Abstract
Technological advances in immunology, protein design, and genetic delivery have unlocked new possibilities for vaccine concepts and delivery technologies that were previously inaccessible. These next-generation vaccine design efforts are particularly promising in their potential to provide solutions to challenging targets for which conventional approaches have proven ineffective—for example, a universal influenza vaccine. In this perspective, we discuss emerging approaches to vaccine design and engineering based on recent insights into immunology, structural biology, computational biology, and immunoengineering. We anticipate that these cutting-edge, interdisciplinary approaches will lead to breakthrough vaccine concepts for ever-evolving and (re)emerging influenza viruses, with important ramifications for global public health.
Keywords: universal influenza vaccine, antigen design, structure-based vaccine, protein engineering, nanoparticle, antigen display, mRNA vaccine, broadly neutralizing antibody
Over the last 100 years, immense efforts have been devoted to understanding influenza viruses, yielding deeper understanding of viral pathogenesis, animal models of infection, innate and adaptive immune responses to the virus, original antigenic sin, immunodominance, and antigenic imprinting. Among a number of achievements, the deployment of influenza vaccines has been the most important milestone and has saved countless lives to date. A major breakthrough in our knowledge of humoral immune responses to influenza virus has been the discovery of broadly neutralizing antibodies (bNAbs) that target structurally and functionally conserved sites of vulnerability on the viral hemagglutinin (HA). Furthermore, several apparently common lineages of bNAbs have been identified across multiple individuals. These findings have instructed us on the developmental pathways that could lead to bNAb responses to influenza and have breathed new life into efforts to design vaccines that protect against a wide variety of virus strains: universal influenza vaccines. These efforts are driven by technological revolutions in several fields that have made possible new approaches to vaccine design and delivery. Here we provide our perspective on the prospects for a next generation of influenza vaccines based on these new approaches and concepts.
NEW CONCEPTS IN TRADITIONAL VACCINE PLATFORMS
Although the majority of universal influenza vaccine efforts are directed at subunit vaccines or alternative platforms, several new concepts in virus manipulation are currently being evaluated in the live-attenuated vaccine format. Orthodox approaches to generate attenuated virus vaccines often involve serial passage of viruses in vitro or in vivo. For example, the first reported live-attenuated influenza vaccine was generated by adapting virus to successively lower temperatures [1]. The resultant cold-adapted virus remains the basis of current commercial live-attenuated influenza vaccines. However, several orthogonal approaches to synthetically attenuate viruses are being explored.
For example, introduction of a conditional defect in influenza HA by engineering its posttranslational processing and maturation was demonstrated as an effective virus attenuation strategy [2]. The engineered HA possesses a proteolytic cleavage site recognized by pancreatic elastase instead of trypsin-like serine proteases, and the resultant virus is highly attenuated while remaining immunogenic. Virus attenuation by genome-wide modifications in codon pair bias, in which adjacent codon usage is deoptimized without introducing nonsynonymous mutations, was initially demonstrated in poliovirus [3] and later proven efficacious in influenza viruses [4]. The precise mechanism of this approach to virus attenuation may relate to inefficient translation of disfavored codon pairs, triggering of innate immune sensors by coincidental increases in the frequency of CpG/UpA dinucleotides [5], or a combination of the two. Incorporation of microRNAs (miRNAs) and their response elements into the viral genome to alter tissue tropism and/or restrict host species range has also been explored as an attenuation strategy [6–8]. Another approach is to recode the influenza genome to include amber codons (TAG) which, under certain circumstances, can be translated as unnatural amino acids (UAAs; eg, Nε-2-azidoethyloxycarbonyl-l-lysine) [9]. The virus with these modifications can only be propagated in engineered cell lines expressing tRNACUA and UAA, yet is fully immunogenic. The error-prone nature of viral RNA-dependent RNA polymerases offers another sophisticated way to attenuate viruses via redirected evolution in sequence space. In a recent example, Coxsackie B3 and influenza viruses were recoded to include codons that are only a single nucleotide substitution away from stop codons [10]. The recoded viruses were highly sensitive to mutagenic conditions and highly attenuated in vivo while retaining their capacity for immune induction. The most advanced candidates using orthogonal viral attenuation methods incorporate specific mutations known to disrupt viral interferon suppression mechanisms, resulting in stronger type I interferon (IFN-I) responses in infected cells [11–13]. It is important to note that these concepts not only make the virus more susceptible to innate immunity and hence attenuate virulence, but also enhance their immunogenicity by providing the transient IFN-β milieu in the absence of other inflammatory cytokines that helps to enhance adaptive immunity [14, 15].
NEW APPROACHES TO ANTIGEN DESIGN
Many recent advances in vaccine development have been driven by structures of antibodies and their targets, often in complex, which inform approaches aimed at eliciting desired responses using designed immunogens [16–20] (Figure 1). In addition to advances in experimental structural biology, methods for computationally modeling and designing proteins are becoming increasingly powerful, enabling rational approaches to highly complex problems in protein engineering [21]. However, there is no single “best” solution for designing immunogens for any disease. Because different pathogens use diverse strategies to evade protective immune responses, vaccine design strategies must be tailored to our knowledge of how protective antibodies function in each disease and why particular antigens may or may not elicit them.
