Abstract
Influenza viruses routinely acquire mutations in their hemagglutinin (HA) and neuraminidase (NA) glycoproteins that abrogate binding of pre-existing antibodies in a process known as antigenic drift. Most human antibodies against HA and NA are directed against epitopes that are hypervariable and not against epitopes that are conserved among different influenza virus strains. Universal influenza vaccines are currently being developed to elicit protective responses against functionally conserved sites on influenza proteins where viral escape mutations can result in large fitness costs [1]. Universal vaccine targets include the highly conserved HA stem domain [2–12], the less conserved HA receptor-binding site (RBS) [13–16], as well as conserved sites on NA [17–19]. One central challenge of universal vaccine efforts is to steer human antibody responses away from immunodominant, variable epitopes and towards subdominant, functionally conserved sites. Overcoming this challenge will require further understanding of the structural basis of broadly neutralizing HA and NA antibody binding epitopes and factors that influence immunodominance hierarchies of human antibody responses.
Keywords: influenza virus, antibody response, immune imprinting, virus evolution, vaccine design
IMMUNODOMINANCE OF THE HEMAGGLUTININ HEAD
Antibodies targeting epitopes in the HA globular head domain can protect animals and humans from influenza virus infections [20]. The majority of these antibodies neutralize by blocking viral attachment to host cells, although other neutralization mechanisms might be in play for some of these antibodies [1, 21]. Infection and vaccination typically elicit strain-specific HA-head antibodies that are often long-lived [22], but these antibodies can become ineffective when viruses acquire antigenic changes in the HA head. Such an example occurred during the 2014–2015 season when a new antigenically drifted H3N2 strain possessing a novel glycosylation site on the HA head caused dramatically reduced vaccine effectiveness [23, 24]. Although most antibodies against the HA head are directed against epitopes adjacent to the conserved HA RBS [25, 26], some antibodies are able to partially mimic the sialic acid receptor and bind to conserved residues within the HA RBS [13, 15, 16, 27, 28]. Hemagglutinin stem antibody responses constitute a small fraction of total anti-influenza virus antibodies in most humans [29]. In contrast to most epitopes on the HA head, the HA stem is less tolerant of change [30–33] and is much more highly conserved across subtypes. Although some anti-HA stem monoclonal antibodies can directly neutralize viruses through inhibiting HA proteolytic processing, pH-induced conformational changes, and viral egress [1, 21], many HA stem antibodies require Fc-mediated effector functions for in vivo protection [34].
HEMAGGLUTININ IMMUNODOMINANCE OF PRIMARY ANTIBODY RESPONSES
Antibodies against highly exposed epitopes on the HA head usually dominate the primary responses against influenza viruses (Figure 1A). Primary H3N2 infections in ferrets elicit high levels of antibodies that are directed towards HA antigenic sites A and B [35, 36], which are located in close proximity to the HA RBS [25]. Likewise, H1N1-infected young children tend to mount antibody responses to epitopes in antigenic sites near the HA RBS [37]. Although steric hindrance or inaccessibility has been suggested to contribute to the immunosubdominance of HA stem antibodies [38], recombinant HA vaccines also fail to elicit high-titer HA stem responses [39]. In fact, cryoelectron tomography has shown that the majority of the HA on influenza virions are indeed available to bind to stem antibodies [40]. Some HA stem antibodies can be polyreactive [38], and it is possible that selection against B-cells specific for HA stem epitopes contributes to HA stem antibody immunosubdominance. Recent data suggest that the fine specificity of influenza virus antibody responses in mice changes over time [41]. Angeletti et al [41] found that (1) antibodies against epitopes near the top of the HA head dominate the early response and (2) antibodies against other epitopes increase later in the response. Given that most studies have only examined a limited range of timepoints, it is likely that shifts in antibody immunodominance dynamics have yet to be fully explored.
