Abstract
Our bodies are colonized by a complex ecosystem of bacteria, unicellular eukaryotes and their viruses that together play a major role in our health. Over the past few years tools derived from the prokaryotic immune system known as CRISPR-Cas have empowered researchers to modify and study organisms with unprecedented ease and efficiency. Here we discuss how various types of CRISPR-Cas systems can be used to modify the genome of gut microorganisms and bacteriophages. CRISPR-Cas systems can also be delivered to bacterial population and programmed to specifically eliminate members of the microbiome. Finally, engineered CRISPR-Cas systems can be used to control gene expression and modulate the production of metabolites and proteins. Together these tools provide exciting opportunities to investigate the complex interplay between members of the microbiome and our bodies, and present new avenues for the development of drugs that target the microbiome.
This article is part of a discussion meeting issue ‘The ecology and evolution of prokaryotic CRISPR-Cas adaptive immune systems’.
Keywords: CRISPR, microbiome, genome engineering, probiotics, phages, homologous recombination
1. Introduction
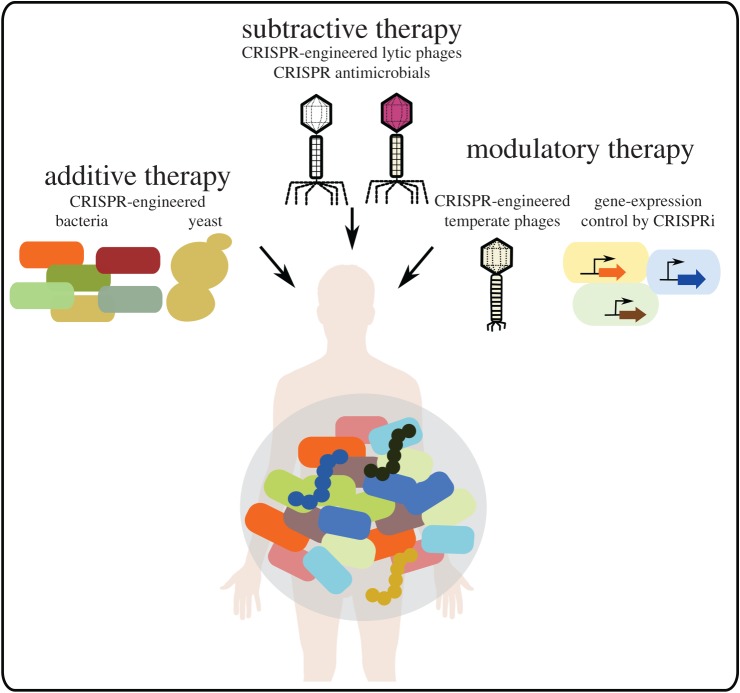
Healthy humans live in a symbiotic relationship with trillions of microorganisms that inhabit the exposed surfaces of our bodies and play an essential role in the maturation of the host-immune response, production of metabolites, brain–gut axis and more (see reviews [1–4]). This close relationship makes our microbiome an interesting target for therapies with the goal to induce desired responses, immunological, metabolic or even neurological in nature. These therapies can be classified into three main types: (i) additive therapies supplementing the host microbiota with individual strains or consortiums of bacterial species, (ii) subtractive therapies aiming to eliminate disease-causing members of the microbiome, and (iii) modulatory therapies aiming to modulate the composition or activity of the endogenous microbiome (see reviews [5,6]). While these therapeutic approaches are still in their infancy, engineered bacteria and viruses can be used to achieve desired outcomes [6]. In this review, we describe how tools derived from the prokaryotic immune system known as clustered regularly interspaced short palindromic repeats (CRISPRs)—and CRISPR-associated (Cas) proteins can be used to modify or eliminate members of the microbiome (figure 1).
Figure 1.
CRISPR approaches to microbiome therapies. CRISPR-Cas systems can be used to engineer designer probiotic strains of bacteria and yeast (additive therapies). CRISPR-Cas systems can also be used to eliminate target bacteria (subtractive therapies), either through the engineering of designer lytic bacteriophages, or through the delivery of CRISPR-Cas systems themselves as antimicrobials. Dead Cas proteins can be used to modify gene expression, and engineered temperate phages can modulate the composition and activity of bacteria in the microbiome (modulatory therapies).
CRISPR-Cas systems are the adaptive immune system of bacteria and archaea [7]. The strong interest in these systems comes from the discovery of a set of diverse RNA-guided nucleases able to destroy target nucleic acid sequences, some DNA and other RNA. The Cas nucleases are guided by CRISPR RNAs (crRNA), produced by transcription and processing of the CRISPR locus: a chromosomal site into which DNA fragments from invading nucleic acids are integrated in between repeats, providing a memory of past infections. Cas proteins associated to CRISPR arrays are very diverse and form the basis of the classification of CRISPR-Cas systems into two classes and six main types [8,9]. Class 1 systems (types I, III and IV) consists of a complex machinery, with several Cas proteins assisting the recognition of foreign nucleic acids and their cleavage. Class 2 systems (types II, V and VI) have a simpler protein architecture with a single effector protein arbitrating both recognition and cleavage. The latter class includes the type II CRISPR-Cas9 system, whose versatility has pushed the limits of genome editing [10]. Some features unique to type II systems are the double-stranded (ds) DNA endonuclease Cas9 and the auxiliary trans-acting crRNA (tracrRNA) [11]. The crRNA and tracrRNA can be fused into a chimeric single guide RNA (sgRNA) further simplifying the use of this system as a tool. More recently, Cas nucleases from other subtypes have been successfully used in a variety of biotechnological applications. These include the Cas12 (Cpf1) DNA endonuclease from type V systems as well as the Cas13 nuclease from type VI systems, which targets RNA rather than DNA [12,13].
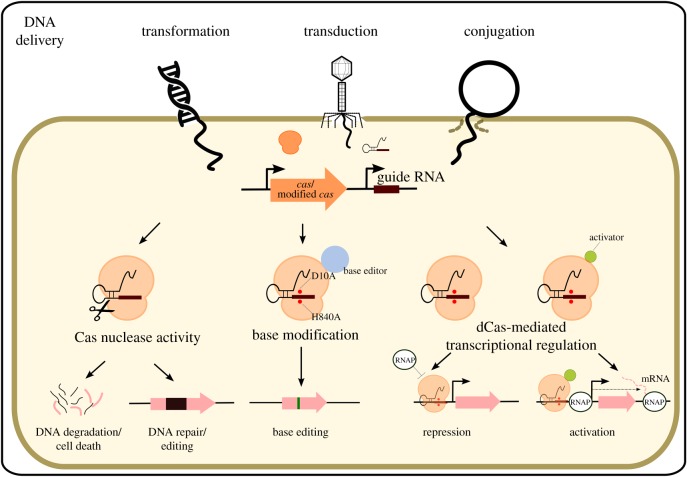
CRISPR-Cas systems are present in approximately 40% of bacteria. Endogenous CRISPR-Cas systems can in some cases be exploited, and engineered CRISPR-Cas systems can otherwise be introduced into target bacteria. These systems can be used to modify the genomes of microbiome-associated or probiotic bacteria, yeast and bacteriophages. They can also be used to kill specific strains based on their sequence without touching the rest of the microbiome (figure 1). Finally, CRISPR-Cas systems can be used to control gene expression without the need to modify the genome. Altogether CRISPR-Cas systems offer a powerful set of tools that will benefit the study of the microbiome and lead to the development of new strategies to modify it (figure 2).
