Abstract
Understanding antigenic variation in influenza virus strains and how the human immune system recognizes strains are central challenges for vaccinologists. Antibodies directed to the 2 major viral surface membrane proteins, hemagglutinin (HA) and neuraminidase (NA), mediate protection against reinfection following natural infection or vaccination, but HA and NA protein sequences in field strains are highly variable. The central questions are how to achieve protective antibody responses in a higher proportion of individuals and how to induce responses with more breadth and durability. Studies using isolation of human monoclonal antibodies followed by structural and functional characterization revealed conserved antigenic sites recognized by broadly cross-reactive antibodies. The antigenic landscape on HA and NA proteins is coming into focus to inform studies of the correlates and mechanisms of immunity. Understanding the antibody determinants of influenza immunity points the way toward development and testing of next-generation vaccines with potential to confer broadly protective immunity.
Keywords: orthomyxoviridae infections; influenza A virus; antibodies; viral; antibodies, neutralizing; hemagglutination inhibition tests; antibody-dependent cell cytotoxicity
Human monoclonal antibody studies have elucidated the structural and genetic basis for molecular recognition of principal protective antigenic sites on influenza HA. This is being used for rational design and testing of experimental vaccines aimed at inducing broadly protective antibodies.
Major antigenic shifts in the influenza virus surface proteins hemagglutinin (HA) and neuraminidase (NA) caused by reassortment of the segmented genome or direct adaptation of an avian virus for human infection and transmission have led to major pandemics in 1898 (suspected H3N8), 1918 (H1N1), 1957 (H2N2), 1968 (H3N2), and 2009 (pH1N1). Point mutations caused by the error-prone viral RNA-dependent RNA polymerase lead to minor antigenic variation in the HA and NA proteins that can be selected on a population basis on a year-to-year basis. The current strategy underlying licensed influenza vaccinations is to adjust the trivalent or quadrivalent HA and NA vaccine components yearly using a best-guess prediction based on worldwide real-time molecular epidemiology studies. In a good year, the vaccine antigens match the dominant circulating strains closely and substantial vaccine effectiveness can be achieved in otherwise healthy populations, but in other years some components of seasonal influenza vaccines confer negligible benefit. Clearly, a better system for dealing with antigenic variation is desirable.
MAINTENANCE OF B-CELL MEMORY
Durability of protective antibody responses following vaccination is a central concern. It is known that serum levels of antiviral antibodies to various microbial antigens exhibit widely differing durations of persistence, varying from years to decades [1], although the mechanisms governing the specific durations are poorly understood. Seasonal subunit (“inactivated”) protein vaccines have been observed to induce serum antibody responses that in some cases are quite short lived, with a duration on the order of months. For convenience, humoral immunity is measured using serum samples containing antibodies, which are secreted by long-lived plasma cells in the bone marrow. Memory B cells, which are not actively secreting antibodies, are present in the lymphoid tissues and in the peripheral blood. The interrelationship of the specificities and function of antibody proteins in the serum and mucosal tissues to the antibody variable genes in naive or memory B cells in blood or tissue, plasma cells in the bone marrow, and plasmablasts in the circulation approximately 1 week following infection or vaccination is poorly understood. Clearly, the frequency of memory B cells in circulation does not correlate exactly with antibody titers for many antigens [1]. Nonhuman primate studies also suggest that plasma cells may persist for a prolonged period of time in the absence of cell division, and in the absence of memory B cells [2].
While persistence of influenza-specific antibodies can be short lived after vaccination, we have found the persistence of memory B cells in the circulation can be extraordinarily long. Most dramatically, in 2007 we isolated B cells from nearly 100-year-old subjects that neutralized the 1918 H1N1 and related early 20th century H1N1 influenza viruses [3–6], even though those viruses had not circulated in the human population for about 5 decades. Intuitively, one might think that maintaining peripheral blood circulation of memory B cells for such a long period, “waiting for” a return of the 1918 influenza virus, would be metabolically costly and inherently inefficient for humans to maintain. However, this persistence of B-cell memory was beneficial when the 2009 H1N1 pandemic occurred (with a virus containing antigenic elements of the 1918 virus) and the extreme elderly exhibited an otherwise unexpected relative protection compared to those without prior exposure to early 20th century H1 viruses [7].
