Abstract
Background
Southern Province, Zambia has experienced a dramatic decline in Plasmodium falciparum malaria transmission in the past decade and is targeted for elimination. Zambia’s National Malaria Elimination Program recommends reactive case detection (RCD) within 140 m of index households to enhance surveillance and eliminate remaining transmission foci.
Methods
To evaluate whether RCD captures local transmission, we genotyped 26 microsatellites from 106 samples collected from index (n = 27) and secondary (n = 79) cases detected through RCD in the Macha Hospital catchment area between January 2015 and April 2016.
Results
Participants from the same RCD event harbored more genetically related parasites than those from different RCD events, suggesting that RCD captures, at least in part, infections related through local transmission. Related parasites clustered in space and time, up to at least 250 m from index households. Spatial analysis identified a putative focal transmission hotspot.
Conclusions
The current RCD strategy detects focal transmission events, although programmatic guidelines to screen within 140 m of index households may fail to capture all secondary cases. This study highlights the utility of parasite genetic data in assessing programmatic interventions, and similar approaches may be useful to malaria elimination programs seeking to tailor intervention strategies to the underlying transmission epidemiology.
Keywords: reactive, case detection, malaria, microsatellites, screen-and-treat, molecular epidemiology
Microsatellite genotyping from Plasmodium falciparum samples collected during reactive case detection in Macha, Zambia, revealed temporal-spatial clustering of genetically related parasites. Relatedness extended at least 250 m from index households, suggesting that the current 140-m screening radius misses secondary cases.
The global decline in malaria between 2000 and 2015 has reinvigorated commitment to elimination [1]. Many countries have malaria elimination targets before 2030, including 8 in southern Africa. Although interventions such as long-lasting insecticide-treated bednets (LLINs), indoor residual spraying, and case management have been and remain important malaria control tools [1], transitioning from control to elimination will necessitate incorporating new strategies capable of permanently interrupting remaining avenues for local transmission [2].
Current elimination strategies recommend variations of active case detection (ACD) in which community health workers (CHWs) screen community members for malaria using rapid diagnostic tests (RDTs) to identify asymptomatically infected individuals who may contribute to ongoing transmission [3, 4]. Despite consensus that reducing the asymptomatic reservoir is theoretically important to elimination by preventing reestablishment of local transmission, the empirical evidence that screen-and-treat initiatives reduce the burden of malaria in practice is inconsistent [5–8]. While simulation analyses and a study from Zambia reported decreases in malaria infection following screen-and-treat efforts [5, 9], 2 cluster-randomized trials in Kenya and Burkina Faso found no difference in the change in malaria burden between villages receiving longitudinal screen-and-treat and villages receiving no intervention [7, 8].
Beyond conflicting evidence of effectiveness, ACD is costly and resource-intensive as an elimination strategy, particularly in low-transmission regions where few individuals have levels of parasitemia above the RDT limit of detection [10]. Therefore, reactive case detection (RCD), a specific type of ACD, has been adopted by numerous control programs in low-transmission settings, focusing screening efforts to geographic transmission “hotspots” rather than the entire community [3]. RCD, initiated after passive detection of an index case at a health facility, involves screening and treating individuals within the index household and in households a prescribed distance from the index household [3, 4]. Multiple studies have reported geographic clustering of malaria cases in such hotspots and noted that individuals living in index case households have a higher likelihood of being infected, supporting RCD as a strategy to disrupt transmission in ecological niches that support malaria transmission [11–19].
In Zambia, RCD was incorporated into the National Malaria Elimination Program (NMEP) in 2011 for regions reporting <1% RDT-positive prevalence, including Southern Province [20, 21]. In 2014, a ground-truth evaluation of RCD in Southern Province revealed that across 26 RCD events, only 32% of the households screened were actually located within the targeted 140-m radius, highlighting the challenges CHWs face in identifying eligible households [22]. Although RCD is theoretically appealing, the inconsistent data regarding ACD’s ability to decrease community-level transmission coupled with the logistical challenges of RCD necessitates validating that RCD is appropriate for the malaria epidemiology of a given region. In particular, it is essential to determine whether clusters of malaria cases are sustained through local transmission or other mechanisms. Whereas local transmission may be effectively targeted through interventions like RCD and focal vector control, eliminating case clusters that arose from household travel or occupational exposure would require different approaches. Distinguishing these scenarios empowers evidence-based prioritization of appropriate interventions tailored to the underlying transmission epidemiology.
Spatial and temporal patterns of malaria cases, sometimes in conjunction with travel data, have been commonly used to draw inferences regarding the epidemiology of local malaria transmission [23–25]. Although they are an important aspect of epidemiological monitoring, spatial and temporal patterns, even when coupled with travel data, cannot reliably distinguish local transmission chains from clusters of unrelated infections. Understanding the contribution of local transmission requires comparing parasite genotypes from infected individuals [26, 27]. To evaluate the ability of RCD to capture local malaria transmission in a region of Southern Province, Zambia, we performed microsatellite genotyping on dried blood spot (DBS) samples collected from index and secondary cases detected through RCD. We characterized the temporal and spatial scales on which genetically related parasites clustered, a proxy for local transmission, and identified a putative focal transmission hotspot.