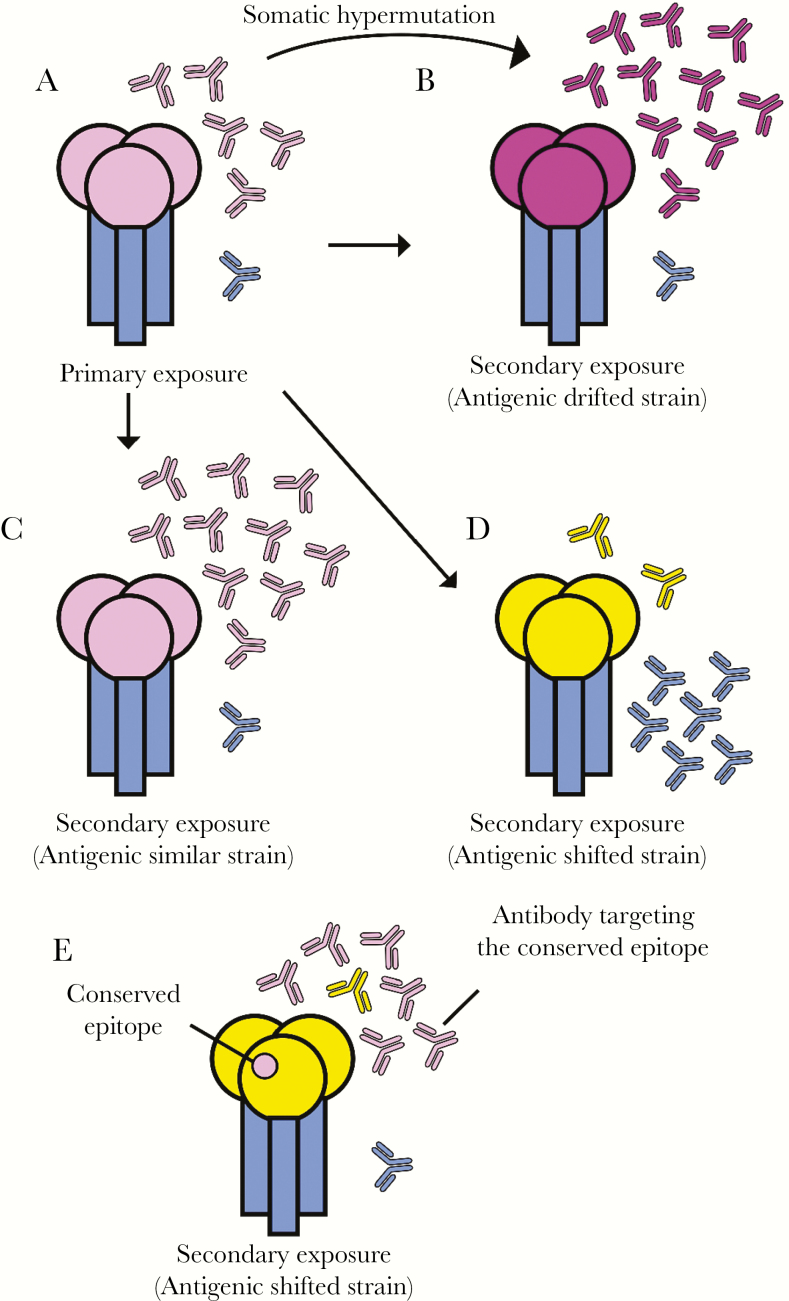
Figure 1.
Immunodominance of primary responses and recall responses against influenza hemagglutinin (HA). (A) The HA head domain (pink) is immunodominant in primary responses, whereas antibodies against the stem domain (blue) are rare. (B–C) Antibodies against the HA head remain dominant after exposure to antigenically similar (B) and antigenically drifted (C) seasonal viral strains. Antibodies elicited by antigenically drifted seasonal influenza virus strains often have high levels of somatic hypermutations that allow recognition of altered epitopes. (D–E) Antibodies against new pandemic viral strains tend to be more dominant initially against the (D) conserved HA stem, and (E) rare conserved epitopes, if any, in the HA head. Memory B cells producing antibodies against these conserved epitopes are preferentially boosted upon exposure to new pandemic viral strains. The color similarity of the HA head domain represents the similarity of the antigenicity in all figure panels.
Almost all immunological studies of influenza virus have been carried out in organisms that make immunoglobulin (Ig)-based humoral responses. To test whether some features of immunodominance are antigen-intrinsic, Altman et al [42] studied immune responses in lampreys that were immunized with influenza virus. Lampreys, a jawless fish, lack Ig genes but encode variable lymphocyte receptors (VLRs), which are an entirely different system of humoral adaptive immunity based on Leu-rich repeats rather than Ig domains. Remarkably, lamprey VLR responses were found to be focused on the same HA epitopes as those that have been observed in mice [42]. The similarity of antibody and VLR responses against HA in mice and lamprey suggest that properties of the HA protein itself contribute to antibody immunodominance hierarchies.
NEW HEMAGGLUTININ STEM-BASED UNIVERSAL VACCINE APPROACHES
Several universal vaccines are being developed to elicit antibodies against the immunosubdominant HA stem. One approach is to generate stable “headless” HA constructs that lack the head domain [10] and, as a result, induce antibody responses exclusively directed against HA stem epitopes [2, 3]. Another approach is sequential immunization with chimeric HAs that express divergent head domains with the goal of refocusing antibody responses towards the HA stem domain [12] (also see Krammer and Palese in this issue). This approach is promising because chimeric HAs selectively recall subdominant HA stem-reactive B-cells in the absence of HA head-reactive immunity. Both of these approaches have shown protection in animal models, but their success in humans will likely depend on their ability to induce protective responses in the context of differing pre-existing immunity in different individuals and age groups.
