISPR acquisition studies in Type I-F systems indicate that alternative PAMs can be explained by a ‘slippage’ event [31,32]. To identify putative slippage events, we analysed the sequences adjacent to GG PAMs and found that 45% of GG PAMs have a canonical GA within three nucleotides of the PAM position, suggesting that the ICP1 acquisition machinery has a propensity to slip (figure 4a).
Figure 4.
Characterizing non-canonical PAMs. (a) The frequency of a canonical GA PAM ± 3 nt from a non-canonical GG PAM across all data sets. (b) Tenfold dilutions of ICP1 engineered to contain a spacer that targets PLE 1 with a non-canonical PAM spotted on V. cholerae PLE (−) or PLE 1 lawns showing the ability of different phage strains to form plaques (dark spots, zones of killing) (left). Sequence context (right) of the region adjacent to the PAM. The protospacer is boxed in purple and PAM is boxed in pink. The consensus canonical PAM GA is bolded and underlined.
We next wanted to determine if these non-canonical PAMs are functional for PLE interference. Since only the newest spacer was sequenced in our high-throughput assay, we could not rule out that multiple spacers were not acquired within the expanded phage CRISPR array. We therefore engineered ICP1 to encode a single spacer reflective of an experimentally acquired spacer with either the canonical PAM or the most common non-canonical PAMs: either a GG or GT (figure 3d), and evaluated plaque formation of the engineered phage on PLE 1 V. cholerae. Despite relying on a non-canonical PAM, we found that ICP1 is able to target those protospacers and overcome PLE, albeit at a lower efficiency than when targeting a protospacer with a canonical GA PAM (figure 4b). Even when no canonical PAM was within ± 3 nt, ICP1 was still able to overcome PLE targeting a protospacer with a GT PAM. As PAM mutations are frequently a source for primed acquisition [33], we tested if the observed residual CRISPR activity was due to further spacer acquisition and interference. We constructed a Cas1 D244A mutation, which disrupts a conserved metal coordinating residue to inhibit spacer acquisition [32] (electronic supplementary material, figure S6), and tested if plaque formation was altered (figure 4b). We observed no difference in the efficiency of plaque formation between the Cas1 mutants and the parental phage, suggesting that the ICP1 CRISPR-Cas system is more tolerant of divergent PAMs during infection than previously characterized [13].
(d). Protospacers in the small chromosome facilitate ICP1 CRISPR-Cas-mediated PLE interference
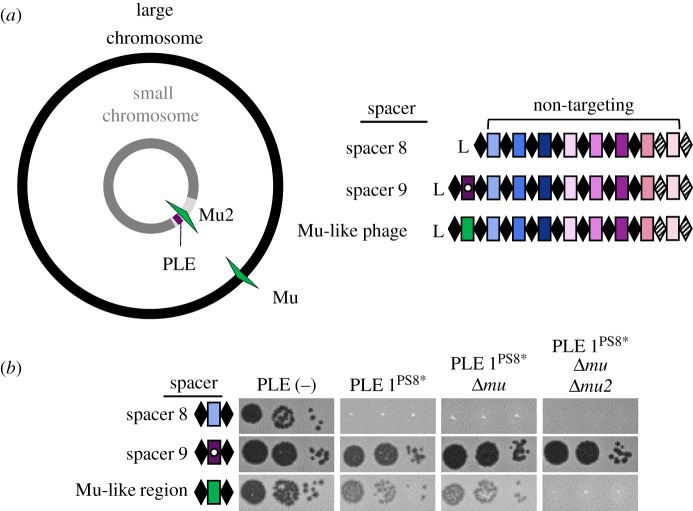
In our spacer acquisition experiment, we identified a subset of spacers that target a mu-like region in the V. cholerae large chromosome (figure 3c), suggesting that CRISPR targeting of the mu-like region was advantageous in overcoming PLE. To test the role of protospacers in the mu-like region in PLE interference, we isolated ICP1 that had acquired a spacer that targets the mu-like region and was able to form plaques on PLE 1PS8* (figure 5). Since assembly of the V. cholerae genome revealed that the mu-like region was present and 100% identical in both chromosomes, presumably due to a duplication of the region on the large chromosome (figure 5a), we wanted to evaluate if targeting the mu-like region per se allowed for plaque formation, or if the chromosomal context was important in allowing for CRISPR-meditated interference with PLE. To test this difference, we generated a single knockout of the mu-like region in the large chromosome and a double knockout in both chromosomes. ICP1 CRISPR-mediated interference with PLE was abolished in the double knockout, however, knocking out the mu-like region in the large chromosome had no effect on ICP1 plaque formation, demonstrating that targeting of the region in the large chromosome is an artefact of the duplication in the small chromosome (figure 5b). These results show that CRISPR targeting of the V. cholerae large chromosome is dispensable for phage overcoming PLE, while targeting the small chromosome is sufficient to overcome PLE activity.