METHODS
Study Region
This study was conducted in the rural catchment area of Macha Hospital, an approximately 2000-km2 region located in Choma District, Southern Province, Zambia, with an estimated 56000 residents, primarily subsistence farmers. This setting is characterized by a tropical savannah climate and experiences peak Plasmodium falciparum malaria during the single rainy season between November and April [28]. Anopheles arabiensis and Anopheles squamosus are the primary and secondary malaria vectors in this region, respectively [28–30]. P. falciparum histidine-rich protein 2 (PfHRP2) based RDT prevalence among actively detected individuals in this region declined from 9.2% in 2008 to <1% in 2013 [31]. The introduction of artemisinin combination therapy with artemether-lumefantrine in 2004 along with LLIN distributions in 2007, 2012, and 2014 may have contributed to the decline in malaria in this setting. LLIN ownership in this region was estimated to be 83% in 2013 [32]. The Zambian NMEP aims to eliminate malaria from the country by 2021, and Southern Province, including the study area, is considered a preelimination setting. For regions like this with low-level malaria transmission, the NMEP recommends RCD whereby CHWs screen using PfHRP2-based RDTs and treat positive individuals living within 140 m of index case households.
Informed Consent
This research was approved by the Institutional Review Board at the Johns Hopkins Bloomberg School of Public Health (Baltimore, Maryland) and the Ethics Review Committee of the Tropical Diseases Research Centre (Ndola, Zambia). All adult participants gave informed consent. All children had parental consent, and children aged 12–15 provided assent.
Enhanced Reactive Case Detection
Following passive detection of PfHRP2-RDT–positive index cases at 14 rural health centers (RHCs) serving the catchment area of Macha Hospital, study team members at Macha Research Trust (MRT) were alerted by text message. Within 1 week of the index case’s presentation at the RHC, study team members accompanied a CHW to screen all individuals living within an expanded RCD radius of 250 m from the index case household using RDTs. RDT-positive cases were offered treatment with artemether-lumefantrine. For all individuals screened, study team members collected a finger-prick blood sample as DBS on filter paper (Whatman 903 Protein Saver Card), administered an epidemiological and demographic survey, and collected global positioning system coordinates for each household. Households within 250 m of the index household were preidentified using satellite imagery to guide the field team in enrolling eligible households. DNA from the DBS samples was extracted using Chelex and screened for P. falciparum using quantitative polymerase chain reaction (qPCR) targeting mitochondrial cytochrome-b at the MRT laboratory in Macha, Zambia [31].
Passively Detected Cases at RHCs
Five of the 14 RHCs closest to Macha Hospital collected finger-prick blood samples as DBS from a convenience sample of passively detected index cases as previously described [33]. DBS samples were transported from the RHCs to MRT. Index cases were linked by name and visit date recorded at the RHC to the epidemiological and demographic survey questionnaire administered to the same individual up to 1 week later during RCD follow-up.
Sample Selection
We selected all RCD events between 1 January 2015 and 18 April 2016 for which at least 2 participants, including the index case, were malaria positive by RDT or qPCR. One hundred six samples (27 index, 79 secondary) across 49 RCD events met our inclusion criteria, and DBS samples were transported to the University of California, San Francisco.
Laboratory Procedures
DBS samples were stored with desiccant at –20° C until transport and processing. Six-millimeter hole-punches from filter cards were extracted using the saponin Chelex method [34] and quantified using var gene acidic terminal sequence (var-ATS) ultra-sensitive qPCR [35]. Samples containing <10 parasites/µL of blood (45/106 samples) underwent selective whole genome amplification (sWGA) to selectively enrich for parasite DNA as previously described, with some modifications (see Supplementary Table 1) [36, 37]. For quality control, we assessed the concordance of alleles and determined the error rate introduced during sWGA by genotyping control samples with and without sWGA. Samples with >10 parasites/µL and post-sWGA samples were amplified at 26 microsatellite loci [38, 39] (Liu et al, unpublished data). PCR products were diluted and sized by denaturing capillary electrophoresis on a Thermo Fisher 3730XL analyzer with GeneScan 400HD ROX size standard (Thermo Fisher Scientific). The resulting electropherograms were analyzed using microSPAT (Murphy et al, unpublished data). Samples with <10 successfully genotyped microsatellites and loci that did not amplify in at least 50% of the samples were excluded from the analysis. For each genotyped sample, we calculated the multiplicity of infection (MOI), or the number of genetically distinct parasite clones, as the second-highest number of alleles present at any locus, accounting for the possibility of false positive allele calls.
Characterizing Relatedness Captured During RCD
For each pair of samples genotyped, pairwise genetic relatedness was determined using a modified identity by state (IBS) metric [40] (Liu et al, unpublished data). Pairwise IBS was calculated from allele similarity between isolates, allowing estimation of genetic relatedness from monoclonal as well as polyclonal samples. For each locus, IBS was estimated by the sum of the product of within host allele frequency (defined as 1 / number of alleles detected) of shared alleles. The overall pairwise IBS was determined as the average of locus-specific estimates under the assumption of independent loci (Liu et al, unpublished data). We calculated the physical distance between all sample pairs as Euclidean distance between households using the R package “geosphere” [41]. Elapsed time between sample pairs was determined from the absolute value of the difference in sample collection dates. Sample pairs were considered related if their pairwise genetic relatedness was ≥0.5, the 96th percentile of pairwise relatedness across all sample pairs. We characterized mean pairwise genetic relatedness as well as the proportion of related samples across bins of increasing physical distance. For each RCD event, we calculated the mean pairwise relatedness between all sample pairs from that event.
