Abstract
In recent years, new genome editing technologies have emerged that can edit the genome of non-human animals with progressively increasing efficiency. Despite ongoing academic debate about the ethical implications of these technologies, no comprehensive overview of this debate exists. To address this gap in the literature, we conducted a systematic review of the reasons reported in the academic literature for and against the development and use of genome editing technologies in animals. Most included articles were written by academics from the biomedical or animal sciences. The reported reasons related to seven themes: human health, efficiency, risks and uncertainty, animal welfare, animal dignity, environmental considerations and public acceptability. Our findings illuminate several key considerations about the academic debate, including a low disciplinary diversity in the contributing academics, a scarcity of systematic comparisons of potential consequences of using these technologies, an underrepresentation of animal interests, and a disjunction between the public and academic debate on this topic. As such, this article can be considered a call for a broad range of academics to get increasingly involved in the discussion about genome editing, to incorporate animal interests and systematic comparisons, and to further discuss the aims and methods of public involvement.
This article is part of a discussion meeting issue ‘The ecology and evolution of prokaryotic CRISPR-Cas adaptive immune systems’.
Keywords: ethics, genome editing, Clustered Regularly Interspaced Palindromic Repeat, human health, environment
1. Introduction
In the past two decades, a host of genome editing technologies have emerged that can edit the genome with progressively increasing efficiency and ease of use. These technologies are based on the use of sequence-specific engineered nucleases, such as zinc finger nucleases (ZFN) [1], meganucleases [2] and transcription activator-like effector nucleases (TALEN) [3]. In more recent years, genome editing was revolutionized by the emergence of clustered regularly interspaced palindromic repeats (CRISPR) and the CRISPR-associated protein 9 (Cas9) [4]. In parallel, new applications of these genome editing technologies have emerged, such as synthetic gene drives, which allow the rapid and super-Mendelian spread of gene alterations within a population or even a species [5,6].
Overall, this new generation of genome editing technologies allows scientists to modify the genomes of non-human animals (from here on: ‘animals’) more precisely than classical transgenesis [7] with comparably fewer off-target effects [8]. Furthermore, engineered nucleases can introduce genetic changes without the use of foreign DNA [9]. These genome editing technologies have a broad range of possible applications in animals, including to increase livestock productivity and disease resistance [10], create new animal models to study human disease [11], protect native species by eradicating invasive species, decrease or even eliminate vector-borne diseases such as malaria, and perhaps even resurrect extinct species [5,12]. Understandably, these technologies and their applications have sparked both excitement and apprehension, raising new questions on ethics and governance and generating significant debate in both academic and public spaces.
Despite this ongoing debate, to our knowledge, no comprehensive overview of the arguments raised in the academic discourse on genome editing in animals exists. Such an overview is a valuable contribution to the academic literature, as it provides insights into patterns of argumentation in the expert debate and can help uncover arguments that go unmentioned or are insufficiently conceptualized. It is particularly salient to study the academic debate because academic experts can have a strong influence on related science and technology policy and governance decisions [13–15]. Moreover, insight into the academic debate is important for understanding whether it differs from the public debate and arguments. For technologies that have a high societal impact, such as genome editing, it is important to identify and bridge potential gaps between the public and academic discourse in the early phases of development.
In this article, we present such a comprehensive overview by reporting the reasons for and against the development and use of genome editing technologies in animals as these have been mentioned in the academic literature. We then critically assess the academic debate and identify perspectives, issues and arguments that are underrepresented in the existing literature.
2. Methods
A systematic review of the reasons that have been given for and against the development and use of new-generation genome editing technologies in animals was conducted. This review was based on the method developed by Strech & Sofaer [16], which can be used to systematically identify reasons and arguments in favour of or against particular (normative or descriptive) positions or claims. This method does not assess the adequacy, quality or normative weight of the reported reasons [16], but enables a systematic collection of all the relevant literature in which opinion, point of view, or position is put forward. Subsequently, it allows for an equally systematic extraction and synthesis of the reasons. It incorporates relevant items from the preferred reporting items for systematic reviews and meta-analyses (PRISMA) statements [17] as well as thematic analysis typical of qualitative research [16].
(a). Search strategy
A literature search of the PubMed, Web of Science, Scopus, CAB Abstracts and Philosopher's Index databases was conducted to find relevant articles. The choice for databases was discussed with experienced librarians; these five databases were selected as they cover a comprehensive area of biomedical, veterinary, and ethics research journals and articles. A search strategy that combined search terms for genome editing, animals (adapted from Hooijmans, Tillema, Leenaars & Ritskes-Hoitinga [18]) and ethics was used (electronic supplementary material, table S1).
(b). Article selection and inclusion criteria
Academic articles or book chapters that were written in English or Dutch and published in 2010 or later were eligible for inclusion. Publications that did not contain a reason for or against the development or use of new-generation genome editing technologies in animals were excluded. Publications that specifically focused on older techniques (e.g. classical transgenesis) were also excluded.
Two researchers independently screened the titles and abstracts and, if applicable, the full texts of the articles. In the case of disagreement about inclusion or exclusion, differences were discussed until consensus was reached. The reference lists of included articles were subsequently screened for additional relevant articles.
(c). Data extraction and analysis
The full text of the selected articles was analysed using a data extraction document (electronic supplementary material, table S2) that was designed prior to start the data extraction to extract data in a systematic way. The contextual data of the included articles, including the discipline of the author(s) and the specific technologies and applications discussed, were also included. Subsequently, all the reasons for and against the development and use of new-generation genome editing technologies in animals were extracted. The reasons that were mentioned in the included articles (reason mentions) were subsequently compared. If different articles mentioned the same reason, these were bundled under the same ‘narrow reason’. Next, a list of narrow reasons was generated: for each narrow reason, we noted which article included that reason and the number of times it was mentioned.
Additionally, the narrow reasons were used to generate an overview of broader themes to which the narrow reasons related. If a narrow reason applied to two themes, the narrow reason was listed under the most applicable theme, as determined by consensus among the researchers. The formulation of both the narrow reasons and themes was an iterative process in which the categories were re-evaluated among all researchers several times to bundle similar narrow reasons together, categorize them and define the themes that best encompassed the narrow reasons.
Finally, an overview of the themes and narrow reasons was created by listing these in a table under the overarching classifications of ‘human-related’, ‘animal-related’ or ‘environment-related’ reasons in order of frequency of appearance. Within each theme, the narrow reasons mentioned in the literature were subcategorized as reasons for or against genome editing in animals; these subcategories were similarly listed in order of frequency of appearance. Where applicable, rebuttals of reasons in favour of genome editing were listed in the subcategory ‘against’ and vice versa.
3. Results
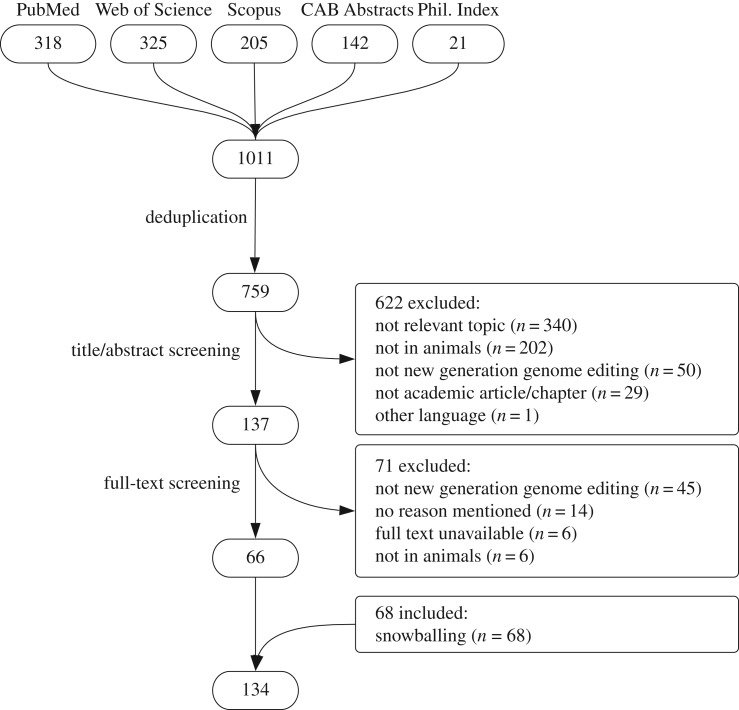
The database searches resulted in a total of 760 unique records. After title/abstract screening, full-text screening, and cross-referencing, 134 articles were included for data extraction and analysis (figure 1).
Figure 1.
Flow chart of article selection and inclusion.
(a). Author affiliation
The included articles were written by professionals working primarily in academic institutions, in a variety of different departments or divisions: biomedical or biological sciences (n = 77/134), animal sciences (n = 30/134), ethics (n = 20/134), philosophy (n = 14/134), biotechnology companies (n = 8/134), governmental organizations (n = 6/134), law (n = 5/134), (bio)engineering (n = 4/134), nutritional or food sciences (n = 3/134), agricultural sciences (n = 3/134), consultancy (n = 2/134), epidemiology (n = 2/134), political sciences (n = 2/134), bioinformatics or computational biology (n = 2/134), psychology (n = 1/134), mathematics (n = 1/134), public and international affairs (n = 1/134) and a private foundation (n = 1/134). In 10/134 articles, no author affiliation was listed (table 1).
Table 1.
Affiliations of the authors of the included manuscripts.
| author affiliation or discipline | N* | references |
|---|---|---|
| biological or (bio)medical sciences | 77 | [3–6,19–91] |
| veterinary medicine or animal sciences | 30 | [10,11,23,24,27,38,47,48,50,54,61,62,67,71,73,77,79,85–88,92–100] |
| ethics | 20 | [6,19,25,34,64,78,80,101–113] |
| philosophy | 14 | [9,20,34,81,114–123] |
| no affiliation or no author listed | 10 | [7,124–132] |
| biotechnology company | 8 | [10,24,73,74,77,79,133,134] |
| governmental organization | 6 | [23,32,83,88,100,135] |
| law | 5 | [12,25,122,136,137] |
| (bio)engineering | 4 | [49,50,77,138] |
| nutritional or food sciences | 3 | [139–141] |
| agricultural sciences | 3 | [100,133,142] |
| consultancy | 2 | [53,115] |
| epidemiology | 2 | [23,111] |
| political sciences | 2 | [32,139] |
| bioinformatics or computational biology | 2 | [32,49] |
| psychology | 1 | [26] |
| mathematics | 1 | [31] |
| public and international affairs | 1 | [143] |
| private foundation | 1 | [6] |
*The numbers add up to more than 134 as various included articles were written by authors with different affiliations or multiple affiliations.
(b). Reasons for and against new-generation genome editing in animals
In total, 115 different reasons were mentioned in the reviewed articles; 67 of these reasons were in favour of and 48 against the development and use of new-generation genome editing in animals. The included articles contained from 1 up to 13 different reasons. The reasons were in response to a broad range of potential applications of genome editing in animals (table 2).
Table 2.
Potential applications of genome editing in animals mentioned in the literature.
| potential application of genome editing in animals | (potential) aim |
|---|---|
| genome editing in general | |
| create an animal model of Parkinson's disease [11] | create animal models of human disease |
| delete an antigen that causes hyperacute rejection in pig-to-human transplantation [51] or inactivating porcine endogenous retroviruses (PERV) to prevent transmission of these viruses to humans [73] | facilitate xenotransplantation from pigs to humans by reducing the chance of immune rejection |
| increase skeletal muscle mass and thereby meat production [48] | increase nutritional value for humans; increase production efficiency in animal farming |
| create a chicken strain with low allergenicity [126] | decrease allergic reactions in humans |
| increase disease resistance to porcine reproductive and respiratory syndrome in livestock [98] | decrease suffering of farm animals; increase production efficiency; reduce use of antibiotics |
| create polled (hornless) cattle [79] | decrease suffering of farm animals (by preventing painful dehorning); decrease costs; increase production efficiency; decrease moral distress of farmers |
| produce poultry in which the embryo's sex can be recognized in the egg, in which genetic males become phenotypical females, or in which male embryos die during early development [100] | decrease suffering of farm animals by preventing the killing of male chicks |
| create the so-called diminished animals in which the ability to sense pain is impaired [78] | decreasing suffering of animals in research and farming |
| revive the woolly mammoth as a major grazing animal in the Arctic [81,91] | curiosity; advance scientific understanding; restore an arctic steppe in the place of the less ecologically rich tundra [139] |
| gene drives | |
| induce mosquito resistance to malaria parasites [29]; induce infertility in mosquitos [101] | reduce the burden of vector-borne diseases |
| reduce fertility or biasing sex towards males in invasive species, creating a population that is not reproductively viable [83] | control or eradicate invasive species |
| increase genetic gain in breeding programmes [10] | increase economic productivity in animal farming |
| change reproductive behaviour of wild animals that give birth to large numbers of offspring, many of which do not survive to adulthood, by decreasing the number of offspring they produce per cycle [114] | prevent wild animal suffering |
These narrow reasons were subsequently categorized into seven broad themes: (1) human health; (2) efficiency; (3) risks and uncertainty; (4) public acceptability; (5) animal welfare; (6) animal dignity and species-specific capacities; (7) environmental considerations (see table 3 in appendix A). In the following sections, the different broad and narrow reasons are discussed in more detail.