Figure 2.
Killing, genome editing or modulation of gene expression by CRISPR-Cas systems. CRISPR-Cas systems can be delivered to target bacteria either in vitro or in vivo through transformation, transduction or conjugation. Cas nucleases induce DNA breaks that can either lead to DNA degradation and cell death, or if the break is repaired to the introduction of mutations. Catalytically dead Cas proteins such as dCas9 can be used to silence genes by blocking the RNA polymerase (RNAP). Dead Cas proteins can also be fused to various protein domains such as activators to induce the expression of genes, or to domains able to modify DNA bases.
2. CRISPR editing of bacteria
Shortly after its discovery and biochemical characterization, the CRISPR-Cas9 system was repurposed to edit eukaryotic and bacterial genomes [14]. It can now be considered as a tool of choice to engineer probiotic strains for additive therapies. Genome editing strategies rely on the use of a guide RNA designed to target a chromosomal sequence of interest where Cas9 will cut. Early bioinformatics studies revealed that bacterial genomes sometimes naturally carry CRISPR-Cas systems that contain guides targeting their own chromosome [15]. In most of these cases clues can be identified showing that the CRISPR-Cas system is inactivated by mutations in the cas genes or CRISPR array, or mutations altering the targeted sequences. A model was thus proposed where the CRISPR-Cas systems sometimes capture self-targeting spacers ‘by mistake’ and can only survive such events if the system is functionally inactivated. The idea that self-targeting of the bacterial chromosome by the CRISPR-Cas system is lethal was also corroborated by reports that bacteria die when endogenous or exogenous CRISPR-Cas systems are programmed to target the chromosome [16–19]. While the primary outcome of self-targeting is cell death, some cells are able to survive through the deletion of large DNA fragments encompassing the target position [18,20–23]. This strategy offers little to no control over the extent of DNA that will be deleted, but could still prove useful in removing undesired genetic elements such as pathogenicity islands or prophages from strains of interest. Lethal self-targeting can also be used to counter-select specific genotypes in complex populations [19,24].
The first evidence that CRISPR-Cas9 could be employed to achieve precise scar-less genome editing in bacteria came from a study in which the Cas9 protein from Streptococcus pyogenes was integrated in the chromosome of Streptococcus pneumoniae, an opportunistic pathogen commonly present in the respiratory tract, sinuses and nasal cavities of healthy carriers [25,26]. In this work, the CRISPR-Cas9 system was programmed to target an antibiotic resistance cassette present in another strain. When the DNA from the first strain was used to transfer the CRISPR-Cas9 system to the second strain through natural transformation, most bacteria died from the activity of the CRISPR-Cas9 system. Nonetheless, a substantial fraction of bacteria survived CRISPR-Cas9 killing through the modification of the targeted position by homologous recombination with the locus present in the donor DNA, which did not carry the antibiotic resistance cassette [26]. This work showed that Cas9 can be used to select for the introduction of mutations at desired positions without the need to leave a selection marker or a scar at the edited position.
While this strategy was easily employed in S. pneumoniae where natural transformation and recombination are efficient, its application to less recombinogenic bacteria such as E. coli requires the use of the phage lambda red recombination system to promote editing and repair of the Cas9-mediated breaks [26]. Many studies have now expanded on this work, making CRISPR-Cas9 editing tools more convenient to use [27–30]. CRISPR-Cas9 editing strategies typically rely on the expression of guide RNAs, Cas9 and the lambda red genes from one or several plasmids. Template DNA can be provided as short single stranded DNA, short or long double stranded DNA (typically PCR products), or cloned on a plasmid. In all cases Cas9 is guided to introduce a break at a position of interest, which leads to cell death unless the target DNA was modified, or unless it can be repaired by recombination with the template DNA. In a different strategy inspired by previous work with the I-SceI nuclease [31], a non-replicative vector can be integrated through homologous recombination into the locus of interest, followed by Cas9 cleavage of the vector backbone leading to recombination and recovery of the desired scar-less mutation [32,33]. In all the strategies above, steps of plasmid curing can also be necessary and are typically achieved by using the temperature sensitive pSC101 origin of replication [34]. When more than one plasmid is required this can be coupled with other strategies such as targeting the second plasmid with a guide RNA or the use of counter-selection markers such as sacB [27–29].
The fairly large number of components involved, and the necessity to clone a guide RNA as well as in some cases a template DNA, can make these strategies more cumbersome than established methods [35]. CRISPR-Cas9 strategies have nonetheless enabled pushing at the limits of what is possible, in particular where scar-less mutations are needed [27]. Of particular interest, a strategy has been devised to perform high-throughput modifications of many positions in parallel [36]. Pools of oligonucleotides designed to carry both a homologous repair cassette and a sgRNA can be cloned on a vector, yielding a library that can be used to perform multiplexed recombineering. Another clever strategy enabled the replacement of large fragments of the E. coli genome with synthetic DNA [37]. In this study, multiple guide RNAs were used simultaneously in the same E. coli cell to cleave two positions in the E. coli chromosome and two positions on a plasmid carrying a synthetic DNA fragment, triggering the replacement of the chromosomal DNA through homologous recombination.
A novel and powerful approach in the field of genome editing is the use of the catalytic dead variant of Cas9 (dCas9) fused to a cytosine deaminase or an adenosine deaminases in order to convert C•G to T•A or A•T to G•C at specific target positions without the need to introduce a DNA break [38–40]. In this strategy, cytosines or adenosines located within a small window 15–25 bp from the protospacer adjacent motif (PAM) are modified, leading to a somewhat random mutational outcome when several C or A are present in the target window. Nonetheless this approach appears to be very efficient and can easily be used to modify many positions in parallel. In a recent study, Kondo and colleagues optimized this strategy for E. coli and were able to modify up to 41 loci simultaneously [41].
CRISPR-Cas9 genome editing tools are already being employed to investigate basic biological questions, as well as in applications such as metabolic engineering [42]. While these applications are not directly related to microbiome engineering, we should keep in mind that E. coli is a gut bacterium that can be used as a probiotic. Probiotic E. coli strains, like Nissle 1917, have been engineered to express antigens [43], antimicrobial compounds [44], enzymes to disperse biofilms, quorum sensing molecules that control pathogen virulence [45], metabolic functions of interest and more [6]. These current efforts to engineer probiotic E. coli strains will certainly benefit from this boon of new tools.
Beyond E. coli, the most commonly used probiotic bacteria are Bifidobacteria and Lactobacilli. Engineered Lactobacilli are being developed by various biotech companies as targeted therapies against a wide range of diseases including oral mucositis, inflammatory bowel disease, viral and bacterial infections [46]. The ability of Bifidobacterium to proliferate in solid tumours offers the possibility to engineer them to produce cancer-suppressing compounds [47]. While CRISPR tools for Bifidobacteria have yet to be developed, Oh and van Pijkeren developed a method to perform genome editing in Lactobacillus reuteri ATCC PTA 6475 [48], a bacteria shown to have interesting immunomodulatory and antimicrobial properties [49,50]. Modifications can be introduced by recombination of a single stranded DNA oligonucleotide mediated by the RecT protein, followed by selection with Cas9. CRISPR-Cas9 mediated genome editing was also recently demonstrated in Lactobacillus plantarum, where putting the recombination template on a plasmid rather than providing it as single stranded DNA led to the best results [51]. Barrangou and colleagues have recently highlighted how CRISPR tools could be used to enhance therapeutic effects of lactic acid bacteria [52]. For instance, researchers are exploring strategies to enhance bile salt hydrolase activity to improve strain survival in the gut, or to modify surface layer-associated proteins to change their immunomodulatory properties.