Characterizing the Temporal and Spatial Boundaries of Malaria Transmission
We investigated the optimal temporal and spatial scales on which to detect related parasites in the study area. Given that the sampling framework inherently oversampled individuals clustered in time and space, we removed all pairwise comparisons between samples from the same RCD event. Using this subset of data corrected for the nonrandom sampling strategy, we binned the remaining pairwise comparisons into 9 space-time categories and calculated the mean genetic relatedness among pairwise comparisons across increasing time and distance.
Identifying Focal Transmission Hotspots
We developed a method to identify regions that sustain local transmission at higher than expected rates, referred to as focal transmission hotspots. We used the R package “sp” [42–44] to draw lines between all pairs of individuals harboring related parasites after removing pairwise comparisons between samples from the same RCD event. We created a grid with 81 equally sized cells across the study region. For each cell, we counted the number of related lines that passed through, generating a count of observed related connections. To calculate an expected number of related connections for each cell under the assumption that connections were random, we generated 1000 sampling replicates of size equal to the total number of related lines, each time sampling from all possible lines connecting all pairs of individuals regardless of relatedness. For each replicate, we summed the number of lines that passed through each cell, generating an expected line count and 95% confidence interval for each cell. We calculated the ratio of observed to expected lines for each cell and exported this file for use in ArcMap (Esri, version 10.5.1) [45].
RESULTS
Genotyping
Of the 26 microsatellite loci amplified, all but locus PolyA had genotyping success rates >50% (Supplementary Figure 2). Locus PolyA was therefore excluded from the analysis. Eighty-five of the 106 extracted samples were successfully genotyped for at least 10 of the 25 microsatellite loci and were included in the analysis. Insufficient coverage was significantly associated with negative RDT result, being a secondary case, and low parasitemia (Supplementary Figure 3). Among the 21 samples with insufficient coverage, 19 underwent sWGA. The relationship between microsatellite coverage and parasitemia is shown in Supplementary Figure 3. Samples excluded from the analysis did not differ significantly from those included in terms of sex, age, self-reported overnight travel, or self-reported LLIN use. In a quality control experiment, there was >99% allele concordance in control samples genotyped pre- and post-sWGA (Supplementary Table 2).
The 85 successfully genotyped samples came from 47 different RCD events. Index (n = 27) and secondary case individuals (n = 58) did not differ significantly with respect to sex, age, self-reported LLIN use, or self-reported overnight travel (Supplementary Table 3). Index cases had a higher proportion of RDT-positive individuals and a lower proportion of sWGA samples compared to secondary cases, as expected given the higher parasite densities generally seen in symptomatic infections (Supplementary Table 3). Among the samples included, the median MOI was 2. MOI did not differ between index and secondary cases (Supplementary Figure 4) or between sWGA (median, 2 [range, 1–3]) and non-sWGA (median, 2 [range, 1–4]) samples.
Relatedness by Physical Distance
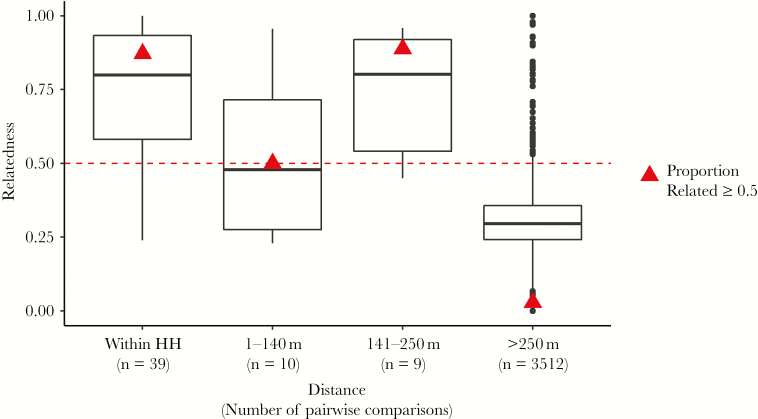
There was no significant difference in mean genetic relatedness between samples collected from within the same household and those 141–250 m apart (Figure 1). Samples collected 1–140 m apart were significantly less related than those within the same household (Student t test, P = .004) and those 141-250 m apart (Student t test, P = .05), but this difference may reflect the small sample size of 10 comparisons in the 1–140 m distance bin. When considered together, samples collected from participants located within 250 m had significantly higher mean genetic relatedness (mean relatedness, 0.71) than participants >250 m apart (mean relatedness, 0.31; Welch t test, P < .001), indicating that screening with a 250-m radius captures genetically related parasites. Among samples from the same RCD event, 78.6% were related, whereas only 2.9% of samples from different RCD events were related (test for difference in proportions, P < .001). The distributions of relatedness among sample pairs from the same RCD event and among those from different RCD events are shown in Supplementary Figure 1. Among the related pairwise comparisons >250 m apart, 60% occurred within 90 days, suggesting that relatedness among physically distant samples is nevertheless clustered temporally. In summary, RCD, both at the 140-m and 250-m radii, captured genetically related cases, suggesting that case clustering is due to local transmission rather than household-level risk factors.