We have found persistence of human B cells in circulation in living survivors not only of the highly virulent 1918 pandemic, but also for those affected by every known pandemic to date. For example, we isolated very potent neutralizing human monoclonal antibodies (mAbs) from middle-aged subjects for the 1957 H2N2 pandemic virus [8], which circulated in humans only from 1957 to 1968. Most human subjects born prior to 1968 have such B cells in peripheral blood. This same type of legacy herd immunity in humans exposed to older influenza strains also has occurred with the H3 viruses, which entered the human population in 1968. Most older adults are immune to H3 viruses that infected humans in the first several decades of H3 circulation. Viruses related to these earlier H3 viruses appear to persist in swine populations in the US and are designated “H3 variant” viruses. We have identified circulating B cells and isolated human mAbs from subjects vaccinated with H3 variant virus experimental vaccines [9, 10], and these antibodies recognize the older H3 human viruses and the current swine H3 variant viruses (but not the more recent seasonal H3 strains) [10]. Children born in the last decade are not immune to the H3 variant viruses, because they have been exposed only to more recent seasonal H3 strains in circulation or recent vaccines, which are antigenically different. When young children lacking prior exposure to older H3 strains are exposed directly to swine infected with H3 variant viruses, they can suffer severe or fatal disease [11, 12]. Thus, there is a general observation that serum titers to many influenza strains can be short lived, but memory B cells often remain in circulation for decades (or life) and their presence may correlate with some level of protection. It is uncertain if vaccination induces as long-lived a circulating memory B-cell response as does natural infection, but likely the durability of vaccine-induced responses is not as durable.
MECHANISMS OF VIRUS NEUTRALIZATION BY ANTIBODIES
Antibodies mediate antiviral effects against replication and disease with diverse mechanisms. The earliest form of inhibition of infection in the virus life cycle is aggregation of viral particles by bivalent cross-linking of 2 virion particles in a process that may progress to large aggregates in the airway lumen. IgM and IgA antibodies exhibit superior performance in this mode of neutralization for 2 reasons. First, they oligomerize to higher-order forms mediated by joining (J) chain protein (using 2 bivalent immunoglobulin molecules for IgA or 10–12 molecules for IgM). Second, polymeric IgM and IgA antibodies are transported actively from the basolateral face of polarized airway epithelial cells to the apical surface and then secreted into the airway lumen, in an active process mediated by the polyimmunoglobulin receptor. Another mechanism of antibody-mediated neutralization is prevention of virus attachment to host sialic acid-bearing receptors on the apical surface of airway cells. While IgM and IgA molecules are actively transported, high concentrations of IgG in respiratory secretions can be achieve by transudation from tissue fluid across the epithelium. The most potent (and validated) serum antibody correlate of protection is IgG blocking of virus attachment to sialic acid receptors, as measured in the hemagglutination inhibition (HAI) assay. This test is the only correlate of protection currently recognized for vaccine licensure by regulatory agencies. Antibodies that bind to (or near) the receptor binding site (RBS) on the globular head domain of the HA molecule block attachment to sialic acid-bearing receptors. The NA protein of some avian influenza strains also has a hemadsorption site that may play a role in this type of activity, and antibodies to that site also may reduce viral attachment for those viruses [13]. N9 NA molecules of recent H7N9 viruses that have crossed from birds to humans have such activity, and the hemadsorption site has been mapped [14, 15]. Like most viral fusion proteins, the HA is a metastable protein that exists on the viral surface in a prefusion state, which is triggered by exposure to low pH conditions in the endosome to switch conformations, facilitating insertion of a hydrophobic fusion peptide into the host membrane.
A third mechanism of neutralization is mediated by antibodies to the stem region that can inhibit the complex conformational changes needed to accomplish virus-cell membrane fusion (ie, they mediate fusion inhibition). Recently, it was demonstrated that the 3 protomers in the HA trimer exhibit dynamic features in the prefusion steady state, prior to conformational switching. Therefore, in the future, it is likely that we will identify antibodies to the HA molecule that bind to alternate surfaces of HA protomers not accessible in the HA visualized in current crystal structures. Also, we suspect there are many antibodies that bridge HA protomers and recognize complex quaternary epitopes, and such antibodies would be expected to reduce the dynamic capacity of the HA trimer and thus the fusogenic capacity of the virus.
A fourth mechanism of neutralizing virus or limiting cell-to-cell spread is to interfere with egress of virus from infected cells. We have identified a human antibody H3v47 that binds to the side of the HA head domain with an epitope in the vestigial esterase (VE) domain, and this antibody exhibits a unique phenotype of neutralizing mechanisms [10, 16]. Although this neutralizing antibody binds to the HA head, it does not block receptor binding (and thus also does not exhibit HAI activity in vitro). Instead, it inhibits virus egress from infected cells. Electron microscopy studies suggested the antibody tethers emerging particles to the cell surface during egress. The antibody also reduces cell-to-cell spread in cell monolayer cultures. These egress and cell-cell fusion inhibition mechanisms reduce transmission within the host and might reduce transmission to other exposed individuals. Because the processes of viral particle scission from the surface of infected cells and the mechanics of cell-cell fusion are so radically different, it is logical to think we could find in the future antibodies that will inhibit one of these processes but not the other, although no such HA-specific antibody has been reported to date. H3v-47 also possesses antibody-dependent cell-mediated cytotoxicity (ADCC) activity.