Figure 5.
ICP1 CRISPR-targeting of the small chromosome facilitates PLE interference. (a) Cartoon (left) of the V. cholerae large and small chromosomes. The superintegron is shown in light grey, the PLE is shown in purple. The two mu-like regions in the large and small chromosome are shown by green arrows. ICP1_2011_A CRISPR variants (right) used to test the role of targeting sites. The internal white circle indicates the PLE 1 targeting spacer. (b) Tenfold dilutions of ICP1 with the spacers indicated spotted on V. cholerae lawns showing the ability of different phage strains to form plaques.
(e). When CRISPR goes off target: going the distance to maintain interference
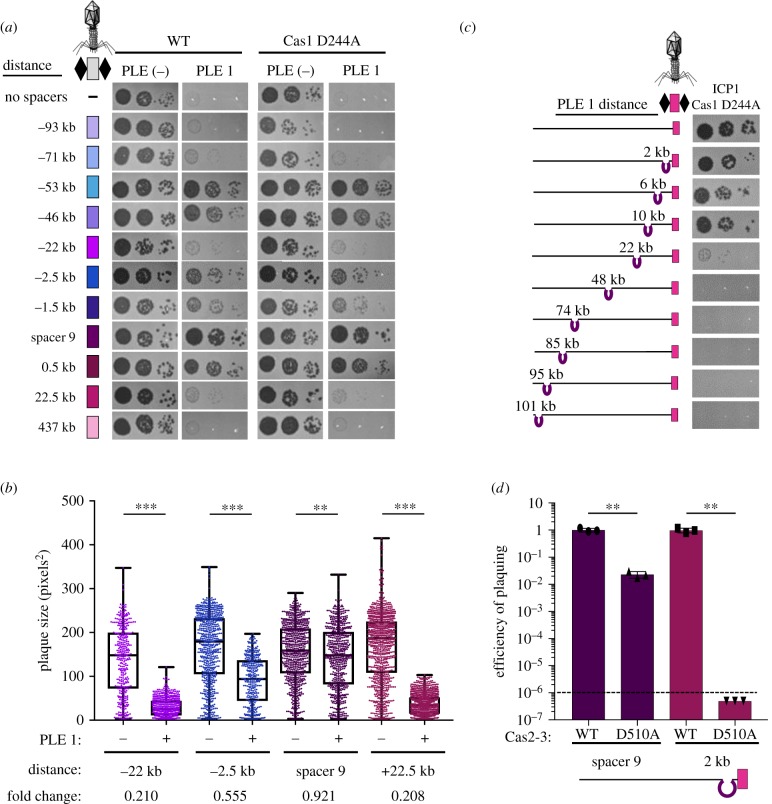
As processivity of Cas2-3 has been demonstrated in vitro [17], we speculated that ICP1 targeting of the small chromosome proximal to PLE interferes with PLE anti-phage activity by the processive degradation of PLE along with the chromosome; however, PLE excises from the chromosome early during ICP1 infection [20]. This timing suggests that CRISPR targeting and Cas2-3 processive degradation of the small chromosome would have to happen prior to PLE excision and would therefore likely be distance dependent. In support of this hypothesis, experimentally acquired spacers mapping to the small chromosome clustered proximal to PLE (figure 3b). To test the impact of targeting at increasing distances from PLE, we engineered ICP1 to possess CRISPR arrays containing only one spacer drawn from the experimental acquisition pool that targets the small chromosome at varying distances away from PLE. We then assayed the ability of this engineered phage to overcome PLE and form plaques (figure 6a). As a positive control, ICP1 engineered with a spacer that targets internal to PLE formed robust and equal plaques on PLE(-) and PLE 1 hosts. In comparison, phage with a spacer that targets far (greater than 400 kb) from PLE were unable to form plaques on PLE 1. Conversely, ICP1 that target a protospacer only 0.5, 1.5 or 2.5 kb from PLE were able to efficiently overcome PLE and form plaques. Phage targeting protospacers at intermediate distances away from PLE (greater than 20 kb) demonstrated weak plaque formation on PLE 1. Surprisingly, we observed that ICP1 with some spacers targeting relatively far from PLE (53 and 46 kb away) were still able to form robust plaques on PLE 1 (figure 6a). While all of the spacers selected for this assay had one perfect protospacer match in the chromosome and have a GA PAM, we identified greater than 100 promiscuous putative target sites