Figure 1.
Genetic relatedness declines after 250 m. Genetic relatedness is shown for pairwise comparisons binned by increasing physical distance between sample pairs (within the same household [HH], samples 1–140 m apart, samples 141–250 m apart, and samples >250 m apart). The number of pairwise comparisons in each physical distance bin is listed below the bin label in parenthesis. Triangles indicate the proportion of comparisons in each category that are related above the 0.5 genetic relatedness threshold, demarcated by the dashed line. Among sample pairs >250 m apart, the mean physical distance was 20.2 km (range, 297 m–46.1 km).
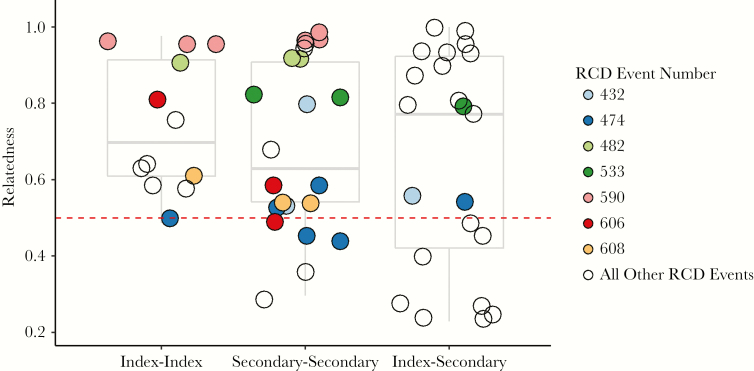
Relatedness Within RCD Events
Among the 56 sample pairs from the same RCD event, mean relatedness was 0.69. There was no significant difference in mean relatedness comparing 2 secondary cases, 2 index cases, or an index and secondary case from the same RCD event (analysis of variance, P = .62; Figure 2). Among the 7 RCD events with comparisons across multiple categories (index–index, secondary–secondary, or index–secondary), there was no difference in relatedness across categories, possibly reflecting the small sample size.
Figure 2.
There were 56 pairwise comparisons between samples from the same RCD event, representing 23 unique RCD events. Each of the 56 comparisons were binned according to whether the comparison was between 2 index cases (index–index), between 2 secondary cases (secondary–secondary), or between an index case and a secondary case (index–secondary) from the same RCD event. Genetic relatedness is shown for each of the 56 comparisons, represented as circles. For 7 RCD events that had comparisons in >1 category (index–index, secondary–secondary, or index–secondary), comparisons are shaded identically. Pairwise comparisons from RCD events for which only 1 category is represented are shown with hollow circles. The distributions of all pairwise comparisons for each of the 3 categories are shown by boxplots.
Relatedness Across Space and Time
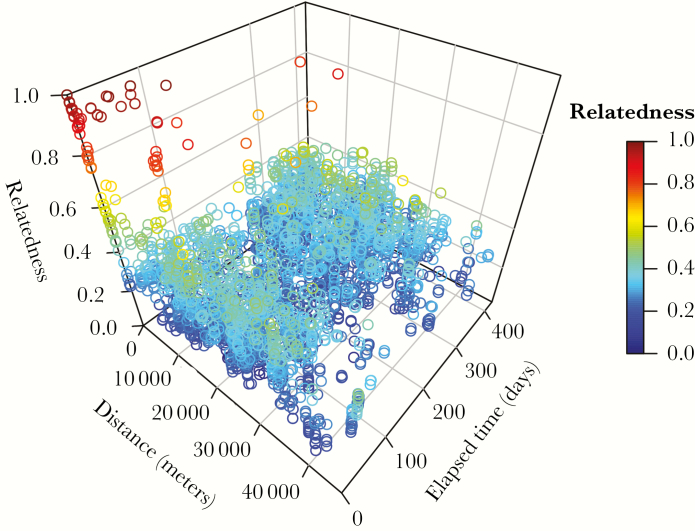
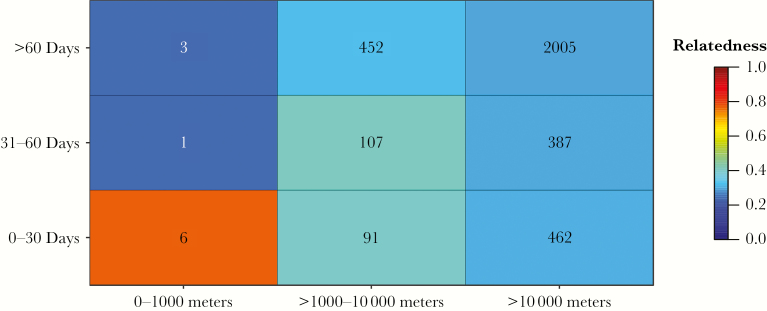
Related comparisons clustered focally in space and time (Figure 3), and pairwise genetic relatedness declined with increasing distance and elapsed time. Ninety-six percent of the 52 most highly related comparisons (related >0.80) were within 10 km and 92% occurred within 90 days (Figure 3), indicating that transmission occurred focally in space and time. Next, to verify that these relationships were not driven by the focal sampling framework, we removed all pairwise comparisons between samples from the same RCD event. Figure 4 illustrates the decline in genetic relatedness across increasing distance and elapsed time after removing these comparisons. After excluding pairs of samples from the same RCD event, most related comparisons occurred within 10 km and 60 days, although the small sample size of comparisons within 1000 m precludes our ability to infer definitive boundaries between 0 and 1000 m. Marginal distributions of relatedness by physical distance and elapsed time are included in Supplementary Figure 5.