IMMUNE HISTORY SHAPES SECONDARY IMMUNE RESPONSES
It has been known since the 1950s that antibodies elicited by primary influenza virus exposures are highly strain-specific, whereas antibodies elicited by secondary exposures with antigenically distinct viral strains tend to be highly cross-reactive with the first strain encountered [43–45] (Figure 1B and C). This observation was originally referred to as “original antigenic sin” [45], and it has been more recently referred to as “antigenic seniority” [46] or “immune imprinting” [47]. Although the mechanisms behind original antigenic sin have yet to be fully elucidated, it is thought that cross-reactive B cells elicited by previous influenza virus exposures are preferentially recalled upon exposure with an antigenically distinct viral strain (Figure 1D).
Several studies suggest that prior seasonal H1N1 exposures influenced the fine-specificity of antibodies elicited against the antigenically distinct 2009 pandemic H1N1 virus in humans [48–53]. In many adults, the 2009 pandemic H1N1 strain preferentially boosted HA stem antibodies [48, 49, 54, 55], likely because this strain possessed a radically different HA head but a similar HA stem compared with previously circulating seasonal H1N1 strains. However, HA stem antibodies were not the only antibody type that was preferentially recalled in humans exposed to the 2009 pandemic H1N1. In some individuals, the 2009 pandemic H1N1 virus elicited antibody responses that were highly focused on rare HA head epitopes that were conserved in seasonal H1N1 strains to which they were exposed in childhood [50–53] (Figure 1E). More importantly, different aged individuals were found to mount antibody responses of different specificities upon exposure to the 2009 pandemic H1N1 virus, due to differences in seasonal H1N1 exposure histories. Age-related differences in antibody specificity appeared to play a role during the 2013–2014 season when a drifted pandemic H1N1 strain acquired an HA mutation in an epitope that was preferentially targeted by middle-aged individuals and, as a consequence, caused a disproportionate amount of disease in this population [51, 56, 57]. Animals sequentially infected with seasonal H1N1 and the 2009 pandemic H1N1 strains produce antibodies that have similar specificities compared with those elicited in humans who were likely sequentially exposed to these viruses [50, 51].
These findings are consistent with a recent study by Gostic et al [47] who used epidemiological data to demonstrate a correlation between the probability of first exposure in early childhood to either a group 1 or group 2 HA and susceptibility to avian H7N9 and H5N1 viral strains, respectively. They found that individuals who were likely exposed to a virus with a group 1 HA in childhood appeared to be protected from H5N1 but susceptible to H7N9, whereas individuals who were likely exposed to a virus with a group 2 HA in childhood appeared to be protected from H7N9 but susceptible to H5N1. Thus, it appears that individuals exposed in childhood to group 1 HAs are more likely to respond well to group 1 HA stem antigens, whereas individuals exposed in childhood to group 2 HAs are more likely to respond well to group 2 HA stem antigens. A deeper understanding of the complexities of human prior exposure and the interplay with universal vaccine candidates will likely be required to design better vaccines and vaccine regimens.
