Abstract
Potentially pandemic strains of influenza pose an undeniable threat to human populations. Therefore, it is essential to develop better strategies to enhance vaccine design and predict parameters that identify susceptible humans. CD4 T cells are a central component of protective immunity to influenza, delivering direct effector function and potentiating responses of other lymphoid cells. Humans have highly diverse influenza-specific CD4 T-cell populations that vary in stimulation history, specificity, and functionality. These complexities constitute a formidable obstacle to predicting immune responses to pandemic strains of influenza and derivation of optimal vaccine strategies. We suggest that more precise efforts to identify and enumerate both the positive and negative contributors of immunity in the CD4 T-cell compartment will aid in both predicting susceptible hosts and in development of vaccination strategies that will poise most human subjects to respond to pandemic influenza strains with protective immune responses.
Keywords: influenza, CD4 T cells, pandemic, vaccine
THE SPECIFICITY OF HUMAN CD4 T CELLS TO INFLUENZA VIRUS
It has become increasingly clear that human CD4 T-cell immunity to influenza has broad viral antigen specificity. In some countries where there are no formal policies for vaccination, immunity is generated primarily by influenza infection. In contrast, within North America, many countries in Europe, and the Western Pacific, where vaccination is recommended (reviewed in [1]), immunity is established by both vaccination and infection. Influenza-specific CD4 T cells have been quantified through approaches such as HLA-class II tetramer staining [2, 3], intracellular cytokine staining [4, 5], cytokine enzyme-linked immunospot (ELISPOTS) [6, 7], or surveyed using epitopes selected with predictive algorithms [8]. Our laboratory has used cytokine ELISPOTS and large peptide libraries to assess the influenza viral protein specificity directly ex vivo in an unbiased and comprehensive manner [9–13], feasible because of the relative small genome size of influenza virus. Collectively, these studies have revealed that human CD4 T cells in circulation are highly diverse and recognize epitopes derived from conserved internal influenza virion proteins such as nucleoprotein (NP) and matrix (M1), as well as the more genetically variable hemagglutinin (HA) and neuraminidase (NA) proteins. Our estimate, based on analyses of a relatively highly vaccinated US population [9], is that influenza A specific CD4 T-cell abundance in circulation is approximately 0.15% of circulating CD4 T cells (range 0.02%–3.6%), when the most abundant viral specificities are summed (Figure 1). The broad specificity of influenza-specific CD4 T cells is due in part to the diversity of HLA class II molecules in humans available to present epitopes, with multiple class II “isotypes” (HLA-DR, HLA-DQ, and HLA-DP), their codominant expression, and heterozygosity in the HLA class II loci [14].
Figure 1.
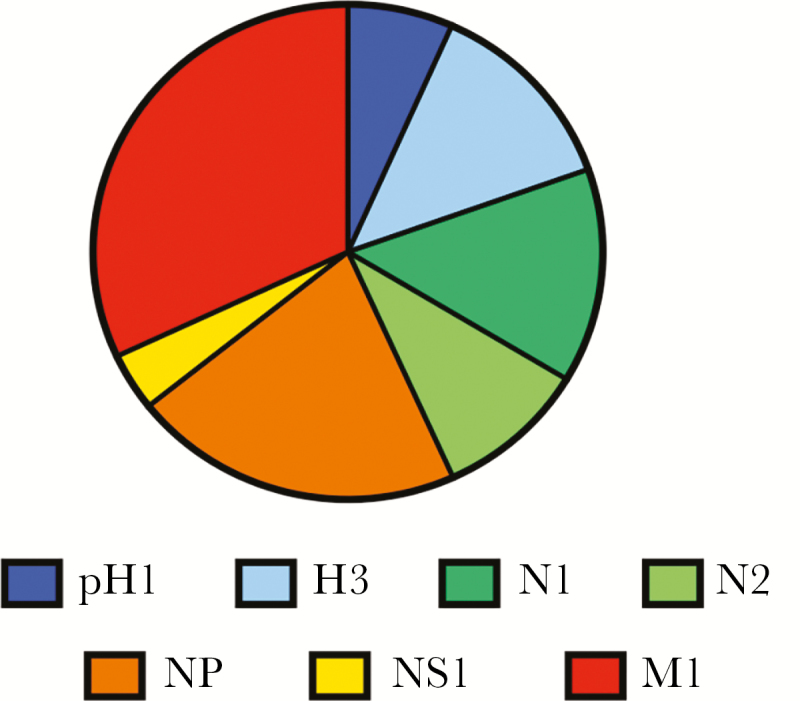
Influenza-specific CD4 T-cell frequencies and specificity in circulating PBMC of healthy adults. Influenza-specific CD4 T-cell frequencies were determined from IFN-γ cytokine ELISPOT assays of circulating PBMC from healthy donors depleted of CD8 and CD56 cells. The range of total influenza-specific CD4 T cells, when the reactivity to HA, NA, NP, NS1, and M1 were summed was 235 to 3570 IFN-γ–producing cells per million CD4 T cells [9]. Based on these frequencies, the influenza-specific CD4 T cells comprise approximately 0.15% of all circulating CD4 T cells, with a range of 0.02%–3.6%. The data on viral specificity are represented as a pie diagram where each slice of the pie depicts the relative fraction of the CD4+ T-cell response dedicated to hemagglutinin (H1, H3), neuraminidase (N1, N2), nucleoprotein (NP), nonstructural protein (NS1), and matrix protein (M1), based on IFN-γ ELISPOT values. The average frequency of IFN-γ–producing cells per million CD4 T cells for pH1 was 6.7%; H3, 12.9%; N1, 14%; N2, 9.6%; NP, 21.1%; NS1, 3.8%; and M1, 31.9%. Abbreviations: ELISPOT, enzyme-linked immunospot; HA, hemagglutinin; IFN-γ, interferon-gamma; NA, neuraminidase; PBMC, peripheral blood mononuclear cells.