Figure 3.
Relatedness declines across space and time. Each hollow circle represents a unique pairwise comparison between 2 samples. Circles are colored and positioned along the vertical axis according to the genetic relatedness between the samples compared. Relatedness of 1.0 (red) indicates that the samples are highly related, and relatedness of 0.0 (blue) indicates that the samples are unrelated. Physical distance and elapsed time between sample pairs are shown on the two horizontal axes.
Figure 4.
Relatedness declines across space and time excluding same reactive case detection (RCD) event comparisons. Pairwise comparisons (excluding those from the same RCD event) were binned into 9 space–time categories represented by the 9 boxes in the matrix. The number in each bin denotes the number of pairwise comparisons in each category. The mean relatedness of the pairwise comparisons in each bin is shown by the color gradient, where relatedness of 1.0 (red) is highly related and relatedness of 0.0 (blue) is unrelated.
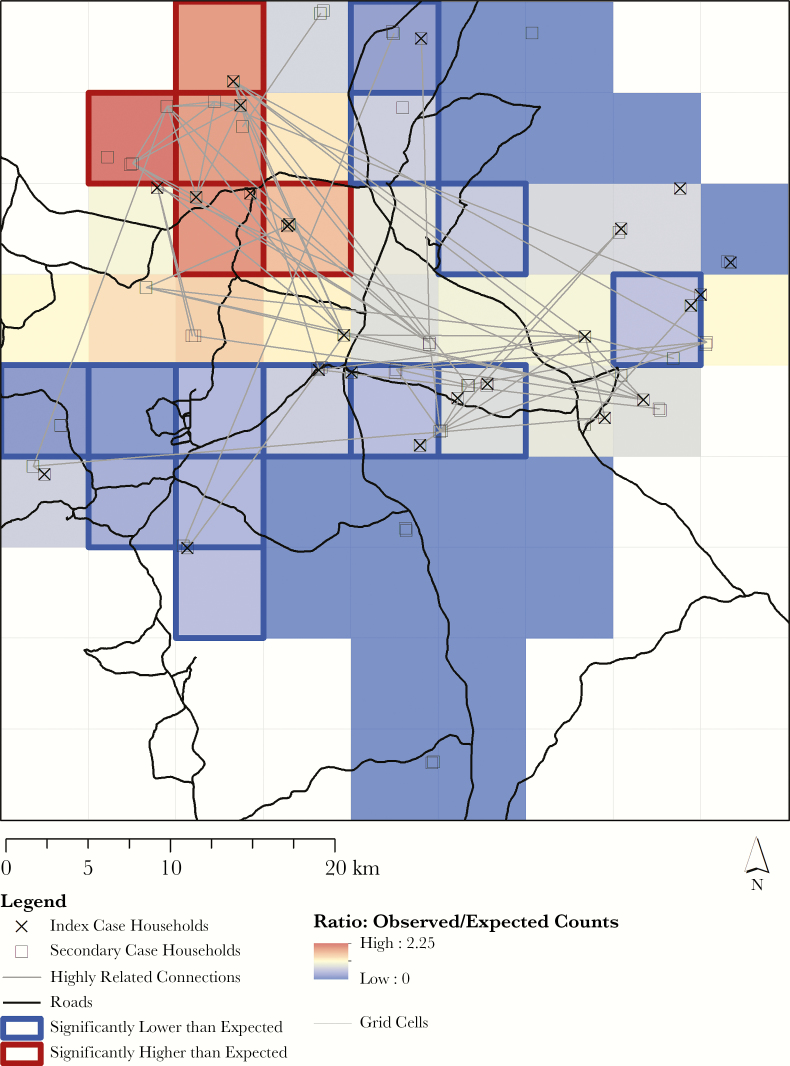
Identifying Focal Transmission Hotspots
We identified a region in the northwest quadrant of our study area as a putative focal transmission hotspot (Figure 5). After removing pairs of samples from the same RCD event and accounting for the spatial distribution and density of households in the analysis, this region had a significantly higher than expected number of related connections. Conversely, 2 regions located in the west-central and north-central areas of our study site had significantly fewer than expected related connections. Parasite genetic data enabled the detection of finer-scale transmission heterogeneities in our study area than could have been detected through epidemiological and spatial patterns alone.
Figure 5.
Observed vs expected counts of highly related connections. A map of the study area is overlaid with 81 equally sized, approximately 5-km grid cells. For each grid cell, the ratio comparing the number of related connections (pairwise comparisons related ≥0.5 after excluding comparisons from the same reactive case detection event) to the number of connections expected is depicted by a color gradient. A ratio > 1 (red) indicates that the cell has more related connections than expected; a ratio < 1 (blue) indicates that the cell has fewer related connections than expected. We observed a similar multiplicity of infection (MOI) comparing individuals living within and outside of focal transmission hotspots (mean MOI, 1.62 among individuals living in focal transmission hotspots vs 1.92 among individuals living outside of focal transmission hotspots; Student t test, P = .28).