Appendix A. Table 3.
Reasons for and against the development and/or use of genome editing technologies in animals.
| n | references | technologies | ||
|---|---|---|---|---|
| human-related reasons | ||||
| human health | ||||
| for (n = 8) | could enhance research by creating better animal model systems of human disease | 35 | [3,4,7,11,19,26,34–51,92–95,108,128,129,133,138–140] | various (ZFN, TALEN, CRISPR); genome editing |
| could improve human health by reducing the burden of vector-borne diseases such as malaria | 35 | [5,6,19–33,52,101–109,114,124–127,135,139,140,143] | gene drives; genetic modification; genome editing | |
| could facilitate xenotransplantation, which could be a solution to the human donor shortage | 26 | [25,37,44,45,48,59–73,94,105,108,111,133,139] | various (ZFN, TALEN, CRISPR) | |
| could expedite research in other species, including non-human primates, which provide more accurate models for human (neurological) disease | 12 | [36,43,55–63,126] | CRISPR | |
| could help to meet the challenge of producing more food more sustainably to ensure the future global population can be fed | 6 | [34,78,79,141–143] | various (ZFN, TALEN); genetic modification | |
| could improve human health through the provision of new medicines and therapies | 4 | [26,126,133,140] | various (TALEN, CRISPR) | |
| could enable genome engineering in non-human primates; this could be considered ethically problematic, but it is much more ethically problematic to watch people die who could be saved | 1 | [57] | CRISPR | |
| could be used to create a chicken strain with low allergenicity | 1 | [126] | CRISPR | |
| against (n = 8) | re simplifying and speeding up the production of new transgenic animal models of human disease: most of such models fail to directly benefit humans; this lack of reproducibility may put human research participants at risk at a later stage | 3 | [53,54,110] | CRISPR |
| re bringing routine genome engineering of non-human primates within reach, which could help identify genetic underpinnings of disease or develop therapies: the moral permissibility of this approach is questionable given available alternatives | 2 | [110,126] | CRISPR | |
| could pose risks to human health if genetic modification is not successful in creating mosquitoes resistant to infections, but instead confers no resistance or actually reduces resistance to the target infection | 2 | [102,109] | genetic modification; gene drives | |
| could disrupt ecosystems, which could be harmful to human populations that depend on them | 2 | [20,143] | gene drives | |
| could be used to re-create species that may become a vector or reservoir for viruses that can be harmful for human beings | 1 | [81] | genetic engineering | |
| re use to create a chicken strain with low allergenicity: there may not be a compelling need for doing so because allergy usually only occurs in children, and alternatives and egg substitutes are available | 1 | [80] | various (ZFN, TALEN, CRISPR) | |
| re use to produce better quality food: little is known about the effects these modified organisms would have on humans when consumed | 1 | [35] | CRISPR | |
| could increase productivity of the livestock sector: this is an undesirable outcome given the negative impact of meat consumption on human health | 1 | [104] | various (ZFN, TALEN, CRISPR) | |
| efficiency | ||||
| for (n = 14) | could be more efficient, versatile, precise, easy to use or accurate than previous editing technologies | 39 | [3,4,6,7,9,33–35,37,40–43,46,49–51,53,56,63,64,69,75,79,82–84,98,105,110,114,126,129,131,133,134,136,140,142] | various (ZFN, TALEN, CRISPR) |
| could lead to advances in scientific understanding or technological advances | 9 | [12,29,64,72,79,102,126,131,140] | various (ZFN, TALEN, CRISPR; genetic modification; active geneticsa) | |
| could be relatively inexpensive in comparison to previous approaches | 9 | [9,17,50,102,108,124,130,133,134] | CRISPR, gene drives | |
| could save costs for the farming industry | 9 | [23,65,72,76,83,84,97,125,141] | various (ZFNs, TALEN, CRISPR) | |
| could accelerate and/or enhance the trait improvement currently accomplished by classic breeding | 8 | [10,45,47,76,96,98,108,130] | various (ZFN, TALEN, CRISPR, gene drives) | |
| could increase production efficiency | 7 | [17,25,46,48,65,82,97] | various (ZFNs, TALEN, CRISPR); genetic modification | |
| could be a potentially efficient and rapid tool to improve important traits in livestock | 3 | [26,96,97] | various (ZFNs, TALEN, CRISPR) | |
| could be a cost-effective strategy to control the transmission of vector-borne diseases | 3 | [6,27,102] | genetic modification, gene drives | |
| could increase economic productivity in animals bred for human consumption | 2 | [97,137] | various (ZFNs, TALEN, CRISPR); genetic engineering | |
| could provide animals with disease resistance, which could reduce the overuse of antibiotics | 2 | [98,139] | various (TALEN, CRISPR) | |
| could be used to eradicate vector-borne diseases in a more efficacious and/or logistically less complex way than other efforts to eliminate these diseases | 2 | [27,28] | gene drives | |
| could be used for pest control, being more precise or effective than other pest management methods such as pesticides | 2 | [20,109] | gene drives | |
| re the possibility of off-target effects: these are fewer and more controlled compared with the mutations that are caused by generally accepted technologies such as conventional breeding | 2 | [80,142] | various (ZFNs, TALEN, CRISPR) | |
| re the possibility of off-target effects: these can be minimized by careful design and testing, and their effects are largely identical to those of the natural processes that continually create variation in the genomes of food animals | 1 | [85] | various (ZFNs, TALEN, CRISPR) | |
| against (n = 1) | could still have inadequate gene targeting efficiency, off-target effects or cause mosaic mutations | 10 | [42,47,54,55,58–60,63,95,116] | various (ZFN, TALEN, CRISPR) |
| risks and uncertainty | ||||
| against (n = 8) | could spread beyond its target population owing to accidental release, cross-breeding or gene flow; this release could have unpredictable ecological consequences | 12 | [20,23,28,35,82,83,88,89,106,109,124,143] | gene drives |
| could introduce off-target mutations into the gene pool and spread these across a species | 5 | [7,20,35,83,143] | gene drives | |
| could have novel features that are unprecedented and unexpected, so the risks and consequences are difficult or even impossible to characterize beforehand | 4 | [20,102,117,143] | various (synthetic biology, gene drives, genetic modification) | |
| could involve risks of deliberate release of (disease carrying [23]) genetically modified mosquitoes to the environment | 2 | [105,117] | synthetic biology (including genome editing), gene drives | |
| could be used to serve the (economic) interests of particular groups with little concern for the general interest | 2 | [20,115] | various (ZFNs, TALEN, CRISPR, gene drives) | |
| could have unexpected effects because our knowledge and understanding of the genetic background of complex traits is incomplete | 1 | [96] | various (ZFNs, TALEN, CRISPR) | |
| could have non-negligible risks because breaches of containment are impossible to rule out and, once released, just a few escaped genetically modified mosquitoes could be capable of spreading transgenes on a global scale | 1 | [22] | gene drives | |
| could benefit humans if used for applications to human disease and agricultural production, however these applications could primarily benefit the current generation, with secondary benefits and potential risks placed upon future generations | 1 | [143] | gene drives | |
| for (n = 7) | re the potential to spread beyond its target population or have unintended consequences: various designs of the gene drive and other containment measures may mitigate these risks | 13 | [5,19,23,25,26,31,32,88,89,102,105,125,131] | gene drives |
| re novel features: could be considered similar to conventional breeding owing to the similarity to natural mutations and the absence of transgenes | 4 | [47,48,80,85] | various (ZFNs, TALEN, CRISPR) | |
| re potential risks: could be researched in a phased approach, allowing sufficient time to evaluate the efficacy and safety of gene drives before regulatory decisions are made on whether they will be suitable for use | 2 | [32,135] | gene drives | |
| re potential for off-target effects with negative effects: genome modification is more precise and consequently has far fewer risks than conventional breeding | 1 | [79] | various (ZFN, TALEN) | |
| re potential risks: it is generally more difficult to prove that something is safe than to find potential risks; the damage of not using a new technique may exceed its potential risks | 1 | [96] | various (ZFNs, TALEN, CRISPR) | |
| re uncertain consequences: these are not in themselves a sufficient reason not to use the technology; the magnitude and likelihood of these risks ought to be thoroughly analysed and balanced against the potential benefits | 1 | [101] | gene drives | |
| re potential risks: these ought to be balanced with the risks and harm caused by the unmodified wild-type | 1 | [23] | gene drives | |
| public acceptability | ||||
| for (n = 6) | could be more acceptable to the public than previous technologies, as no foreign DNA is introduced | 4 | [9,33,96,97] | various (ZFNs, TALEN, CRISPR) |
| could increase the chance of a publicly justified policy permitting genome editing | 1 | [9] | CRISPR | |
| could be less controversial than using pesticides for pest control | 1 | [20] | gene drives | |
| could impact community members who have not consented to the release of genetically modified mosquitoes; however, this may be justifiable if the public health benefits of the trial for the community are important enough | 1 | [102] | genetic modification | |
| could be used in field trials with genetically modified animals while respecting the interests of community members if community advisory boards and a community authorization process are used | 1 | [107] | gene drives | |
| re public resistance: could lead to resistance when modified mosquitoes cross borders to communities who did not agree with this; however, various designs of the gene drive may prevent this, enabling local communities to make local decisions | 1 | [31] | gene drives | |
| against (n = 1) | could lead to public resistance | 6 | [12,22,89,102,108,126] | gene drives; genetic modification; genome editing, genetic engineering |
| animal-related reasons | ||||
| animal welfare | ||||
| for (n = 13) | could decrease animal suffering in dairy farming by creating dehorned cattle, preventing invasive and painful dehorning | 10 | [9,19,78–80,85,96,126,139,140] | various (ZFNs, TALEN, CRISPR) |
| could counter welfare problems by creating the so-called diminished animals in which the ability to sense pain is impaired | 8 | [78,112,115,116,119–121,137] | genome editing (CRISPR); genetic engineering/modification | |
| could increase animal health and welfare by providing animals with disease resistance | 8 | [78,80,96,98,126,133,135,139] | various (ZFNs, TALEN, CRISPR) | |
| could increase adaptations to different environmental conditions | 2 | [19,137] | various (ZFNs, TALEN, CRISPR); genetic engineering | |
| could be used to prevent the killing of day-old male chicks | 2 | [100,126] | CRISPR; genetic modification | |
| re the possible creation of animals with welfare problems: if they have a life worth living we cannot say that they are worse off owing to the genetic modification, for if they had not been created with genetic modification, they would not have existed at all | 2 | [115,118] | genetic modification | |
| re off-target effects: could result in fewer off-target effects than previous techniques, which could improve welfare of genetically modified animals | 1 | [9] | CRISPR | |
| could reduce the numbers of animals used to create model organisms compared to traditional methods, which typically sacrifice many animals before achieving the desired genotype and phenotype | 1 | [110] | CRISPR | |
| could remove known harmful recessive alleles that impair fertility or health and in that sense repair accumulated damage in the genome of breeding animals | 1 | [96] | various (ZFNs, TALEN, CRISPR) | |
| could prevent wild animal suffering by using genome editing to change reproductive behaviour; the harm that would be prevented by doing so would outweigh the harm of developing and testing these strategies | 1 | [114] | CRISPR | |
| could lead us to ignore the predicament of the animal and to accept negative effects on animal welfare for the sake of other goals; however, this concern may be addressed by using less drastic gene drive designs and using these to promote animal welfare | 1 | [9] | gene drives | |
| re applications that would permit even greater intensification of farming resulting in decreased animal welfare: this seems unlikely given recent trends of companies to improve animal welfare | 1 | [78] | various (ZFNs, TALEN, CRISPR) | |
| could be a humane method to eliminate invasive species | 1 | [6] | gene drives | |
| against (n = 11) | could result in off-target mutations or unintended effects, which could negatively affect animal health | 6 | [9,80,90,104,139,143] | various (ZFNs, TALEN, CRISPR) |
| could contribute to animal suffering by perpetuating the use of animals in research | 5 | [9,36,53,108,110] | genome editing; CRISPR | |