The diversity and abundance of influenza-specific CD4 T cells in most humans might initially suggest that CD4 T-cell function is not a limiting factor in protective immunity to influenza. With many CD4 T cells in many humans dedicated to highly conserved internal virion proteins, one might predict that there should be sufficient cross-reactive CD4 T cells to provide protection against even novel and potentially pandemic strains of influenza. If true, then vaccine efforts should logically focus on the compartments of the adaptive response that are clearly lacking, such as B cells that produce broadly protective antibodies. However, recent data on vaccine responses, as well as a body of experimental evidence dissecting the complex functionality of CD4 T cells, argue that this assessment is almost certainly incorrect.
Critical Effector Functions of CD4 T cells
Because of the importance of neutralizing antibody in protection against influenza [15–17], there has been increasing emphasis on exploring the subset of CD4 T cells termed T follicular helper cells (Tfh) that promote the generation and maintenance of the germinal center reaction and the production of high affinity, class-switched immunoglobulin (Ig) (reviewed in [18–20]). Characterized by the expression of the chemokine receptor CXCR5 within the lymph node during an immune response to infection or vaccination, Tfh cells migrate toward the T:B border of secondary lymphoid tissue where they form conjugates to antigen-specific B cells that have internalized antigen and display antigenic peptides in association with host MHC class II molecules. These cognate interactions between antigen-specific B cells and CD4 T cells provide help for B-cell expansion and survival, and promote affinity maturation in the elicited antibody response. The resulting germinal center response is essential for long-lived B-cell memory and persistence of antibody secreting cells [21, 22]. The cognate CD4 T-cell help to B cells includes localized secretion of cytokines, such as interleukin-4 (IL-4) and IL-21, and direct cell contact-mediated costimulation via cell surface proteins expressed on B cells and CD4 T cells, such as CD40 and CD40L, respectively. Human circulating CD4 T cells corresponding to Tfh have been identified and quantified in recent studies (reviewed in [18, 23]). Subsets of these CXCR5-positive Tfh cells, distinguished by expression of chemokine receptors CXCR3 and CCR6, demonstrate a superior capacity to help B cells become antibody-producing cells in vitro via cognate interactions and their emergence into the blood after vaccination correlates with a productive antibody response [24, 25]. After resolution of the response, they are thought to represent circulating memory Tfh cells that can be quickly mobilized in future responses. The precise relationship between the CD4 T cells that emerge into the blood after vaccination versus those that persist in the draining lymph node is not clear at the moment. There are efforts to sample these sites of the germinal center response in situ, using such methods as fine-needle aspiration [26]. These approaches would permit much more precise insight into the evolution of the germinal center response over time, including a detailed understanding of the responding B-cell and CD4 T-cell receptor repertoire.
CD4 T cells also play key functions distinct from delivery of B-cell help (reviewed in [27–29]). They are important for protective immunity conveyed by CD8 T cells, enhancing their priming, expansion, and establishment of long-lived memory. Recent studies suggest that they also contribute to the development of effector CD8 T cells that have cytotoxic potential and express proteins important for homing and extravasation, all features required for protective immunity to influenza. An additional, potentially critical function of influenza-specific CD4 T-cell memory during infection is the ability to accelerate recruitment of innate effectors to the lung, blunting virus replication. A final discreet function of CD4 T cells that has been increasingly validated is cytotoxicity that has the potential to directly eliminate infected cells (reviewed in [30–33]), and as has been recently demonstrated directly ex vivo [34].
Links Between Specificity and Function of CD4 T Cells in Influenza
The diversity in epitope specificity and functional roles of CD4 T cells in protective immunity to influenza raises critical questions. Do most CD4 T cells convey similar functions? If not, does the viral antigen protein specificity of CD4 T cells influence their functionality? For example, can we predict which CD4 T cells have the ability to recruit innate effectors to the lung, mediate cytotoxicity, or provide help for antibody responses based on their viral antigen specificity? Does the functional potential of virus-specific CD4 T cells depend on the events that originally primed them — for example infection versus vaccination? Another critical issue is whether the highly inflammatory environment and abundant levels and persistence of viral proteins after infection together give the elicited CD4 T cells priority for persistence into memory. If only some subsets of memory CD4 T cells convey particular functions, which of these subsets are a limiting factor in protective responses to infection from novel potentially pandemic strains of influenza or robust responses to vaccine? Finally, do CD4 T cells of some viral protein specificities or phenotypic characteristics compete with or antagonize the effector functions of others? At present, we have only limited understanding of these issues.
Due to the clear role that neutralizing antibodies play in immunity to influenza, our laboratory has explored the role of CD4 T-cell viral protein specificity in provision of help for antibody responses to vaccines and infection. We used an experimental mouse model to address whether enhanced representation of CD4 T-cell memory could potentiate antibody responses to influenza infection. Using synthetic peptides previously identified to be coimmunodominant to generate CD4 T-cell memory independently of B-cell memory, mice were subsequently challenged with influenza virus. These studies revealed, first, that despite the abundance of CD4 T cells elicited by infection in the primary response [35–38], the presence of CD4 T-cell memory accelerated the influenza-specific B-cell response. Second, and somewhat surprisingly, these studies also revealed an inseparable linkage of specificity in the provision of CD4 help to antigen-specific B cells [39]. Mice with CD4 T-cell memory to NP demonstrated an enhanced antibody response to NP but not HA, while those with HA-specific memory CD4 T cells exhibited an accelerated antibody response to HA, a phenotype associated with lower viral titers in the lungs. We interpret this important result to mean that HA-specific memory CD4 T cells, but not other CD4 T-cell specificities, can potentiate production of early neutralizing antibodies that can diminish the yield of replicating virus.