DISCUSSION
Microsatellite genotyping from RCD participants demonstrated that focal malaria transmission was ongoing between January 2015 and April 2016 in a region of Southern Province, Zambia, targeted for elimination. Although spatial clustering of malaria cases has been previously described [11–14], this is, to our knowledge, the first study to genotype P. falciparum identified during RCD in southern Africa to attribute case clustering to local transmission. Using genetic methods to clarify the mechanism through which clusters of malaria cases arise has important implications for programmatic planning, enabling evidence-based decisions regarding effective control tools suited to a region’s unique malaria epidemiology. This study demonstrates that RCD captures genetically related infections and underscores the added value of genetic data to understanding the underlying malaria transmission epidemiology of a given region beyond what can be inferred through spatial and temporal patterns alone.
Our data, implicating local transmission as a contributor to the sustained malaria burden, highlights the importance of addressing the underlying transmission biology and ecology that perpetuate malaria in this region. These results suggest that local transmission in this region extends up to 250 m from the index household, and possibly beyond. Therefore, the Zambian NMEP’s recommended 140-m screening radius at 1 timepoint likely fails to capture infected individuals who may be capable of sustaining transmission after RCD. However, performing RCD across the entire landscape of local transmission may not be feasible or efficient, as previously suggested [46, 47]. Exploring additional options, such as repeated visits or coupling focal vector control with RCD may be useful to interrupt local transmission in this and similar settings. With the goal of identifying regions where transmission-disrupting interventions such as focal vector control could have the largest impact, we describe a novel approach for detecting focal transmission hotspots. Focal transmission hotspots may be ideal locations to implement additional transmission-disrupting interventions in conjunction with RCD. This proposed method for identifying focal transmission hotspots has not yet been rigorously assessed and will require expanded evaluation before being considered a reliable indicator for transmission foci.
The RCD sampling framework in this study is not the ideal dataset to delineate the spatial and temporal scales of local transmission. We did not genotype every index and secondary case between January 2015 and April 2016, but rather selected samples that came from RCD events where at least 2 individuals tested positive for malaria by RDT or qPCR. Given the nonrandom inclusion criteria and somewhat sparse sampling of this study, the spatial and temporal boundaries of local transmission we can infer—10 km and 60–90 days—require additional validation. Despite our inability to unequivocally identify the spatial and temporal boundaries of local transmission, our data strongly suggest that local transmission occurs focally in space and time. Temporal and spatial clustering was observed regardless of whether sample pairs from the same RCD event were included or excluded, indicating that our sampling framework did not solely determine the results. Our observations are consistent with simulation analyses from the region that suggested a 500-m RCD radius would identify 77% of additional cases [47]. Similarly, our observation that transmission occurs temporally for up to 60–90 days is corroborated by 24-SNP (single-nucleotide polymorphism) molecular barcode data suggesting ongoing transmission by highly related subsequent infections within index households 30 and 90 days after the initial RCD intervention [48]. Defining exact boundaries of local transmission would necessitate sampling longitudinally from a spatially representative framework.
We inferred local transmission from pairwise genetic relatedness. Although we are unable to confidently assert that any sample pairs harboring related parasites represented direct transmission from one to another, it is improbable that we would see genetic relatedness values >0.5 unless the individuals were connected in some manner through transmission. Our 0.5 threshold represented the 96th percentile of all pairwise relatedness values in the study, indicating how rare these related events are. Despite not having the granularity of connecting individual transmission events, the observed related infections likely indicate direct or indirect local transmission.
The ability to obtain genotypes from asymptomatic infections, the diverse, multiallelic loci used, and the use of a similarity metric capable of comparing infections with varying MOI contributed to the success of this study in a setting of low-density, polyclonal infections. However, there is room for further improvement of these methods. For example, we were unable to assign meaning to categories of genetic relatedness values <0.5, given the low precision of characterizing how relatedness across transmission generations decays for these markers in the context of polyclonal infections.
Characterizing the contribution of local transmission to sustained malaria is critical to programs transitioning from control to elimination. Knowledge of whether cases arise through travel, occupational exposure, or locally acquired transmission can guide intervention priorities. We discerned signatures of local malaria transmission using 106 DBS samples and a relatively low-tech genotyping method. Similar approaches could be valuable to other researchers and program leaders, particularly in resource-limited settings, interested in characterizing local transmission to guide malaria elimination.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We are grateful to the community of Macha, Zambia, and to the individuals who participated in this research.