DIFFERENCES BETWEEN INFLUENZA GROUP 1 AND GROUP 2 HEMAGGLUTININ STEMS
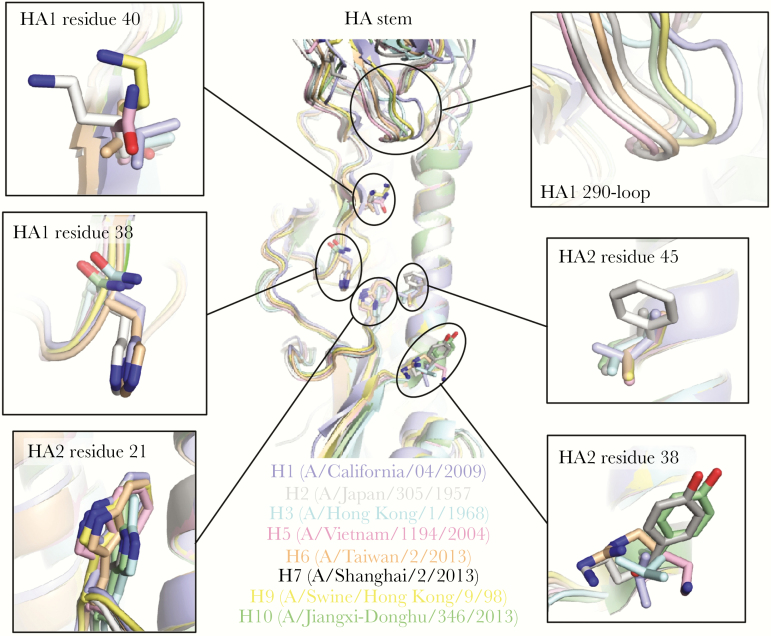
Although a few HA stem-reactive antibodies can target both group 1 and 2 HAs [27, 58–64], many are group specific [58, 63–70]. Several structural features are conserved within, but not across, group 1 or group 2 HAs. For example, the N-glycosylation site at HA1 Asn38 is highly conserved in group 2, but it is not present in group 1 HAs (Figure 2). In addition, the orientation and positioning of HA2 Trp21 differ between group 1 and group 2 HAs (Figure 2). A higher variability can be observed in other structural features in the HA stem and proximal regions, such as HA2 residue 38 and HA1 290-loop that are often contacted by stem-reactive antibodies [58–60, 65, 69, 71]. Subtype-specific variation can also be observed. For example, whereas HA1 Ile45 is highly conserved across group 1 and 2 HAs, HAs of the human H2 subtype have HA1 Phe45 instead (Figure 2). Some of these features have been associated with limiting the breadth of HA stem-reactive antibodies. The most well known example is perhaps the group 2-specific N-glycosylation site at HA1 Asn38 [72], which restricts the approach angle for antibodies to the highly conserved epitope in the stem of group 2 HAs [60]. Due to the structural difference in group 1 and group 2 HAs, it is not surprising that the germline usage of group 1-specific HA stem antibodies also seems to have a different preference than that of group 2-specific HA stem antibodies [73]. Most group 1-specific HA stem-binding antibodies utilize VH1-69 germline [64–66, 69, 70, 74], whereas group 2-specific HA stem-binding antibodies utilize a more diverse set of germlines, such as VH1-2 [64], VH3-53 [64], VH1-3 [67], and VH1-18 [75]. Nonetheless, studies have shown that the breadth of broadly neutralizing antibodies (bnAbs) can be increased during memory B cell evolution [59, 62, 70]. For example, although FI6 and MEDI8852 are both cross-group anti-HA stem antibodies, their germline versions (VH3-30 and VH6-1, respectively) only react with group 1 HAs [59, 61]. In fact, VH6-1 has been proposed to be a germline that encodes a multidonor class of bnAbs [63]. Therefore, it is possible for an HA group-specific anti-HA stem antibody to evolve a cross-group breadth through affinity maturation.
Figure 2.
Structural variation in the hemagglutinin (HA) stem and neighboring regions. Structures of HA protein from subtypes that have caused human infection are aligned by the helix A (HA2 residues 38–55) in the stem region: H1 (PDB 3LZG) [53], H2 (PDB 3KU5) [93], H3 (PDB 4FNK) [16], H5 (PDB 4BGW) [94], H6 (PDB 4XKD) [95], H7 (PDB 4LN6) [96], H9 (PDB 1JSD) [97], H10 (PDB 4XQ5) [98]. Zoomed-in views for several structural features of interest are shown. PDB, Protein Data Bank.
Recent studies from the Vaccine Research Center at the National Institutes of Health have elucidated several classes of cross-group anti-HA stem antibodies that were commonly observed in individuals after H5N1 (group 1) or H7N9 (group 2) vaccinations [63, 64]. An additional observation is that the anti-HA stem antibodies from H5N1-vaccinated individuals are primarily group 1-specific, whereas most antistem antibodies from H7N9-vaccinated individuals can react with both group 1 and group 2 HAs [64]. Such results suggest that there is a higher chance to induce cross-group anti-HA stem antibodies from a group 2 HA stem-based immunogen than from a group 1 HA stem-based immunogen. Nonetheless, most HA stem-based immunogen designs to date [76] are based on group 1 HAs (H1 [2, 3, 5, 6, 9] and H5 [4]), although some success has come from group 2 based HAs (H3 [7, 8]). Head-to-head comparison should be performed in the future between group 2 HA stem-based and group 1 HA stem-based immunogens to see whether one is superior to the other in inducing cross-group antistem antibodies. It will also be important to establish whether sequential vaccinations with group 1 HA stem vaccines followed by group 2 HA stem vaccines elicit different types of antibodies compared with sequential vaccinations with group 2 HA stem vaccines followed by group 1 HA stem vaccines. Throughout all of these studies, it will be crucial to take into account vaccinees’ year of birth and the potential effects of HA imprinting from early childhood influenza virus exposures.