Our studies of the human response to influenza vaccination agree with and extend this concept of linked CD4 T cell–B-cell antigen specificity to vaccination. Although licensed vaccines are quantified only for HA, inactivated vaccines also contain the membrane protein NA and internal viral proteins such as M1 and NP [40, 41]. Therefore, these vaccines will recruit CD4 T cells of many protein specificities. In 2 separate studies, we have tracked expansion of human CD4 T cells after vaccination, using cytokine ELISPOTS and large peptide pools derived from discrete viral proteins, bypassing the need for antigen processing by antigen-presenting cells in culture. When CD4 T-cell responses were tracked over time, expansion of CD4 T cells specific for peptide epitopes within HA, but not NP, were positively correlated with the neutralizing anti-HA antibody response [11, 12].
We speculate that the importance of viral antigen specificity in the ability of recruited CD4 T cells to facilitate the neutralizing antibody response is dependent on the nature of the antigen/complex internalized by HA-specific B cells and that during the course of influenza infection or vaccination, this complex does not include proteins such as NP. If true, there are 2 implications of this work. First, a portion of the influenza-specific CD4 T-cell repertoire, particularly those dedicated to internal virion proteins such as NP and polymerase, may be irrelevant to antibody production during the course of infection or vaccination. Second, CD4 T-cell help for production of the protective antibodies specific for HA and NA may cross-help each other as they cluster together in lipid rafts [42] and may have persistent interactions after infection or vaccination. These issues need to be experimentally explored.
THE CHALLENGE TO SUCCESSFUL VACCINATION AND PROTECTION FROM NOVEL PANDEMIC STRAINS OF INFLUENZA
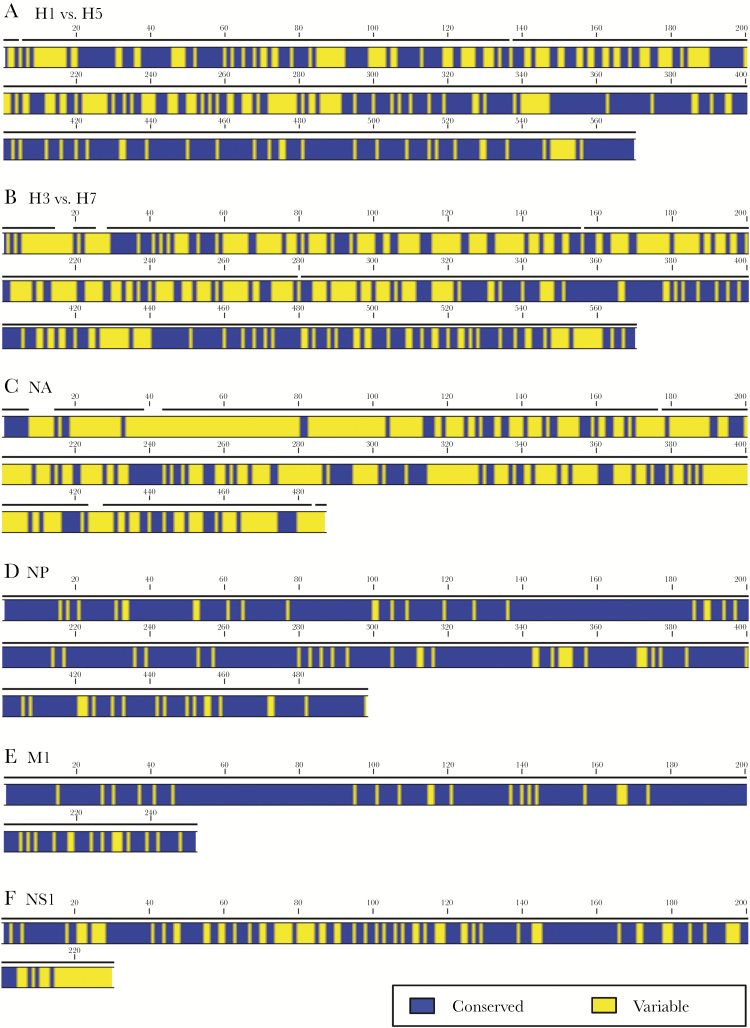
The preceding findings regarding the link between HA specificity of CD4 T cells and antibody responses are of particular importance when considering the ability of circulating CD4 memory to provide help for production against novel and potentially pandemic strains of influenza. Figure 2 shows a comparison of the degree of sequence identity among influenza proteins (HA, NA, NP, nonstructural protein [NS1], and M1) derived from seasonal strains and potentially avian pandemic strains. The HA sequence comparisons (Figure 2A and 2B) are organized into pairs with the seasonal strain aligned with the genetically closest avian strain, where blue represents sequence identity. Sequence identity among internal virion proteins for all seasonal and avian strains of influenza A is also shown. In these illustrations, NP, NA, and NS1 from 3 recently circulating seasonal strains of influenza (an H3N2 and 2 H1N1 viruses circulating within the last 30 years) were aligned with their counterparts in avian H5N1 and H7N9 viruses isolated in humans. It is clear from this comparison that M1 and NP (Figure 2D and 2E) offer far more regions of sequence identity than do HA proteins. Across avian and human viruses, the NA protein also has very dissimilar amino acid sequences (Figure 2C). Thus, if adequate help for neutralizing antibody is sought during responses to vaccination, and CD4 T cells specific for M1 and NP cannot provide it, the abundance of HA-specific CD4 T cells for avian strains will likely be limiting if drawn exclusively from the memory pool established from seasonal influenza H1N1 or H3N2 viruses. We have shown that there are H7-reactive CD4 T cells in humans never exposed to H7N9, enriched in reactivity toward HA2-derived epitopes shared by H7 and H3, suggesting that these CD4 T cells were generated by exposure to seasonal viruses. Although detectable, these H7 cross-reactive cells are of quite low abundance and, in most individuals, inadequate to promote B-cell responses with single, unadjuvanted vaccines [13].