for these spacers which would bring the chromosomal target much closer to PLE 1 (electronic supplementary material, table S7), which may explain these phage's ability to overcome PLE. To test if spacer acquisition had a role in plaque formation, we engineered the chromosomal targeting phage in a Cas1-deficient background and assayed for plaque formation on the PLE 1 host. Despite being unable to acquire spacers (electronic supplementary material, figure S6), the Cas 1 = deficient phage retained the same plaquing phenotype. We quantified the weaker plaque formation observed when ICP1 targets 2.5 kb and greater than 20 kb away from PLE 1 by measuring plaque size compared to PLE (−) V. cholerae (figure 6b). As compared to phage with PLE internal and PLE proximal spacers, phage with chromosomal spacers targeting greater than 20 kb away from PLE had significantly limited plaque size; however, even phage with a chromosomal spacer that is proximal to PLE has an approximately 50% smaller plaque size when compared to plaques on a PLE (−) host. These results indicate that some PLE-mediated anti-phage activity is retained when CRISPR-Cas is directed at increasing distances from PLE in the small chromosome, but direct targeting of PLE is still required for maximizing phage fitness.
Figure 6.
The interference potential of spacers directed to the small chromosome is dependent on the proximity to PLE. (a) Tenfold dilutions of ICP1 engineered with a spacer that targets the small chromosome or PLE 1 +/− Cas1 spotted on lawns of V. cholerae showing the ability of different phage strains to form plaques. Spacer 9 is the same as spacer 9 in ICP1_2011_A. The distance of the chromosomal protospacer from the PLE 1 integration site is indicated. (b) Plaque size of ICP1 variants plated with V. cholerae. The distance and colour scheme correspond to the spacers tested in (a). The fold change in average size of a plaque on a PLE (+) host compared to a PLE (−) host is indicated at the bottom. Significance was determined by Mann–Whitney U-test, **p < 0.005, ***p < 0.0001. (c) Tenfold dilutions of ICP1 engineered with a chromosomal protospacer spotted on lawns of V. cholerae harbouring PLE in different locations in the small chromosome. (d) Efficiency of plaquing of phage engineered to contain a spacer that is internal to PLE 1 (spacer 9) or targeting the small chromosome 2 kb away from PLE 1 (same as in (c), cartoon below graph) with the WT or helicase dead (D510A) Cas2-3 allele. The dashed line indicates the limit of detection. Significance was determined by T-test, **p < 0.005.