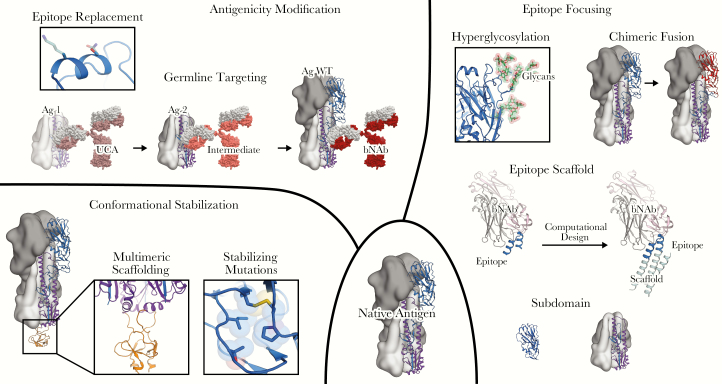
Figure 1.
Structural manipulations of vaccine antigens. Design starts from a native antigen (lower center), which may not have the desired antigenic and biophysical profile. Conformational stabilization (lower left) can be achieved by genetic fusion to a multimeric scaffold protein, or by mutations that stabilize a desired conformational state. Epitope focusing (right) can be achieved by diverse strategies, including subdomain-based engineering, hyperglycosylation, epitope scaffolding, and the design of chimeric antigens. Antigenicity can be more directly modified for other desired purposes (upper left). For example, antigens can be engineered to selectively engage B cells expressing germline precursors to a desired broadly neutralizing antibody. Abbreviations: Ag, antigen; bNAb, broadly neutralizing antibody; UCA, unmutated common ancestor; WT, wild type.
Many antigens adopt multiple conformational states throughout the life cycle of the pathogen, either as part of their natural function or as an explicit strategy for immune evasion. For such antigens, stabilization of a desired antigenic state is a conceptually straightforward approach for improving the quality of antibody responses. A common problem encountered when expressing antigens outside their native context is that they fail to adopt the correct quaternary structure (eg, the ectodomains of enveloped virus glycoproteins expressed without their native transmembrane domains). Genetic fusion to multimeric protein scaffolds with higher stability has proven to be a general method for quaternary stabilization of such antigens. This is usually performed using protein complexes with symmetries matching that of the antigen, and can range from small scaffolds such as the foldon trimerization domain from T4 fibritin [22, 23] to larger multimers such as ferritin [24] that display multiple copies of the oligomeric antigen. While often required for the production of stable antigens, heterologous multimerization domains generally do not address sources of instability unrelated to quaternary structure. For example, many viral glycoproteins that mediate host-virus membrane fusion are present on virions in a metastable prefusion state that transitions through transient fusion intermediates to an energetically more stable postfusion state [25]. This type of conformational heterogeneity can be problematic if the metastable prefusion state is desired antigenically [22, 26, 27], and it can be exaggerated when antigens are recombinantly expressed and purified in isolation. With detailed knowledge of desired and undesired structural states, more precise techniques in protein engineering and design can be used to favor or disfavor specific conformations of target proteins. Common strategies include designing disulfide bonds to immobilize linked domains [28–32], improved hydrophobic packing to stabilize a particular conformational state [22, 33], proline mutations to constrain conformational flexibility, and proline/glycine mutations to discourage helix formation [32, 34, 35]. In one well-characterized example, the Fab of a potent, prefusion-specific neutralizing antibody against the fusion glycoprotein of respiratory syncytial virus (RSV F) was used in combination with a foldon trimerization domain at the C terminus to maintain F in its metastable prefusion conformation and prevent irreversible transition to the postfusion state. After solving the structure of the prefusion state in complex with the Fab [36], mutations that introduced a disulfide bond and cavity-filling hydrophobic side chains were made to F to stabilize the protein in this conformation. The designed prefusion-stabilized antigen, DS-Cav1, elicits much stronger neutralizing antibody responses than postfusion F [22] and is currently being evaluated in a phase I clinical trial (NCT03049488). Additional prefusion-stabilized RSV F antigens have been generated by introducing proline mutations in between discontinuous helices that form a continuous helix upon transition to the postfusion structure [34], and this approach has also been used to stabilize other viral glycoproteins such as the spike (S) proteins of Middle East respiratory syndrome coronavirus (MERS CoV), severe acute respiratory syndrome (SARS) CoV, and human CoV HKU-1 [35]. For HIV, combining disulfide and proline mutations has been shown to stabilize soluble versions of the Env glycoprotein in trimeric, native-like, prefusion conformations [32]. Extensions of this design strategy have been generalized across the Env glycoproteins of many different strains and have become an invaluable tool for vaccine development and the study of humoral responses against the virus [37–39]. These stabilization strategies have proven immensely useful for generating conformationally homogeneous, antigenically intact molecules; continued development and application of these methods will be applicable in many vaccine design contexts.