Antibodies to NA also inhibit virus replication by multiple mechanisms, although to date these mechanisms are less well studied and fewer human mAbs to NA have been isolated than to HA. NA possesses an enzymatic site that cleaves host cell sialic acid, which is thought to facilitate release of particles from infected cells without attaching back to the cell of origin. Zanamivir is a small molecule drug that inhibits influenza virus by binding to the active site of the NA protein, thus inhibiting viral cleavage of sialic acid on cells and preventing virus egress. Some antibodies to NA bind in or near the active enzymatic site, preventing sialic acid cleavage in the same way that zanamivir does [17].
ANTIGENIC SITES RECOGNIZED BY INHIBITORY ANTIBODIES
The antigenic landscape on the surface of HA and NA can be described in terms of structural domains, antigenic sites, and epitopes. HA can be divided into 2 major domains, the head and stem. Both of the protein subunits have been expressed as separate soluble protein domains, and indeed the design and production of “headless” stem antigens is the basis for a number of current “universal influenza” candidate vaccine programs, discussed below. Within these 2 domains, some sites on the surface of these proteins are more immunogenic than others, and antibodies that bind sufficiently near each other to compete for binding can be organized into clones recognizing a major antigenic site. Within these sites, the specific features of the HA or NA antigen to which 1 antibody binds is termed an epitope.
There are several historical and overlapping nomenclatures that designate particular sites on the HA molecule for antibody recognition. The conventional numbering of HA residues differs between the 2 subtypes within influenza A, including group 1 HAs such as H1 and H5 and group 2 HAs such as H3 and H7, leading to conventions of “H1 numbering” and “H3 numbering”. Also, antigenic sites have been designated by mapping onto the H1N1 A/Puerto Rico/8/34 (A/PR/8/34) HA with vocabulary describing Sa, Sb, Ca1, Ca2, and Cb sites [18], or H3 strains with A, B, C, D, E sites [19, 20]. Although these site designations have utility, we have found that mAbs rarely recognize a single site designated by this type of nomenclature. For instance, the epitope of the 1918 H1 HA-specific antibody 2D1 extends to residues beyond the conventionally defined antigenic site to sites Sb and Ca1 [5]. The crystal structure of the 1918 influenza mAb 1F1 demonstrates that 1F1 interacts with residues within Sa, Sb, and Ca2 and also reaches into the HA RBS [6].
A more consistent and inclusive nomenclature for HA antigenic sites and epitopes within them is needed. With more recent structure-function studies, it has been helpful to designate structural features recognized by particular antibodies. Such designations are particularly helpful for antibodies recognizing 4 features surrounding the RBS, including elements designated the 130-loop, 150-loop, 190-helix, and 220-loop, which are named based on the numbered amino acid positions in the primary sequence of the HA. Residues in these loops and helices are hypervariable, and this mutability in field strains underlies much of the antigenic drift that is observed over time in influenza. Genetic drift in influenza, unlike many other RNA viruses, is directional in nature [21]. Antigenic variation trends in the general direction of the genetic drift, but the antigenic relationships tend to cluster, and transition from one antigenic cluster may result from relatively low numbers of amino acids changes [22]. The regions in and around the RBS form an intragenic network that is maintained by the simultaneous requirements for sialic acid receptor binding and the need to escape immune selection pressure from neutralizing antibodies [23]. Changes in HA receptor specificity (from the α2,3 sialic acid linkage preferred in avian influenza viruses to the α2,6 linkage in human transmissible strains) is a factor that can drive the selection of variant HA head domains. Broadly neutralizing antibodies have to accommodate HA changes associated with shift in receptor specificity when they occur [24].
NEUTRALIZATION OF INFLUENZA BY ANTIBODIES RECOGNIZING SITES IN OR NEAR THE RBS
Human B-cell and serum antibody studies have shown that the most frequent cell response is directed to the influenza HA globular head. The most potently inhibiting human mAbs also are directed to the head domain, especially to the RBS. Likely, this immunodominance of the head domain is due to the fact that the structural elements of the HA head are highly exposed on the virion surface and very accessible to B-cell receptors and thus antibody recognition. Many of these antigenic features are hydrophilic in nature and project articulated structures into the solute, accounting for their antigenicity. The majority of the HA head domain surface also is highly mutable, allowing viral escape from antibody recognition. Many antibodies recognize complex quaternary structures on HA, including epitopes formed by more than 1 HA protomer [25].