Other bacteria of interest include the Clostridia, a diverse class of bacteria that include strains of industrial interest but also many commensals of the gut microbiome, of which a few—and most notoriously Clostridium difficile—can be opportunistic pathogens. Several reports have demonstrated the use of CRISPR tools to modify species of biotechnological interest including Clostridium acetobutylicum, Clostridium beijerinckii and Clostridium cellulolyticum [53–55]. CRISPR-Cas9 tools have now also enabled the engineering of C. difficile [56,57]. Note that non-toxigenic Clostridia, including non-toxigenic C. difficile, form part of the normal human gut microbiome [58] and could potentially be engineered as interesting probiotics. Clostridium butyricum MIYAIRI 588 has notably been developed as a probiotic against C. difficile infections [59], and could likely be engineered using the CRISPR tools developed for other Clostridium species.
Finally, CRISPR-Cas9 tools have also been developed for Staphylococcus aureus [60–62], an opportunistic pathogen commonly found on the skin [63]. Similarly to Clostridiae, Staphylococci can either be beneficial commensals or pathogens depending on the genetic makeup of specific strains. In addition to these microbiome-associated bacteria, CRISPR-Cas9 tools have been developed for other bacterial species including Bacillus subtilis [64]. In the future the catalogue of bacteria that can be engineered with CRISPR tools will likely continue to expand, enabling an increasing number of applications.
3. CRISPR editing of unicellular eukaryotes
While less studied than bacteria, commensal protozoans and fungi are highly prevalent in healthy populations [65,66], and some yeast can be used as probiotics in additive therapies. Most famously Saccharomyces boulardii was isolated by the French scientist Henri Boulard in 1923 with the purpose of controlling the symptoms of diarrhoea and is now commonly used as a probiotic. This yeast strain has been engineered with CRISPR-Cas9 to introduce various modifications, including the introduction of an exogenous metabolic pathway and the production of human lysozyme [67]. Saccharomyces boulardii has also been engineered with the help of CRISPR for the development of oral vaccines [68]. More generally, many CRISPR-Cas9 editing strategies have been developed in Saccharomyces cerevisiae [69,70] as well as microbiome-associated fungi, including the opportunistic pathogen Candida albicans [71,72]. Along the same line, CRISPR tools developed for protozoan parasites like Plasmodium falciparum [73,74] could likely be adapted to commensal protozoans like Blastocystis.
4. CRISPR editing of bacteriophages
Phages have been applied to cure bacterial infections, with many reported successes in various animal models of lung, skin or gut infection. Engineered phages present interesting subtractive therapeutic opportunities to treat infectious diseases and target the microbiome. In particular, phage adsorption elements like tail fibres and tail tips can be engineered to modify their host range [75]. Temperate phages can be engineered to remove potential virulence factors and turned into lytic phages to limit the risks associated with their use in phage therapy [76]. Phages can further be modified to disperse biofilm [77], encode antimicrobial proteins [78] or other functions of interest. Temperate phages that can be stably maintained in the bacterial cell as prophage or plasmid can be engineered during their lysogenic cycle using the tools described above. However, the modification of lytic phages is particularly challenging as they never reside as a stable genetic element in the cell and antibiotic selection markers cannot be used.
Strategies to edit lytic bacteriophages with CRISPR tools have now been developed. A guide RNA is designed to target the phage genome and a genetic modification of interest is typically cloned on a vector with homology arms to promote recombination with the phage DNA. Shortly after viral DNA entry, Cas nucleases cleave the target sequence and the lesion is repaired through recombination with the provided template, resulting in the edited phage. Only edited phages can then form plaques on bacteria carrying the CRISPR-Cas9 system. This strategy was first demonstrated using the type I-E CRISPR-Cas system of E. coli to engineer phage T7 [79], followed shortly thereafter by a demonstration that the CRISPR-Cas9 system from Streptococcus thermophilus could be used to edit virulent phages of this bacterium [80]. These techniques have now been extended to the engineering of Lactococcus lactis phage p2 [81], of phage T4 in E. coli [82] and phage vB_BsuP-Goe1 in Bacillus subtilis [83]. In one example, the type III CRISPR-Cas10 system from Staphylococcus epidermidis has also been used to edit staphylococcal phages [84]. Note that lytic phages can also be cloned and engineered in yeast [75], where CRISPR-Cas9 tools are readily available.
Beyond the possible use of natural or engineered bacteriophages to cure infections caused by specific pathogens, the recent description of their role as key components of the microbiome will likely open the way to new phage-based therapies [85]. Not only can phages alter the structure of the microbiome by infecting specific species, but they can also alter the genotype and phenotype of the bacteria they infect through horizontal gene transfer and lysogeny. As such, phages likely contribute to the maintenance of the intestinal homeostasis either in health or in disease (dysbiosis) [86]. The use of temperate phages to influence the composition and phenotype of bacteria in the microbiome could thus be viewed as an interesting modulatory therapeutic strategy, but a better understanding of these complex ecological interactions will be needed for the development of such therapies.
5. CRISPR antimicrobials
Besides their use to directly kill target bacteria, phages can be used as DNA delivery vectors. Plasmids carrying a phage packaging signal, known as phagemids, can be used to deliver various effector DNA circuits to target bacterial populations. Phagemids can be packaged into phage particles in the presence of a helper phage that carries all the elements necessary for the production of functional capsids that are missing from the phagemid DNA [87]. In addition, the helper phage can be modified in order to block packaging of its DNA [88]. The M13 phagemid was used to deliver various toxins or restriction enzymes to E. coli [89–91]. The Pf3 phage has also been used to a deliver a restriction enzyme, and successfully treat a Pseudomonas infection in mice [78].
CRISPR-Cas systems themselves can be delivered to populations of bacteria using this strategy with the purpose of specifically eliminating bacteria carrying target sequences in their genome. This strategy has already been demonstrated in E. coli [92] and in S. aureus [93]. In the first study, a plasmid carrying Cas9 and guide RNAs targeting antibiotic resistance genes were injected into bacterial populations using the M13 phagemid system. Efficient cell death was observed as expected when the target gene was present. In the second study, a phagemid based on Staphylococcus phage phiNM1 was constructed by cloning its packaging site on a plasmid carrying a CRISPR-Cas9 system. This phagemid was then tested against various antibiotic resistance genes and virulence factors. Both reports demonstrated the possibility of using CRISPR-Cas systems to eliminate a specific target bacterial genotype in a mixed population, both in vitro and in vivo, testing a wax worm infection model in the former case and a mouse skin colonization model in the second one. These studies also investigated the outcome of targeting a plasmid rather than the chromosome. Cas9 cleavage of a target plasmid leads to cell survival and plasmid loss. Note however that in cases where the plasmid carries a toxin–antitoxin addiction system, the cells will die as a consequence of plasmid loss.