| could result in secondary complications that are bad for animal welfare (e.g. increased muscle growth could lead to increased rates of caesarean sections, leg problems or breathing complications) | 3 | [80,96,104] | various (ZFNs, TALEN, CRISPR) | |
| could be used for applications that would permit even greater intensification of farming; this outcome would be undesirable | 3 | [36,104,116] | various (ZFNs, TALEN, CRISPR); genome engineering | |
| could be used to the decrease animal suffering (by creating polled cattle or diminished animals); however, there are alternatives to doing so (e.g. by improving animals’ environments) | 2 | [80,118] | various (ZFNs, TALEN, CRISPR) | |
| could be combined with somatic cell nuclear transfer (SCNT) cloning to deliver the nuclease-mediated genetic alterations, which are associated with embryonic losses, postnatal death and birth defects | 2 | [95,97] | various (ZFNs, TALEN, CRISPR) | |
| could bring routine genome editing of non-human primates within reach; this use of the technologies may substantially diminish these organisms' welfare and quality of life | 1 | [110] | CRISPR | |
| re use to prevent wild animal suffering: the complexity of ecosystems, the unpredictability of climate change and the indeterminacy of human behaviour leave us with too little confidence that this aim will be successful | 1 | [122] | genome editing | |
| re use to create diminished animals that lack the affective dimension of pain: no proof of concept experiment has been done on farm animals and conducting these experiments would itself cause suffering | 1 | [116] | genetic engineering | |
| re use to revive extinct species: the revived animals may end up suffering either as a result of the processes used or because of their particular genomic variations | 1 | [12] | genetic engineering | |
| re use to revive extinct species: the re-created species may become a vector or reservoir for viruses that can be harmful for other animals | 1 | [81] | genetic engineering | |
| animal dignity and species-specific characteristics | ||||
| against (n = 9) | could be objectionable because it instrumentalizes animals by using them as mere objects to serve human purposes | 5 | [36,64,81,104,115] | various (ZFNs, TALEN, CRISPR) |
| could be used to revive extinct species or create gene-edited pets, but it is questionable if physiological limits should be altered or animals should be exploited for unimportant human purposes like entertainment | 3 | [12,126,128] | various (TALEN, CRISPR) | |
| could impinge on animals' dignity as altering the genome of an animal is a failure to acknowledge its dignity or prevents the animal from living according to its instinct | 3 | [36,96,111] | various (ZFNs, TALEN, CRISPR) | |
| could affect the ‘telos’ (the essence and purpose of a creature) if they are genetically altered to the point where they lose the behaviour that characterizes that animal | 3 | [80,120,137] | various (ZFNs, TALEN, CRISPR); genetic modification | |
| could expedite transgenesis in other species, including non-human primates, which likely occupy a level of moral status that would obligate us to protect them from being used in this way or to allow it only in extremely exceptional circumstances | 2 | [53,110] | CRISPR | |
| could create diminished animals to decrease animal suffering, but this is an inappropriate response to the historical wronging of agricultural animals; we have a duty to repair these wrongs | 2 | [116,119] | genome editing (CRISPR mentioned) | |
| could only be rightfully done if the permissibility of genome editing in research is evaluated for each species on its own merits | 1 | [36] | CRISPR | |
| could be used to facilitate xenotransplantation, which could be considered ethically untenable as it compromises species boundaries | 1 | [64] | CRISPR | |
| could be viewed as the initiation of increasingly imbalanced power distribution between humans and animals | 1 | [80] | various (ZFNs, TALEN, CRISPR) | |
| for (n = 11) | re breaching species norms if used for animal diminishment: species norms are only indirectly morally significant, as a generally useful guide to evaluating animal welfare | 2 | [118,119] | genome editing (CRISPR) |
| re violating animal dignity or integrity: such arguments focus only on respect for individual animals, they ultimately cannot justify an objection that is based on a species-norm, as is the case in the discussion on enhancement | 2 | [115,118] | genetic modification | |
| re use to create diminished animals, which could be said to harm these animals as their species-typical essence would be changed: as the literature about human disability has taught us, we should not assume that ‘disabilities’ caused by diminishment make animals worse off | 2 | [78,112] | various (ZFNs, TALEN, CRISPR; genetic engineering) | |
| re violating rights, violating dignity or wrongly instrumentalizing: genome editing determines which individual will come into existence rather than modifying existing individuals, making it hard to say how its rights could have been infringed, its dignity violated, or even that it has been wrongly instrumentalized | 1 | [119] | genome editing (CRISPR mentioned) | |
| re breaching the sanctity of the lives of mosquitoes by making them go extinct: neither existing mosquitoes nor the species holistically bear a significant degree of moral status | 1 | [101] | gene drives | |
| re impinging on an animal's dignity by making them serve better as objects for human use: the Kantian concept of dignity cannot be applied to animals, for this concept is tied to prerequisite conditions that animals do not possess | 1 | [113] | genetic engineering | |
| re use to modify an animal's telos or nature: this could be morally acceptable if the animals are made less miserable or happier as one does not morally wrong the telos by changing it; only individuals can be wronged | 1 | [121] | genetic engineering | |
| could be used to prevent additional violations to animal rights, which would be preferable to the status quo, even on an account that considers raising animals for human consumption impermissible | 1 | [78] | various (ZFNs, TALEN, CRISPR) | |
| re impinging on an animal's integrity or dignity and thereby harming it even if welfare is improved: what is good for an individual must in some way resonate with that individual; what is good for it cannot diverge from its welfare | 1 | [78] | various (ZFNs, TALEN, CRISPR) | |
| re impact on the ‘telos’ of an animal: the animal's telos can still be respected if it is provided with an environment that fits its altered genetic predispositions | 1 | [78] | various (ZFNs, TALEN, CRISPR) | |
| re impact on the ‘telos’ of an animal: the idea that there is some ‘true essence’ of a species is mistaken as behaviours and tendencies change over time, making it hard to see why this should be seen as morally problematic | 1 | [78] | various (ZFNs, TALEN, CRISPR) | |
| environment-related reasons | ||||
| environmental considerations | ||||
| against (n = 10) | could have unknown negative effects on ecosystems | 13 | [6,7,20,28,34,35,82,83,106,108,126,139,143] | gene drives |
| could cross moral limits by exceeding the extent to which humans breach natural boundaries or act out of hubris; nature/life cannot be completely manufactured or planned and we ought to acknowledge their unpredictability | 4 | [12,36,115,117] | various (ZFNs, TALEN, CRISPR, synthetic biology) | |
| could constitute an unnatural interference with nature | 2 | [100,115] | various (ZFNs, TALEN, CRISPR); genetic modification | |
| could be used to revive extinct species, for which there may no longer be a niche | 2 | [12,81] | genetic engineering | |
| could be used to revive extinct species, which might diminish the desire to protect existing species | 2 | [12,81] | genetic engineering | |
| re use to revive extinct species: genome editing will fail to genuinely recreate species while preserving their species identity | 2 | [12,123] | genome editing; genetic engineering | |
| could disrupt the natural order; although this order should not hold an intrinsic moral value, deleting genetic diversity could carry risks by deleting traits that are advantageous | 1 | [105] | gene drives | |
| could lead to increased productivity of the livestock section, which is not desirable given the negative impact of this sector on the environment (e.g. greenhouse gas production and water and land pollution) | 1 | [104] | various (ZFNs, TALEN, CRISPR) | |
| could be used to control certain invasive species; if this succeeds, this could become a trojan horse to legitimate the eradication of other species without questioning to whom or what they are harmful | 1 | [20] | CRISPR, gene drives | |
| could be more transformative, uncontrollable and ecologically damaging than organisms modified to contain self-limiting genes | 1 | [107] | gene drives | |
| for (n = 9) | could enable ecological conservation by eradicating invasive species or reviving extinct species | 8 | [5,12,21,31,91,103,124,143] | active geneticsa; gene drives; genetic engineering |
| could help to develop and support more sustainable agricultural models | 4 | [5,31,32,105] | gene drives | |
| re potential to be considered unnatural or alike ‘playing god’: it is unclear what is meant by naturalness; furthermore, there is no reason to accept that the natural is necessarily good and the unnatural necessarily bad | 2 | [78,111] | various (ZFNs, TALEN, CRISPR); genetic engineering | |
| could contribute to reducing the environmental impact of animal production | 1 | [96] | various (ZFNs, TALEN, CRISPR) | |
| could protect threatened species and reduce invasive species to conserve the natural and cultural world for future generations, which could be imperative from an intergenerational justice perspective | 1 | [143] | gene drives | |
| re potential of driving mosquitoes to extinction being considered ‘playing god’ or displaying hubris: there may be sufficient reasons—such as saving many lives—that may justify improving the given | 1 | [101] | gene drives | |
| could be used to control agricultural pests; this may be a more environmentally sound control method than using insecticides | 1 | [23] | gene drives | |
| could be used to revive extinct species, which would be just; because humans killed extinct species and have the power to revive them, there is a duty to do so | 1 | [12] | genetic engineering | |
| re ecological risks created by using gene drives to prevent wild animal suffering by using genome editing to change reproductive behaviour: these risks may be offset by modifying other features of the ecosystem, too | 1 | [114] | CRISPR, gene drives | |
aGenetic manipulations in which a ‘genetic element is copied from one chromosome to the identical insertion site on the sister chromosome using cas9 and guide RNA elements’ [21].
(i). Human-related reasons
Human health
Most reasons in favour of genome editing in animals concerned its potential to improve human health. First, these hoped-for improvements included using gene drives to reduce the burden of vector-borne diseases [5,6,19–33,101–109,114,124–127,135,139,140,143], either by suppressing or eradicating insect populations [21,101] or inducing vector resistance to disease pathogens [22,101]. At the same time, however, some authors noted that gene drives could pose risks to human health if they disrupted ecosystems on which humans are dependent [20,143], or if modified mosquitoes did not confer resistance—or if they actually reduced instead of increased resistance to the target infection [102,109].
Second, various authors noted that genome editing in animals could enhance research in animal systems by creating better animal models of human disease [3,4,7,11,19,26,34–52,92–95,108,128,129,133,138–140], which could ultimately benefit human health, for example, by leading to the creation of new medicines and therapies [26,126,133,140]. At the same time, it was argued that there is a lack of reproducibility of animal findings in humans [53,54,110], which could put human research participants at risk at a later stage of the research [110].
In a similar way, authors argued that genome editing could expedite research in other species, including non-human primates, which could provide more accurate models for human (neurological) disease [36,43,55–63,126]. The permissibility of this approach was questioned, however, given available alternatives such as using organoids or stem cell models of disease [110] or using animal models of smaller animals such as mice [126]. It was mentioned that although genome editing in non-human primates could be considered ethically problematic, it would be even more ethically problematic to let humans die who could be saved [57].
Third, genome editing in animals could provide a solution to the long-standing shortage of human organ donors by facilitating xenotransplantation from pigs into humans [26,39,47,50,51,64–77,93,111,130,133,135,139,140], either by reducing the chance of immune rejection in xenotransplantation [37,44,45,48,60,62,63,66,67,70–72,94,105,108,133,139] or by decreasing the risk of transmission of porcine pathogens such as porcine endogenous virus (PERV) [26,39,51,64,66,68,73,74,76,77,130,135,140]. It was mentioned that this solution should be compared to alternative solutions to this problem in terms of resource allocation and prioritization [105].