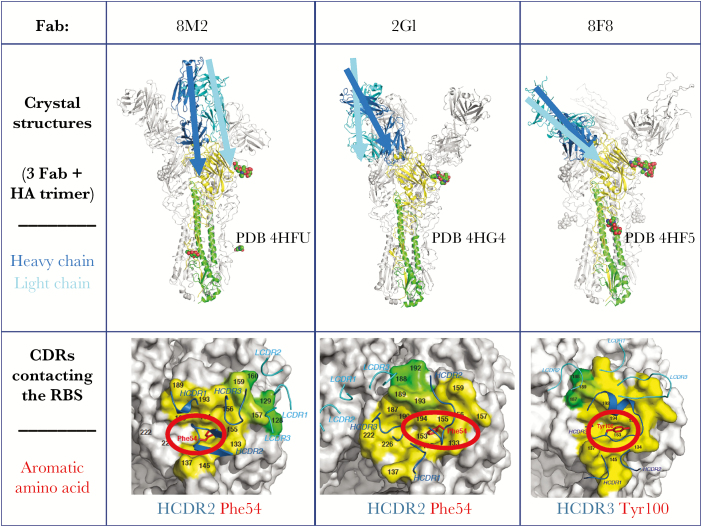
The most potent neutralizing antibodies recognize the RBS on HA, and many of the amino acids in this site are highly conserved in order to preserve sialic acid binding function. The RBS is a simple structural domain, comprising a shallow pocket surrounded by hypervariable loops and helices. The paratope region on antibodies that forms the antigen combining surface is formed by a surface of 6 hypervariable loops (designated complementarity determining regions [CDRs]). The loops encoded by some antibody variable region genes interact optimally with the pocket that serves as the RBS. The CDR2 or CDR3 loops of the heavy chain of some antibodies reach into the RBS and form direct contacts with conserved residues in the base of the RBS. Because the contact residues in the RBS are highly conserved (to maintain sialic acid binding), only certain residues on the antibody CDRs satisfy the requirement for interaction through typical bond formation. Therefore, several canonical modes of interaction of the influenza RBS with particular amino acids in antibody CDRs have been recognized. One of the most common modes of interaction is the presentation of an aromatic residue, typically Phe or Tyr, on the tip of a CDR inserting into the RBS, creating pi-pi interactions (attractive, noncovalent interactions between aromatic rings) between the antibody and HA [8] (Figure 1). This interaction is interesting because the protein structure of the antibody CDR mimics the interaction of the sialic acid receptor with HA. This mimicry occurs by the insertion of a large hydrophobic amino acid into the RBS with a backbone carbonyl group, making an interaction similar to that of the sialic acid carboxylate on host receptors. In these instances, the role of the CDR is principally to position the hydrophobic amino acid correctly.
Figure 1.
Interaction of aromatic residues on the tip of antibody HCDR2 or HCDR3 with the influenza A H2 hemagglutinin receptor binding site (RBS). Top, Crystal structures of H2 HA with each of 3 human neutralizing antibody Fabs are shown (3 Fabs are associated with each trimer). One of the Fabs is colored in blue (heavy chain) and cyan (light chain), and the approximate axis of orientation of that chain is indicated. The corresponding HA1 is shown in yellow and the HA2 in green for 1 protomer, with N-linked glycans that are observed in the crystal structure in spheres. Bottom, Footprints of the Fabs on the surface representation of H2 HA are indicated, with heavy chain contacts in yellow and light chain contacts in green. Complementarity determining region (CDR) loops are indicated by ribbons. The aromatic residue on HCDR2 or HCDR3 that inserts into the RBS is highlighted in red. Abbreviation: ASP, aspartate. Based on data in Xu et al [8].
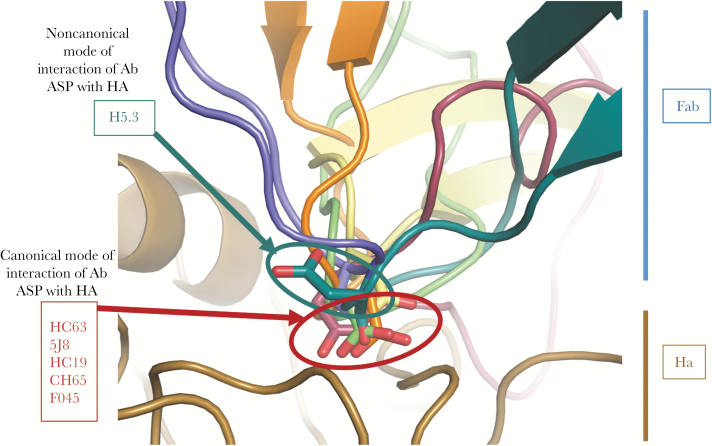
A second canonical mode of interaction of antibodies and the RBS is the presentation of an aspartate residue on the tip of a CDR, which mediates interaction with the RBS because of favorable charge interactions with amino acids in the HA protein. If an aspartic acid is positioned properly on the tip of a CDR, backbone atoms mimic the acetamido groups of the receptor and a carboxylic acid mimics the carboxylate of sialic acid. A large number of mAbs with an aspartate in the proper interacting position (or dipeptide with an aspartic acid hydrophobic motif) have been identified in cocrystal structures of antibodies with HA [26]. Several potent H1-specific neutralizing antibodies have been studied in detail in this regard, including 5J8 [27, 28] and CH65 [29, 30]. One of the interesting observations from those studies is that the binding pose of antibodies during interaction with the RBS determines the breadth of recognition of diverse H1 strains. The combination of 5J8 and CH65 probably would cover all H1 strains, because of the differing angles of approach to the RBS [31]. Simple presentation of an aspartate residue in this position is not sufficient to satisfy the requirements for interaction. For example, we isolated mAb H5.3 that neutralizes influenza and possesses an aspartate in the correct position in a CDR3, but a cocrystal structure of the complex revealed that the CDR does not use the aspartate to interact in the typical manner [32, 33] (Figure 2). Antibodies that interact with the canonical aromatic or aspartate residue have been described, but also at least 1 antibody is described that mediates both interactions, an antibody designated F045-092 [34, 35]. This antibody has a long (23-residue) HCDR3 that interacts with the RBS in a manner that mimics sialic acid [35]. The F045-092 HCDR3 creates a hydrogen bond between the Fab main chain using the Tyr100b to interact with a residue on HA, and the carboxylate side chain of the antibody Asp100e residue closely aligns with the carboxylate of sialic acid that would be found in the same position and binds to