In yet another study, it was proposed to use temperate phages rather than phagemids in order to introduce CRISPR-Cas systems in E. coli [94]. The CRISPR array was programmed to cure plasmids carrying antibiotic resistance genes, thereby sensitizing bacteria to antibiotics. In this work an additional trick was played: a lytic phage was engineered to carry sequences matching the guide RNAs encoded by the CRISPR array. This phage could then be used to kill bacteria that did not carry a functional CRISPR-Cas system and ensure the fixation of the CRISPR prophage in the population.
CRISPR-based antimicrobials offer the possibility to develop novel subtractive therapies, enabling the killing of bacteria based on their sequence without disturbing the rest of the microbiota. We now understand increasingly how different strains of a given bacterial species can have profoundly different effects on our health. The accessory genome that differentiates bacterial strains includes genomic islands, prophages and plasmids that impact the interaction of bacteria with each other and with their host. These elements frequently include virulence factors, toxins and antibiotic resistance genes [95,96]. CRISPR antimicrobials could become a powerful tool both to study the effect of specific strains by removing them from the microbiome, and to eliminate undesired strains as a therapeutic strategy. Note that many bacteria carry active CRISPR-Cas systems of their own that can also be harnessed to trigger self-targeting and cell death. The different approaches to CRISPR antimicrobials and the associated challenges have been reviewed in more detail elsewhere [97,98].
6. Controlling gene expression with CRISPR
Beyond the targeted elimination of strains and the genetic modification of commensals, CRISPR-Cas systems can also be engineered to modulate gene expression. These engineered CRISPR-Cas systems could be used to modulate the activity of bacteria used as probiotics in additive strategies, or could directly be delivered to the resident bacteria of the microbiome through transduction or conjugation (figure 2).
The Cas9 protein carries an HD nuclease domain and a RuvC nuclease domain, each cleaving the target DNA on a different strand [10]. Mutation of the catalytic residues abolishes DNA restriction while maintaining strong on-target binding. Binding of dCas9 to promoter sequences strongly inhibits the initiation of transcription, while binding inside transcribed regions can inhibit transcription elongation [99,100]. Note that dCas9 is only able to efficiently block the running RNA polymerase when it is guided by an RNA that binds to the non-template (coding) strand of DNA. The level of complementarity between the guide RNA and the target can be used to control the rate at which RNA polymerase ‘kicks out’ dCas9 from the target and completes transcription [33]. This mechanism can be used to precisely and robustly reduce gene expression by defined relative amounts, offering a powerful tool to modulate the physiology of target bacteria.
Gene silencing with dCas9, also known as CRISPRi, is comparatively much easier to perform than Cas9-mediated genome editing in bacteria. This has led to the rapid and broad adoption of this technology in a wide range of bacteria, including pathogenic streptococci [100,101], Pseudomonas [102], Staphyloccoci [103], Mycobacteria [104] and Mycoplasma [105], as well as a large number of bacterial species of industrial interest. Directly relevant to this review, dCas9 repression has also been performed in microbiome-associated bacteria like Lactococcus lactis [106] and Bacteroides thetaiotaomicron, where it was used to alter the bacterial metabolic capacity and its resistance to antimicrobial peptides [107]. In the latter work, the CRISPRi repression was also shown to be functional in the mouse gut.
It is also possible to repurpose dCas9 as a transcriptional activator, in a strategy also known as CRISPRa. In a first proof of concept, dCas9 was fused to the omega subunit of the RNA polymerase, yielding moderate activation [100]. In a recent study, activator domains were recruited to the dCas9 ribonucleic complex using a more elaborate strategy. Activator domains were fused to the MS2 coat protein, which binds to a MS2 hairpin itself fused to the guide RNA [108]. Several candidate activators could easily be tested in this manner and strong activation was obtained with SoxS, which could further be optimized to yield up to a 50-fold increase in the expression of a target reporter gene. The authors further showed how this design could be used to activate silent metabolic pathways. Note however that this strategy is very sensitive to the distance between the target and the promoter. Binding needs to occur within a narrow window roughly 60–90 bases upstream of the transcription start site. When no PAM is available in the desired range, it might not be possible to use this tool without further efforts to modify PAM specificity [109].
One appealing feature of CRISPRi is how it can easily be scaled to high-throughput genetic screens. Arrayed libraries of guide RNAs have already proven very useful to investigate the function of essential genes in B. subtilis and S. pneumoniae [101,110]. Potential caveats of using dCas9 were discovered in a recent study in which a pool of guide RNAs was used to target approximately 105 positions in the chromosome of E. coli [111]. This study revealed that dCas9 can cause off-target effects at positions with as little as 9 nt of homology to the guide RNA. The same study also revealed an unexpected toxicity of dCas9 in E. coli. Among other important design rules, this work highlighted the importance of fine-tuning the concentration of dCas9 to avoid this toxicity while maintaining strong on-target repression. Using such an optimized expression cassette and relying on the information gathered by multiple guides targeting the same gene can enable powerful screens to be performed using dCas9 [112,113].
Other types of CRISPR-Cas systems can also be used to silence genes. In type I systems, the Cas3 nuclease is responsible for DNA degradation. In its absence, the multiprotein complex known as Cascade will bind target sequences without introducing any DNA damage. This can be used to block gene expression in the same manner as dCas9 [114,115]. The dead variants of the Cas12 (Cpf1) protein from type V system can also be used to block gene expression in bacteria, as already demonstrated with Cas12 proteins of various origins [116–118]. Finally gene silencing can also be achieved at the RNA level directly using the Cas13 protein from type VI systems, which acts as a RNA-guided RNAse [12,119]. Note however that Cas13 displays a non-specific RNAse activity once it has found its target. This triggers collateral RNA damage that is likely toxic to the cell and was seen to reduce cell growth in E. coli [12]. This collateral activity makes it impractical to silence genes in bacteria, but variants of Cas13 might be identified in the future that only carry a specific RNAse activity. Technologies have also been developed to edit RNA with a deactivated Cas13 fused to a base modification domain [120].
CRISPRi and CRISPRa have also been adapted to eukaryotic cells and—of particular relevance to this review—to Saccharomyces cerevisiae [121], where it could help in the development of probiotic yeast. Note that while dCas9 itself can efficiently block transcription in bacteria, CRISPRi in eukaryotic cells requires the fusion of dCas9 to effector domains.
7. Challenges and future prospects
Over the past few years, we have seen the application of CRISPR tools to organisms as diverse as Firmicutes, Proteobacteria, yeasts and human cells. The fact that the Cas9 protein from S. pyogenes worked easily in so many different cellular backgrounds was surprising to many. This bodes well for the future applications of CRISPR technologies to the extremely diverse set of organisms that compose our microbiome, even though Cas9 from different origins, or even other Cas proteins, might be better suited than others to specific organisms.