Another major challenge in antigen design is the development of strategies to elicit broadly protective responses against antigenically variable viruses such as influenza or HIV. Many approaches are currently being evaluated in preclinical research, and a few are beginning to enter clinical trials. One intuitively appealing approach is to focus immune responses on epitopes that are conserved across multiple virus strains, subtypes, clades, or species. This concept is supported by the observation that bNAbs against many pathogens target precisely these epitopes [40]. However, design strategies must overcome the presence of less conserved, distracting epitopes that are either strain-specific or incapable of eliciting protective responses [41, 42]. Inspired by a widespread viral immune evasion mechanism, physically masking antigen surfaces (such as the immunodominant head domain of influenza HA) using hyperglycosylation has been shown to redirect antibody responses to desired epitopes in some cases [43–45].
Subdomain-based approaches that remove elements containing undesired epitopes from the antigen entirely have also been widely employed to focus immune responses. In some cases, simple elimination of undesired portions of the antigen, usually along domain boundaries, can be used to produce minimal antigens that are both structurally and antigenically intact [46–49]. In other cases more complex manipulations such as novel loop connections and stabilizing mutations have been required; “headless” or “stabilized stem” HAs lacking the immunodominant head region are a successful example that have been shown to induce responses to the immunologically subdominant stem with broader cross-reactivity [29, 33, 50–52]. Similarly, radical deletions, loop connections, and stabilizing mutations were used to produce a minimal version of the CD4 binding site of HIV Env called eOD [53] that has proven to be a robust platform for further engineering, as discussed below. A more extreme variation on the subdomain approach known as epitope-focused vaccine design has been used to generate antigens that present only specific, well-defined epitopes known to be targeted by neutralizing antibodies. Several examples have demonstrated that computational protein design can be used to incorporate such epitopes into unrelated scaffold proteins with high structural fidelity. These “epitope scaffolds” bind to their target antibodies with high affinity and, upon immunization, induce antibodies that bind to the native antigen and in some cases neutralize the pathogen [54–58]. In another flavor of epitope-focused vaccine design, conserved linear epitopes targeted by neutralizing antibodies, such as the fusion peptide of HIV Env, can be presented as antigens on multivalent display platforms in a structure-agnostic manner [59, 60]. While epitope-focused vaccine design offers a conceptual framework for targeting a desired epitope with high precision, inducing high levels of neutralizing antibodies with minimal, epitope-based antigens has been challenging and there is a risk of immune evasion as this approach typically targets a single vulnerable epitope. Combining epitope-focused design with continued exploration and optimization of multivalent presentation platforms, discussed below, could help address these issues.
The controlled use of antigenic variation is another strategy for highlighting a conserved region for immune responses. For example, heterologous prime-boost strategies have been used to administer antigens that significantly vary in off-target regions with undesired epitopes while keeping the desired epitope invariant across all antigens to maximize the response against that region [59, 61–63]. This process can be enhanced by designing antigens optimized for this purpose, as exemplified by chimeric HA antigens in which the head region of HA from a seasonally circulating virus is replaced with the corresponding region of exotic subtypes that most humans have never encountered [63]. Through a combination of low-level preexisting immunity against conserved stem epitopes and sequential administration of antigens with distinct head regions for which no preexisting immunity exists, this strategy aims to maximize stem-directed responses and is currently being evaluated in clinical trials (NCT03300050).
While targeting conserved epitopes is a promising strategy for eliciting broadly protective responses, differences in surrounding motifs or epitope conformations between strains can impede cross-reactive immunity. In influenza, this is particularly true for the receptor-binding site (RBS), as many mutations near the RBS significantly limit the breadth of all known cross-reactive antibodies against this region [64–66]. Stem-directed responses are inherently broader due to the higher conservation of this region of HA, yet it remains challenging to elicit strong anti-stem responses that span group 1 and group 2 influenza A viruses, or ideally protect against both influenza A and B viruses. To achieve universal protection from such a diverse collection of viruses, epitope-focused strategies must be optimized to cater towards maximal breadth.
A different approach to inducing bNAbs by immunization, referred to as germline-targeting or lineage-based vaccine design, has been inspired by the ever-growing collection of bNAbs against HIV and influenza. The identification of structural and genetic constraints on bNAb development that are commonly found across multiple individuals, enabled by high-throughput antibody sequencing technologies, provides a system for classifying bNAbs and has made the idea of targeting particular classes a viable option for vaccine design. The central concept of lineage-based vaccine design is that designing antigens that strongly bind to inferred unmutated precursors of known classes of bNAbs has the potential to drive direct elicitation of bNAb responses [67, 68]. This strategy is most established in the context of HIV vaccine research, where it is primarily (though not only [61, 62]) focused on eliciting bNAbs against the CD4 binding site of HIV Env [49, 62, 69–71]. A leading example is the eOD-GT8 antigen, which has been extensively engineered to have high affinity to the unmutated common ancestor of VRC01-class bNAbs. eOD-GT8, when displayed in multivalent fashion on a self-assembling protein scaffold, has successfully activated target precursor B cells in knock-in mouse models [62, 72] and has recently advanced into proof-of-concept studies in humans (NCT03547245). There is significant room to explore the application of this strategy to viruses other than HIV, and particularly for influenza where there is growing knowledge of multidonor classes of bNAbs that could be targeted [73–76].