HA using a similar network of hydrogen bonds. (There are several numbering schemes for amino acids in antibodies based upon variable regions; here the designations are according to the Kabat numbering scheme). C05 is a particularly interesting antibody with a very long HCDR3 that interacts with the RBS and avoids contacting the more variable residues around the RBS [36]. This antibody possesses what must surely be a minimum interacting region, thus reducing the exposure to loss of binding caused by HA protein variability. Nevertheless, even C05 is not a “universal” antibody for all influenza strains because of variability in the RBS in field strains. It should be noted that the residues in the HA 220-loop differ across HA subtypes and species. For example, residues 226 and 228 usually are leucine or serine in the human H2 or H3 subtype viruses, but these residues are glutamine or glycine in viruses of the human H1 or some avian subtypes. Likewise, residues 190 and 225 typically are aspartate in H1 human influenza strains, but these positions have glutamine or glycine residues in most H2 or H3 human influenza strains.
Figure 2.
Canonical or noncanonical modes of binding of human antibodies with aspartate (Asp) residues at the tip of complementarity determining regions (CDRs) interacting with the hemagglutinin (HA) receptor binding site (RBS). The CDRH3 of monoclonal antibody H5.3 (teal, top) is inserted into the HA (gold, bottom) receptor binding site. The Asp at the tip of the H5.3 CDRH3 (teal) is oriented away from the RBS, unlike the Asp at the tips of CDRH3 in HC63 (blue, 1KEN), 5J8 (orange, 4M5Z), HC19 (gray, 2VIR), CH65 (green, 3SM5), F045-092 (4O5I, burgundy), which are inserted into the receptor binding site, mimicking the carboxylate group of sialic acid. Based on data in Winarski et al [33].
The side of the HA head domain also contains conserved elements that are the targets of neutralizing antibodies, especially when those antibodies are directed to the VE domain [16]. Influenza type C HA protein has a region containing a 9-O-acetylesterase domain within the HA-esterase fusion glycoprotein that cleaves the host receptor to facilitate viral budding. A similar VE domain in influenza A and B viruses has been defined based on approximately 50% structural homology with the functional VE domain. In influenzas A and B, the VE domain does not cleave the receptor during budding, rather the neuraminidase protein serves this function in those viruses. The VE domain is an interesting antigenic target because the amino acid residues in the domain are very conserved within influenza A subtypes. The VE domain-specific mAb H3v47, discussed above, is interesting in that it neutralizes H3 viruses mostly by the inhibition of virus egress from cells.
STEM ANTIBODIES
In recent years, the influenza vaccine development field has been reenergized by the rediscovery of stem-reactive antibodies [37–40]. Neutralizing antibodies with unusual breadth have been isolated and characterized, engendering hope for design of broadly protective or “universal” vaccines. Several prototype human mAbs, for example CR6261 or F10 that unexpectedly recognized both H1 and H5 HA molecules, were described in 2009 and, since that time, many stem antibodies with broad cross-reactivity have been described. A number of mAbs are being developed and tested as therapeutic molecules in clinical trials, including CR6261 and CR8020 (NCT02371668 and NCT01938352) and MEDI8852 (NCT02603952). In general, stem antibodies can achieve broad cross-reactivity, including recognition of both group 1 and 2 influenza A viruses, such as by the stem antibody FI6 [41]. Many stem antibodies exhibit low potency in virus neutralization tests compared to head domain antibodies, and the protective and therapeutic effects for many stem antibodies in preclinical animal models is associated with ADCC activity [42]. Investigators also have used structure-based design to develop small protein mimics of antibodies that bind to the stem region in a manner analogous to CR6261 [43, 44].
It is not clear that the relatively weak neutralizing activity coupled with ADCC activity will be sufficient for therapy in humans as monotherapy using human mAbs. Nevertheless, the concept of the stem as a broad and protective antigen has important implications for understanding natural heterosubtypic immunity and design of broadly protective vaccines. Based on structural studies of stem-antibody complexes, investigators have designed novel immunogens to focus the immune response on the stem, using headless HA constructs, chimeric HA molecules with rare subtype head domains and the H1 stem, or other strategies [45–50]. Stem vaccines will be tested in clinical trials. The rationale for focusing on the stem region is that it not only is relatively conserved across subtypes, but also the stem domain evolves more slowly under immune pressure than the head domain [51].