Figure 2.
Sequence alignment of closely related influenza A virus strains. The sequences of influenza A viral proteins were aligned using CLC sequence Viewer 7.7 (Qiagen, Aarhus) to show sequence identity. HA alignments for H1N1 A/California/07/09 and H5N1 A/Vietnam/1203/04 (A) and H3N2 A/ New York/384/05 HA and H7N9 A/Anhui/1/2013 (B) are shown in pairs, based on the genetic relatedness. The sequence relatedness of NA (C) NP (D), M1 (E), and NS1 (F) are also shown where sequences from gene segments from 5 viruses (H1N1 A/Caledonia/20/99H1N1, A/California/07/09, H5N1 A/Vietnam/1203/04, H3N2 A/ New York/384/05, and H7N9 A/Anhui/1/2013) were aligned against each other. Dark gray segments represent sequence identity while light gray segments represent areas of any sequence disparity across the viral proteins that were compared. Abbreviations: HA, hemagglutinin; M1, matrix protein; NA, neuraminidase; NP, nucleoprotein; NS1, nonstructural protein.
An unusual pandemic strain emerged in 2009 that expressed a novel HA protein originally encoded within a classic swine influenza strain [43–46]. However, because this HA was an H1 protein, there was significant (approximately 80%) sequence identity with recently circulating seasonal H1N1 HAs. We [11] and others [47, 48] found that vaccination of humans with a monovalent pH1N1 vaccine successfully elicited neutralizing antibody responses, despite very little B-cell memory to draw from, suggesting that the memory CD4 T cells from the cross-reactive CD4 T-cell pool were able to facilitate a primary B-cell response to unique pH1 B-cell epitopes or to expand rare cross reactive HA-specific B cells. Our studies revealed that in these vaccinated human subjects, expansion of CD4 T cells specific for conserved HA epitopes was the best correlate of the neutralizing antibody response [11]. We suspect that the high degree of CD4 T-cell cross-reactivity between the pH1N1 and the previous seasonal H1N1 strain may be responsible for the better antibody response to pH1N1 vaccination compared to avian-influenza–derived vaccines and may also have served to temper disease progression during the initial spread of this virus. Completely novel strains of influenza of avian origin are likely to present a more substantial hurdle for protective immunity. The potential for cross-reactive CD4 T-cell recognition of these pandemic strains in protection will depend on the abundance of CD4 T cells that provide the most critical function to early protective responses in the lung and robust antibody responses.
UNKNOWNS AND THE WAY FORWARD
There have been investigators who have speculated that it is not a matter of “if” but “when” the next influenza pandemic will occur. To guard against this, efforts by many scientists have advocated for derivation of vaccination strategies that will elicit broadly neutralizing antibodies that can recognize many strains of influenza virus (reviewed in [49–53]). Clearly, this is an important priority. However, our perspective is that protective immunity to novel and pandemic strains of influenza would also be enhanced by vaccination strategies that more fully leverage all of the effector functions of CD4 T cells. There are many unknowns at present regarding the utility of CD4 T cells to promote broadly protective immunity to novel and potentially pandemic strains of influenza. Development of the most effective vaccines requires that the significant gaps in knowledge be addressed.
The central unknown is the type(s) of CD4 T-cell effector function that are most needed to enhance protective immunity or to modulate severity of disease. Insight into this issue is necessary to devise strategies to enhance their representation in susceptible hosts. The complexity of the influenza-specific repertoire in humans, distinguished by expression of cell surface molecules, transcription factors, cytokine potential, and antigen specificity, makes this goal extremely challenging. Although there are many circulating CD4 T cells specific for influenza in humans, it is not known what fraction of these participate in the response to vaccination or infection. Even for those CD4 T cells that are drawn into the response, it is likely that only some CD4 T-cell subsets are truly limiting, constituting a bottleneck in the response. Below we divide the consideration of influenza-specific CD4 T cells based on the site in which effector function is conveyed.