Financial support. This research was financially supported by the National Institutes of Health (5T32AI007417 to J.C.P. as a trainee on the Molecular and Cellular Basis of Infectious Diseases Training Grant in the Department of Molecular Microbiology and Immunology at the Johns Hopkins Bloomberg School of Public Health and 5U19AI089680 to the Southern and Central African International Centers of Excellence in Malaria Research). This work was supported by the Bloomberg Philanthropies. G.C. received a postdoctoral fellowship award from the Johns Hopkins Malaria Research Institute. A. W. is funded by a Career Award at the Scientific Interface by the Burroughs Wellcome Fund. B. G. is a Chan Zuckerberg Biohub investigator.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Genetic Epidemiology of Malaria Conference, Hixton, United Kingdom, June 2018; 67th Annual Meeting of the American Society of Tropical Medicine and Hygiene, 28 October–1 November 2018, New Orleans, Louisiana.
References
- 1. Bhatt S, Weiss DJ, Cameron E, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 2015; 526:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Moonen B, Cohen JM, Snow RW, et al. Operational strategies to achieve and maintain malaria elimination. Lancet 2010; 376:1592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sturrock HJ, Novotny JM, Kunene S, et al. Reactive case detection for malaria elimination: real-life experience from an ongoing program in Swaziland. PLoS One 2013; 8:e63830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. World Health Organization. A framework for malaria elimination. Geneva, Switzerland: WHO, 2017. [Google Scholar]
- 5. Kern SE, Tiono AB, Makanga M, et al. Community screening and treatment of asymptomatic carriers of Plasmodium falciparum with artemether-lumefantrine to reduce malaria disease burden: a modelling and simulation analysis. Malar J 2011; 10:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sutcliffe CG, Kobayashi T, Hamapumbu H, et al. Reduced risk of malaria parasitemia following household screening and treatment: a cross-sectional and longitudinal cohort study. PLoS One 2012; 7:e31396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tiono AB, Ouédraogo A, Ogutu B, et al. A controlled, parallel, cluster-randomized trial of community-wide screening and treatment of asymptomatic carriers of Plasmodium falciparum in Burkina Faso. Malar J 2013; 12:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Halliday KE, Okello G, Turner EL, et al. Impact of intermittent screening and treatment for malaria among school children in Kenya: a cluster randomised trial. PLoS Med 2014; 11:e1001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Larsen DA, Bennett A, Silumbe K, et al. Population-wide malaria testing and treatment with rapid diagnostic tests and artemether-lumefantrine in southern Zambia: a community randomized step-wedge control trial design. Am J Trop Med Hyg 2015; 92:913–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rogawski ET, Congpuong K, Sudathip P, et al. Active case detection with pooled real-time PCR to eliminate malaria in Trat province, Thailand. Am J Trop Med Hyg 2012; 86:789–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bejon P, Williams TN, Liljander A, et al. Stable and unstable malaria hotspots in longitudinal cohort studies in Kenya. PLoS Med 2010; 7:e1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bousema T, Drakeley C, Gesase S, et al. Identification of hot spots of malaria transmission for targeted malaria control. J Infect Dis 2010; 201:1764–74. [DOI] [PubMed] [Google Scholar]
- 13. Pinchoff J, Henostroza G, Carter BS, et al. Spatial patterns of incident malaria cases and their household contacts in a single clinic catchment area of Chongwe District, Zambia. Malar J 2015; 14:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Björkman A, Cook J, Sturrock H, et al. Spatial distribution of falciparum malaria infections in Zanzibar: implications for focal drug administration strategies targeting asymptomatic parasite carriers. Clin Infect Dis 2017; 64:1236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Eijk AM, Ramanathapuram L, Sutton PL, et al. What is the value of reactive case detection in malaria control? A case-study in India and a systematic review. Malar J 2016; 15:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stresman GH, Kamanga A, Moono P, et al. A method of active case detection to target reservoirs of asymptomatic malaria and gametocyte carriers in a rural area in Southern Province, Zambia. Malar J 2010; 9:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Littrell M, Sow GD, Ngom A, et al. Case investigation and reactive case detection for malaria elimination in northern Senegal. Malar J 2013; 12:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stresman GH, Baidjoe AY, Stevenson J, et al. Focal screening to identify the subpatent parasite reservoir in an area of low and heterogeneous transmission in the Kenya highlands. J Infect Dis 2015; 212:1768–77. [DOI] [PubMed] [Google Scholar]
- 19. Aidoo EK, Afrane YA, Machani MG, et al. Reactive case detection of Plasmodium falciparum in western Kenya highlands: effective in identifying additional cases, yet limited effect on transmission. Malar J 2018; 17:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zambia National Malaria Elimination Centre. National Malaria Elimination Strategic Plan 2017–2021: moving from accelerated burden reduction to malaria elimination in Zambia. Lusaka: Zambia National Malaria Elimination Centre, 2017. [Google Scholar]