ESCAPE MUTATIONS TO HEMAGGLUTININ STEM ANTIBODIES
An important consideration for antiviral and vaccine development is the potential emergence of escape mutants. The HA head can tolerate a lot more mutations, additional or changing glycosylation sites, or even insertions, when compared with the HA stem [30–33]. As a result, anti-HA head antibodies are more prone to escape than anti-HA stem antibodies [77]. Nevertheless, strong escape mutations to the anti-HA stem antibodies have been isolated [66, 67, 71, 75, 78–80]. Chai et al [80] have shown that both decrease in antibody-binding affinity and enhancing membrane fusion can contribute to escape from anti-HA stem antibodies. However, not all attempts to isolate strong escape mutants to the anti-HA stem antibodies have been successful. Doud et al [77] used deep mutational scanning to systematically search for escape mutations against 2 HA stem-binding bnAbs, namely FI6v3 [59] and C179 [78], but only weak escape mutants were found. Likewise, escape mutants to HA stem-binding bnAb CR6261 were only identified after extensive passaging [66]. It remains to be resolved how strong escape can be readily identified in some studies but not in others. It is possible that certain anti-HA stem antibodies are more prone to result in viral escape. It should also be noted that different studies often use different viral strains to search for escape mutants. The escape profiles of anti-HA stem antibodies may also vary among strains and subtypes, as suggested by the differential ability of A/California/7/2009 (H1N1) and A/Perth/16/2009 (H3N2) to escape from anti-HA stem antibody 39.29 [80]. Likewise, whereas only weak escape mutants to C179 were identified in A/WSN/1933 (H1N1) [77], complete escape mutants were identified in A/Suita/1/1989 (H1N1) and A/Izumi/5/1965 (H2N2) [78]. In fact, such a phenomenon of strain- or subtype-specific mutational effects has been described in the study of the HA RBS, where the tolerability to certain mutations differs between subtypes [81] or even among strains within a given subtype [82]. Several anti-HA stem antibodies are undergoing clinical trials as therapeutics [83], and an increasing amount of resources is being invested in the development of an HA stem-based universal vaccine [84]. Therefore, a comprehensive understanding of possible escape mutations is desirable to minimize unwanted surprises. Furthermore, because combining multiple antibodies can potentially minimize the emergence of escape mutants [85–87], a universal vaccine where escape is minimized will likely require elicitation of polyclonal responses targeting different HA epitopes.
MOVING FORWARD
Although current influenza HA stem-based universal vaccine candidates are promising, the development of a universal vaccine will likely be an iterative process, and a better understanding of the dynamics of immunodominance in humans will be essential for improving such vaccines. In the case of human immunodeficiency virus, a great deal has also been learned about the development of broadly neutralizing antibody responses by studying antibody-virus coevolution from the time of infection [88–90]. The analogous situation in influenza is more challenging, because it requires following individuals from birth in longitudinal studies and defining how immunodominance changes over the course of a response and from response to response. These longitudinal cohort studies have the potential to answer fundamental questions about (1) what antibody specificities dominate the plasmablast response versus B-cell memory and (2) which lineages are recalled in the response to an antigenically drifted strain. More importantly, these studies will also allow us to explore differences in responses elicited by infection and vaccination. Some studies suggest that B cells recalled in response to vaccination have a reduced ability to undergo somatic mutation relative to those recalled by an infection [91]. Although titers elicited by vaccination in adults exhibit modest waning [92], we know very little about the longevity of responses elicited in children.
Another major challenge will be to develop standardized assays to detect antibodies against different HA and NA epitopes. The standard assays used to select vaccine strains almost exclusively detect antibodies that bind the HA head and block viral attachment to cellular receptors. New assays to measure antibody functions such as HA stem binding and neutralization, NA inhibition, and Fc-mediated effector engagement need to be developed. Dissecting the contribution of different epitopes to protection in universal vaccine trials will allow us to precisely determine which epitopes are targeted in different individuals and whether viral escape is occurring at particular epitopes.
CONCLUSIONS
The current generation of universal vaccine candidates are the product of decades of work across multiple disciplines and represent the first attempt to control influenza immunodominance to elicit long-lived, protective responses against conserved sites. Understanding and manipulating immunodominance will be the crux of continued progress towards universal influenza immunity.
Notes
Financial support. I. A. W. received funding from National Institutes of Health (NIH) Grant R56 AI127371. S. E. H. received funding from NIH Grants 1R01AI113047, 1R01AI108686, and HHSN272201400005C. S. E. H. holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund.