To control for differences in spacer sequences, we also varied the location of the PLE and tested the ability of ICP1 with a single spacer targeting the small chromosome to interfere with PLE 1. Following ICP1-mediated transduction, PLE 1 integrates into a V. cholerae repeat in the new host [12]. We collected a pool of PLE 1 transductants where PLE was integrated at varying distances from the chromosomal protospacer and challenged these strains with ICP1. As a control, we determined that all of the tested PLE 1 V. cholerae hosts were susceptible to ICP1 CRISPR-Cas interference when ICP1 possessed a PLE internal spacer (electronic supplementary material, figure S7a). Consistent with our earlier finding, PLE integrated at an increasing distance away from the protospacer was less susceptible to ICP1-encoded CRISPR interference (figure 6c).
As Cas2-3 has been demonstrated to translocate in vitro, we next wanted to see if the indirect inhibition of PLE by spacers that target the small chromosome was due to Cas2-3 processivity. We constructed a Cas2-3 helicase dead variant by mutating the conserved DExx helicase motif II (D510A) [16] (electronic supplementary material, figure S7b) and tested the ability of ICP1 to form plaques on PLE 1 V. cholerae. ICP1 Cas2-3 D510A engineered with a spacer that targets internal to PLE was still able to form plaques on a PLE (+) host, although both the efficiency of plaquing and plaque size were negatively impacted by the helicase mutation (figure 6d; electronic supplementary material, figure S7c). Conversely, when ICP1 targets the V. cholerae small chromosome 2 kb away from PLE, the helicase activity of ICP1 Cas2-3 was absolutely essential for PLE interference and plaque formation (figure 6d; electronic supplementary material, figure S7c). These findings are the first direct demonstration of functional Cas2-3 processivity in vivo and support our model of indirect targeting (figure 7).
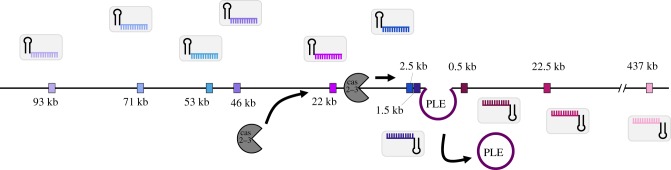
Figure 7.
Model of race between ICP1 Cas2-3 processive degradation of the V. cholerae chromosome and ICP1-mediated PLE excision. Csy complexes (grey boxes) with crRNAs (coloured) search for a complementary protospacer (coloured rectangles, experimentally assessed in figure 6a). Cas2-3 (dark grey) is recruited to the protospacer and processively degrades the DNA towards PLE (purple). ICP1 is able to form plaques when Cas2-3 degrades PLE before PLE excises from the chromosome, which occurs within 5 min of ICP1 infection.
4. Discussion
Our results reveal that the latest front in the ongoing arms race between contemporary isolates of epidemic V. cholerae and its predator ICP1 necessitate the persistence of the ICP1-encoded Type I-F CRISPR-Cas system to counter PLE-mediated anti-phage activity (figure 1). By using a high-throughput spacer acquisition assay, we gained insight into the full range of spacers that can combat PLE. Interestingly, our experimental findings on acquisition and interference do not reflect the rather limited diversity of spacers that ICP1 maintains against PLE in nature. These results highlight that not all spacers are equally proficient for interference, and that coupled analysis of these competing mobile genetic elements from nature reveals the evolutionary benefits of a particular complement of spacers more so than laboratory-based studies. Despite a lack of clear evidence indicating where the ICP1-encoded CRISPR-Cas system originated, it serves as a tractable model through which we can examine the biology of an endogenous Type I-F CRISPR-Cas system against its cognate foe.