VIRUS ESCAPE FROM ANTIBODY RECOGNITION AND NEUTRALIZATION
Two principal mechanisms underlie the ability of influenza viruses to drift antigenically, thus escaping recognition by particular antibody clones. The virus uses a viral RNA-dependent RNA polymerase for replication, which is error prone and introduces missense mutations that can cause minor structural changes. First, altering the backbone or side chain configuration of residues in the antibody epitope can eliminate binding. Second, acquisition of a glycosylation site in or near an antigenic site can shield epitopes from antibody recognition. This glycan-mediated inhibition of antibody binding can occur in both the HA head domain, including near the RBS, and in the stem domain. Stem domain glycans regulate group 1 versus group 2 influenza A subtype specificity of stem antibodies to a large degree.
One of the curious observations with influenza is that although there are 4 or 5 major antigenic sites for neutralizing antibodies near the RBS, the virus still can drift antigenically year to year on a population basis. If each of the sites is recognized equally, then statistically it is highly unlikely that a virus could simultaneously acquire escape mutations in all of the major antigenic sites. Therefore, it has been unclear in the past how influenza actually accomplishes steady antigenic drift on a population basis. In many cases, mutations facilitating glycosylation at the apex of HA accumulate as drift occurs, obscuring some neutralizing determinants. Recent detailed studies also have shown that, over time, a single antigenic site on the HA head can achieve a sufficient level of immunodominance that escape in that one site can mediate viral escape from polyclonal responses [52].
HAI AS A BIOMARKER OF NEUTRALIZATION
The most potently neutralizing and protective HA antibodies block attachment of virion particles to sialic acid-bearing receptors on host epithelial cells. This process can be mimicked by blocking attachment of virus to sialic acid on the surface of animal red blood cells in vitro, and thus blocking hemagglutination. The HAI test is the only test currently accepted by regulatory authorities as a correlate of immunity for vaccine studies and licensure. Antibodies to the VE domain appear to be multifunctional, with neutralizing activity that is associated with egress inhibition and ADCC activity [16]. Antibodies binding below the head domain may inhibit conformational changes needed to accomplish fusion of the viral and cell endosomal membranes. It is difficult to use such alternate mechanisms of virus inhibition as laboratory endpoints in clinical trials, because currently there are no validated test versions of these assays that have been accepted by regulatory agencies.
NONNEUTRALIZING MECHANISMS OF VIRUS INHIBITION
Additional inhibitory mechanisms occur in vivo, which are mediated by the antibody Fragment crystallizable (Fc) region. Variations in sequence or glycosylation state in the immunoglobulin Fc region modulate binding of antibodies to Fc receptors on innate immune cells and affect their ability to induce Fc receptor-mediated innate cell signaling. The Fc region also can activate circulating soluble immune components, including complement made in the liver and distributed systemically. A large number of Fc variants that modulate binding to and activity induced by Fc receptors has been identified and reported in recent years. Variation in immunoglobulin isotype (IgM, IgD, IgG, IgA, IgE) or the subclasses of IgG (IgG1, IgG2, IgG3, or IgG4) are the principal naturally occurring physiologic drivers of multifunctional differences, but Fc mutants also have been engineered. IgG3 typically is the most active isotype in many Fc-mediated activities, while IgG4 is often silent. Over 80 single or multiple amino acid polymorphisms have been described to modulate interaction with various Fc receptors. The interaction also can be regulated by small glycan modifications. Recombinant immunoglobulins can be produced in altered production cell lines (such as Chinese hamster ovary cells expressing afucosylated proteins) to increase activities like ADCC. Many Fc-mediated functions have been described for influenza, including ADCC, antibody-dependent cellular phagocytosis, and antibody-dependent complement deposition [16, 53, 54]. Vaccine formulations and adjuvants can modulate the types of antibodies induced in active vaccination schemes, but the pattern of isotypes induced in a population is difficult to control precisely. Also, there are polymorphisms in the FcγRIIIa receptor that modulate ADCC functionality.