Potential conflicts of interest. S. E. H. has received consultancy fees from Lumen, Novavax, and Merck for work unrelated to this report. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Krammer F, Palese P. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 2015; 14:167–82. [DOI] [PubMed] [Google Scholar]
- 2. Impagliazzo A, Milder F, Kuipers H, et al. . A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015; 349:1301–6. [DOI] [PubMed] [Google Scholar]
- 3. Yassine HM, Boyington JC, McTamney PM, et al. . Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 2015; 21:1065–70. [DOI] [PubMed] [Google Scholar]
- 4. Valkenburg SA, Mallajosyula VV, Li OT, et al. . Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci Rep 2016; 6:22666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wohlbold TJ, Nachbagauer R, Margine I, Tan GS, Hirsh A, Krammer F. Vaccination with soluble headless hemagglutinin protects mice from challenge with divergent influenza viruses. Vaccine 2015; 33:3314–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mallajosyula VV, Citron M, Ferrara F, et al. . Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci U S A 2014; 111:E2514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mallajosyula VV, Citron M, Ferrara F, et al. . Hemagglutinin sequence conservation guided stem immunogen design from influenza A H3 subtype. Front Immunol 2015; 6:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bommakanti G, Citron MP, Hepler RW, et al. . Design of an HA2-based Escherichia coli expressed influenza immunogen that protects mice from pathogenic challenge. Proc Natl Acad Sci U S A 2010; 107:13701–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bommakanti G, Lu X, Citron MP, et al. . Design of Escherichia coli-expressed stalk domain immunogens of H1N1 hemagglutinin that protect mice from lethal challenge. J Virol 2012; 86:13434–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steel J, Lowen AC, Wang TT, et al. . Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 2010; 1:e00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krammer F, Hai R, Yondola M, et al. . Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J Virol 2014; 88:3432–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krammer F, Pica N, Hai R, Margine I, Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 2013; 87:6542–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whittle JR, Zhang R, Khurana S, et al. . Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 2011; 108:14216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmidt AG, Xu H, Khan AR, et al. . Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A 2013; 110:264–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmidt AG, Therkelsen MD, Stewart S, et al. . Viral receptor-binding site antibodies with diverse germline origins. Cell 2015; 161:1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ekiert DC, Kashyap AK, Steel J, et al. . Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012; 489:526–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen YQ, Wohlbold TJ, Zheng NY, et al. . Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell 2018; 173:417–429.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu WC, Lin CY, Tsou YT, Jan JT, Wu SC. Cross-reactive neuraminidase-inhibiting antibodies elicited by immunization with recombinant neuraminidase proteins of H5N1 and pandemic H1N1 influenza A viruses. J Virol 2015; 89:7224–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krammer F, Fouchier RAM, Eichelberger MC, et al. . NAction! How can neuraminidase-based immunity contribute to better influenza virus vaccines? mBio 2018; 9:e02332–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Coudeville L, Bailleux F, Riche B, Megas F, Andre P, Ecochard R. Relationship between haemagglutination-inhibiting antibody titres and clinical protection against influenza: development and application of a bayesian random-effects model. BMC Med Res Methodol 2010; 10:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brandenburg B, Koudstaal W, Goudsmit J, et al. . Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS One 2013; 8:e80034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu X, Tsibane T, McGraw PA, et al. . Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 2008; 455:532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zimmerman RK, Nowalk MP, Chung J, et al. . 2014–2015 influenza vaccine effectiveness in the United States by vaccine type. Clin Infect Dis 2016; 63:1564–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zost SJ, Parkhouse K, Gumina ME, et al. . Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci U S A 2017; 114:12578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koel BF, Burke DF, Bestebroer TM, et al. . Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 2013; 342:976–9. [DOI] [PubMed] [Google Scholar]
- 26. Popova L, Smith K, West AH, et al. . Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS One 2012; 7:e41895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCarthy KR, Watanabe A, Kuraoka M, et al. . Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity 2018; 48:174–184.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr, Wilson IA. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 2013; 20:363–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sui J, Sheehan J, Hwang WC, et al. . Wide prevalence of heterosubtypic broadly neutralizing human anti-influenza A antibodies. Clin Infect Dis 2011; 52:1003–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heaton NS, Sachs D, Chen CJ, Hai R, Palese P. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc Natl Acad Sci U S A 2013; 110:20248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doud MB, Bloom JD. Accurate measurement of the effects of all amino-acid mutations on influenza hemagglutinin. Viruses 2016; 8:E155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thyagarajan B, Bloom JD. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. eLife 2014; 3:e03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu NC, Young AP, Al-Mawsawi LQ, et al. . High-throughput profiling of influenza A virus hemagglutinin gene at single-nucleotide resolution. Sci Rep 2014; 4:4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med 2014; 20:143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981; 289:366–73. [DOI] [PubMed] [Google Scholar]