Co-culture studies competing phage against CRISPR-Cas proficient bacterial hosts demonstrated that mutational escape by phage is limited by bacterial populations that have heterogenous CRISPR arrays [34]. Here, we see that PLE 1 is highly conserved over time, even when co-circulating with CRISPR proficient ICP1. In the light of previous suggestions, the diversity of CRISPR arrays in ICP1 populations may limit the success of PLE escape mutants. Surprisingly, however, we see very little diversity in the spacer composition of ICP1 CRISPR arrays with the same minimal spacers being conserved in phage circulating for over 8 years (figure 1b). Likewise, CRISPR-proficient ICP1 isolated from nature always encoded more than one spacer against PLE, which would be expected to limit CRISPR escape mutations. It may be that there is limited room for genetic drift in the PLE genome, permitting ICP1 to streamline its CRISPR array, keeping only the most efficient spacers while also maintaining an advantageous genome size.
Akin to studies of bacterial Type I-F CRISPR-Cas mediated interference with plasmid transformation and conjugation [35], we similarly see that the spacer sequence and quantity of spacers in the array have a role in ICP1's ability to abolish PLE spread (figure 2). This may be due to differences in crRNA abundance or stability, or sequence-dependent subtleties that dictate interference potential, as has been proposed previously [36]. Despite spacer 9's improved interference with PLE mobilization compared to spacer 8, we still observed a slight defect in plaque size when comparing engineered phage with only spacer 9 relative to a PLE (−) host (figure 6b), suggesting that even this improved spacer alone is not sufficient to fully overcome PLE-mediated anti-phage activity. By encoding a seemingly redundant set of spacers targeting PLE, ICP1 increases its ability to overcome PLE and limit PLE spread in the environment. Additionally, multi-site targeting of V. cholerae PLE by ICP1 CRISPR-Cas may contribute to the modular evolution observed between PLE variants and dictate which PLEs are circulating within and between epidemics [12].
As expected, the majority of spacers acquired in our high-throughput acquisition assay directly target PLE (figure 3a). Analysis of natural ICP1 isolates recovered from cholera patient stool samples shows that the phage-encoded CRISPR-Cas system recognizes a GA PAM (electronic supplementary material, figure S4) which, although atypical for Type I-F systems [37], has been confirmed through single mutations to a C in both positions [13]. Notably, we found that ICP1 was able to incorporate spacers that targeted non-canonical PAMs (figure 3d) and that these spacers can suffice for PLE interference (figure 4b). In comparison to another high-throughput spacer acquisition assay in a Type I-F system, which found greater than 90% of all protospacers flanked by the canonical PAM [31], it appears that the phage-encoded system is less discriminating with only 70% of protospacers flanked by the expected PAM. However, targeting a protospacer with a non-canonical PAM reduced the ability of ICP1 to form plaques compared to the canonical PAM (figure 4b). As such, in nature ICP1 targeting a protospacer with a non-canonical PAM would not be able to completely interfere with PLE and thus would be selected against. This hypothesis is additionally supported by the observation that very few non-canonical PAM protospacers were associated with indirect targets in the small chromosome. As these chromosomal spacers are themselves less proficient for interference (figure 6a,b), the added disadvantage of targeting a protospacer with a non-canonical PAM likely tips the balance in favour of PLE, possibly explaining the lower abundance of these spacers in our selection experiments.
Despite the presence of spacers that target the V. cholerae large chromosome in the high-throughput spacer acquisition assay (figure 3c), we show that targeting this chromosome is dispensable for CRISPR interference of PLE and is likely an artefact of a duplication event of the mu-like region in the strain used in our assays (figure 5). Interestingly, two of the natural ICP1 isolates contain a spacer that targets a gene on the V. cholerae large chromosome (figure 1b). We speculate that this spacer was acquired from a V. cholerae strain possessing a duplication or rearrangement that is not represented in currently sequenced isolates, in which the protospacer was in the small chromosome proximal to PLE, allowing the phage to overcome PLE activity. However, this spacer does not seem to be maintained in the phage population, likely due to diminished PLE interference relative to PLE-direct spacers as we experimentally observed.