- 21. Larsen DA, Chisha Z, Winters B, et al. Malaria surveillance in low-transmission areas of Zambia using reactive case detection. Malar J 2015; 14:465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Searle KM, Hamapumbu H, Lubinda J, et al. Southern Africa International Centers of Excellence for Malaria Research Evaluation of the operational challenges in implementing reactive screen-and-treat and implications of reactive case detection strategies for malaria elimination in a region of low transmission in southern Zambia. Malar J 2016; 15:412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sturrock HJ, Roberts KW, Wegbreit J, Ohrt C, Gosling RD. Tackling imported malaria: an elimination endgame. Am J Trop Med Hyg 2015; 93:139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruktanonchai NW, DeLeenheer P, Tatem AJ, et al. Identifying malaria transmission foci for elimination using human mobility data. PLoS Comput Biol 2016; 12:e1004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reiner RC, Le Menach A, Kunene S, et al. Mapping residual transmission for malaria elimination. Elife 2015; 4:e09520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mi Choi K, Kyoung Choi Y, Kang YA, et al. Study of the genetic discrimination between imported and autochthonous cases of malaria in South Korea. J Travel Med 2011; 18:63–6. [DOI] [PubMed] [Google Scholar]
- 27. Rice BL, Golden CD, Anjaranirina EJ, Botelho CM, Volkman SK, Hartl DL. Genetic evidence that the Makira region in northeastern Madagascar is a hotspot of malaria transmission. Malar J 2016; 15:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kent RJ, Thuma PE, Mharakurwa S, Norris DE. Seasonality, blood feeding behavior, and transmission of Plasmodium falciparum by Anopheles arabiensis after an extended drought in southern Zambia. Am J Trop Med Hyg 2007; 76:267–74. [PMC free article] [PubMed] [Google Scholar]
- 29. Stevenson JC, Pinchoff J, Muleba M, et al. Southern Africa International Centers of Excellence in Malaria Research Spatio-temporal heterogeneity of malaria vectors in northern Zambia: implications for vector control. Parasit Vectors 2016; 9:510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fornadel CM, Norris LC, Glass GE, Norris DE. Analysis of Anopheles arabiensis blood feeding behavior in southern Zambia during the two years after introduction of insecticide-treated bed nets. Am J Trop Med Hyg 2010; 83:848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laban NM, Kobayashi T, Hamapumbu H, et al. Southern Africa International Centers of Excellence for Malaria Research Comparison of a PfHRP2-based rapid diagnostic test and PCR for malaria in a low prevalence setting in rural southern Zambia: implications for elimination. Malar J 2015; 14:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pinchoff J, Hamapumbu H, Kobayashi T, et al. Southern Africa International Centers of Excellence for Malaria Research Factors associated with sustained use of long-lasting insecticide-treated nets following a reduction in malaria transmission in southern Zambia. Am J Trop Med Hyg 2015; 93:954–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Searle KM, Katowa B, Kobayashi T, et al. Southern Africa International Centers of Excellence for Malaria Research Distinct parasite populations infect individuals identified through passive and active case detection in a region of declining malaria transmission in southern Zambia. Malar J 2017; 16:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg 1995; 52:565–8. [DOI] [PubMed] [Google Scholar]
- 35. Hofmann N, Mwingira F, Shekalaghe S, Robinson LJ, Mueller I, Felger I. Ultra-sensitive detection of Plasmodium falciparum by amplification of multi-copy subtelomeric targets. PLoS Med 2015; 12:e1001788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oyola SO, Ariani CV, Hamilton WL, et al. Whole genome sequencing of Plasmodium falciparum from dried blood spots using selective whole genome amplification. Malar J 2016; 15:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sundararaman SA, Plenderleith LJ, Liu W, et al. Genomes of cryptic chimpanzee Plasmodium species reveal key evolutionary events leading to human malaria. Nat Commun 2016; 7:11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anderson TJ, Haubold B, Williams JT, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol 2000; 17:1467–82. [DOI] [PubMed] [Google Scholar]
- 39. Anderson TJ, Su XZ, Bockarie M, Lagog M, Day KP. Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology 1999; 119(Pt 2):113–25. [DOI] [PubMed] [Google Scholar]
- 40. Jacquard A, Charlesworth B, Charlesworth D.. The genetic structure of populations. Berlin: Springer, 2012. [Google Scholar]
- 41. Hijmans RJ. Geosphere: spherical trigonometry 2017. https://cran.r-project.org/package=geosphere. Accessed 18 April 2018.
- 42. Bivand RS, Pebesma E, Gómez-Rubio V.. Applied spatial data analysis with R. New York: Springer, 2013. [Google Scholar]
- 43. Pebesma E, Bivand RS. Classes and methods for spatial data: the sp package 2005. https://cran.r-project.org/web/packages/sp/vignettes/intro_sp.pdf. Accessed 15 March 2018.
- 44. R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2018. https://www.r-project.org/. Accessed 22 June 2018. [Google Scholar]
- 45. Esri. ArcGIS desktop: release 10.5.1. Redlands, CA: Environmental Systems Research Institute, 2017. [Google Scholar]
- 46. Deutsch-Feldman M, Hamapumbu H, Lubinda J, et al. Efficiency of a malaria reactive test-and-treat program in southern Zambia: a prospective, observational study. Am J Trop Med Hyg 2018; 98:1382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Searle KM, Shields T, Hamapumbu H, et al. Efficiency of household reactive case detection for malaria in rural southern Zambia: simulations based on cross-sectional surveys from two epidemiological settings. PLoS One 2013; 8:e70972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Searle KM, Pringle JC, Hamapumbu H, et al. Evaluating the efficiency of reactive case detection to achieve malaria elimination in rural southern Zambia using follow-up household visit and parasite genotyping [abstract 1326]. In: Program and Abstracts of the Proceedings of the 66th Annual Meeting of the American Society of Tropical Medicine and Hygiene, Baltimore, MD, 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.