Fourth, genome editing could help to meet the challenge of producing more food more sustainably to ensure that the future human population can be fed [34,78,79,141,142], for example, by increasing skeletal muscle mass and thereby meat production. Concurrently, it was mentioned that little is known about the effects these modified organisms would have on humans when consumed [35] and that it could be undesirable to increase meat production given the negative impact of meat consumption on human health [104].
Finally, the authors noted that genome editing could be used to create a chicken strain with low allergenicity, which could benefit humans with egg allergies [126]. On the other hand, authors mentioned that there may not be a compelling need to produce such chickens because the allergy usually only occurs in children and because alternatives and egg substitutes are available [80]. Finally, some authors noted that if genome editing were used to revive extinct species (also known as de-extinction), the re-created species could potentially be harmful to humans if it became a vector or reservoir for viruses [81].
Efficiency
Many reasons in favour of genome editing in animals mentioned the efficiency of these techniques. First, it was argued that genome editing could be a potentially efficient and rapid tool to improve important traits in livestock [26,96,97], which could increase production efficiency [19,48,70,96,115,133,139], for example, by achieving a higher meat yield [19,48,70,96,139]. Various authors argued that genome editing using engineered nucleases (ZFN, TALEN or CRISPR) was more efficient, versatile, precise, easy to use or accurate than previous genetic technologies [3,4,6,7,9,33–35,37,40–43,46,49–51,53,56,63,64,69,75,79,82–84,98,105,110,114,126,129,131,133,134,136,140,142]. On the other hand, it was argued that genome editing technologies could still have inadequate gene targeting efficiency and cause off-target effects or mosaic mutations [76], particularly in non-human primates [42,47,54,55,58–60,63,95,116]. Other authors mentioned that these off-target effects could be identical to those of natural processes that continually create variation in the genomes of food animals [85], and that they could be fewer and more controlled than the mutations caused by generally accepted technologies such as conventional breeding [80,142]. Finally, it was suggested that off-target effects could be minimized by careful design [85].
Second, authors compared the efficiency of these technologies to alternative strategies in which genome editing was not used. It was argued that genome editing could facilitate quicker or more effective trait improvement than classic breeding [10,47,79,85,94,99,140,142]. For gene drives, it was mentioned that this technology could be more efficacious than other approaches at eliminating vector-borne diseases [27,28] or than other pest management methods such as pesticides [20,109].
Third, it was argued that these technologies could lead to advances in scientific understanding [12,21,26,33,69,75,81,102,134] or to technological advances [12]. Authors also mentioned that genome editing could reduce the overuse of antibiotics in farm animals by providing these animals with disease resistance [98,139].
Fourth, issues of cost were addressed. It was mentioned that CRISPR could be relatively inexpensive in comparison to both previous genetic technologies [9,26,34,110,114,132,142], other pest management techniques such as insecticides [20,23] and traditional sterile insect methods [23], and that it could increase economic productivity in animals bred for human consumption [97,137]. Moreover, authors mentioned that genome editing could save costs for the farming industry by providing animals with disease resistance [70,75,86,98,126] or by transferring polled genes to horned cattle, obviating the need for expensive dehorning [19,79,87,96]. Finally, gene drives could be a cost-effective strategy for controlling the transmission of vector-borne diseases [6,27,109].
Risks and uncertainty
Other reasons given for or against the use of genome editing technologies concerned their potential risks and uncertainties.
For gene drives, the risks addressed primarily related to an accidental or deliberate release of gene drive organisms. It was mentioned that the genes drive could spread beyond their target population [35,83,143] owing to accidental release [20,23,28,82,88,89,106,124], horizontal transfer [28,109,143], cross-breeding [20] or gene flow [20], with unpredictable ecological consequences. Authors noted that it could be impossible to rule out breaches of containment, which would constitute a non-negligible risk as release of just a few gene drive organisms could cause the transgenes to spread on a global scale [22]. Authors also mentioned that gene drive organisms could be released deliberately, exposing the public and the environment to risk [105,117], particularly if these organisms were engineered to carry diseases rather than prevent them [105]. The potential for off-target mutations affecting the gene drive was mentioned as another risk [7,20,35,83]; guide RNA could, for example, mutate over time and consequently target an unintended part of the genome [7].
Several authors mentioned potential ways to mitigate these risks. Various designs of the gene drive and other containment measures could mitigate unintended consequences or the risk that the change would spread beyond the target population [5,19,23,25,26,31,32,88,89,102,105,125,131]. Authors also suggested that gene drives could be researched in a phased approach, allowing sufficient time to evaluate the efficacy and safety of gene drive organisms before regulatory decisions are made about whether they are suitable for widespread use [32,135]. Furthermore, it was argued that these potential negative consequences are not in themselves a sufficient reason not to use gene drives; the magnitude and likelihood of these risks ought to be analysed thoroughly and balanced against the potential benefits [101] as well as the risks and harm caused by the unmodified wild-type animal [23].
For genome editing in general, the uncertainty involved in assessing potential consequences of genome editing technologies was stressed. It was argued that the risks or consequences of genome editing technologies could be difficult or even impossible to characterize beforehand, given their novel features [20,102,117,143] and our incomplete knowledge and understanding of the genetic background of complex traits [96]. With respect to applications of genome editing in animal farming, on the other hand, it was argued that genome editing could be considered similar to conventional breeding because the created modifications are comparable to natural mutations and no transgenes are involved [47,48,80,85]. Although genome editing could result in off-target effects with potential negative consequences, it was argued that genome editing is more precise and therefore has fewer risks than conventional breeding and consequently should be generally regarded as safe [79]. Some authors also argued that it is generally more difficult to prove that something is safe than to find potential risks; the damage of not using a new technique may exceed its potential risks [96].
Finally, it was mentioned that genome editing could be used to serve the (economic) interests of particular groups, such as the agriculture or food industry [20], with little concern for the public interest [20,115]. Additionally, applications of gene drives to human disease and agricultural production could primarily benefit the current generation, with secondary benefits and potential risks placed upon future generations; it was argued that this may not be acceptable from a standpoint of intergenerational equity given the irreversibility and uncertainties inherent to the deployment of gene drives [143].
Public acceptability
Other human-related reasons in favour of or against genome editing in animals concerned public acceptance or rejection of the technologies. Some authors argued that the new generation of genome editing technologies might be more acceptable to the public than previous technologies because no foreign DNA is introduced into the animal [9,33,96,97]. It was mentioned that this could consequently increase the chance of a publicly justified policy [9]. It was also mentioned that the public might consider gene drive applications in agriculture less controversial than using pesticides for pest control [20].
By contrast, it was argued that some uses of genome editing could generate public resistance to the technologies [12,22,89,102,108,126], for example, if public funds were used to bring back extinct species [12] or if genetically modified mosquitoes were to cross borders to other countries that did not support their release [22,89,102]. Other authors asserted that the latter concern could be mitigated by using gene drive designs that could enable local communities to make decisions concerning their own local environments [31]. While authors acknowledged that it would not be possible to seek consent from all humans who could potentially be impacted by the release of genome-edited mosquitoes, it was argued that release could nonetheless be justified if the public health benefits of the trial are important enough for the community [102]. It was suggested that one way to conduct field trials with genetically modified animals while respecting the interests of community members is to use community advisory boards and a community authority [107].
(ii). Animal-related reasons
Animal welfare
Reasons related to animal welfare were used to argue both in favour of and against genome editing in different types of animals.
First, it was argued that genome editing could decrease the suffering of farm animals. For example, genome editing could be used to prevent the killing of day-old male chicks [100,126] by enabling the production of poultry in which the embryo's sex can be recognized in the egg, in which genetic males become phenotypical females or in which male embryos die during early development. Authors also suggested that genome editing could be used to repair accumulated damage in the genome of breeding animals by removing harmful recessive alleles that impair animal fertility and health [96]. Additionally, genome editing could be used to create hornless cattle, which would not require the painful dehorning that is commonly performed in the farming industry to protect both cows and farmers from injury [9,19,78–80,85,96,126,139,140]. At the same time, it was mentioned that this goal could be accomplished in other ways too; instead of creating polled animals, the rearing environment of cattle could be improved to prevent accidents, horn covers could be used, or dehorning could be performed under anaesthesia [80,118].
Other authors emphasized the potential use of genome editing to increase animal health and welfare by making animals resistant to diseases [78,80,96,98,126,133,135,139] or better able to adapt to environmental conditions [19,137]. By contrast, it was argued that such uses of genome editing would enable even greater intensification of farming, for example, by generating polled or disease resistant animals that could be kept at higher density [36,104,116]. While these authors noted that any intensification of farming would decrease animal welfare, others questioned the likelihood of this outcome given recent trends of companies improving animal welfare [78].
Some authors considered the possible use of genome editing to counter welfare problems of farm animals by creating the so-called diminished animals with an impaired ability to sense pain [78,112,115,116,119–121,137]. In response, the authors noted that there is no proof-of-concept experiment for such an application in farm animals and argued that conducting these experiments would itself cause suffering [116]. Lastly, authors noted that if farm animals were edited to improve production efficiency, some of these genome modifications could result in secondary complications that are bad for animal welfare [80,96,104]; increased muscle growth, for example, could lead to increased rates of Caesarean sections, leg problems or breathing complications.
Second, it was argued that genome editing could be used to decrease the suffering of research animals, for example, by decreasing the occurrence of unwanted genetic effects [53] and reducing the number of animals [110] used to create animal model systems compared to traditional methods [110]. On the other hand, it was argued that, if genome editing were to be widely used, this decrease in suffering per experiment would be offset by the overall increase in the numbers of transgenic animals used in research [36,53]; in this way, genome editing could contribute to animal suffering by perpetuating their continued use in research [9,36,53,108]. Moreover, it was mentioned that genome editing could bring routine genome editing of non-human primates within reach, which could substantially diminish these organisms' welfare and quality of life [110].
Third, it was mentioned that genome editing might decrease the suffering of many species of wild animals, for example, by changing the reproductive behaviour of prey animals in ways that reduce their high infant mortality rate [114]. It was argued that the harm that would be prevented by doing so would outweigh the harm inflicted on animals during development and testing of these strategies [114]. On the other hand, authors argued that scientists cannot be confident enough that this strategy will successfully decrease wild animal suffering given the complexity of ecosystems, the unpredictability of climate change and the indeterminacy of human behaviour [122]. With regards to reviving extinct species, it was mentioned that these animals could end up suffering as a result of the processes used or because of their genomic variations [12], and that revived species could threaten other animals if they become a vector or reservoir for viruses [81].
Finally, it was argued that genome editing could affect animal welfare in several other ways. Authors noted that genome editing could decrease animal welfare if somatic cell nuclear transfer (SCNT) cloning were used to deliver the nuclease-mediated modifications; SCNT is associated with embryonic losses, postnatal death and birth defects [95,97]. Authors also mentioned that genome editing could result in off-target mutations or unintended effects, which could negatively affect animal health [9,80,90,104,139]. Others argued that genome editing using engineered nucleases could result in fewer off-target effects than previous techniques [9]. Furthermore, the so-called non-identity problem was raised in the context of creating genetically modified animals; if these animals have a life worth living, one cannot conclude that they are worse off, even if they have welfare problems, for they would not have existed if they had not been genetically modified [115,118].
With regard to gene drives, it was mentioned that this technology could be a humane method to eliminate invasive species [6]. On the other hand, it was argued that such applications could lead humans to ignore the predicament of the animal and to accept negative effects on animal welfare for the sake of other goals [9], although this risk could be prevented by using less drastic gene drive designs and using them to promote animal welfare (for instance, by driving disease resistance into wild populations) [9].
Animal dignity and species-specific capacities
Several authors argued that (applications of) genome editing are undesirable not because they might harm the welfare of these animals, but because they might be harmed in other ways. First, it was argued that genome editing instrumentalizes animals by using them as mere objects to serve human purposes [36,64,81,104,115], whereas these animals have intrinsic value [104], and in any case prospective human benefits should not be used to justify harm to animals [36]. For particular applications such as reviving extinct species or creating genome-edited pets, authors argued that it could be inappropriate to alter physiological limits [126,128] or to exploit the animals for unimportant human purposes like entertainment [12]. Additionally, it was mentioned that genome editing could be viewed as the initiation of increasingly imbalanced power distribution between humans and animals [80]. On the other hand, some authors argued that genome editing could prevent additional violations to animal rights, which should be considered preferable to the status quo, even on an account that considers raising animals for human consumption to be impermissible [78].