CRISPR targeting of the V. cholerae small chromosome can overcome PLE, but our results suggest a model in which there is a limit to the distance over which processive Cas2-3 degradation can occur to reach the PLE prior to excision (figure 7), an action which occurs within 5 min of ICP1 infection that is directed by an early expressed ICP1 protein [20]. The limit of processivity appears to be around a distance of 23 kb (figure 6a,c), at which point either Cas2-3 is unable to continue to process along the V. cholerae chromosome or PLE excises before interference occurs. In vitro studies of Cas3 from Type I-E systems have demonstrated Cas3 translocation velocities of 89–300 bases per second and average processivities between 12 and 19 kb [38,39], however, the functional role and limitations of processivity in vivo are not known. Our results are the first to demonstrate that Cas2-3 is processive in vivo, with over 22 kb from a distal chromosomal protospacer over which the ICP1 CRISPR-Cas can maintain activity to overcome PLE when Cas2-3 has the ability to translocate along DNA (figure 6d). As this event must occur within 5 min of ICP1 initiating infection, the estimated processivity of ICP1 Cas2-3 is within the range of what has been reported for Type I-E Cas3, which is especially remarkable given the complexity of the crowded intracellular environment compared to simplified in vitro systems.
In comparison to other Cas nucleases like Cas9, which introduces a single double-stranded break [40,41], Cas2-3 degrades DNA as it translocates away from the protospacer [17], making it more likely to destroy and thus interfere with its target. In fact, we see that the helicase dead Cas2-3 is less able to overcome PLE even when directly targeting the anti-phage island, suggesting that the processive degradation of PLE contributes to interference (figure 6d). Similarly, this predicted advantage may account for the increased prevalence of Type I systems for phage defence [42]. In the context of the battle between ICP1 and PLE, this processivity permits interference even with an indirect CRISPR target and has important implications for harnessing CRISPR-Cas in biotechnology and medicine. Since the characterization of the ICP1-encoded CRISPR-Cas system, phage engineered with CRISPR-Cas systems to target virulent, antibiotic-resistant bacteria have been assayed for therapeutic applications [43,44], showing the value of innovating from natural systems to overcome disparate biological problems.
Supplementary Material
Supplementary Material
Acknowledgements
The authors are especially thankful to ICDDR,B hospital and laboratory staff for their support and would like to thank Shirajum Monira, Kazi Zillur Rahman, Fatema-tuz Johura, Marzia Sultana and Monika Sultana in particular. The authors thank members of the Seed lab for critical feedback and thoughtful discussion regarding this manuscript.
Ethics
The collection of cholera patient stools was approved by the ICDDR,B institutional review board. All samples were deidentified and written informed consent was obtained from adult participants and from the guardians of children.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material. The sequencing data have been deposited in the Sequence Read Archive database under accession codes SRR7826356, SRR7826357, SRR7826358, SRR7827053, SRR7827054 and SRR7827055.
Authors' contributions
A.C.M., K.N.L. and K.D.S. carried out the molecular laboratory work. A.A. developed and implemented tools for sequence data analysis. M.A. coordinated the collection of clinical specimens. A.C.M., M.A. and K.D.S. conceived of the study. All authors participated in data analysis. A.C.M. and K.D.S. wrote the manuscript with input from all authors and all authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This research was funded by the National Institute of Allergy and Infectious Diseases grant number R01AI127652 (K.D.S.). A.C.M. received support from the Kathleen L. Miller Fellowship from the Henry Wheeler Center for Emerging and Neglected Diseases. K.N.L. received support from the National Science Foundation Graduate Research Fellowship Program. K.D.S. is a Chan Zuckerberg Biohub Investigator. M.A. of ICDDR,B gratefully acknowledges the following donors which provide unrestricted support: Government of the People's Republic of Bangladesh, Global Affairs Canada (GAC), Swedish International Development Cooperation Agency (SIDA), and the Department for International Development, UK Aid.