ANTIBODY-DEPENDENT ENHANCEMENT
Antibody-dependent enhancement (ADE) has been reported in vitro for a number of viruses, including dengue viruses and more recently Ebola virus. This activity typically is mediated by Fc engagement of an Fc receptor on a replication-competent cell for the virus of interest, causing increased viral entry. It is not clear at this point whether medically significant ADE occurs in humans. During the 2009 H1N1 pandemic, Argentina suffered the highest incidence of severe influenza including deaths in otherwise healthy young adults. Investigators showed that severe cases had preexisting serum antibodies that appeared to cross-react with, but did not protect against, 2009 H1N1 influenza virus in adults [55]. The studies also showed C4d deposition in lung sections of fatal cases, a marker of complement activation mediated by immune complexes. This study raised the concern that nonneutralizing antibodies might contribute to pathogenesis of severe influenza. Others have developed a swine model of influenza vaccine-associated enhanced respiratory disease (VAERD) using an H1N1 mismatched virus challenge. Whole inactivated H1N2 (human-like) virus vaccine (WIV-H1N2) appeared to cause enhanced pneumonia and disease in pigs after pandemic H1N1 virus challenge [56], although the design of additional control treatments in these types of studies is warranted to clarify if this effect is real and reproducible. It is uncertain whether or not these porcine studies pertain to human immunity. Others have investigated the role of previous conventional vaccination on vaccine effectiveness in humans and observed reduced effectiveness with history of repeated vaccination in some cases [57], although substantial heterogeneity is observed in such studies. In general, previous infection or vaccination with influenza is protective in humans.
POTENTIAL FOR USING MONOCLONAL ANTIBODIES AS PREVENTION OR THERAPY
Antibodies are increasingly used as medical interventions, especially in the fields of cancer and autoimmunity. Given the strong safety history of antibodies in humans, and the increasing effectiveness of antibodies in immunotherapy settings, it is logical to think that antibodies could be deployed as biological drugs to prevent or treat influenza infection. The challenges are that the most potent antibodies are to the head domain, which is hypervariable, while antibodies to the stem are broader but reduced in potency and may require ADCC, which is unproven as a correlate of immunity. The cost of mAbs has been prohibitive historically, but the cost is dropping in the industry by using new engineering methods. Also, new methods for delivering cDNAs expressing antibody genes in vivo using mRNA, DNA or adeno-associated virus vectors are being developed for other applications. An antibody discovery and development approach could leverage the isolation of ultrapotent, broadly cross-reactive antibodies for human use in the next decade. A combination of several antibodies seems attractive to provide redundant mechanisms of protection, to reduce the risk of escape mutants, and to achieve synergy in mechanisms of action.
Notes
Financial support. This work was supported by National Institutes of Health, National Institute of Allergy and Infectious Diseases (grant number U19 AI117905); and Department of Health and Human Services (contract number HHSN272201400024C).
Potential conflict of interest. J. E. C. has served as a consultant for Sanofi, Novavax, and Takeda, is on the Scientific Advisory Boards of CompuVax, GigaGen, and Meissa Vaccines, is a recipient of previous unrelated research grants from Moderna and Sanofi, and is founder of IDBiologics, Inc. The author has submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med 2007; 357:1903–15. [DOI] [PubMed] [Google Scholar]
- 2. Hammarlund E, Thomas A, Amanna IJ, et al. Plasma cell survival in the absence of B cell memory. Nat Commun 2017; 8:1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krause JC, Tumpey TM, Huffman CJ, et al. Naturally occurring human monoclonal antibodies neutralize both 1918 and 2009 pandemic influenza A (H1N1) viruses. J Virol 2010; 84:3127–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu X, Tsibane T, McGraw PA, et al. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 2008; 455:532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 2010; 328:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsibane T, Ekiert DC, Krause JC, et al. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog 2012; 8:e1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dawood FS, Jain S, Finelli L, et al. ; Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. New Engl J Med 2009; 360:2605–15. [DOI] [PubMed] [Google Scholar]
- 8. Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr, Wilson IA. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 2013; 20:363–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keitel WA, Jackson LA, Edupuganti S, et al. ; VTEU H3N2v Vaccine Study Work Group Safety and immunogenicity of a subvirion monovalent unadjuvanted inactivated influenza A(H3N2) variant vaccine in healthy persons ≥18 years old. J Infect Dis 2015; 212:552–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bangaru S, Nieusma T, Kose N, et al. Recognition of influenza H3N2 variant virus by human neutralizing antibodies. JCI Insight 2016; 1:pii: e86673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jhung MA, Epperson S, Biggerstaff M, et al. Outbreak of variant influenza A(H3N2) virus in the United States. Clin Infect Dis 2013; 57:1703–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lindstrom S, Garten R, Balish A, et al. Human infections with novel reassortant influenza A(H3N2)v viruses, United States, 2011. Emerg Infect Dis 2012; 18:834–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Laver WG, Colman PM, Webster RG, Hinshaw VS, Air GM. Influenza virus neuraminidase with hemagglutinin activity. Virology 1984; 137:314–23. [DOI] [PubMed] [Google Scholar]
- 14. Webster RG, Air GM, Metzger DW, et al. Antigenic structure and variation in an influenza virus N9 neuraminidase. J Virol 1987; 61:2910–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nuss JM, Air GM. Transfer of the hemagglutinin activity of influenza virus neuraminidase subtype N9 into an N2 neuraminidase background. Virology 1991; 183:496–504. [DOI] [PubMed] [Google Scholar]