While lineage-based approaches may enable elicitation of very specific classes of antibodies, designed antigens can be tailored to bias the specificity of antibody response patterns at a polyclonal level as well. In the case of complex antigens with a variety of epitopes that could mediate neutralization or protection, approaches that combine multiple sequences from diverse strains into a consensus or genetic-mosaic sequence by presenting epitopes from all included strains have the potential to induce broad responses. For influenza, this concept has been evaluated using “computationally optimized broadly reactive antigens” [77, 78]. The design strategy focuses on the head region of HA with the intention of maximizing the breadth of hemagglutination inhibition (HAI) titers, and has been successful at improving cross-reactivity within individual subtypes of influenza. Similar approaches have been used in HIV vaccine development to produce Env gp140 trimers that are composed of a variety of sequences from diverse [79], and a polyclonal-centered strategy has been recently combined with lineage-based vaccine design [71]. In that case, germline-targeting mutations were added to two distinct epitopes on a SOSIP trimer to activate the unmutated common ancestors of two classes of bNAbs, a strategy that was validated in knock-in mice.
Looking ahead, the coordinated design and coadministration of multiple antigens could be a promising approach to increasing breadth against viruses exhibiting extreme structural and antigenic diversity, such as influenza and HIV. In particular, designing antigens that are specifically intended to be codelivered or multimerically displayed may be a promising avenue for future investigation.
ANTIGEN DISPLAY AND DELIVERY PLATFORMS
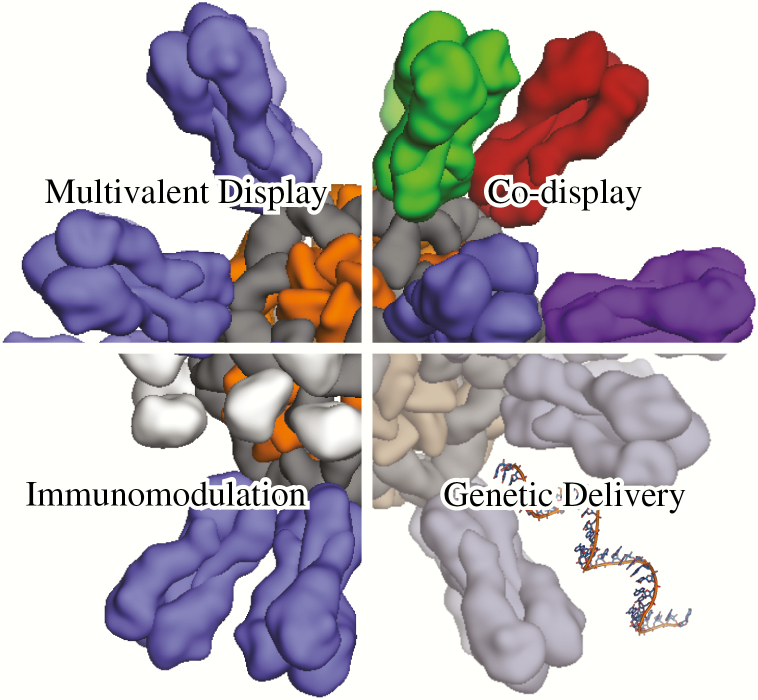
The manner in which antigens are presented to the immune system profoundly influences the ensuing response [80]. Recent technological advances have enabled new approaches to antigen display and delivery that are increasing the potency and durability of vaccine responses and optimizing the balance between humoral and cellular immunity (Figure 2). We focus here on two such technologies: self-assembling proteins as multivalent antigen display platforms and genetic immunization via mRNA vaccines.
Figure 2.
Antigen display and delivery methods. Antigen presentation on self- assembling nanoparticles (either natural or designed) has proven to be a robust method for enhancing humoral responses (upper left). Codisplay of related antigens (antigenic variants, colored uniquely) on individual nanoparticles offers a customizable strategy for altering the profile of the vaccine-induced antibody response (upper right). Immunomodulation can be achieved by colocalizing antigens and immunomodulatory molecules (silver blobs) on the same nanoparticle (lower left). Genetic delivery involves administration of a nucleic acid to produce immunogens in situ, offering a versatile strategy for vaccine production that may also improve immunogenicity (lower right).
Multivalent antigen display platforms induce potent antibody responses by presenting repetitive arrays of antigen, which drives robust activation of B cells through B cell receptor (BCR) clustering [80–82]. Much of the work in this area has focused on the repetitive display of haptens and peptide antigens using a number of different platforms, including classic carrier proteins (eg, keyhole limpet hemocyanin [83]), virus-like particles [84], liposomes [85], and synthetic polymers [86]. Several general principles have emerged from this work, including the observation that higher epitope density correlates with more potent responses [87] and that the physical size of the immunogen or formulation affects antigen biodistribution in vivo [88], although in ways that may be specific to each material or platform. A significant limitation of early display platforms was their inability to scaffold complex antigens such as the intact ectodomains of viral glycoproteins. The discovery that nonviral self-assembling proteins such as ferritin and lumazine synthase can form monodisperse immunogens that present large, complex antigens has recently moved the field beyond this limitation [24, 69]. A small set of such proteins has been used to present a wide variety of antigens, including those derived from influenza [24, 33], HIV [49, 70, 72, 89–92], and Epstein-Barr virus [48]. In these studies, multivalent display consistently and robustly enhanced the potency of the antigen-specific response. In some cases, self-assembling scaffolds were successfully combined with antigens designed to focus the response on specific epitopes and therefore improve neutralization potency or breadth [33, 44, 48]. A first-in-class clinical trial of a self-assembling nanoparticle-based influenza vaccine has recently been started (NCT03186781).