References
- 1.Parikka KJ, Le Romancer M, Wauters N, Jacquet S. 2017. Deciphering the virus-to-prokaryote ratio (VPR): insights into virus-host relationships in a variety of ecosystems. Biol. Rev. 92, 1081–1100. ( 10.1111/brv.12271) [DOI] [PubMed] [Google Scholar]
- 2.Dy RL, Richter C, Salmond GPC, Fineran PC. 2014. Remarkable mechanisms in microbes to resist phage infections. Annu. Rev. Virol. 1, 307–331. ( 10.1146/annurev-virology-031413-085500) [DOI] [PubMed] [Google Scholar]
- 3.Hille F, Richter H, Wong SP, Bratovič M, Ressel S, Charpentier E. 2018. The biology of CRISPR-Cas: backward and forward. Cell 172, 1239–1259. ( 10.1016/j.cell.2017.11.032) [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. ( 10.1126/science.1138140) [DOI] [PubMed] [Google Scholar]
- 5.Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30, 1335–1342. ( 10.1038/emboj.2011.41) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koonin EV, Makarova KS. 2017. Mobile genetic elements and evolution of CRISPR-Cas systems: all the way there and back. Genome Biol. Evol. 9, 2812–2825. ( 10.1093/gbe/evx192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shmakov S, Sitnik V, Makarova K, Wolf Y, Severinov K, Koonin E. 2017. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. MBio. 8, e01397-17 ( 10.1128/mBio.01397-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-Cas. MBio. 1, e00227-10 ( 10.1128/mBio.00227-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. 2012. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12, 177–186. ( 10.1016/j.chom.2012.06.003) [DOI] [PubMed] [Google Scholar]
- 10.Novick RP, Ram G. 2016. The floating (pathogenicity) island: a genomic dessert. Trends Genet. 32, 114–126. ( 10.1016/j.tig.2015.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Box AM, McGuffie MJ, O'Hara BJ, Seed KD. 2015. Functional analysis of bacteriophage immunity through a Type I-E CRISPR-Cas system in Vibrio cholerae and its application in bacteriophage genome engineering. J. Bacteriol. 198, 578–590. ( 10.1128/JB.00747-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Hara BJ, Barth ZK, McKitterick AC, Seed KD. 2017. A highly specific phage defense system is a conserved feature of the Vibrio cholerae mobilome. PLoS Genet. 13, e1006838 ( 10.1371/journal.pgen.1006838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seed KD, Lazinski DW, Calderwood SB, Camilli A. 2013. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature 494, 489–491. ( 10.1038/nature11927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haurzitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329, 1355–1358. ( 10.1126/science.1192272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richter C, Fineran PC. 2013. The subtype I-F CRISPR–Cas system influences pathogenicity island retention in Pectobacterium atrosepticum via crRNA generation and Csy complex formation. Biochem. Soc. Trans. 41, 1468–1474. ( 10.1042/BST20130151) [DOI] [PubMed] [Google Scholar]
- 16.Fagerlund RD, et al. 2017. Spacer capture and integration by a type I-F Cas1–Cas2-3 CRISPR adaptation complex. Proc. Natl Acad. Sci. USA 114, E5122–E5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rollins MF, et al. 2017. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc. Natl Acad. Sci. USA 114, E5113–E5121. ( 10.1073/pnas.1620722114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellas CM, Anesio AM, Barker G. 2015. Analysis of virus genomes from glacial environments reveals novel virus groups with unusual host interactions. Front. Microbiol. 6, 656 ( 10.3389/fmicb.2015.00656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chénard C, Wirth JF, Suttle CA. 2016. Viruses infecting a freshwater filamentous cyanobacterium (Nostoc sp.) encode a functional CRISPR array and a proteobacterial DNA polymerase B. MBio. 7, e00667-16 ( 10.1128/mBio.00667-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKitterick AC, Seed KD. 2018. Anti-phage islands force their target phage to directly mediate island excision and spread. Nat. Commun. 