- 16. Bangaru S, Zhang H, Gilchuk IM, et al. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat Commun 2018; 9:2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen YQ, Wohlbold TJ, Zheng NY, et al. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 2018; 173:417–29.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gerhard W, Yewdell J, Frankel ME, Webster R. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 1981; 290:713–7. [DOI] [PubMed] [Google Scholar]
- 19. Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981; 289:366–73. [DOI] [PubMed] [Google Scholar]
- 20. Wilson IA, Cox NJ. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol 1990; 8:737–71. [DOI] [PubMed] [Google Scholar]
- 21. Volz EM, Koelle K, Bedford T. Viral phylodynamics. Plos Comput Biol 2013; 9:e1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith DJ, Lapedes AS, de Jong JC, et al. Mapping the antigenic and genetic evolution of influenza virus. Science 2004; 305:371–6. [DOI] [PubMed] [Google Scholar]
- 23. Wu NC, Thompson AJ, Xie J, et al. A complex epistatic network limits the mutational reversibility in the influenza hemagglutinin receptor-binding site. Nat Commun 2018; 9:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu NC, Grande G, Turner HL, et al. In vitro evolution of an influenza broadly neutralizing antibody is modulated by hemagglutinin receptor specificity. Nat Commun 2017; 8:15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Knossow M, Gaudier M, Douglas A, et al. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology 2002; 302:294–8. [DOI] [PubMed] [Google Scholar]
- 26. Lee PS, Wilson IA. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol 2015; 386:323–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hong M, Lee PS, Hoffman RM, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol 2013; 87:12471–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE Jr. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol 2011; 85:10905–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whittle JR, Zhang R, Khurana S, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 2011; 108:14216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmidt AG, Therkelsen MD, Stewart S, et al. Viral receptor-binding site antibodies with diverse germline origins. Cell 2015; 161:1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crowe JE., Jr Principles of broad and potent antiviral human antibodies: insights for vaccine design. Cell Host Microbe 2017; 22:193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thornburg NJ, Nannemann DP, Blum DL, et al. Human antibodies that neutralize respiratory droplet transmissible H5N1 influenza viruses. J Clin Invest 2013; 123:4405–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Winarski KL, Thornburg NJ, Yu Y, Sapparapu G, Crowe JE Jr, Spiller BW. Vaccine-elicited antibody that neutralizes H5N1 influenza and variants binds the receptor site and polymorphic sites. Proc Natl Acad Sci U S A 2015; 112:9346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol 2011; 85:11048–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee PS, Ohshima N, Stanfield RL, et al. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 2014; 5:3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ekiert DC, Kashyap AK, Steel J, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012; 489:526–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Throsby M, van den Brink E, Jongeneelen M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 2008; 3:e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sui J, Hwang WC, Perez S, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 2009; 16:265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ekiert DC, Bhabha G, Elsliger MA, et al. Antibody recognition of a highly conserved influenza virus epitope. Science 2009; 324:246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 1993; 67:2552–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Corti D, Voss J, Gamblin SJ, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011; 333:850–6. [DOI] [PubMed] [Google Scholar]
- 42. Leon PE, He W, Mullarkey CE, et al. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc Natl Acad Sci U S A 2016; 113:E5944–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whitehead TA, Chevalier A, Song Y, et al. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol 2012; 30:543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fleishman SJ, Whitehead TA, Ekiert DC, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science 2011; 332:816–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Steel J, Lowen AC, Wang TT, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio 2010; 1:pii: e00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Impagliazzo A, Milder F, Kuipers H, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015; 349:1301–6. [DOI] [PubMed] [Google Scholar]
- 47. Yassine HM, Boyington JC, McTamney PM, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 2015; 21:1065–70. [DOI] [PubMed] [Google Scholar]
- 48. Valkenburg SA, Mallajosyula VV, Li OT, et al. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci Rep 2016; 6:22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hai R, Krammer F, Tan GS, et al. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol 2012; 86:5774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 2013; 87:6542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kirkpatrick E, Qiu X, Wilson PC, Bahl J, Krammer F. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci Rep 2018; 8:10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Linderman SL, Chambers BS, Zost SJ, et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013-2014 influenza season. Proc Natl Acad Sci U S A 2014; 111:15798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med 2014; 20:143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol 2004; 172:5598–605. [DOI] [PubMed] [Google Scholar]
- 55. Monsalvo AC, Batalle JP, Lopez MF, et al. Severe pandemic 2009 H1N1 influenza disease due to pathogenic immune complexes. Nat Med 2011; 17:195–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Khurana S, Loving CL, Manischewitz J, et al. Vaccine-induced anti-HA2 antibodies promote virus fusion and enhance influenza virus respiratory disease. Sci Transl Med 2013; 5:200ra114. [DOI] [PubMed] [Google Scholar]
- 57. Belongia EA, Skowronski DM, McLean HQ, Chambers C, Sundaram ME, De Serres G. Repeated annual influenza vaccination and vaccine effectiveness: review of evidence. Expert Rev Vaccines 2017; 16:1–14. [DOI] [PubMed] [Google Scholar]