Second, it was argued that genome editing could be an affront to an animal's dignity [96] or could prevent the animal from living according to its instincts [111]. On the other hand, it was argued that the Kantian concept of dignity cannot be applied to animals, for it is tied to prerequisite conditions, such as the ability to exert self-determination or to be a moral agent, that animals do not possess [113]. Likewise, it was argued that it does not make sense to propose that genome editing could impinge on an animal's dignity and thereby harm that animal even if its welfare is improved, because what is good for an individual must in some way resonate with that individual [78]. Similarly, it was argued that dignity-related arguments ultimately cannot justify an objection that is based on a species norm rather than on respect for individual animals, as is the case in the discussion on enhancement [115,118]. Finally, authors noted that because genome editing could determine which individual comes into existence, it could be hard to say that its rights were infringed, its dignity violated, or even that it was wrongly instrumentalized because it would otherwise not exist [119].
Third, it was argued that genome editing could affect the telos (the essence and purpose) of an animal [80] if they are genetically altered to the point where they lose the behaviour that makes them that particular animal [120], for example, if genome editing were used to create diminished animals [137]. In response, it was argued that the idea that there is a ‘true essence’ of a species is mistaken, as behaviours and tendencies change over time [78]; furthermore, the telos of a creature could still be respected by providing it with an environment that fits its altered genetic predispositions [78]. Moreover, it was argued that it could be morally acceptable to modify an animal's telos if the animal was made less miserable or indeed happier because only an individual animal, not its telos, can be harmed [121].
With regard to species-specific considerations, it was argued that genome editing could expedite transgenesis in non-human primates, which likely occupy a level of moral status that would obligate us to protect them from being used in this way [110] or to allow it only in extremely exceptional circumstances [53]. It was also mentioned that genome editing could only be rightfully done if its permissibility were evaluated for each species on its own merits [36]. With regard to mosquitoes, it was mentioned that using gene drives to drive them to extinction could breach the sanctity of their lives, however, it was argued that neither existing mosquitoes (that will not die nor suffer, but merely fail to reproduce), nor the species holistically (for which it could not be considered clear that they possess relevant cognitive capacities) bear a significant degree of moral status [101].
Finally, objections were made to specific applications of genome editing. It was argued that although genome editing could increase animal welfare by facilitating diminishment, this result would be an inappropriate response to the systematic wronging [119] or inappropriate valuation [116] of agricultural animals, whereas we have a duty of reparation to members of this historically wronged group [119]. Authors also mentioned that genome editing could facilitate xenotransplantation, which might be considered ethically untenable because it compromises species boundaries and treats animals as re-designable systems for human use [64]. On the other hand, it was argued that species norms (which could also be breached if genome editing were used for animal diminishment) are only indirectly morally significant as a generally useful guide to evaluating animal welfare [118,119]. Similarly, it was mentioned that ‘disabilities’ caused by diminishment, which could affect the species-typical essence of these animals, would not necessarily make these animals worse off, as the literature on human disabilities has taught us [78,112].
(iii). Environment-related reasons
Environmental considerations
Environmental considerations were mostly used to argue against genome editing. One line of argument pursued the potential impacts of genome-edited animals on ecosystems. Authors argued that both genome-edited organisms [28,139] and gene drive organisms [6,7,20,28,34,35,82,83,106,108,126,139,143] could have unknown negative effects on ecosystems. It was mentioned that gene drive organisms could be more transformative, uncontrollable and ecologically damaging than other genome-edited organisms that contain self-limiting genes [107], particularly if gene drives were used to eradicate species [7,34,35,83,108,139]. By eradicating a species, gene drives could disrupt the positive contributions of these species in native ecosystems [89], for example, by eliminating the food source of another species [7,34,83] or promoting the proliferation of invasive pests [7,34]. By contrast, it was argued that genome editing could enable ecological conservation [21,143] and save endangered native species [5,103,124] if used to eradicate invasive species [5,31,103,124] or revive ecological proxies of extinct species [12,91]. It was argued that using gene drives to protect threatened species and reduce invasive species could conserve the natural and cultural world for future generations, possibly rendering its use imperative from an intergenerational justice perspective [143].
Authors also argued that genome editing could impact the environment in other ways. On the one hand, it was reasoned that using genome editing to increase the productivity of livestock could be undesirable given the negative impact of farming on the environment, for example, through greenhouse gas production and water and land pollution [104]. On the other hand, genome editing could perhaps contribute to reducing the environmental impact of animal production, for example, by decreasing the amount of phosphate pollution [96]. Similarly, authors noted that using gene drives to control agricultural pests could be a more environmentally sound control method than using insecticides [23] and that gene drives could help scientists to develop and support more sustainable agricultural models [5,31,32,105], for example, by editing populations of resistant species to become vulnerable to pesticides and herbicides again [5,32,105].
Authors raised several environmental considerations in response to specific proposed applications of genome editing, in particular reviving extinct species. On the one hand, it was argued that reviving extinct species could be just; because humans caused the extinction and have the power to revive them, they may have a duty to do so [12]. On the other hand, it was mentioned that in some cases there may no longer be a niche for a particular revived species [12,81], and as a result the revived species may do substantial environmental damage if it is released or escapes into the environment. Reviving animals could also diminish the desire to protect existing species [12,81]. Finally, it was mentioned that genome editing will fail to genuinely recreate species because there would not be a reproductive nor spatio-temporal relationship between the resurrected animal and other members of its species [12,123]. In response to the ecological damage that could result from using genome editing to change the reproductive behaviour of wild animals to prevent suffering, it was mentioned that such damage could be offset by modifying other features of the ecosystem, too [114].
Finally, it was argued that genome editing could cross moral limits if humans were to use it to breach natural boundaries or to act out of hubris [12,36,115,117], as nature and life should not be completely manufactured or planned and we should acknowledge their unpredictability [12,115]. Some authors noted that genome editing might in itself constitute an unnatural interference with nature [100,115]. Authors also argued that while the natural order might not hold an intrinsic moral value, deleting genetic diversity risks eliminating advantageous traits [105]. In response, authors noted that it is unclear what is meant by ‘naturalness’ [78,111]. Furthermore, the natural is not necessarily good and the unnatural is not necessarily bad [78,111]. Similarly, it was argued that although it could be said that using genome editing could amount to ‘playing God’ or displaying hubris, there may be sufficient reasons—such as saving many lives—to justify improving the given [101]. For gene drives, it was mentioned that the use of this technology to control certain invasive species, if successful, could become a Trojan horse to legitimize the eradication of other species without questioning to whom or what they are harmful [20].
4. Discussion
To the best of our knowledge, this review constitutes the first systematic review of reasons for and against development and use of new-generation genome editing technologies in non-human animals as reported in the academic literature. Our review shows that a wide and diverse range of reasons is brought forward and provides a descriptive overview of these reasons, offering a starting point for subsequent further research and normative analysis [16].
Importantly, many arguments mentioned in this review are not reasons for or against all uses of genome editing in animals. Instead, they point to possible conditions for the responsible use of these technologies. For example, the fact that genetically modified (non-gene drive) mosquitoes could potentially cause negative consequences by spreading the modified gene beyond the target population, could lead to the requirement that, among other conditions, a first trial site be geographically isolated, such as an island [102]. Our review also underlines that different ethical considerations apply to different applications of genome editing in animals. From this point of view, the question is not whether genome editing in animals is ethically acceptable, but whether there are conditions under which it can be ethically employed.
In what follows, we make four additional observations about the academic debate, and suggest areas for future research and analysis. In particular, we note a low disciplinary diversity in the authors shaping the academic debate, a scarcity of systematic comparisons of potential consequences of using these technologies, underrepresented or missing concerns, especially regarding animal interests, and a disjunction between the public and academic debate on this topic. We elaborate on these observations below.
(a). The academic literature lacks disciplinary diversity
Our findings provide insight into who is shaping the academic debate on the use of gene editing technologies in non-human animals. As table 1 illustrates, while authors from different backgrounds are involved in this debate, the large majority are (mostly biomedical or veterinary) scientists, investigating the technical feasibility of different applications of genome editing in animals. On the one hand, a concern for ethics on the part of scientists is important and encouraging. On the other hand, it shows that authors working in ethics, philosophy and the social sciences are underrepresented. This low disciplinary diversity is particularly problematic as the debate moves from discussions of technical feasibility to (potential) real-world applications, in which academic experts will likely influence policy and regulatory decisions [14,144]. To critically assess the applications of genome editing in animals from different perspectives, multidisciplinary and proactive evaluation of the technologies and their ethical and societal implications—for example, through ethics parallel research [145,146]—is essential. Ethics parallel research entails an ethical evaluation of emerging technologies in parallel with—or even in advance of—the developing science, allowing scientists and ethicists to co-shape innovation processes and governance in an ethically sound way during the development of the technology [145].
(b). Few articles include systematic comparisons
Our findings also illuminate the characteristics of the specific reasons addressed in the literature. While many reasons related to potential harms and benefits, surprisingly few articles engaged in a systematic comparison of the harms and benefits of the proposed application of genome editing compared to alternatives. This is noteworthy, as such systematic comparisons are necessary to draw conclusions about what would result in the best overall consequences. Such an analysis could draw on the principles of proportionality and subsidiarity. According to the principle of proportionality, potential benefits should be balanced against potential harms or risks; those that argue in favour of or against (applications of) genome editing in animals ought to present an explicit comprehensive overview of the benefits, harms and risks in question and argue why the harms outweigh the benefits or vice versa. The principle of subsidiarity entails that a policy should only be adopted if there is no less harmful policy that would achieve the same result. This principle suggests that applications of genome editing ought to be compared to alternative policies in terms of potential harms and benefits, including the—often forgotten—benefits and harms of the status quo, including the costs of inaction. In the case of gene drives, for example, potential ecological damage resulting from their use is a pressing concern, warranting a thorough inventory of related risks and harms. When weighing those, the principle of subsidiarity requires us—among other things—to balance the possible ecological damage of using gene drives to eradicate vector-borne diseases with the deaths that are now caused by these diseases and the ecological damage of using pesticides. This kind of analysis is consistent with calls from the scientific community to integrate comparative assessment of harms, risks and benefits into the regulatory framework [147,148]. Yet where some scientific reports define benefits in narrow economic terms, the principle of subsidiarity requires a broad definition of and metric for benefits.
(c). Underrepresented or missing concerns
Given that this review concerns genome editing in animals, it is remarkable how few animal-related reasons have been put forward; most reasons for or against the use of genome editing in animals rest on human-related grounds. Little of the biomedical literature considered the welfare of (research) animals; for example, articles that mentioned off-target effects seldom considered whether these effects could have an impact on animal welfare. Similarly, there was relatively little reflection on species-specific considerations. Although the moral status and interests of non-human primates were brought up [53,110,126], the moral status of other animals was rarely mentioned. Given that accounts of moral status are generally founded in sentience [149] and consciousness, the interests of other animals appear worthy of more attention within this debate.
On a related note, while the relationship between humans and animals was brought up in several reasons, particularly those related to animal dignity, this relationship was never framed in terms of human virtues [150]. Such an analysis might ask, for example, who we become when we use and alter animals in certain ways. Indeed, when it comes to ethical theory, we note that the most frequently reported reasons—to a large extent originating from biomedical literature—were consequentialist in nature, i.e. focusing on potential (positive or negative) outcomes of using genome editing technology in animals for human health, animal welfare or ecosystems. While an initial emphasis on consequentialism is consistent with general argumentative patterns around new and emerging science and technologies [151], other ethical theories are relevant to this debate and will also be necessary to understand and engage with public attitudes and concerns.