9, 2348 ( 10.1038/s41467-018-04786-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naser IB, Hoque MM, Nahid MA, Rocky MK, Faruque SM. 2017. Analysis of the CRISPR-Cas system in bacteriophages active on epidemic strains of Vibrio cholerae in Bangladesh. Sci. Rep. 7, 14880 ( 10.1038/s41598-017-14839-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seed KD, et al. 2011. Evidence of a dominant lineage of Vibrio cholerae-specific lytic bacteriophages shed by cholera patients over a 10-year period in Dhaka, Bangladesh. MBio 2, e00334-10 ( 10.1128/mBio.00334-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angermeyer A, Das MM, Singh DV, Seed KD. 2018. Analysis of 19 highly conserved Vibrio cholerae bacteriophages isolated from environmental and patient sources over a twelve-year period. Viruses 10, 299 ( 10.3390/v10060299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. ( 10.1101/gr.215087.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 ( 10.1371/journal.pone.0112963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalia AB, Lazinski DW, Camilli A. 2014. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. MBio 5, e01028-13 ( 10.1128/mBio.01028-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazinski DW, Camilli A. 2013. Homopolymer tail-mediated ligation PCR: a streamlined and highly efficient method for DNA cloning and library construction. Biotechniques 54, 25–34. ( 10.2144/000113981) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 ( 10.1186/gb-2009-10-3-r25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cock PJA, et al. 2009. Biopython : freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. ( 10.1093/bioinformatics/btp163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barker A, Clark CA, Manning PA. 1994. Identification of VCR, a repeated sequence associated with a locus encoding a hemagglutinin in Vibrio cholerae O1. J. Bacteriol. 176, 5450–5458. ( 10.1128/jb.176.17.5450-5458.1994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staals RHJ, Jackson SA, Biswas A, Brouns SJJ, Brown CM, Fineran PC. 2016. Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR-Cas system. Nat. Commun. 7, 12853 ( 10.1038/ncomms12853) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vorontsova D, et al. 2015. Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery. Nucleic Acids Res. 43, 10 848–10 860. ( 10.1093/nar/gkv1261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Erp PBG, Jackson RN, Carter J, Golden SM, Bailey S, Wiedenheft B. 2015. Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res. 43, 8381–8391. ( 10.1093/nar/gkv793) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Houte S, et al. 2016. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388. ( 10.1038/nature17436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter C, et al. 2014. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 42, 8516–8526. ( 10.1093/nar/gku527) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xue C, Seetharam AS, Musharova O, Severinov K, Brouns SJJ, Severin AJ, Sashital DG. 2015. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res. 43, 10 831–10 847. ( 10.1093/nar/gkv1259) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. ( 10.1099/mic.0.023960-0) [DOI] [PubMed] [Google Scholar]
- 38.Redding S, et al. 2015. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell 163, 854–865. ( 10.1016/j.cell.2015.10.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown MW, et al. 2017. Assembly and translocation of a CRISPR-Cas primed acquisition complex. Cell 175, 934–946. ( 10.1016/j.cell.2018.09.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garneau JE, et al. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. ( 10.1038/nature09523) [DOI] [PubMed] [Google Scholar]
- 41.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 337, 816–821. ( 10.1126/science.1225829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova KS, et al. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. ( 10.1038/nrmicro3569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yosef I, Manor M, Kiro R, Qimron U. 2015. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl Acad. Sci. USA 112, 7267–7272. ( 10.1073/pnas.1500107112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bikard D, et al. 2014. Exploiting CRISPR-cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32, 1146–1150. ( 10.1038/nbt.3043) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material. The sequencing data have been deposited in the Sequence Read Archive database under accession codes SRR7826356, SRR7826357, SRR7826358, SRR7827053, SRR7827054 and SRR7827055.