While CRISPR-Cas systems seem easily portable between species, one key consideration is that the outcome of DNA cleavage by Cas nucleases can be very different depending on the type and efficiency of the DNA repair pathways present in different organisms. As an example, Cas9 cleavage in the chromosome of mammalian or yeast cells leads to DNA repair through homologous recombination or non-homologous end joining, while the main outcome of efficient Cas9 cleavage in bacterial genomes is cell death. This is likely owing to the absence or poor efficiency of non-homologous end joining (NHEJ) systems in most bacterial species. Among bacteria that naturally carry NHEJ systems, Cas9 breaks could be repaired by NHEJ in some species but not others [55,122,123]. Efforts to import NHEJ systems into bacteria that lack them have yielded limited success [20,123,124]. Different bacterial species also carry more or less efficient homologous recombination systems. Bacteria that can undergo natural transformation and efficient homologous recombination can easily be modified using CRISPR-Cas9 tools, while other bacteria like E. coli require the use of an exogenous recombination system [26]. This heterogeneity in DNA repair capabilities between organisms means that specific tools and strategies need to be designed for different species. CRISPR tools that do not rely on DNA cleavage will likely be more easily portable. These include base editing and CRISPRi strategies.
One of the main obstacles for the broad adoption of these technologies to manipulate members of the microbiome is the difficulty of introducing exogenous DNA in many bacterial species. Some bacteria are more or less amenable to techniques such as electroporation, conjugation or transduction. Many bacteria also carry restriction systems that destroy incoming DNA. Others might not be able to replicate plasmid DNA if the origin of replication is not carefully chosen. Finally, bacteria that cannot be easily grown in the laboratory are obviously hard to engineer. These obstacles are not specific to CRISPR technologies and researchers have found ways to engineer many bacterial species through the construction of dedicated vectors. CRISPR tools thus need to be redesigned to fit the requirements of individual target species. Finding more standardized approaches and more universal tools to deliver DNA to bacteria would go a long way to accelerate research and engineering of a diverse set of bacteria. In a recent study, Peters and colleagues used conjugation and transposition for DNA delivery and integration in the bacterial chromosome to facilitate the use of CRISPRi in many species [125]. Beyond the use of CRISPR tools in vitro, conjugation as well as phage delivery systems enable the delivery of DNA directly in vivo and could be used for the development of novel therapies. Both transduction and conjugation also open the possibility of targetting non-culturable bacteria directly in their natural environment. Phage particles can be used to deliver DNA efficiently into specific bacterial strains, however this specificity is such that dedicated phage capsids will likely have to be engineered for each therapeutic indication. The host range of bacteriophage capsids can be extended through the modification of the proteins that interact with the bacterial envelope such as the tail fibres [75]. In a recent study, Qimron and colleagues employed a directed evolution approach to extend the host range of bacteriophage particles for DNA transduction [126]. This type of approach will be critical to engineer delivery vectors for a wide range of target bacteria, but whether transduction or conjugation can efficiently reach the right bacteria in the complex spatial structure of the microbiome largely remains to be investigated.
The increasing knowledge of the profound impact that the microbiome has on human health is driving the development of novel therapeutic avenues to treat infectious disease, metabolic disease, immune disease and even neurological disorders. However, the outcome of current therapies that target the microbiome is often uncertain owing to our limited understanding of the complex ecological interactions that occur within microbial communities and with the host immune system. CRISPR tools provide exciting strategies not only to study the biology of microbes, but also to elucidate their role within complex communities and drive the development of novel therapies.
Data accessibility
This article has no additional data.
Competing interest
D.B. is a board member and shareholder of Eligo Bioscience, a company developing CRISPR-based therapies targeting the microbiome.
Funding
This work was supported by the European Research Council (ERC) under the Europe Union's Horizon 2020 research and innovation program (grant agreement no. [677823]); the French Government's Investissement d'Avenir program; Laboratoire d'Excellence ‘Integrative Biology of Emerging Infectious Diseases’ [ANR-10-LABX-62-IBEID]; the Pasteur-Weizmann consortium and the Bill and Melinda Gates Foundation - BMGF Grand Challenge grant no. OPP1141338.
References
- 1.Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R. 2018. Current understanding of the human microbiome. Nat. Med. 24, 392–400. ( 10.1038/nm.4517) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barko PC, McMichael MA, Swanson KS, Williams DA. 2018. The gastrointestinal microbiome: a review. J. Vet. Intern. Med. 32, 9–25. ( 10.1111/jvim.14875) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharon G, Sampson TR, Geschwind DH, Mazmanian SK. 2016. The central nervous system and the gut microbiome. Cell 167, 915–932. ( 10.1016/j.cell.2016.10.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martens EC, Neumann M, Desai MS. 2018. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol. 16, 457 ( 10.1038/s41579-018-0036-x) [DOI] [PubMed] [Google Scholar]
- 5.Bober JR, Beisel CL, Nair NU. 2018. Synthetic biology approaches to engineer probiotics and members of the human microbiota for biomedical applications. Annu. Rev. Biomed. Eng. 20, 277–300. ( 10.1146/annurev-bioeng-062117-121019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mimee M, Citorik RJ, Lu TK. 2016. Microbiome therapeutics—advances and challenges. Adv. Drug Deliv. Rev. 105, 44–54. ( 10.1016/j.addr.2016.04.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marraffini LA. 2015. CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61. ( 10.1038/nature15386) [DOI] [PubMed] [Google Scholar]
- 8.Koonin EV, Makarova KS, Zhang F. 2017. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin Microbiol. 37, 67–78. ( 10.1016/j.mib.2017.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova KS, et al. 2015. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13, 722–736. ( 10.1038/nrmicro3569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. ( 10.1126/science.1225829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. ( 10.1038/nature09886) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abudayyeh OO, et al. 2016. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 ( 10.1126/science.aaf5573) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zetsche B, et al. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. ( 10.1016/j.cell.2015.09.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrangou R, Doudna JA. 2016. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941. ( 10.1038/nbt.3659) [DOI] [PubMed] [Google Scholar]
- 15.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 26, 335–340. ( 10.1016/j.tig.2010.05.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. 2012. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12, 177–186. ( 10.1016/j.chom.2012.06.003) [DOI] [PubMed] [Google Scholar]