- 36. Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981; 289:373–8. [DOI] [PubMed] [Google Scholar]
- 37. Koel BF, Mögling R, Chutinimitkul S, et al. . Identification of amino acid substitutions supporting antigenic change of influenza A(H1N1)pdm09 viruses. J Virol 2015; 89:3763–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Andrews SF, Huang Y, Kaur K, et al. . Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med 2015; 7:316ra192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nachbagauer R, Choi A, Izikson R, Cox MM, Palese P, Krammer F. Age dependence and isotype specificity of influenza virus hemagglutinin stalk-reactive antibodies in humans. MBio 2016; 7:e01996–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harris AK, Meyerson JR, Matsuoka Y, et al. . Structure and accessibility of HA trimers on intact 2009 H1N1 pandemic influenza virus to stem region-specific neutralizing antibodies. Proc Natl Acad Sci U S A 2013; 110:4592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Angeletti D, Gibbs JS, Angel M, et al. . Defining B cell immunodominance to viruses. Nat Immunol 2017; 18:456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Altman MO, Bennink JR, Yewdell JW, Herrin BR. Lamprey VLRB response to influenza virus supports universal rules of immunogenicity and antigenicity. eLife 2015; 4:e07467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davenport FM, Hennessy AV. A serologic recapitulation of past experiences with influenza A; antibody response to monovalent vaccine. J Exp Med 1956; 104:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davenport FM, Hennessy AV, Francis T Jr. Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J Exp Med 1953; 98:641–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Francis T. On the doctrine of original antigenic sin. Proc Am Philos Soc 1960; 104:572–8. [Google Scholar]
- 46. Lessler J, Riley S, Read JM, et al. . Evidence for antigenic seniority in influenza A (H3N2) antibody responses in southern China. PLoS Pathog 2012; 8:e1002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gostic KM, Ambrose M, Worobey M, Lloyd-Smith JO. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 2016; 354:722–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wrammert J, Koutsonanos D, Li GM, et al. . Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 2011; 208:181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li GM, Chiu C, Wrammert J, et al. . Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A 2012; 109:9047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li Y, Myers JL, Bostick DL, et al. . Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med 2013; 210:1493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Linderman SL, Chambers BS, Zost SJ, et al. . Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013-2014 influenza season. Proc Natl Acad Sci U S A 2014; 111:15798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang KY, Rijal P, Schimanski L, et al. . Focused antibody response to influenza linked to antigenic drift. J Clin Invest 2015; 125:2631–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 2010; 328:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pica N, Hai R, Krammer F, et al. . Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc Natl Acad Sci U S A 2012; 109:2573–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thomson CA, Wang Y, Jackson LM, et al. . Pandemic H1N1 influenza infection and vaccination in humans induces cross-protective antibodies that target the hemagglutinin stem. Front Immunol 2012; 3:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Petrie JG, Parkhouse K, Ohmit SE, Malosh RE, Monto AS, Hensley SE. Antibodies against the current influenza A(H1N1) vaccine strain do not protect some individuals from infection with contemporary circulating influenza A(H1N1) virus strains. J Infect Dis 2016; 214:1947–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Epperson S, Blanton L, Kniss K, et al. . Influenza activity - United States, 2013-14 season and composition of the 2014-15 influenza vaccines. MMWR Morb Mortal Wkly Rep 2014; 63:483–90. [PMC free article] [PubMed] [Google Scholar]
- 58. Dreyfus C, Laursen NS, Kwaks T, et al. . Highly conserved protective epitopes on influenza B viruses. Science 2012; 337:1343–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Corti D, Voss J, Gamblin SJ, et al. . A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011; 333:850–6. [DOI] [PubMed] [Google Scholar]
- 60. Lang S, Xie J, Zhu X, Wu NC, Lerner RA, Wilson IA. Antibody 27F3 broadly targets influenza A group 1 and 2 hemagglutinins through a further variation in VH1-69 antibody orientation on the HA stem. Cell Rep 2017; 20:2935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kallewaard NL, Corti D, Collins PJ, et al. . Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 2016; 166:596–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fu Y, Zhang Z, Sheehan J, et al. . A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve. Nat Commun 2016; 7:12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Joyce MG, Wheatley AK, Thomas PV. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 2016; 166:609–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Andrews SF, Joyce MG, Chambers MJ, et al. . Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci Immunol 2017; 2:eaan2676. [DOI] [PubMed] [Google Scholar]
- 65. Ekiert DC, Bhabha G, Elsliger MA, et al. . Antibody recognition of a highly conserved influenza virus epitope. Science 2009; 324:246–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Throsby M, van den Brink E, Jongeneelen M, et al. . Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 2008; 3:e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Friesen RH, Lee PS, Stoop EJ, et al. . A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U S A 2014; 111:445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nakamura G, Chai N, Park S, et al. . An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe 2013; 14:93–103. [DOI] [PubMed] [Google Scholar]