(d). Disjunction between the expert and public debate
Academic experts have made significant calls for public engagement with and debate about genome editing [4,28,70,126,152,153], particularly with regard to the possible use of gene drives [5,6,20,32,83,88,109,140]. A study commissioned by the United Kingdom's Royal Society explores public perceptions and the reasoning behind them [154]. In both this study and the academic debate more generally, considerable weight is given to the potential for genetically modified animals to improve human health or (negatively) impact ecosystems [154]. However, other public concerns regarding genome editing technologies are thus far underrepresented in the academic literature, including the public concern for equity of access to the potential benefits of genome editing technologies, questions about the just distribution of governmental funding of genome editing compared with other investments, and concerns about the commercialization of genome editing technologies. With regard to commercialization, members of the public have raised the worry that businesses could prioritize profit-making over the public good and could fail to provide a balanced representation of the benefits and risks of these technologies [154]. The fact that these concerns are largely absent from the academic debate on genome editing in animals is particularly significant given ongoing calls for public engagement and raises interesting questions that relate to a broader discussion about what the rationale, form and aim of public engagement should be. If the goals of such engagement are not merely to inform the public, but also to address societal challenges and to allow the public to be involved in shaping technological developments together with other stakeholders, then issues regarding commercialization, distributive justice and access to the benefits of genome editing technologies are worthy of more attention in the academic literature.
(e). Limitations
This systematic review provides a comprehensive overview of the reasons brought forward in the academic debate on genome editing in animals. The articles presented were included after a thorough screening of the academic literature on the topic by two independent reviewers, based on a search strategy that was guided by experienced librarians. Nonetheless, this review has several limitations.
First, given the focus on relatively new genome editing technologies and a large amount of literature on this topic, this review included articles published between 2010 and 2018. We recognize that arguments raised previously, in different contexts or in older but related debates, may be relevant for the current discussion of genome editing. Second, a systematic review of this kind always involves reporting bias; a different group of researchers could have selected or grouped the included reasons in a different way. Third, we could not systematically perform a quality assessment of the included literature, as there is no screening instrument to assess the quality of normative papers or the reasons mentioned. Finally, we note that it was beyond the scope of this paper to assess the scientific validity of the reasons and different applications of genome editing discussed in the included articles.
5. Conclusion
Genome editing has a broad range of possible applications in research animals, farm animals and wild animals. Despite an ongoing academic debate on this topic, this study is the first comprehensive overview of this debate. Our article provides a systematic review of the reasons for and against the development and use of genome editing technologies in animals as reported in the academic literature. We identified 67 different reasons for and 48 different reasons against genome editing in animals, which related to human health, efficiency, risks and uncertainty, animal welfare, animal dignity, environmental considerations and public acceptability. Our findings illuminate several key features of the academic debate thus far, including a low disciplinary diversity in the contributing professionals, a scarcity of systematic comparisons of potential consequences of using these technologies, an underrepresentation of animal interests, and a disjunction between the public and academic debate on this topic.
As such, our article can be considered a call for professionals from a wide range of disciplines to become involved in the academic discussion about genome editing in non-human animals. We also suggest that this ongoing debate seek to incorporate animal interests, systematically compare applications of these technologies using the principles of proportionality and subsidiarity, and further research the range of concerns uncovered through public engagement. Proactive and multidisciplinary collaboration can both advance these technological developments and the academic discourse about them, allowing us to go beyond rhetoric of promises or fears and positioning their ethical analysis in real-world practices [145,155].
Supplementary Material
Supplementary Material
Acknowledgements
We thank Professor Dr Niels Geijsen for his comments on this work.
Data accessibility
This article has no additional data.
Authors' contributions
N.G., K.R.J. and A.L.B. were responsible for designing the article concept. N.G. and K.R.J. performed the data collection and analysis. N.G. was responsible for drafting and revision of the manuscript, to which K.R.J., S.H., J.J. and A.L.B. provided critical intellectual input and revisions. All authors approved the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This project was supported by the division of Applied and Engineering Sciences of The Netherlands Organisation for Scientific Research (NWO; project no. 15804). J.J.'s work is funded by a grant from the John Templeton Foundation; S.H.'s work is funded by a grant from the British Academy's ‘Tackling the UK's International Challenges Programme: Knowledge Frontiers’.
References
- 1.Carroll D. 2008. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 15, 1463–1468. ( 10.1038/gt.2008.145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pâques F, Duchateau P. 2007. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr. Gene Ther. 7, 49–66. ( 10.2174/156652307779940216) [DOI] [PubMed] [Google Scholar]
- 3.Joung JK, Sander JD. 2013. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell. Biol. 14, 49–55. ( 10.1038/nrm3486.TALENs) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doudna JA, Charpentier E. 2014. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096 ( 10.1126/science.1258096) [DOI] [PubMed] [Google Scholar]
- 5.Esvelt KM, Smidler AL, Catteruccia F, Church GM. 2014. Concerning RNA-guided gene drives for the alteration of wild populations. Elife 3, e03401 ( 10.7554/eLife.03401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emerson C, James S, Littler K, Randazzo F. 2017. Principles for gene drive research. Science 358, 1135–1136. ( 10.1126/science.aap9026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ledford H. 2015. CRISPR, the disruptor. Nature 522, 20–24. ( 10.1038/522020a) [DOI] [PubMed] [Google Scholar]
- 8.Sovová T, Kerins G, Demnerová K, Ovesná J. 2017. Genome editing with engineered nucleases in economically important animals and plants: state of the art in the research pipeline. Curr. Issues Mol. Biol. 21, 41–62. ( 10.21775/cimb.021.041) [DOI] [PubMed] [Google Scholar]
- 9.Schultz-Bergin M. 2018. Is CRISPR an ethical game changer? J. Agric. Environ. Ethics 31, 219–238. ( 10.1007/s10806-018-9721-z) [DOI] [Google Scholar]
- 10.Gonen S, Jenko J, Gorjanc G, Mileham AJ, Whitelaw CBA, Hickey JM. 2017. Potential of gene drives with genome editing to increase genetic gain in livestock breeding programs. Genet. Sel. Evol. 49, 1–14. ( 10.1186/s12711-016-0280-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma D, Liu F. 2015. Genome editing and its applications in model organisms. Genomics, Proteomics Bioinforma. 13, 336–344. ( 10.1016/j.gpb.2015.12.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherkow JS, Greely HT. 2013. What if extinction is not forever? Science 340, 32–33. ( 10.1126/science.1113442) [DOI] [PubMed] [Google Scholar]
- 13.Ambrus M, Arts K, Hey E, Raulus H (eds). 2014. The role of ‘experts’ in international and European decision-making processes: advisors, decision makers or irrelevant actors? Cambridge, UK: Cambridge University Press. [Google Scholar]
- 14.Hartley S. 2016. Policy masquerading as science: an examination of non-state actor involvement in European risk assessment policy for genetically modified animals. J. Eur. Public Policy 23, 276–295. ( 10.1080/13501763.2015.1049196) [DOI] [Google Scholar]
- 15.Pielke RA. 2007. The honest broker: making sense of science in policy and politics. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 16.Strech D, Sofaer N. 2012. How to write a systematic review of reasons. J. Med. Ethics 38, 121–126. ( 10.1136/medethics-2011-100096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moher D, Liberati A, Tetzlaff J, Altman DG. 2009. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6, e1000097 ( 10.1371/journal.pmed.1000097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooijmans CR, Tillema A, Leenaars M, Ritskes-Hoitinga M. 2010. Enhancing search efficiency by means of a search filter for finding all studies on animal experimentation in PubMed. Lab. Anim. 44, 170–175. ( 10.1258/la.2010.009117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll D, Charo RA. 2015. The societal opportunities and challenges of genome editing. Genome Biol. 16, 242 ( 10.1186/s13059-015-0812-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Courtier-Orgogozo V, Morizot B, Boëte C. 2017. Agricultural pest control with CRISPR-based gene drive: time for public debate. Sci. Soc. 18, 878–880. ( 10.15252/embr.201744205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gantz VM, Bier E. 2016. The dawn of active genetics. Bioessays 38, 50–63. ( 10.1002/bies.201500102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall JM. 2010. The Cartagena Protocol and genetically modified mosquitoes. Nat. Biotechnol. 28, 896–897. ( 10.1038/nbt0910-896) [DOI] [PubMed] [Google Scholar]
- 23.Benedict MQ, Burt A, Capurro ML, De Barro P, Handler AM, Hayes KR, Marshall JM, Tabachnick WJ, Adelman ZN. 2018. Recommendations for laboratory containment and management of gene drive systems in arthropods. Vector Borne Zoonotic Dis. 18, 2–13. ( 10.1089/vbz.2017.2121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown DM, Alphey LS, Mckemey A, Beech C, James AA. 2014. Criteria for identifying and evaluating candidate sites for open-field trials of genetically engineered mosquitoes. Vector Borne Zoonotic Dis. 14, 291–299. ( 10.1089/vbz.2013.1364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charo RA, Greely HT. 2015. CRISPR critters and CRISPR cracks. Am. J. Bioeth. 15, 11–17. ( 10.1080/15265161.2015.1104138) [DOI] [PubMed] [Google Scholar]
- 26.Ju X-D, Xu J, Sun ZS. 2018. CRISPR editing in biological and biomedical investigation. J. Cell. Biochem. 119, 52–61. ( 10.1002/jcb.26154) [DOI] [PubMed] [Google Scholar]
- 27.Eckhoff PA, Wenger EA, Godfray HCJ, Burt A. 2017. Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. Proc. Natl Acad. Sci. USA 114, E255–E264. ( 10.1073/pnas.1611064114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabrieli P, Smidler A, Catteruccia F. 2014. Engineering the control of mosquito-borne infectious diseases. Genome Biol. 15, 535 ( 10.1186/s13059-014-0535-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl Acad. Sci. USA 112, E6736–E6743. ( 10.1073/pnas.1521077112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kistler KE, Vosshall LB, Matthews BJ. 2016. Genome-engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. 11, 51–60. ( 10.1016/j.celrep.2015.03.009.Genome-engineering) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noble C, et al. 2016. Daisy-chain gene drives for the alteration of local populations. bioRxiv, 57307. ( 10.1101/057307) [DOI]