Based on our own and other’s data, a functional subset that is almost certainly a limiting factor in pandemic vaccine efficacy is helper CD4 T cells that recognize avian HA proteins. CD4 Tfh cells are critical in the germinal center response that produces high affinity, class-switched antibodies. As we have discussed earlier, there is good evidence that broad-based “prepandemic priming” of human subjects can poise them for a productive future antibody response to avian HA [12]. These data argue that in most human subjects, there are insufficient CD4 T cells of the correct epitope specificity to optimally enable a neutralizing antibody response to novel avian-derived viruses. Although in our own work, an intact H5 protein of a distinct clade was used to establish CD4 T-cell memory, some might have the concern that such priming would lead to a B-cell response to a second encounter with a related strain of HA that is preferentially focused on the first clade. To overcome this worry, novel HA constructs could be pursued. For example, several groups have designed vaccines composed of stalk only, HA2 domains of HA [54–56], or modified head domains that are less immunogenic [57]. These constructs could elicit CD4 T cells recognizing conserved epitopes in avian HA proteins that could be recalled upon infection or vaccination. The added benefit of these constructs is that they are also likely to elicit B cells whose surface Ig recognize conserved epitopes in the HA stalk domain. Protein constructs derived from avian HA proteins from viruses that are currently of concern, such as H5, H7, and H9, could be combined in a single vaccine. Upon peripheral vaccination, sufficient levels of circulating CD4 T cells specific for epitopes within these avian HA proteins could be established to potentiate or accelerate the antibody response to infection. Such CD4 T-cell memory could also limit the dose of vaccine or adjuvants needed for successful vaccination in the case of an emerging pandemic.
In addition to help for antibody responses, it would be extremely valuable to leverage the capabilities of other effector functions of CD4 T cells, particularly those that are delivered in the respiratory tract.
With regard to these effector functions, there is a dearth of knowledge regarding the availability and specificity of lung-localized CD4 T cells in adults and children. Using animal models of infection, it is known that intranasal infection generates a robust CD4 T-cell response detectable in both the local draining lymph node and in the respiratory tract [58–60]. Despite the magnitude of the initial response in the respiratory tract, most antigen-specific CD4 T cells in the lung decay rather quickly, typically within 3–4 weeks [59, 61]. However, it is possible that these encounters through infection establish niches of such memory CD4 T cells, either within the respiratory tract or in the periphery [62–65]. If memory CD4 T cells with lung homing potential exist in most humans, we need the capacity to quantify them, interrogate their functional capabilities (including whether they are protective or pathogenic), and then develop strategies to selectively boost the needed effector function. Activities of CD4 T cells, such as cytolysis or production of cytokines or chemokines that initiate the host antiviral response, can theoretically be enhanced.
A number of studies have suggested that vaccination strategies that initiate responses in the respiratory tract may be the most efficacious, and have included attenuated viruses, nanoparticles, and antibody-antigen conjugates (reviewed in [66, 67]). Intranasal protein-based or attenuated vaccines that are aimed at selective recruitment or amplification of lung-localized CD4 T cells with broad cross-reactivity to diverse influenza viruses might not only allow persistence of tissue resident influenza specific memory cells but might also establish cells in the periphery with lung homing preferences. Recent studies have suggested that priming of CD4 T cells via lung-draining dendritic cells leads to development of lung homing potential of the elicited CD4 cells [68] and that intranasal vaccines can seed cells in peripheral lymph nodes as well as the lung [69]. It is possible that these peripheral CD4 T cells may have preferential and early access to the respiratory tract upon influenza infection. We do not yet have precise chemokine receptor profiles and adequate knowledge of other markers expressed by circulating CD4 T cells that predict their ability to be recruited into the respiratory tract early after infection. Viral antigen specificity within the vaccines may be important as well. In addition to selection of viral protein specificities that are shared across influenza virus strains, strategies that focus on influenza proteins with abundant expression during infection, such as HA, NA, M1, NS1, and NP [70] may also be important in order to recruit a broad and diverse repertoire of CD4 T cells that will be rapidly recalled during infection. HA- and NA-specific CD4 T cells could provide help for lung-localized antibody production, while CD4 T cells specific for antigens such as M1, NP, and NS1 could provide protection via mechanisms distinct from help for antibody responses, such as cytokine production or direct cytotoxicity.
Here, we have suggested 2 distinct modes of vaccination — one that focuses on priming hosts for adequate CD4 T-cell help for production of HA-specific neutralizing antibody responses and the other for provision of localized effector function to the lung. It is important to consider whether these goals can be met by heterogeneous inactivated influenza vaccines, a single-protein vaccine, or will require vaccination strategies with complex and well-characterized viral components, perhaps administered through different routes of vaccination. With regard to systemic immunity, we would advocate for a more broad-based vaccination strategy that includes efforts to promote antibody responses to the increasingly better-understood targets and mechanisms of protective antibodies, including HA, NP, and NA. However, even for this straightforward goal, recruitment of many viral specificities into the CD4 T-cell response to vaccination may have unpredictable consequences. Our own studies have suggested that CD4 T cells specific for NP have less Tfh potential than CD4 T cells specific for HA [71] and that some CD4 T-cell viral specificities are enriched for cytotoxic potential [9] or interferon-gamma (IFN-γ) production, which can have immunosuppressive effects [72]. Also, infection can elicit regulatory CD4 T cells [73] and, in humans, these Treg may be enriched for CD4 T cells of some viral specificities [74]. Therefore, the immunogenicity of multicomponent vaccines, relative to single-component vaccines, and the functionality of the CD4 T cells elicited needs to be explicitly evaluated. Additionally, we suggest that it would be beneficial to complement these peripherally administered vaccines with strategies that specifically recruit CD4 T cells that deliver effector function into the respiratory tract. Such strategies may result in seeding broadly distributed and long-lived memory CD4 T cells that can be rapidly recruited into the lung during infection. Critical considerations for this goal likely include administration of antigen via intranasal inoculation, inclusion of influenza proteins robustly expressed early during infection, targeting to the most efficacious lung-derived antigen-presenting cell, via engagement of specific lectin receptors on antigen presenting cell [75–77]. Also of potential value would be development of viral antigen constructs, such as self-replicating RNA, that promote antigen persistence [78, 79], based on the studies of natural infection that suggest antigen persistence is an important parameter for effector T-cell function in the lung [62, 80].