Self-assembling proteins have several advantages over other multivalent antigen presentation platforms [82]. Seamless integration of the antigen and self-assembling scaffold via genetic fusion—an approach unique to self-assembling protein scaffolds—facilitates immunogen production because, for recombinant antigens, production of both the antigen and the scaffold can be accomplished in a single molecule. Alternatively, simple, robust, and gentle protein-based conjugation systems such as SpyCatcher/SpyTag [93] enable rapid prototyping of many different antigens on self-assembling protein scaffolds in an approach that has been dubbed “plug-and-display” [94, 95]. Because most functional proteins fold and assemble to well-defined and unique low energy states, self-assembling protein immunogens tend to be highly ordered and monodisperse, making them easier to characterize for quality and potency than more heterogeneous materials. As noted above, self-assembling scaffolds can also stabilize antigens in their native oligomeric state. Furthermore, the fact that self-assembling proteins are genetically encoded opens up the possibility of deploying potent nanoparticle immunogens via genetic immunization (eg, viral vectors or nucleic acid vaccines; see below).
However, unlike polymeric systems or liposomes, a potential liability of using self-assembling proteins as scaffolds is that the resultant immunogens tend to induce antibody responses against the scaffolds themselves [24, 96]. While this has generally not prevented the elicitation of potent antigen-specific responses, it remains to be seen whether engineering mechanisms to dampen or avoid anti-scaffold responses into these systems [97, 98] might further improve antigen-specific, protective responses.
Looking ahead, the limited number of existing self-assembling protein scaffolds and the fact that their structural properties are essentially fixed severely constrains the design space that may be explored. Recently, new computational methods have been developed for designing novel self-assembling proteins with atomic-level accuracy [99–103]. These methods have enabled the generation of new protein scaffolds in which structural features such as size, shape, number of subunits, and the location and orientation of displayed antigen can be systematically and precisely controlled. We expect such studies to improve our understanding of the structural correlates of immunogenicity and generate potent new vaccine candidates. The first examples of the use of these computationally designed self-assembling proteins as vaccine scaffolds have recently been reported [95, 104]. In both cases, highly ordered, monodisperse immunogens notable for their extreme stability were found to induce potent immune responses.
An emerging concept in antigen presentation and delivery that exploits several advantages of recombinant approaches is the codisplay of multiple antigenic variants on a single continuous surface [92, 105]. For antigenically variable targets such as influenza HA and HIV Env, codisplay could provide a mechanism for improving the breadth of vaccine responses by focusing them on conserved epitopes known to be targeted by bNAbs. The hypothesis is that antigen presentation in a heterogeneous array would alter the hierarchy of B-cell activation by providing a selective advantage to cross-reactive B cells targeting conserved epitopes through nanoparticle-mediated BCR clustering and potentially bivalent BCR ligation. In contrast, B cells expressing BCRs with narrow, strain-specific binding would not receive the advantages offered by the particulate antigen and would be relatively poorly activated. While this type of “mosaic antigen array” does not exist in nature, it can be generated on the surface of self-assembling protein scaffolds, as reported in a recent study in which multiple monomeric HA receptor binding domains (RBDs) were displayed on ferritin to generate a series of antigenically variable nanoparticle immunogens [106]. Intriguingly, the breadth of the immune response induced by these immunogens correlated with the complexity of the mosaic antigen array (ie, the number of antigenically distinct RBDs displayed on the nanoparticle immunogen), and was superior to that induced by cocktails of nanoparticles individually displaying each of the RBDs. It will be interesting to determine whether this concept is applicable to other antigenically diverse, “hard-to-hit” targets like HIV. Given suitable scaffolds, codisplay could also be used to present heteromeric protein complexes (eg, the human cytomegalovirus glycoprotein pentamer) or multiple proteins that associate on pathogen surfaces (eg, herpesvirus gB and gH/gL). In these contexts, codisplay could simplify vaccine manufacturing and characterization in addition to offering the enhanced potency that is typically obtained through multivalent display.
Immunogens targeting certain types of cells induce stronger immune responses than nontargeted counterparts [88, 107]. New approaches to modulating antigen trafficking, uptake, or transport by manipulating size, surface chemistry, or through accessory molecules could enhance antigen availability in lymph nodes for cognate immune cells (ie, B and T cells). Accessory molecules could also be used to potentiate immune responses by exploiting immune signaling pathways. For example, complement C3d, a classical opsonin, is recognized by complement receptors 2 (CR2, CD21) expressed on B cells and follicular dendritic cells. On B cells, CR2 forms a trimolecular complex with CD19 and CD81 (TAPA-1), and signaling through this complex amplifies and compensates for low levels of BCR signaling [108, 109]. Therefore, targeting this trimolecular complex may act as a molecular adjuvant [110]. The key for successful targeting/adjuvanting is to colocalize the targeting/adjuvanting molecules on the same surface where the vaccine antigens are located. This should provide maximal enhancement of the antigen-specific immune response and mitigate potential off-target effects [111]. While the recent development of highly customizable nanoparticle platforms capable of codisplaying multiple molecules has enabled new approaches to modulating targeting or immune signaling, these concepts are still in their infancy and need to be rigorously tested. Nevertheless, in the near future, we anticipate that it will become possible to reconstitute immunological synapses with tailored functions by codisplaying a variety of costimulatory, inhibitory, regulatory, accessory, and immunomodulatory molecules in addition to vaccine antigen all on the same surface to maximize or fine-tune immune stimulation by design.