- 17.Edgar R, Qimron U. 2010. The Escherichia coli CRISPR system protects from lambda lysogenization, lysogens, and prophage induction. J. Bacteriol. 192, 6291–6294. ( 10.1128/JB.00644-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vercoe RB, et al. 2013. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 9, e1003454 ( 10.1371/journal.pgen.1003454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. 2013. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio 5, e00928-13 ( 10.1128/mBio.00928-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui L, Bikard D. 2016. Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic Acids Res. 44, 4243–4251. ( 10.1093/nar/gkw223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selle K, Klaenhammer TR, Barrangou R. 2015. CRISPR-based screening of genomic island excision events in bacteria. Proc. Natl Acad. Sci. USA 112, 8076–8081. ( 10.1073/pnas.1508525112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan J, Wang W, Sun B. 2017. Chromosomal targeting by the Type III-A CRISPR-Cas system can reshape genomes in Staphylococcus aureus. mSphere 2, e00403-17 ( 10.1128/mSphere.00403-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stout EA, Sanozky-Dawes R, Goh YJ, Crawley AB, Klaenhammer TR, Barrangou R. 2018. Deletion-based escape of CRISPR-Cas9 targeting in Lactobacillus gasseri. Microbiology 164, 1098–1111. ( 10.1099/mic.0.000689) [DOI] [PubMed] [Google Scholar]
- 24.Hullahalli K, Rodrigues M, Palmer KL. 2017. Exploiting CRISPR-Cas to manipulate Enterococcus faecalis populations. Elife 6, e26664 ( 10.7554/eLife.26664) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiser JN, Ferreira DM, Paton JC. 2018. Streptococcus pneumoniae: transmission, colonization and invasion. Nat. Rev. Microbiol. 16, 355–367. ( 10.1038/s41579-018-0001-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31, 233–239. ( 10.1038/nbt.2508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zerbini F, et al. 2017. Large scale validation of an efficient CRISPR/Cas-based multi gene editing protocol in Escherichia coli. Microb. Cell Fact. 16, 68 ( 10.1186/s12934-017-0681-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y, Chen B, Duan CL, Sun BB, Yang JJ, Yang S. 2015. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514. ( 10.1128/AEM.04023-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ronda C, Pedersen LE, Sommer MOA, Nielsen AT. 2016. CRMAGE: CRISPR optimized MAGE recombineering. Sci. Rep. 6, 19452 ( 10.1038/srep19452) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reisch CR, Prather KLJ. 2015. The no-SCAR (Scarless Cas9 Assisted Recombineering) system for genome editing in Escherichia coli. Sci. Rep. 5, 15096 ( 10.1038/srep15096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pósfai G, Kolisnychenko V, Bereczki Z, Blattner FR. 1999. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 27, 4409–4415. ( 10.1093/nar/27.22.4409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Cheng Q-X, Liu A-M, Zhao G-P, Wang J. 2017. A novel and efficient method for bacteria genome editing employing both CRISPR/Cas9 and an antibiotic resistance cassette. Front. Microbiol. 8, 812 ( 10.3389/fmicb.2017.00812) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vigouroux A, Oldewurtel E, Cui L, Bikard D, van Teeffelen S. 2018. Tuning dCas9's ability to block transcription enables robust, noiseless knockdown of bacterial genes. Mol. Syst. Biol. 14, e7899 ( 10.15252/msb.20177899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hashimoto-Gotoh T, Sekiguchi M. 1977. Mutations of temperature sensitivity in R plasmid pSC101. J. Bacteriol. 131, 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645. ( 10.1073/pnas.120163297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garst AD, et al. 2017. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 35, 48–55. ( 10.1038/nbt.3718) [DOI] [PubMed] [Google Scholar]
- 37.Wang K, Fredens J, Brunner SF, Kim SH, Chia T, Chin JW. 2016. Defining synonymous codon compression schemes by genome recoding. Nature 539, 59–64. ( 10.1038/nature20124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. 2017. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471. ( 10.1038/nature24644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. 2016. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. ( 10.1038/nature17946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishida K, et al. 2016. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729 ( 10.1126/science.aaf8729) [DOI] [PubMed] [Google Scholar]
- 41.Banno S, Nishida K, Arazoe T, Mitsunobu H, Kondo A. 2018. Deaminase-mediated multiplex genome editing in Escherichia coli. Nat. Microbiol. 3, 423–429. ( 10.1038/s41564-017-0102-6) [DOI] [PubMed] [Google Scholar]
- 42.Liang L, Liu R, Garst AD, Lee T, Nogué VSI, Beckham GT, Gill RT. 2017. CRISPR EnAbled Trackable genome Engineering for isopropanol production in Escherichia coli. Metab. Eng. 41, 1–10. ( 10.1016/j.ymben.2017.02.009) [DOI] [PubMed] [Google Scholar]
- 43.Konieczny MPJ, Suhr M, Noll A, Autenrieth IB, Alexander Schmidt M. 2000. Cell surface presentation of recombinant (poly-) peptides including functional T-cell epitopes by the AIDA autotransporter system. FEMS Immunol. Med. Microbiol. 27, 321–332. ( 10.1111/j.1574-695X.2000.tb01446.x) [DOI] [PubMed] [Google Scholar]
- 44.Hwang IY, Koh E, Wong A, March JC, Bentley WE, Lee YS, Chang MW. 2017. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 8, 15028 ( 10.1038/ncomms15028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duan F, March JC. 2010. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc. Natl Acad. Sci. USA 107, 11 260–11 264. ( 10.1073/pnas.1001294107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh B, Mal G, Marotta F. 2017. Designer probiotics: paving the way to living therapeutics. Trends Biotechnol. 35, 679–682. ( 10.1016/j.tibtech.2017.04.001) [DOI] [PubMed] [Google Scholar]
- 47.Fujimori M. 2006. Genetically engineered bifidobacterium as a drug delivery system for systemic therapy of metastatic breast cancer patients. Breast Cancer 13, 27–31. ( 10.2325/jbcs.13.27) [DOI] [PubMed] [Google Scholar]
- 48.Oh JH, van Pijkeren JP. 2014. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 42, e131 ( 10.1093/nar/gku623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spinler JK, Taweechotipatr M, Rognerud CL, Ou CN, Tumwasorn S, Versalovic J. 2008. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe 14, 166–171. ( 10.1016/j.anaerobe.2008.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin YP, Thibodeaux CH, Peña JA, Ferry GD, Versalovic J. 2008. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm. Bowel Dis. 14, 1068–1083. ( 10.1002/ibd.20448) [DOI] [PubMed] [Google Scholar]
- 51.Leenay RT, Vento JM, Shah M, Martino ME, Leulier F, Beisel CL. 2018. Streamlined, recombinase-free genome editing with CRISPR-Cas9 in Lactobacillus plantarum reveals barriers to efficient editing. bioRxiv 352039 ( 10.1101/352039) [DOI] [PubMed] [Google Scholar]
- 52.Hidalgo-Cantabrana C, O'Flaherty S, Barrangou R. 2017. CRISPR-based engineering of next-generation lactic acid bacteria. Curr. Opin Microbiol. 37, 79–87. ( 10.1016/j.mib.2017.05.015) [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Zhang ZT, Seo SO, Choi KJ, Lu T, Jin YS, Blaschek HP. 2015. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J. Biotechnol. 200, 1–5. ( 10.1016/j.jbiotec.2015.02.005) [DOI] [PubMed] [Google Scholar]