To fully leverage the influenza-specific CD4 T cells in the human host for protective immunity to both seasonal and pandemic strains of influenza, the available repertoire needs to be better defined and key subsets that participate in the response to infection and vaccination quantified. There have been tremendously powerful experimental tools developed in the past decades to analyze human CD4 T cells. These include definition of a vast array of cell-surface markers that are coupled with highly specific monoclonal antibodies, a greater understanding of chemokines and chemokine receptors, more detailed understanding of transcriptional regulators of CD4 T-cell subsets, and experimental techniques such as multiparameter flow cytometry, single-cell transcriptomics, and analytical tools to handle the complex data sets that emerge from these techniques. Coupled with development of human vaccine studies and human infection challenge models, gaining insight into the CD4 T-cell correlates of protection and strategies to enhance their representation in vulnerable hosts should be within reach within the next few years.
Notes
Acknowledgements. I would like to thank Katherine Richards for assistance with the manuscript and for preparing Figure 1. I would also like to thank Dr. Ajitanju Rattan for preparation of Figure 2.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant number HHSN272201400005C).
Potential conflicts of interest. The author reports no potential conflicts of interest. The author has submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Palache A, Oriol-Mathieu V, Fino M, Xydia-Charmanta M;. Influenza Vaccine Supply Task Force Seasonal influenza vaccine dose distribution in 195 countries (2004–2013): Little progress in estimated global vaccination coverage. Vaccine 2015; 33:5598–605. [DOI] [PubMed] [Google Scholar]
- 2. Roti M, Yang J, Berger D, Huston L, James EA, Kwok WW. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J Immunol 2008; 180:1758–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Uchtenhagen H, Rims C, Blahnik G, et al. . Efficient ex vivo analysis of CD4+ T-cell responses using combinatorial HLA class II tetramer staining. Nat Commun 2016; 7:12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weaver JM, Yang H, Roumanes D, et al. . Increase in IFNγ(-)IL-2(+) cells in recent human CD4 T cell responses to 2009 pandemic H1N1 influenza. PLoS One 2013; 8:e57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Savic M, Dembinski JL, Kim Y, et al. . Epitope specific T-cell responses against influenza A in a healthy population. Immunology 2016; 147:165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babon JA, Cruz J, Orphin L, et al. . Genome-wide screening of human T-cell epitopes in influenza A virus reveals a broad spectrum of CD4(+) T-cell responses to internal proteins, hemagglutinins, and neuraminidases. Hum Immunol 2009; 70:711–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayward AC, Wang L, Goonetilleke N, et al. ; Flu Watch Group Natural T cell-mediated protection against seasonal and pandemic influenza. Results of the Flu Watch Cohort Study. Am J Respir Crit Care Med 2015; 191:1422–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Assarsson E, Bui HH, Sidney J, et al. . Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J Virol 2008; 82:12241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Richards KA, Treanor JJ, Nayak JL, Sant AJ. Overarching immunodominance patterns and substantial diversity in specificity and functionality in the circulating human influenza A and B virus-specific CD4+ T-cell repertoire. J Infect Dis 2018; 218:1169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Richards KA, Topham D, Chaves FA, Sant AJ. Cutting edge: CD4 T cells generated from encounter with seasonal influenza viruses and vaccines have broad protein specificity and can directly recognize naturally generated epitopes derived from the live pandemic H1N1 virus. J Immunol 2010; 185:4998–5002. [DOI] [PubMed] [Google Scholar]
- 11. Nayak JL, Fitzgerald TF, Richards KA, Yang H, Treanor JJ, Sant AJ. CD4+ T-cell expansion predicts neutralizing antibody responses to monovalent, inactivated 2009 pandemic influenza A(H1N1) virus subtype H1N1 vaccine. J Infect Dis 2013; 207:297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nayak JL, Richards KA, Yang H, Treanor JJ, Sant AJ. Effect of influenza A(H5N1) vaccine prepandemic priming on CD4+ T-cell responses. J Infect Dis 2015; 211:1408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richards KA, Nayak J, Chaves FA, et al. . Seasonal influenza can poise hosts for CD4 T-Cell immunity to H7N9 avian influenza. J Infect Dis 2015; 212:86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trowsdale J. HLA genomics in the third millennium. Curr Opin Immunol 2005; 17:498–504. [DOI] [PubMed] [Google Scholar]
- 15. Treanor J, Wright PF. Immune correlates of protection against influenza in the human challenge model. Dev Biol (Basel) 2003; 115:97–104. [PubMed] [Google Scholar]
- 16. de Jong JC, Palache AM, Beyer WE, Rimmelzwaan GF, Boon AC, Osterhaus AD. Haemagglutination-inhibiting antibody to influenza virus. Dev Biol 2003; 115:63–73. [PubMed] [Google Scholar]
- 17. Rimmelzwaan GF, McElhaney JE. Correlates of protection: novel generations of influenza vaccines. Vaccine 2008; 26 (Suppl 4):D41–4. [DOI] [PubMed] [Google Scholar]
- 18. Hale JS, Ahmed R. Memory T follicular helper CD4 T cells. Front Immunol 2015; 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vinuesa CG, Linterman MA, Yu D, MacLennan IC. Follicular helper T cells. Annu Rev Immunol 2016; 34:335–68. [DOI] [PubMed] [Google Scholar]
- 20. Fazilleau N, Mark L, McHeyzer-Williams LJ, McHeyzer-Williams MG. Follicular helper T cells: lineage and location. Immunity 2009; 30:324–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qi H. T follicular helper cells in space-time. Nat Rev Immunol 2016; 16:612–25. [DOI] [PubMed] [Google Scholar]