Immunization with mRNA for in situ translation has many potential advantages as a method of antigen delivery. Like other approaches to genetic immunization, such as DNA vaccines [112] and viral vectors [113], the production of antigen from a delivered mRNA mimics the replication of infectious agents in three key ways. First, antigen is provided continuously for a period of time, in contrast to the bolus of antigen delivered by protein-based subunit vaccines. Several recent studies have demonstrated that prolonged antigen exposure can improve the magnitude and quality of vaccine-induced responses [114–117]. Second, antigen is synthesized, posttranslationally modified, processed, and presented to B cells and T cells the same way it is during natural infection, including antigen presentation on MHC class I, which results in stronger cellular responses than are typically achieved with subunit vaccines [118]. Third, various types of antigens can be expressed, including cytoplasmic, transmembrane, or secreted proteins as well as engineered antigens that are highly immunogenic yet incompatible with the manufacture of live or inactivated whole-pathogen vaccines because they do not support viral replication (eg, prefusion-stabilized RSV F [119]). A major advantage that mRNA vaccines have over other genetic immunization platforms is that there is no risk of infection or genome integration, resulting in a better safety profile. Manufacturing can be done synthetically, eliminating the need for cell culture and potential contamination by animal products. Furthermore, manufacturing is rapid and for the most part independent of sequence, making this vaccine modality particularly well-suited to the development of rapid response platforms to protect against periodic outbreaks or pandemics. Finally, unlike viral vectors [120], antivector immunity tends not to be an issue with existing delivery platforms, which could help enable repeated dosing.
While early approaches to genetic immunization focused on DNA vaccines and viral vectors due to concerns about mRNA instability and immunogenicity, recent technological breakthroughs have made mRNA a plausible commercial vaccine modality. Decades of investigation into the basic biology of mRNA have defined sequence elements (eg, 5′ and 3′ UTRs [121, 122]) and optimization strategies (eg, codon optimization [123]) that enable modulation of intracellular stability and translational efficiency. Improvements in potency have also been obtained by minimizing the intrinsic immunogenicity of nonendogenous mRNA, which can trigger innate immune responses that suppress translation of the encoded antigen. For example, the related discoveries that mRNAs containing modified nucleosides such as pseudouridine are weaker activators of RNA-sensing pattern recognition receptors [124] and that they therefore yield higher levels of translated protein for a longer period of time in cells and in vivo [125] were major steps forward. The subsequent observation that mRNA preparations free of double-stranded RNA contaminants further reduce innate immune activation and increase translation efficiency [126] has made high-level expression of delivered antigen without undesired immune activation possible. Antigen-encoding viral replicons are an alternative approach to obtaining high-level antigen expression [127]. In addition to antigen, the self-amplifying mRNA encodes a viral replicase that makes copies of the mRNA vector itself [128], leading to robust antigen production and potent immune responses from very small doses of mRNA. One potential drawback of self-amplifying mRNAs is that they intrinsically activate innate immunity, which, while likely contributing to their potency, eliminates control over this important aspect of mRNA vaccine design and limits the duration of antigen production.
Delivery systems that can transport nucleic acids into the cytoplasm are required for mRNA vaccines to be effective. While ex vivo loading of dendritic cells is being pursued for cancer vaccines [129], direct immunization with formulated mRNA will likely be the only viable approach for infectious disease in the near future. Although substantial progress has been made, biologics delivery generally remains an unsolved problem and many strategies are being evaluated preclinically. To date, there are at least 3 delivery platforms under clinical investigation for mRNA vaccines, including uncomplexed mRNAs mixed with mRNA-protamine complexes that stimulate innate immunity through TLR7 signaling [130, 131], mRNA-decorated cationic lipid nanoemulsions [132], and mRNA-encapsidating lipid nanoparticles [118, 133–135]. The latter systems were pioneered for intravenous delivery of siRNA therapeutics, which results primarily in delivery to the liver. However, intradermal, intramuscular, and subcutaneous administration of mRNA vaccines formulated in lipid nanoparticles results in robust local expression at the injection site [136]. Encouragingly, mRNA vaccines have provided impressive results in preclinical animal models.
Data are just beginning to emerge from phase I clinical trials of mRNA vaccines for infectious diseases. A clinical trial of an mRNA/mRNA-protamine complex vaccine encoding the rabies virus glycoprotein induced neutralizing antibodies and virus-specific T cells and demonstrated an acceptable safety profile [137]. However, the potency and durability of the immune response were lower than in preclinical animal models and required the use of needle-free injection devices. Similarly, an mRNA vaccine with modified nucleosides formulated in lipid nanoparticles encoding influenza HA induced HAI titers in humans that were above the protective threshold and similar to conventional, commercially available influenza vaccines, yet below the levels induced in animal models [133]. These early data, while encouraging, suggest that additional improvements to mRNA vaccines will be required to realize the full potential of this promising delivery platform. Better delivery systems and innovative approaches to triggering innate immunity without sacrificing antigen expression are two areas where substantial improvements could be made. Improving the design of the encoded antigens—for example, using the self-assembling nanoparticle immunogens discussed earlier—is also likely to enhance performance.