- 54.Li Q, et al. 2016. CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol. J. 11, 961–972. ( 10.1002/biot.201600053) [DOI] [PubMed] [Google Scholar]
- 55.Xu T, Li Y, Shi Z, Hemme CL, Li Y, Zhu Y, Van Nostrand JD, He Z, Zhou J. 2015. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl. Environ. Microbiol. 81, 4423–4431. ( 10.1128/AEM.00873-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McAllister KN, Bouillaut L, Kahn JN, Self WT, Sorg JA. 2017. Using CRISPR-Cas9-mediated genome editing to generate C. difficile mutants defective in selenoproteins synthesis. Sci. Rep. 7, 14672 ( 10.1038/s41598-017-15236-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S, Hong W, Dong S, Zhang Z-T, Zhang J, Wang L, Wang Y. 2018. Genome engineering of Clostridium difficile using the CRISPR-Cas9 system. Clin. Microbiol. Infect. 24, 1095–1099. ( 10.1016/j.cmi.2018.03.026) [DOI] [PubMed] [Google Scholar]
- 58.Rousseau C, Poilane I, De Pontual L, Maherault A-C, Le Monnier A, Collignon A. 2012. Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin. Infect. Dis. 55, 1209–1215. ( 10.1093/cid/cis637) [DOI] [PubMed] [Google Scholar]
- 59.Woo TDH, et al. 2011. Inhibition of the cytotoxic effect of Clostridium difficile in vitro by Clostridium butyricum MIYAIRI 588 strain. J. Med. Microbiol. 60, 1617–1625. ( 10.1099/jmm.0.033423-0) [DOI] [PubMed] [Google Scholar]
- 60.Chen W, Zhang Y, Yeo W-S, Bae T, Ji Q. 2017. Rapid and efficient genome editing in Staphylococcus aureus by using an engineered CRISPR/Cas9 system. J. Am. Chem. Soc. 139, 3790–3795. ( 10.1021/jacs.6b13317) [DOI] [PubMed] [Google Scholar]
- 61.Penewit K, Holmes EA, McLean K, Ren M, Waalkes A, Salipante SJ. 2018. Efficient and scalable precision genome editing in Staphylococcus aureus through conditional recombineering and CRISPR/Cas9-mediated counterselection. MBio 9, e00067-18 ( 10.1128/mBio.00067-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Q, Jiang Y, Shao L, Yang P, Sun B, Yang S, Chen D. 2017. CRISPR/Cas9-based efficient genome editing in Staphylococcus aureus. Acta Biochim. Biophys. Sin. 49, 764–770. ( 10.1093/abbs/gmx074) [DOI] [PubMed] [Google Scholar]
- 63.Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. ( 10.1056/NEJM199808203390806) [DOI] [PubMed] [Google Scholar]
- 64.Altenbuchner J. 2016. Editing of the Bacillus subtilis genome by the CRISPR-Cas9 system. Appl. Environ. Microbiol. 82, 5421–5427. ( 10.1128/AEM.01453-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chabé M, Lokmer A, Ségurel L. 2017. Gut protozoa: friends or foes of the human gut microbiota? Trends Parasitol. 33, 925–934. ( 10.1016/j.pt.2017.08.005) [DOI] [PubMed] [Google Scholar]
- 66.Huseyin CE, O'Toole PW, Cotter PD, Scanlan PD. 2017. Forgotten fungi—the gut mycobiome in human health and disease. FEMS Microbiol. Rev. 41, 479–511. ( 10.1093/femsre/fuw047) [DOI] [PubMed] [Google Scholar]
- 67.Liu J-J, et al. 2016. Metabolic engineering of probiotic Saccharomyces boulardii. Appl. Environ. Microbiol. 82, 2280–2287. ( 10.1128/AEM.00057-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bagherpour G, Ghasemi H, Zand B, Zarei N, Roohvand F, Ardakani EM, Azizi M, Khalaj V. 2018. Oral administration of recombinant Saccharomyces boulardii expressing ovalbumin-CPE fusion protein induces antibody response in mice. Front. Microbiol. 9, 723 ( 10.3389/fmicb.2018.00723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343. ( 10.1093/nar/gkt135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stovicek V, Holkenbrink C, Borodina I. 2017. CRISPR/Cas system for yeast genome engineering: advances and applications. FEMS Yeast Res. 17, fox030 ( 10.1093/femsyr/fox030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shapiro RS, et al. 2018. A CRISPR–Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat. Microbiol. 3, 73–82. ( 10.1038/s41564-017-0043-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vyas VK, Barrasa MI, Fink GR. 2015. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv. 1, e1500248 ( 10.1126/sciadv.1500248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wagner JC, Platt RJ, Goldfless SJ, Zhang F, Niles JC. 2014. Efficient CRISPR-Cas9–mediated genome editing in Plasmodium falciparum. Nat. Methods 11, 915–918. ( 10.1038/nmeth.3063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grzybek M, Golonko A, Górska A, Szczepaniak K, Strachecka A, Lass A, Lisowski P. 2018. The CRISPR/Cas9 system sheds new lights on the biology of protozoan parasites. Appl. Microbiol. Biotechnol. 102, 4629–4640. ( 10.1007/s00253-018-8927-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ando H, Lemire S, Pires DP, Lu TK. 2015. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 1, 187–196. ( 10.1016/j.cels.2015.08.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kilcher S, Studer P, Muessner C, Klumpp J, Loessner MJ. 2018. Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc. Natl Acad. Sci. USA 115, 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu TK, Collins JJ. 2007. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl Acad. Sci. USA 104, 11 197–11 202. ( 10.1073/pnas.0704624104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hagens S, Habel A, Ahsen U, Gabain A, Bläsi U. 2004. Therapy of experimental pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob. Agents Chemother. 48, 3817–3822. ( 10.1128/AAC.48.10.3817-3822.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kiro R, Shitrit D, Qimron U. 2014. Efficient engineering of a bacteriophage genome using the type I-E CRISPR-Cas system. RNA Biol. 11, 42–44. ( 10.4161/rna.27766) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martel B, Moineau S. 2014. CRISPR-Cas: an efficient tool for genome engineering of virulent bacteriophages. Nucleic Acids Res. 42, 9504–9513. ( 10.1093/nar/gku628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lemay M-L, Tremblay DM, Moineau S. 2017. Genome engineering of virulent lactococcal phages using CRISPR-Cas9. ACS Synth. Biol. 6, 1351–1358. ( 10.1021/acssynbio.6b00388) [DOI] [PubMed] [Google Scholar]
- 82.Tao P, Wu X, Tang W-C, Zhu J, Rao V. 2017. Engineering of bacteriophage T4 genome using CRISPR-Cas9. ACS Synth. Biol. 6, 1952–1961. ( 10.1021/acssynbio.7b00179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schilling T, Dietrich S, Hoppert M, Hertel R. 2018. A CRISPR-Cas9-based toolkit for fast and precise in vivo genetic engineering of Bacillus subtilis phages. Viruses 10, 241 ( 10.3390/v10050241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bari SMN, Walker FC, Cater K, Aslan B, Hatoum-Aslan A. 2017. Strategies for editing virulent staphylococcal phages using CRISPR-Cas10. ACS Synth. Biol. 6, 2316–2325. ( 10.1021/acssynbio.7b00240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dalmasso M, Hill C, Ross RP. 2014. Exploiting gut bacteriophages for human health. Trends Microbiol. 22, 399–405. ( 10.1016/j.tim.2014.02.010) [DOI] [PubMed] [Google Scholar]
- 86.Manrique P, Bolduc B, Walk ST, Oost J, Vos WM, Young MJ. 2016. Healthy human gut phageome. Proc. Natl Acad. Sci. USA 113,10 400–10 405. ( 10.1073/pnas.1601060113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dotto GP, Enea V, Zinder ND. 1981. Functional analysis of bacteriophage f1 intergenic region. Virology 114, 463–473. ( 10.1016/0042-6822(81)90226-9) [DOI] [PubMed] [Google Scholar]
- 88.Russel M, Kidd S, Kelley MR. 1986. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene 45, 333–338. ( 10.1016/0378-1119(86)90032-6) [DOI] [PubMed] [Google Scholar]
- 89.Hagens S, Bläsi U. 2003. Genetically modified filamentous phage as bactericidal agents: a pilot study. Lett. Appl. Microbiol. 37, 318–323. ( 10.1046/j.1472-765X.2003.01400.x) [DOI] [PubMed] [Google Scholar]