- 22. Dufaud CR, McHeyzer-Williams LJ, McHeyzer-Williams MG. Deconstructing the germinal center, one cell at a time. Curr Opin Immunol 2017; 45:112–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ueno H. Human circulating T follicular helper cell subsets in health and disease. J Clin Immunol 2016; 36 (Suppl 1):34–9. [DOI] [PubMed] [Google Scholar]
- 24. Spensieri F, Borgogni E, Zedda L, et al. . Human circulating influenza-CD4+ ICOS1+IL-21+ T cells expand after vaccination, exert helper function, and predict antibody responses. Proc Natl Acad Sci U S A 2013; 110:14330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bentebibel SE, Lopez S, Obermoser G, et al. . Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med 2013; 5:176ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patricia D’Souza M, Allen MA, Baumblatt JAG, et al. ; Lymph Node Webinar Contributors Innovative approaches to track lymph node germinal center responses to evaluate development of broadly neutralizing antibodies in human HIV vaccine trials. Vaccine 2018; 36:5671–7. [DOI] [PubMed] [Google Scholar]
- 27. Devarajan P, Jones MC, Kugler-Umana O, Vong AM, Xia J, Swain SL. Pathogen recognition by CD4 effectors drives key effector and most memory cell generation against respiratory virus. Front Immunol 2018; 9:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zens KD, Farber DL. Memory CD4 T cells in influenza. Curr Top Microbiol Immunol 2015; 386:399–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sant AJ, Richards KA, Nayak J. Distinct and complementary roles of CD4 T cells in protective immunity to influenza virus. Curr Opin Immunol 2018; 53:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Juno JA, van Bockel D, Kent SJ, Kelleher AD, Zaunders JJ, Munier CM. Cytotoxic CD4 T cells-friend or foe during viral infection? Front Immunol 2017; 8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Devarajan P, Bautista B, Vong AM, McKinstry KK, Strutt TM, Swain SL. New insights into the generation of CD4 memory may shape future vaccine strategies for influenza. Front Immunol 2016; 7:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brown DM, Lampe AT, Workman AM. The differentiation and protective function of cytolytic CD4 T cells in influenza infection. Front Immunol 2016; 7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takeuchi A, Saito T. CD4 CTL, a cytotoxic subset of CD4+ T cells, their differentiation and function. Front Immunol 2017; 8:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marshall NB, Vong AM, Devarajan P, et al. . NKG2C/E marks the unique cytotoxic CD4 T cell subset, ThCTL, generated by influenza infection. J Immunol 2017; 198:1142–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nayak JL, Richards KA, Chaves FA, Sant AJ. Analyses of the specificity of CD4 T cells during the primary immune response to influenza virus reveals dramatic MHC-linked asymmetries in reactivity to individual viral proteins. Viral Immunol 2010; 23:169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Richards KA, Chaves FA, Krafcik FR, Topham DJ, Lazarski CA, Sant AJ. Direct ex vivo analyses of HLA-DR1 transgenic mice reveal an exceptionally broad pattern of immunodominance in the primary HLA-DR1-restricted CD4 T-cell response to influenza virus hemagglutinin. J Virol 2007; 81:7608–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Richards KA, Chaves FA, Sant AJ. Infection of HLA-DR1 transgenic mice with a human isolate of influenza a virus (H1N1) primes a diverse CD4 T-cell repertoire that includes CD4 T cells with heterosubtypic cross-reactivity to avian (H5N1) influenza virus. J Virol 2009; 83:6566–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Richards KA, DiPiazza AT, Rattan A, Knowlden ZAG, Yang H, Sant AJ. Diverse epitope specificity, immunodominance hierarchy, and functional avidity of effector CD4 T Cells established during priming is maintained in lung after influenza A virus infection. Front Immunol 2018; 9:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Alam S, Knowlden ZA, Sangster MY, Sant AJ. CD4 T cell help is limiting and selective during the primary B cell response to influenza virus infection. J Virol 2014; 88:314–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Co MD, Orphin L, Cruz J, et al. . In vitro evidence that commercial influenza vaccines are not similar in their ability to activate human T cell responses. Vaccine 2009; 27:319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. García-Cañas V, Lorbetskie B, Bertrand D, Cyr TD, Girard M. Selective and quantitative detection of influenza virus proteins in commercial vaccines using two-dimensional high-performance liquid chromatography and fluorescence detection. Anal Chem 2007; 79:3164–72. [DOI] [PubMed] [Google Scholar]
- 42. Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology 2011; 411:229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garten RJ, Davis CT, Russell CA, et al. . Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009; 325:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Smith GJ, Vijaykrishna D, Bahl J, et al. . Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009; 459:1122–5. [DOI] [PubMed] [Google Scholar]
- 45. Itoh Y, Shinya K, Kiso M, et al. . In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 2009; 460:1021–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009; 459:931–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Greenberg ME, Lai MH, Hartel GF, et al. . Response to a monovalent 2009 influenza A (H1N1) vaccine. N Engl J Med 2009; 361:2405–13. [DOI] [PubMed] [Google Scholar]
- 48. Clark TW, Pareek M, Hoschler K, et al. . Trial of 2009 influenza A (H1N1) monovalent MF59-adjuvanted vaccine. N Engl J Med 2009; 361:2424–35. [DOI] [PubMed] [Google Scholar]
- 49. Nachbagauer R, Palese P. Development of next generation hemagglutinin-based broadly protective influenza virus vaccines. Curr Opin Immunol 2018; 53:51–7. [DOI] [PubMed] [Google Scholar]