In addition to these technologies, the goal of single-injection subunit vaccines continues to drive the exploration of controlled release formulations based on synthetic or biological polymers [117, 138, 139]. For more detail, the reader is referred to excellent recent reviews [86, 88].
CONCLUDING REMARKS—TOWARDS A UNIVERSAL INFLUENZA VACCINE
One hundred years after the 1918 Spanish flu pandemic, influenza remains a massive burden to public health. In recent years, methods developed for immunogen design have resulted in significant preclinical progress towards a universal influenza vaccine. Even though such a vaccine has not yet been deployed, many recently developed approaches could nonetheless provide substantial improvements in breadth of protection over existing licensed vaccines. In aiming for universal protection, it will be vital to determine the best possible ways to redirect immune responses towards conserved regions of influenza antigens, including the RBS and stem of HA and possibly more unexplored regions on the neuraminidase and M2 proteins. New approaches to immunogen design, enabled by the development of new technologies, will be needed to achieve the breadth of responses against these conserved regions required to provide universal protection. Better knowledge of convergent antibody lineages and additional experience designing antigens that target them to give rise to bNAbs is expected to improve current approaches. Rigorous exploration of new methods for antigen display using self-assembling scaffolds has the potential to further amplify responses targeting these conserved regions, and the ability to design novel self-assembling protein platforms should make it possible to tailor them to maximize the potency and durability of humoral responses. These challenges and opportunities represent a novel space to explore in conjunction with antigen design and the development of delivery technologies to induce CD8+ T cell responses or mucosal immunity in addition to targeted antibody responses. With respect to influenza in particular, it will be essential to understand how preexisting immunity influences responses to novel immunogens, and how the vaccine design process can best accommodate these effects.
The novel strategies and technologies developed in response to the challenges presented by a universal influenza vaccine will be applicable to the structure-based design of vaccines for a wide range of pathogens. It is an incredibly exciting time for the field of vaccine development, and we predict a structure-based, design-driven renaissance that recalls or even surpasses the original golden age of vaccines.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgment. The authors thank B. Hartman (Vaccine Research Center) for help with manuscript preparation.
Financial support. This work was supported by the Intramural Research Programs of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health (M. K.); and by a generous gift from the Open Philanthropy Project (N. P. K. and D. E.).
Potential conflicts of interest. M. K. is a named inventor of several patents and patent applications describing vaccine design and vaccine display technologies filed by US Department of Health and Human Services. D. E. and N. P. K. are named inventors of several patents and patent applications describing designed protein nanoparticle-based vaccine delivery technologies filed by University of Washington. N. P. K. is an academic co-founder, shareholder, and chair of the scientific advisory board of Icosavax, Inc. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- (50 selected references, with numbers in Text maintained. The full list of references is in the Supplementary Material, accessible online) [Google Scholar]
- 1. Maassab HF. Adaptation and growth characteristics of influenza virus at 25 degrees c. Nature 1967; 213:612–4. [DOI] [PubMed] [Google Scholar]
- 2. Stech J, Garn H, Wegmann M, Wagner R, Klenk HD. A new approach to an influenza live vaccine: modification of the cleavage site of hemagglutinin. Nat Med 2005; 11:683–9. [DOI] [PubMed] [Google Scholar]
- 3. Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science 2008; 320:1784–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mueller S, Coleman JR, Papamichail D, et al. . Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol 2010; 28:723–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tulloch F, Atkinson NJ, Evans DJ, Ryan MD, Simmonds P. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. Elife 2014; 3:e04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe 2008; 4:239–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Perez JT, Pham AM, Lorini MH, Chua MA, Steel J, tenOever BR. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat Biotechnol 2009; 27:572–6. [DOI] [PubMed] [Google Scholar]
- 8. Langlois RA, Albrecht RA, Kimble B, et al. . MicroRNA-based strategy to mitigate the risk of gain-of-function influenza studies. Nat Biotechnol 2013; 31:844–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Si L, Xu H, Zhou X, et al. . Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science 2016; 354:1170–3. [DOI] [PubMed] [Google Scholar]
- 10. Moratorio G, Henningsson R, Barbezange C, et al. . Attenuation of RNA viruses by redirecting their evolution in sequence space. Nat Microbiol 2017; 2:17088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Du Y, Xin L, Shi Y, et al. . Genome-wide identification of interferon-sensitive mutations enables influenza vaccine design. Science 2018; 359:290–6. [DOI] [PubMed] [Google Scholar]
- 12. Talon J, Salvatore M, O’Neill RE, et al. . Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc Natl Acad Sci U S A 2000; 97:4309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mössler C, Groiss F, Wolzt M, Wolschek M, Seipelt J, Muster T. Phase I/II trial of a replication-deficient trivalent influenza virus vaccine lacking NS1. Vaccine 2013; 31:6194–200. [DOI] [PubMed] [Google Scholar]
- 14. Prchal M, Pilz A, Simma O, et al. . Type I interferons as mediators of immune adjuvants for T- and B cell-dependent acquired immunity. Vaccine 2009; 27(Suppl 6):G17–20. [DOI] [PubMed] [Google Scholar]