- 90.Moradpour Z, Sepehrizadeh Z, Rahbarizadeh F, Ghasemian A, Yazdi MT, Shahverdi AR. 2009. Genetically engineered phage harbouring the lethal catabolite gene activator protein gene with an inducer-independent promoter for biocontrol of Escherichia coli. FEMS Microbiol. Lett. 296, 67–71. ( 10.1111/j.1574-6968.2009.01620.x) [DOI] [PubMed] [Google Scholar]
- 91.Westwater C, Kasman LM, Schofield DA, Werner PA, Dolan JW, Schmidt MG, Norris JS. 2003. Use of genetically engineered phage to deliver antimicrobial agents to bacteria: an alternative therapy for treatment of bacterial infections. Antimicrob. Agents Chemother. 47, 1301–1307. ( 10.1128/AAC.47.4.1301-1307.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Citorik RJ, Mimee M, Lu TK. 2014. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32, 1141–1145. ( 10.1038/nbt.3011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. 2014. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32, 1146–1150. ( 10.1038/nbt.3043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yosef I, Manor M, Kiro R, Qimron U. 2015. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl Acad. Sci. USA 112, 7267–7272. ( 10.1073/pnas.1500107112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393. ( 10.1111/j.1574-6976.2008.00136.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Davies EV, Winstanley C, Fothergill JL, James CE. 2016. The role of temperate bacteriophages in bacterial infection. FEMS Microbiol. Lett. 363, fnw015 ( 10.1093/femsle/fnw015) [DOI] [PubMed] [Google Scholar]
- 97.Bikard D, Barrangou R. 2017. Using CRISPR-Cas systems as antimicrobials. Curr. Opin. Microbiol. 37, 155–160. ( 10.1016/j.mib.2017.08.005) [DOI] [PubMed] [Google Scholar]
- 98.Pursey E, Sünderhauf D, Gaze WH, Westra ER, Houte S. 2018. CRISPR-Cas antimicrobials: challenges and future prospects. PLoS Pathog. 14, e1006990 ( 10.1371/journal.ppat.1006990) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim W. 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. ( 10.1016/j.cell.2013.02.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. 2013. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437. ( 10.1093/nar/gkt520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu X, et al. 2017. High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Mol. Syst. Biol. 13, 931 ( 10.15252/msb.20167449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tan SZ, Reisch CR, Prather KLJ. 2018. A robust CRISPR interference gene repression system in Pseudomonas. J. Bacteriol. 200, e00575-17 ( 10.1128/JB.00575-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao C, Shu X, Sun B. 2017. Construction of a gene knockdown system based on catalytically inactive (“Dead”) Cas9 (dCas9) in Staphylococcus aureus. Appl. Environ. Microbiol. 83, e00291-17 ( 10.1128/AEM.00291-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Choudhary E, Thakur P, Pareek M, Agarwal N. 2015. Gene silencing by CRISPR interference in mycobacteria. Nat. Commun. 6, 6267 ( 10.1038/ncomms7267) [DOI] [PubMed] [Google Scholar]
- 105.Mariscal AM, et al. 2018. Tuning gene activity by inducible and targeted regulation of gene expression in minimal bacterial cells. ACS Synth. Biol. 7, 1538–1552. ( 10.1021/acssynbio.8b00028) [DOI] [PubMed] [Google Scholar]
- 106.Berlec A, Škrlec K, Kocjan J, Olenic M, Štrukelj B. 2018. Single plasmid systems for inducible dual protein expression and for CRISPR-Cas9/CRISPRi gene regulation in lactic acid bacterium Lactococcus lactis. Sci. Rep. 8, 1009 ( 10.1038/s41598-018-19402-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mimee M, Tucker AC, Voigt CA, Lu TK. 2015. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 1, 62–71. ( 10.1016/j.cels.2015.06.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dong C, Fontana J, Patel A, Carothers JM, Zalatan JG. 2018. Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun. 9, Article number 2489 ( 10.1038/s41467-018-04901-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hu JH, et al. 2018. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63. ( 10.1038/nature26155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peters JM, et al. 2016. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165, 1493–1506. ( 10.1016/j.cell.2016.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cui L, Vigouroux A, Rousset F, Varet H, Khanna V, Bikard D. 2018. A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nat. Commun. 9, 1912 ( 10.1038/s41467-018-04209-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rousset F, Cui L, Siouve E, Depardieu F, Bikard D. 2018. Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. bioRxiv. 26, 308916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang T, Guan C, Guo J, Liu B, Wu Y, Xie Z, Zhang C, Xing X-H. 2018. Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nat. Commun. 9, 2475 ( 10.1038/s41467-018-04899-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Luo ML, Mullis AS, Leenay RT, Beisel CL. 2015. Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 43, 674–681. ( 10.1093/nar/gku971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rath D, Amlinger L, Hoekzema M, Devulapally PR, Lundgren M. 2015. Efficient programmable gene silencing by Cascade. Nucleic Acids Res. 43, 237–246. ( 10.1093/nar/gku1257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang X, Wang J, Cheng Q, Zheng X, Zhao G, Wang J. 2017. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov. 3, 17018 ( 10.1038/celldisc.2017.18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim SK, Kim H, Ahn W-C, Park K-H, Woo E-J, Lee D-H, Lee S-G. 2017. Efficient transcriptional gene repression by type V-A CRISPR-Cpf1 from Eubacterium eligens. ACS Synth. Biol. 6, 1273–1282. ( 10.1021/acssynbio.6b00368) [DOI] [PubMed] [Google Scholar]
- 118.Leenay RT, et al. 2016. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol. Cell 62, 137–147. ( 10.1016/j.molcel.2016.02.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Abudayyeh OO, et al. 2017. RNA targeting with CRISPR–Cas13. Nature 550, 280–284. ( 10.1038/nature24049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cox DBT, et al. 2017. RNA editing with CRISPR-Cas13. Science 358, 1019–1027. ( 10.1126/science.aaq0180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gilbert LA, et al. 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. ( 10.1016/j.cell.2013.06.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tong YJ, Charusanti P, Zhang LX, Weber T, Lee SY. 2015. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth. Biol. 4, 1020–1029. ( 10.1021/acssynbio.5b00038) [DOI] [PubMed] [Google Scholar]
- 123.Bernheim A, Calvo-Villamañán A, Basier C, Cui L, Rocha EPC, Touchon M, Bikard D. 2017. Inhibition of NHEJ repair by type II-A CRISPR-Cas systems in bacteria. Nat. Commun. 8, 2094 ( 10.1038/s41467-017-02350-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Su T, Liu F, Gu P, Jin H, Chang Y, Wang Q, Liang Q, Qi Q. 2016. A CRISPR-Cas9 assisted non-homologous end-joining strategy for one-step engineering of bacterial genome. Sci. Rep. 6, 37895 ( 10.1038/srep37895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peters JM, et al. 2019. Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nature Microbiol. 4, 244 ( 10.1038/s41564-018-0327-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yosef I, Goren MG, Globus R, Molshanski-Mor S, Qimron U. 2017. Extending the host range of bacteriophage particles for DNA transduction. Mol. Cell 66, 721–728. ( 10.1016/j.molcel.2017.04.025) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.