- 50. Krammer F. Strategies to induce broadly protective antibody responses to viral glycoproteins. Expert Rev Vaccines 2017; 16:503–13. [DOI] [PubMed] [Google Scholar]
- 51. Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 2015; 14:167–82. [DOI] [PubMed] [Google Scholar]
- 52. Sautto GA, Kirchenbaum GA, Ross TM. Towards a universal influenza vaccine: different approaches for one goal. Virol J 2018; 15:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Paules CI, Marston HD, Eisinger RW, Baltimore D, Fauci AS. The pathway to a universal influenza vaccine. Immunity 2017; 47:599–603. [DOI] [PubMed] [Google Scholar]
- 54. Lu Y, Welsh JP, Swartz JR. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc Natl Acad Sci U S A 2014; 111:125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yassine HM, Boyington JC, McTamney PM, et al. . Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 2015; 21:1065–70. [DOI] [PubMed] [Google Scholar]
- 56. Impagliazzo A, Milder F, Kuipers H, et al. . A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015; 349:1301–6. [DOI] [PubMed] [Google Scholar]
- 57. Eggink D, Goff PH, Palese P. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J Virol 2014; 88:699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Knudson CJ, Weiss KA, Hartwig SM, Varga SM. The pulmonary localization of virus-specific T lymphocytes is governed by the tissue tropism of infection. J Virol 2014; 88:9010–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. DiPiazza A, Laniewski N, Rattan A, Topham DJ, Miller J, Sant AJ. CD4 T Cell Epitope Specificity and Cytokine Potential Are Preserved as Cells Transition from the Lung Vasculature to Lung Tissue following Influenza Virus Infection. J Virol 2018; 92:pii: e00377-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Román E, Miller E, Harmsen A, et al. . CD4 effector T cell subsets in the response to influenza: heterogeneity, migration, and function. J Exp Med 2002; 196:957–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Turner DL, Bickham KL, Thome JJ, et al. . Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol 2014; 7:501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim TS, Hufford MM, Sun J, Fu YX, Braciale TJ. Antigen persistence and the control of local T cell memory by migrant respiratory dendritic cells after acute virus infection. J Exp Med 2010; 207:1161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Takamura S. Niches for the long-term maintenance of tissue-resident memory T cells. Front Immunol 2018; 9:1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Iijima N, Iwasaki A. T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science 2014; 346:93–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zens KD, Chen JK, Farber DL. Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight 2016; 1:pii: e85832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Clemens EB, van de Sandt C, Wong SS, Wakim LM, Valkenburg SA. Harnessing the power of T cells: the promising hope for a universal influenza vaccine. Vaccines 2018; 6:pii: E18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Takamura S. Persistence in temporary lung niches: a survival strategy of lung-resident memory CD8+ T cells. Viral Immunol 2017; 30:438–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mikhak Z, Strassner JP, Luster AD. Lung dendritic cells imprint T cell lung homing and promote lung immunity through the chemokine receptor CCR4. J Exp Med 2013; 210:1855–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ciabattini A, Pettini E, Fiorino F, Prota G, Pozzi G, Medaglini D. Distribution of primed T cells and antigen-loaded antigen presenting cells following intranasal immunization in mice. PLoS One 2011; 6:e19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kummer S, Flöttmann M, Schwanhäusser B, et al. . Alteration of protein levels during influenza virus H1N1 infection in host cells: a proteomic survey of host and virus reveals differential dynamics. PLoS One 2014; 9:e94257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Leddon SA, Richards KA, Treanor JJ, Sant AJ. Abundance and specificity of influenza reactive circulating memory follicular helper and non-follicular helper CD4 T cells in healthy adults. Immunology 2015; 146:157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mellor AL, Lemos H, Huang L. Indoleamine 2,3-dioxygenase and tolerance: Where are we now? Front Immunol 2017; 8:1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Betts RJ, Prabhu N, Ho AW, et al. . Influenza A virus infection results in a robust, antigen-responsive, and widely disseminated Foxp3+ regulatory T cell response. J Virol 2012; 86:2817–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Su LF, Del Alcazar D, Stelekati E, Wherry EJ, Davis MM. Antigen exposure shapes the ratio between antigen-specific Tregs and conventional T cells in human peripheral blood. Proc Natl Acad Sci U S A 2016; 113:E6192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Macri C, Dumont C, Johnston AP, Mintern JD. Targeting dendritic cells: a promising strategy to improve vaccine effectiveness. Clin Transl Immunology 2016; 5:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Monteiro JT, Lepenies B. Myeloid C-type lectin receptors in viral recognition and antiviral immunity. Viruses 2017; 9:pii: E59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Geijtenbeek TB, Gringhuis SI. C-type lectin receptors in the control of T helper cell differentiation. Nat Rev Immunol 2016; 16:433–48. [DOI] [PubMed] [Google Scholar]
- 78. Démoulins T, Englezou PC, Milona P, et al. . Self-replicating RNA vaccine delivery to dendritic cells. Methods Mol Biol 2017; 1499:37–75. [DOI] [PubMed] [Google Scholar]
- 79. Tews BA, Meyers G. Self-replicating RNA. Methods Mol Biol 2017; 1499:15–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. McKinstry KK, Strutt TM, Bautista B, et al. . Effector CD4 T-cell transition to memory requires late cognate interactions that induce autocrine IL-2. Nat Commun 2014; 5:5377. [DOI] [PMC free article] [PubMed] [Google Scholar]