- 32.Oye KA, et al. 2014. Biotechnology. Regulating gene drives. Science. 345, 626–628. ( 10.1126/science.1254287) [DOI] [PubMed] [Google Scholar]
- 33.Reid W, O'brochta DA. 2016. Applications of genome editing in insects. Curr. Opin. Insect Sci. 13, 43–54. ( 10.1016/j.cois.2015.11.001) [DOI] [PubMed] [Google Scholar]
- 34.Caplan AL, Parent B, Shen M, Plunkett C. 2015. No time to waste – the ethical challenges created by CRISPR. EMBO Rep. 16, 1421–1426. ( 10.15252/embr.201541337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau V, Davie JR. 2017. The discovery and development of the CRISPR system in applications in genome manipulation. Biochem. Cell Biol. 95, 203–210. ( 10.1139/bcb-2016-0159) [DOI] [PubMed] [Google Scholar]
- 36.Greenfield A. 2017. Editing mammalian genomes: ethical considerations. Mamm. Genome 28, 388–393. ( 10.1007/s00335-017-9702-y) [DOI] [PubMed] [Google Scholar]
- 37.Cribbs AP, Perera SMW. 2017. Science and bioethics of CRISPR-Cas9 gene editing: an analysis towards separating facts and fiction. Yale J. Biol. Med. 90, 625–634. [PMC free article] [PubMed] [Google Scholar]
- 38.Eaton SL, Wishart TM. 2017. Bridging the gap: large animal models in neurodegenerative research. Mamm. Genome 28, 324–337. ( 10.1007/s00335-017-9687-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meier RPH, Muller YD, Balaphas A, Morel P, Pascual M, Seebach JD, Buhler LH. 2017. Xenotransplantation: back to the future? Transpl. Int. 31, 465–477. ( 10.1111/tri.13104) [DOI] [PubMed] [Google Scholar]
- 40.Singh V, Braddick D, Dhar PK. 2017. Exploring the potential of genome editing CRISPR-Cas9 technology. Gene 599, 1–18. ( 10.1016/j.gene.2016.11.008) [DOI] [PubMed] [Google Scholar]
- 41.Waddington SN, Privolizzi R, Karda R, O'Neill HC. 2016. A broad overview and review of CRISPR-Cas technology and stem cells. Curr. Stem Cell Reports 2, 9–20. ( 10.1007/s40778-016-0037-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Musunuru K. 2017. The hope and hype of CRISPR-Cas9 genome editing: a review. JAMA Cardiol. 2, 914–919. ( 10.1001/jamacardio.2017.1713) [DOI] [PubMed] [Google Scholar]
- 43.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. ( 10.1016/j.cell.2014.05.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heidenreich M, Zhang F. 2015. Applications of CRISPR-Cas systems in neuroscience. Nat. Rev. Neurosci. 2, 147–185. ( 10.1038/nrn.2015.2.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm IE, Alstrup AKO, Luo Y. 2016. Genetically modified pig models for neurodegenerative disorders. J. Pathol. 238, 267–287. ( 10.1002/path.4654) [DOI] [PubMed] [Google Scholar]
- 46.Mashimo T. 2014. Gene targeting technologies in rats: zinc finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats. Dev. Growth Differ. 56, 46–52. ( 10.1111/dgd.12110) [DOI] [PubMed] [Google Scholar]
- 47.Petersen B, Niemann H. 2015. Molecular scissors and their application in genetically modified farm animals. Transgenic Res. 24, 381–396. ( 10.1007/s11248-015-9862-z) [DOI] [PubMed] [Google Scholar]
- 48.Qian L, et al. 2015. Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci. Rep. 5, 1–13. ( 10.1038/srep14435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. 2014. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. ( 10.1016/j.cell.2013.04.025.One-Step) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao J, et al. 2014. Efficient bi-allelic gene knockout and site-specific knock-in mediated by TALENs in pigs. Sci. Rep. 4, 6926 ( 10.1038/srep06926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao J, Huang J, Zhao J. 2016. Genome editing revolutionize the creation of genetically modified pigs for modeling human diseases. Hum. Genet. 135, 1093–1105. ( 10.1007/s00439-016-1710-6) [DOI] [PubMed] [Google Scholar]
- 52.Boete C. 2011. Scientists and public involvement: a consultation on the relation between malaria, vector control and transgenic mosquitoes. Trans. R. Soc. Trop. Med. Hyg. 105, 704–710. ( 10.1016/j.trstmh.2011.08.006) [DOI] [PubMed] [Google Scholar]
- 53.Combes RD, Balls M. 2014. Every silver lining has a cloud: the scientific and animal welfare issues surrounding a new approach to the production of transgenic animals. Altern. Lab. Anim. 42, 137–145. [DOI] [PubMed] [Google Scholar]
- 54.Chen Y, et al. 2015. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum. Mol. Genet. 24, 3764–3774. ( 10.1093/hmg/ddv120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo X, Li M, Su B. 2016. Application of the genome editing tool CRISPR/Cas9 in non-human primates. Zool. Res. 37, 214–219. ( 10.13918/j.issn.2095-8137.2016.4.214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stouffer RL, Woodruff TK. 2017. Nonhuman primates: a vital model for basic and applied research on female reproduction, prenatal development, and women's health. ILAR J. 58, 281–294. ( 10.1093/ilar/ilx027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willyard C. 2016. New models: gene-editing boom means changing landscape for primate work. Nat. Med. 22, 1200–1202. ( 10.1038/nm1116-1200) [DOI] [PubMed] [Google Scholar]
- 58.Chen Y, Niu Y, Ji W. 2016. Genome editing in nonhuman primates: approach to generating human disease models. J. Intern. Med. 280, 246–251. ( 10.1111/joim.12469) [DOI] [PubMed] [Google Scholar]
- 59.Jennings CG, et al. 2016. Opportunities and challenges in modeling human brain disorders in transgenic primates. Nat. Neurosci. 19, 1123–1130. ( 10.1038/nn.4362) [DOI] [PubMed] [Google Scholar]
- 60.Guo X, Li XJ. 2015. Targeted genome editing in primate embryos. Cell Res. 25, 767–768. ( 10.1038/cr.2015.64) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu H, et al. 2014. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell 14, 323–328. ( 10.1016/j.stem.2014.01.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Niu Y, et al. 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156, 836–843. ( 10.1016/j.cell.2014.01.027) [DOI] [PubMed] [Google Scholar]
- 63.Tu Z, Yang W, Yan S, Guo X, Li XJ. 2015. CRISPR/Cas9: a powerful genetic engineering tool for establishing large animal models of neurodegenerative diseases. Mol. Neurodegener. 10, 1–8. ( 10.1186/s13024-015-0031-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fung RKF, Kerridge IH. 2016. Gene editing advance re-ignites debate on the merits and risks of animal to human transplantation. Intern. Med. J. 46, 1017–1022. ( 10.1111/imj.13183) [DOI] [PubMed] [Google Scholar]
- 65.Burlak C. 2015. Xenotransplantation literature update, November–December 2014. Xenotransplantation 22, 80–83. ( 10.1111/xen.12158) [DOI] [PubMed] [Google Scholar]
- 66.De Salvatore S, Segreto A, Chiusaroli A, Congiu S, Bizzarri F. 2015. Role of xenotransplantation in cardiac transplantation. J. Card. Surg. 30, 111–116. ( 10.1111/jocs.12454) [DOI] [PubMed] [Google Scholar]
- 67.Zeyland J, Hryhorowicz M, Nowak-Terpiłowska A, Jura J, Slomnsko R, Smorąg Z, Gajda B, Lipiński D. 2018. The production of UL16-binding protein 1 targeted pigs using CRISPR technology. 3Biotech 8, 70 ( 10.1007/s13205-018-1107-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mourad NI, Gianello P. 2017. Gene editing, gene therapy, and cell xenotransplantation: cell transplantation across species. Curr. Transplant. Reports 4, 193–200. ( 10.1007/s40472-017-0157-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peterson A. 2017. CRISPR: express delivery to any DNA address. Oral Dis. 23, 5–11. ( 10.1111/odi.12487) [DOI] [PubMed] [Google Scholar]
- 70.Carroll D. 2014. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439. ( 10.1146/annurev-biochem-060713-035418) [DOI] [PubMed] [Google Scholar]
- 71.Butler JR, et al. 2016. Silencing porcine genes significantly reduces human-anti-pig cytotoxicity profiles: an alternative to direct complement regulation. Transgenic Res. 25, 751–759. ( 10.1007/s11248-016-9958-0) [DOI] [PubMed] [Google Scholar]
- 72.Lutz AJ, et al. 2013. Double knockout pigs deficient in N-glycolylneuraminic acid and Galactose α-1,3-Galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation 20, 27–35. ( 10.1111/xen.12019) [DOI] [PubMed] [Google Scholar]
- 73.Niu D, et al. 2018. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9 Dong. Science 357, 1303–1307. ( 10.1126/science.aan4187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cooper DKC, Ekser B, Ramsoondar J, Phelps C, Ayares D. 2016. The role of genetically engineered pigs in xenotransplantation research. J. Pathol. 238, 288–299. ( 10.1002/path.4635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reyes LM, et al. 2018. Creating class I MHC null pigs using gRNA and the Cas9 endonuclease. 193, 5751–5757. ( 10.4049/jimmunol.1402059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salomon DR. 2016. A CRISPR way to block PERVs — engineering organs for transplantation. N. Eng. J. Med. 374, 1089–1091. ( 10.1056/NEJMcibr1515623) [DOI] [PubMed] [Google Scholar]
- 77.Yang L. 2015. Genome-wide inactivation of porcine endogenous retroviruses. Science 350, 1101–1104. ( 10.1126/science.aad1191) [DOI] [PubMed] [Google Scholar]
- 78.Shriver A, Mcconnachie E. 2018. Genetically modifying livestock for improved welfare: a path forward. J. Agric. Environ. Ethics 31, 161–180. ( 10.1007/s10806-018-9719-6) [DOI] [Google Scholar]
- 79.Tan F, Carlson DF, Walton MW, Fahrenkrug SC, Hacket PB. 2012. Precision editing of large animal genomes. Adv. Genet.80, 37–97. ( 10.1016/B978-0-12-404742-6.00002-8) [DOI] [Google Scholar]
- 80.Ishii T. 2017. Genome-edited livestock: ethics and social acceptance. Anim. Front. 7, 24–32. ( 10.2527/af.2017.0115) [DOI] [Google Scholar]
- 81.Martinelli L, Oksanen M, Siipi H. 2014. De-extinction: a novel and remarkable case of bio-objectification. Croat. Med. J. 55, 423–427. ( 10.3325/cmj.2014.55.423.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shinwari ZK, Tanveer F, Khali AT. 2017. Ethical issues regarding CRISPR-mediated genome editing. Cris. Syst. Emerg. Technol. Appl. 26, 103–110. ( 10.21775/9781910190630.09) [DOI] [PubMed] [Google Scholar]
- 83.Webber BL, Raghu S, Edwards OR. 2015. Opinion: is CRISPR-based gene drive a biocontrol silver bullet or global conservation threat? Proc. Natl Acad. Sci. USA 112, 10 565–10 567. ( 10.1073/pnas.1514258112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miano JM, Zhu QM, Lowenstein CJ. 2016. A CRISPR path to engineering new genetic mouse models for cardiovascular research. Arterioscler. Thromb. Vasc. Biol. 36, 1058–1075. ( 10.1161/ATVBAHA.116.304790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carroll D, Van Eenennaam AL, Taylor JF, Seger J, Voytas DF. 2016. Regulate genome-edited products, not genome editing itself. Nat. Biotechnol. 34, 477–479. ( 10.1038/nbt.3566) [DOI] [PubMed] [Google Scholar]
- 86.Whitworth KM, et al. 2015. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 34, 20–22. ( 10.1038/nbt.3434) [DOI] [PubMed] [Google Scholar]
- 87.Carlson DF, Lancto CA, Zang B, Kim ES, Walton M, Oldeschulte D, Seabury C, Sonstegard TS, Fahrenkrug SC. 2016. Production of hornless dairy cattle from genome-edited cell lines. Nat. Biotechnol. 34, 479–481. ( 10.1038/nbt.3560) [DOI] [PubMed] [Google Scholar]
- 88.Akbari OS, et al. 2016. Safeguarding gene drive experiments in the laboratory. Science 349, 927–929. ( 10.1126/science.aac7932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Esvelt KM, Gemmell NJ. 2017. Conservation demands safe gene drive. PLoS Biol. 15, 1–8. ( 10.1371/journal.pbio.2003850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carroll D. 2011. Genome engineering with zinc-finger nucleases. Genetics 188, 773–782. ( 10.1534/genetics.111.131433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shapiro B. 2017. Pathways to de-extinction: how close can we get to resurrection of an extinct species? Funct. Ecol. 31, 996–1002. ( 10.1111/1365-2435.12705) [DOI] [Google Scholar]
- 92.Pavlovic G, Brault V, Sorg T, Hérault Y. 2014. Generation and use of transgenic mice in drug discovery. In In vivo models for drug discovery (eds Vela JM, Maldonado M, Hamon M), pp. 131–148. New York, NY: Wiley Blackwell. [Google Scholar]