- 69. Sui J, Hwang WC, Perez S, et al. . Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 2009; 16:265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pappas L, Foglierini M, Piccoli L, et al. . Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature 2014; 516:418–22. [DOI] [PubMed] [Google Scholar]
- 71. Wu NC, Wilson IA. A Perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J Mol Biol 2017; 429:2694–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Julien JP, Lee PS, Wilson IA. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol Rev 2012; 250:180–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Andrews SF, McDermott AB. Shaping a universally broad antibody response to influenza amidst a variable immunoglobulin landscape. Curr Opin Immunol 2018; 53:96–101. [DOI] [PubMed] [Google Scholar]
- 74. Kashyap AK, Steel J, Oner AF, et al. . Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci U S A 2008; 105:5986–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ekiert DC, Friesen RH, Bhabha G, et al. . A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 2011; 333:843–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu NC, Wilson IA. Structural insights into the design of novel anti-influenza therapies. Nat Struct Mol Biol 2018; 25:115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Doud MB, Lee JM, Bloom JD. How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat Commun 2018; 9:1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Okuno Y, Isegawa Y, Sasao F, Ueda S. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 1993; 67:2552–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Henry Dunand CJ, Leon PE, Kaur K, et al. . Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J Clin Invest 2015; 125:1255–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chai N, Swem LR, Reichelt M, et al. . Two escape mechanisms of influenza a virus to a broadly neutralizing stalk-binding antibody. PLoS Pathog 2016; 12:e1005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wu NC, Xie J, Zheng T, et al. . Diversity of functionally permissive sequences in the receptor-binding site of influenza hemagglutinin. Cell Host Microbe 2017; 21:742–753.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wu NC, Thompson AJ, Xie J, et al. . A complex epistatic network limits the mutational reversibility in the influenza hemagglutinin receptor-binding site. Nat Commun 2018; 9:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sparrow E, Friede M, Sheikh M, Torvaldsen S, Newall AT. Passive immunization for influenza through antibody therapies, a review of the pipeline, challenges and potential applications. Vaccine 2016; 34:5442–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Erbelding EJ, Post DJ, Stemmy EJ, et al. . A universal influenza vaccine: the strategic plan for the national institute of allergy and infectious diseases. J Infect Dis 2018; 218:347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bakker AB, Marissen WE, Kramer RA, et al. . Novel human monoclonal antibody combination effectively neutralizing natural rabies virus variants and individual in vitro escape mutants. J Virol 2005; 79:9062–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Keeffe JR, Van Rompay KK, Olsen PC, et al. . A combination of two human monoclonal antibodies prevents Zika virus escape mutations in non-human primates. Cell Rep 2018; 25:1385–1394.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mendoza P, Gruell H, Nogueira L, et al. . Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 2018; 561:479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bonsignori M, Liao HX, Gao F, et al. . Antibody-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol Rev 2017; 275:145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Landais E, Murrell B, Briney B, et al. . HIV envelope glycoform heterogeneity and localized diversity govern the initiation and maturation of a V2 apex broadly neutralizing antibody lineage. Immunity 2017; 47:990–1003.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rantalainen K, Berndsen ZT, Murrell S, et al. . Co-evolution of HIV envelope and apex-targeting neutralizing antibody lineage provides benchmarks for vaccine design. Cell Rep 2018; 23:3249–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ellebedy AH, Jackson KJ, Kissick HT, et al. . Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat Immunol 2016; 17:1226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Petrie JG, Ohmit SE, Johnson E, Truscon R, Monto AS. Persistence of antibodies to influenza hemagglutinin and neuraminidase following one or two years of influenza vaccination. J Infect Dis 2015; 212:1914–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Xu R, McBride R, Paulson JC, Basler CF, Wilson IA. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J Virol 2010; 84:1715–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Xiong X, Coombs PJ, Martin SR, et al. . Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 2013; 497:392–6. [DOI] [PubMed] [Google Scholar]
- 95. Tzarum N, de Vries RP, Zhu X, et al. . Structure and receptor binding of the hemagglutinin from a human H6N1 influenza virus. Cell Host Microbe 2015; 17:369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J Virol 2013; 87:12433–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ha Y, Stevens DJ, Skehel JJ, Wiley DC. H5 avian and H9 swine influenza virus haemagglutinin structures: possible origin of influenza subtypes. EMBO J 2002; 21:865–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhang H, de Vries RP, Tzarum N, et al. . A human-infecting H10N8 influenza virus retains a strong preference for avian-type receptors. Cell Host Microbe 2015; 17:377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]