- 15. Trinchieri G. Type I interferon: friend or foe? J Exp Med 2010; 207:2053–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kwong PD. What are the most powerful immunogen design vaccine strategies? A structural biologist’s perspective. Cold Spring Harb Perspect Biol 2017; 9:pii: a029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rappuoli R, Bottomley MJ, D’Oro U, Finco O, De Gregorio E. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J Exp Med 2016; 213:469–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burton DR. Antibodies, viruses and vaccines. Nat Rev Immunol 2002; 2:706–13. [DOI] [PubMed] [Google Scholar]
- 19. Burton DR. What are the most powerful immunogen design vaccine strategies? reverse vaccinology 2.0 shows great promise. Cold Spring Harb Perspect Biol 2017; 9:pii: a030262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Graham BS. Advances in antiviral vaccine development. Immunol Rev 2013; 255:230–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang PS, Boyken SE, Baker D. The coming of age of de novo protein design. Nature 2016; 537:320–7. [DOI] [PubMed] [Google Scholar]
- 22. McLellan JS, Chen M, Joyce MG, et al. . Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 2013; 342:592–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 2004; 303:1866–70. [DOI] [PubMed] [Google Scholar]
- 24. Kanekiyo M, Wei CJ, Yassine HM, et al. . Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 2013; 499:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harrison SC. Viral membrane fusion. Virology 2015; 479-480:498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ngwuta JO, Chen M, Modjarrad K, et al. . Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 2015; 7:309ra162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanders RW, van Gils MJ, Derking R, et al. . HIV-1 Vaccines. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 2015; 349:aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang P, Gorman J, Geng H, et al. . Interdomain stabilization impairs CD4 binding and improves immunogenicity of the HIV-1 envelope trimer. Cell Host Microbe 2018; 23:832–44.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Impagliazzo A, Milder F, Kuipers H, et al. . A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015; 349:1301–6. [DOI] [PubMed] [Google Scholar]
- 30. Lee PS, Zhu X, Yu W, Wilson IA. Design and structure of an engineered disulfide-stabilized influenza virus hemagglutinin trimer. J Virol 2015; 89:7417–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Godley L, Pfeifer J, Steinhauer D, et al. . Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 1992; 68:635–45. [DOI] [PubMed] [Google Scholar]
- 32. Sanders RW, Vesanen M, Schuelke N, et al. . Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 2002; 76:8875–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yassine HM, Boyington JC, McTamney PM, et al. . Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 2015; 21:1065–70. [DOI] [PubMed] [Google Scholar]
- 34. Krarup A, Truan D, Furmanova-Hollenstein P, et al. . A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun 2015; 6:8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pallesen J, Wang N, Corbett KS, et al. . Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 2017; 114: E7348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McLellan JS, Chen M, Leung S, et al. . Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 2013; 340:1113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sanders RW, Moore JP. Native-like ENV trimers as a platform for HIV-1 vaccine design. Immunol Rev 2017; 275:161–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Joyce MG, Georgiev IS, Yang Y, et al. . Soluble prefusion closed DS-SOSIP.664-Env trimers of diverse HIV-1 strains. Cell Rep 2017; 21:2992–3002. [DOI] [PubMed] [Google Scholar]
- 39. Rutten L, Lai YT, Blokland S, et al. . A universal approach to optimize the folding and stability of prefusion-closed HIV-1 envelope trimers. Cell Rep 2018; 23:584–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 2012; 337:183–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ward AB, Wilson IA. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol Rev 2017; 275:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krammer F, Smith GJD, Fouchier RAM, et al. . Influenza. Nat Rev Dis Primers 2018; 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eggink D, Goff PH, Palese P. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J Virol 2014; 88:699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Duan H, Chen X, Boyington JC, et al. . Glycan masking focuses immune responses to the HIV-1 CD4-binding site and enhances elicitation of VRC01-class precursor antibodies. Immunity 2018; 49:301–11.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chandramouli S, Ciferri C, Nikitin PA, et al. . Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody. Nat Commun 2015; 6:8176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Song L, Xiong D, Kang X, et al. . An avian influenza A (H7N9) virus vaccine candidate based on the fusion protein of hemagglutinin globular head and Salmonella typhimurium flagellin. BMC Biotechnol 2015; 15:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McGuire AT, Hoot S, Dreyer AM, et al. . Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med 2013; 210:655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kanekiyo M, Bu W, Joyce MG, et al. . Rational design of an Epstein-Barr virus vaccine targeting the receptor-binding site. Cell 2015; 162:1090–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McGuire AT, Gray MD, Dosenovic P, et al. . Specifically modified ENV immunogens activate B-cell precursors of broadly neutralizing HIV-1 antibodies in transgenic mice. Nat Commun 2016; 7:10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bommakanti G, Citron MP, Hepler RW, et al. . Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci U S A 2010; 107:13701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.