- 93.Petersen B, Frenzel A, Lucas-Hahn A, Herrmann D, Hassel P, Klein S, Ziegler M, Hadeler KG, Niemann H. 2016. Efficient production of biallelic GGTA1 knockout pigs by cytoplasmic microinjection of CRISPR/Cas9 into zygotes. Xenotransplantation 23, 338–346. ( 10.1111/xen.12258) [DOI] [PubMed] [Google Scholar]
- 94.Wang X, et al. 2016. Multiplex gene editing via CRISPR/Cas9 exhibits desirable muscle hypertrophy without detectable off-target effects in sheep. Sci. Rep. 6, 1–11. ( 10.1038/srep32271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Whitelaw CBA, Sheets TP, Lillico SG, Telugu BP. 2016. Engineering large animal models of human disease. J. Pathol. 238, 247–256. ( 10.1002/path.4648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eriksson S, Jonas E, Rydhmer L, Rocklinsberg H. 2018. Invited review: Breeding and ethical perspectives on genetically modified and genome edited cattle. J. Dairy Sci. 101, 1–17. ( 10.3168/jds.2017-12962) [DOI] [PubMed] [Google Scholar]
- 97.Bhat SA, Malik AA, Ahmad SM, Shah RA, Ganai NA, Shafi SS, Shabir N. 2017. Advances in genome editing for improved animal breeding: a review. Vet. World 10, 1361–1366. ( 10.14202/vetworld.2017.1361-1366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Reiner G. 2016. Genetic resistance - an alternative for controlling PRRS? Porc. Heal. Manag. 2, 27 ( 10.1186/s40813-016-0045-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hickey JM, Bruce C, Whitelaw A, Gorjanc G. 2016. Promotion of alleles by genome editing in livestock breeding programmes. J. Anim. Breed. Genet. 133, 83–84. ( 10.1111/jbg.12206) [DOI] [PubMed] [Google Scholar]
- 100.Leenstra F, Munnichs G, Beekman V, Van Den Heuvel-Vromans E, Aramyan L, Woelders H. 2011. Killing day-old chicks? Public opinion regarding potential alternatives. Anim. Welf. 20, 37–45. [Google Scholar]
- 101.Pugh J. 2016. Driven to extinction? The ethics of eradicating mosquitoes with gene-drive technologies. J. Med. Ethics J. Inst. Med. Ethics 42, 578–581. ( 10.1136/medethics-2016-103462) [DOI] [PubMed] [Google Scholar]
- 102.Resnik DB. 2014. Ethical issues in field trials of genetically modified disease-resistant mosquitoes. Dev. World Bioeth. 14, 37–46. ( 10.1111/dewb.12011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kaebnick GE. 2017. The spectacular garden: where might de-extinction lead? Hastings Cent. Rep. 47, S60–S64. ( 10.1002/hast.754) [DOI] [PubMed] [Google Scholar]
- 104.Benz-Schwarzburg J, Ferrari A. 2016. Super-muscly pigs trading ethics for efficiency. Issues Sci. Technol. 32, 29–32. [Google Scholar]
- 105.Camporesi S, Cavaliere G. 2016. Emerging ethical perspectives in the clustered regularly interspaced short palindromic repeats genome-editing debate. Per. Med. 13, 575–586. ( 10.2217/pme-2016-0047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lunshof J. 2015. Regulate gene editing in wild animals. Nature 521, 127 ( 10.1038/521127a) [DOI] [PubMed] [Google Scholar]
- 107.Neuhaus CP. 2018. Community engagement and field trials of genetically modified insects and animals. Hastings Cent. Rep. 48, 25–36. ( 10.1002/hast.808) [DOI] [PubMed] [Google Scholar]
- 108.Neuhaus CP, Caplan AL. 2017. Genome editing: bioethics shows the way. PLoS Biol. 15, e2001934 ( 10.1371/journal.pbio.2001934) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Resnik DB. 2017. Ethics of community engagement in field trials of genetically modified mosquitoes. Dev. World Bioeth. 375, 135–143. ( 10.1111/dewb.12147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Neuhaus CP. 2017. Ethical issues when modelling brain disorders in non-human primates. J. Med. Ethics 44, 323–327. ( 10.1136/medethics-2016-104088) [DOI] [PubMed] [Google Scholar]
- 111.Manesh SB, Samani RO, Manesh SB. 2014. Ethical issues of transplanting organs from transgenic animals into human beings. Cell J. 16, 353–360. [PMC free article] [PubMed] [Google Scholar]
- 112.Shriver A. 2015. Would the elimination of the capacity to suffer solve ethical dilemmas in experimental animal research? Curr. Top. Behav. Neurosci. 19, 117–132. ( 10.1007/7854_2014_318) [DOI] [PubMed] [Google Scholar]
- 113.Heeger R. 2015. Dignity only for humans? A controversy. In The Cambridge handbook of human dignity: interdisciplinary perspectives, pp. 541–545. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 114.Johannsen K. 2017. Animal rights and the problem of r-strategists. Ethical Theory Moral Pract. 20, 333–345. ( 10.1007/S10677-016-9774-X) [DOI] [Google Scholar]
- 115.Bovenkerk B, Nijland HJ. 2017. The pedigree dog breeding debate in ethics and practice: beyond welfare arguments. J. Agric. Environ. Ethics 30, 387–412. ( 10.1007/s10806-017-9673-8) [DOI] [Google Scholar]
- 116.Schultz-Bergin M. 2014. Making better sense of animal disenhancement: a reply to Henschke. Nanoethics 8, 101–109. ( 10.1007/s11569-014-0190-1) [DOI] [Google Scholar]
- 117.Ahteensuu M. 2017. Synthetic biology, genome editing, and the risk of bioterrorism. Sci. Eng. Ethics 23, 1541–1561. ( 10.1007/s11948-016-9868-9) [DOI] [PubMed] [Google Scholar]
- 118.Palmer C. 2011. Animal disenhancement and the non-identity problem: a response to Thompson. Nanoethics 5, 43–48. ( 10.1007/s11569-011-0115-1) [DOI] [Google Scholar]
- 119.Schultz-Bergin M. 2017. The dignity of diminished animals: species norms and engineering to improve welfare. Ethical Theory Moral Pract. 20, 843–856. ( 10.1007/s10677-017-9828-8) [DOI] [Google Scholar]
- 120.Noll S. 2013. Broiler chickens and a critique of the epistemic foundations of animal modification. J. Agric. Environ. Ethics 26, 273–280. ( 10.1007/s10806-011-9362-y) [DOI] [Google Scholar]
- 121.Rollin BE. 2015. Telos, conservation of welfare, and ethical issues in genetic engineering of animals. Curr. Top. Behav. Neurosci. 19, 99–116. ( 10.1007/7854_2014_279) [DOI] [PubMed] [Google Scholar]
- 122.Delon N, Purves D. 2018. Wild animal suffering is intractable. J. Agric. Environ. Ethics. 31, 239–260. ( 10.1007/s10806-018-9722-y) [DOI] [Google Scholar]
- 123.Jebari K. 2016. Should extinction be forever? Philos. Technol. 29, 211–222. ( 10.1007/s13347-015-0208-9) [DOI] [Google Scholar]
- 124.Venkatraman V. 2016. Turning point: Kevin Esvelt. Nature 536, 117 ( 10.1038/nj7614-117a) [DOI] [PubMed] [Google Scholar]
- 125.2015. Defensive drives. Nature 527, 275–276. ( 10.1038/527275b) [DOI] [PubMed] [Google Scholar]
- 126.Reardon S. 2016. Welcome to the CRISPR zoo. Nature 531, 160–163. ( 10.1038/531160a) [DOI] [PubMed] [Google Scholar]
- 127.Callaway E. 2015. Mosquitoes engineered to pass down genes that would wipe out their species. Nature 1–2. ( 10.1038/nature.2015.18974) [DOI] [Google Scholar]
- 128.Cyranoski D. 2015. Gene-edited pigs to be sold as pets. Nature 526, 18 ( 10.1038/nature.2015.18448) [DOI] [PubMed] [Google Scholar]
- 129.Ledford H. 2016. CRISPR: gene editing is just the beginning. Nature 531, 156–169. ( 10.1038/531156a) [DOI] [PubMed] [Google Scholar]
- 130.Reardon S. 2015. New life for pig-to-human transplants. Nature 527, 152–154. ( 10.1038/527152a) [DOI] [PubMed] [Google Scholar]
- 131.Graham DM. 2016. Putting the brakes on CRISPR-Cas9 gene drive systems. Lab. Anim. 45, 47 ( 10.1038/laban.941) [DOI] [PubMed] [Google Scholar]
- 132.Knox M. 2014. The gene genie. Sci. Am. 311, 42–46. ( 10.1038/scientificamerican1214-42) [DOI] [PubMed] [Google Scholar]
- 133.West J, Gill WW. 2016. Genome editing in large animals. J. Equine Vet. Sci. 41, 1–6. ( 10.1016/j.jevs.2016.03.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. 2010. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646. ( 10.1038/nrg2842) [DOI] [PubMed] [Google Scholar]
- 135.Fears R, Ter Meulen V. 2017. How should the applications of genome editing be assessed and regulated? Elife 6, e26295 ( 10.7554/eLife.26295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Webber P. 2014. Does CRISPR-Cas open new possibilities for patents or present a moral maze? Nat. Biotechnol. 32, 331–333. ( 10.1038/nbt.2843) [DOI] [PubMed] [Google Scholar]
- 137.Fox D. 2010. Retracing liberalism and remaking nature: designer children, research embryos, and featherless chickens. Bioethics 24, 170–178. ( 10.1111/j.1467-8519.2008.00707.x) [DOI] [PubMed] [Google Scholar]
- 138.Perez-Pinera P, Ousterout D, Gersbach C. 2012. Advances in targeted genome editing. Curr. Opin Chem. Biol. 16, 268–277. ( 10.1016/j.cbpa.2012.06.007.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hefferon KL, Herring RJ. 2017. The end of the GMO? Genome editing, gene drives and new frontiers of plant technology. Rev. Agrar. Stud. 7, 1–32. [Google Scholar]
- 140.Barrangou R, Doudna JA. 2016. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34, 933–941. ( 10.1038/nbt.3659) [DOI] [PubMed] [Google Scholar]
- 141.Buttriss JL. 2011. Feeding the planet: an unprecedented confluence of pressures anticipated. Nutr. Bull. 36, 235–241. ( 10.1111/j.1467-3010.2011.01894.x) [DOI] [Google Scholar]
- 142.Laible G, Wei J, Wagner S. 2015. Improving livestock for agriculture—technological progress from random transgenesis to precision genome editing heralds a new era. Biotechnol. J. 10, 109–120. ( 10.1002/biot.201400193) [DOI] [PubMed] [Google Scholar]
- 143.Kuzma J, Rawls L. 2016. Engineering the wild: gene drives and intergenerational equity. Jurimetrics J. Law Sci. Technol. 56, 279 ( 10.3868/s050-004-015-0003-8) [DOI] [Google Scholar]
- 144.Ambrus M, Arts K, Hey E, Raulus H. 2014. The role of experts in international and European decision-making processes: setting the scene. In The role of ‘experts’ in international and European decision-making processes: advisors, decision makers or irrelevant actors? (eds Hey E, Raulus H, Arts K, Ambrus M), pp. 1–16. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 145.Jongsma KR, Bredenoord AL, Lucivero F. 2018. Digital medicine: an opportunity to revisit the role of bioethicists. Am. J. Bioeth. 18, 69–70. ( 10.1080/15265161.2018.1498952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Van Delden JJ, Bredenoord AL. 2015. Future challenges for bioethicists: regenerative medicine. In Global bioethics: what for? 20th anniversary of UNESCO's bioethics program (ed. Solinis G.), pp. 137–141. Paris, France: UNESCO Publishing. [Google Scholar]
- 147.Advisory Committee on Releases to the Environment (ACRE). 2007. Managing the footprint of agriculture: towards a comparative assessment of risks and benefits for novel agricultural systems. Available online: https://webarchive.nationalarchives.gov.uk/20080604145150/http://www.defra.gov.uk/Environment/acre/fsewiderissues/pdf/acre-wi-final.pdf.
- 148.Scientific Committee on Health and Environmental Risks (SCHER), Scientific Committee on Emerging and Newly Identified Health Risks (SCENIHR) & Scientific Committee on Consumer Safety (SCCS). 2013. Making risk assessment more relevant for risk management. ( 10.2772/34776) [DOI]
- 149.Degrazia D. 2008. Moral status as a matter of degree? South. J. Philos. 46, 181–198. ( 10.1111/j.2041-6962.2008.tb00075.x) [DOI] [Google Scholar]
- 150.Hursthouse R. 2006. Applying virtue ethics to our treatment of the other animals. In The practice of virtue: classic and contemporary readings in virtue ethics (ed. Welchman J.), pp. 136–155. Indianapolis, IN: Hackett Publishing Company. [Google Scholar]
- 151.Swierstra T, Rip A. 2007. Nano-ethics as NEST-ethics: patterns of moral argumentation about new and emerging science and technology. Nanoethics 1, 3–20. ( 10.1007/s11569-007-0005-8) [DOI] [Google Scholar]
- 152.Sarewitz D. 2015. CRISPR: science can't solve it. Nature 522, 413–414. ( 10.1038/522413a) [DOI] [PubMed] [Google Scholar]
- 153.Burall S. 2018. Rethink public engagement for gene editing. Nature 555, 438–439. ( 10.1038/d41586-018-03269-3) [DOI] [PubMed] [Google Scholar]
- 154.Van Mil A, Hopkins H, Kinsella S.2017. Potential uses for genetic technologies: dialogue and engagement research conducted on behalf of the Royal Society Findings Report. Available online: https://royalsociety.org/~/media/policy/projects/gene-tech/genetic-technologies-public-dialogue-hvm-full-report.pdf .
- 155.Lucivero F. 2016. Promises, expectations and visions: on appraising the plausibility of socio-technical futures. Basel, Switzerland: Springer International Publishing. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.