Abstract
The year 2018 marked the 100th anniversary of the deadliest event in human history. In 1918–1919, pandemic influenza spread globally and caused an estimated 50–100 million deaths associated with unexpected clinical and epidemiological features. The descendants of the 1918 virus continue to circulate as annual epidemic viruses causing significant mortality each year. The 1918 influenza pandemic serves as a benchmark for the development of universal influenza vaccines. Challenges to producing a truly universal influenza vaccine include eliciting broad protection against antigenically different influenza viruses that can prevent or significantly downregulate viral replication and reduce morbidity by preventing development of viral and secondary bacterial pneumonia. Perhaps the most important goal of such vaccines is not to prevent influenza, but to prevent influenza deaths.
Keywords: influenza, pandemic, pathogenesis, vaccine
In 1918, the world experienced the deadliest single event in recorded human history [1, 2]—the sudden emergence of an influenza virus of extraordinary lethality, unprecedented in more than a millennium of influenza pandemic observation (Figure 1). In considering the rationale for and the desired characteristics of so-called “universal” influenza vaccines, we must, before any other consideration, look back from the vantage point of 2018 to that century-old tragedy and ask: What are we trying to prevent, and how do we expect a vaccine to prevent it?
Figure 1.
The 1918 pandemic caused so many deaths, so quickly, that in some hospitals bodies were stacked up layers deep; hasty burials, and burials in mass graves, were common.
The novel 1918 pandemic virus killed an estimated 50–100 million people within the span of a year [3], which would equate, given the same degree of lethality, to as many as 400 million or more deaths today. Preventing a repeat of such an unimaginable disaster is a litmus test, and the ultimate standard, for any universal influenza vaccine.
But pandemic vaccine prevention is only one part of what must be achieved with a putative “universal” influenza vaccine. Over time, nonpandemic influenza viruses can be just as deadly, or more so. The 1918 H1N1 pandemic influenza A virus (IAV) was a founder virus that introduced a new and deadly viral era [4]. A century later we are still living in this era: All of the influenza A viruses that have circulated in humans since 1918, including H1N1, H2N2, and H3N2 viruses, are genetic descendants of the original 1918 virus, including new pandemic viruses derived by reassortment (“antigenic shift”) with other IAVs in 1957 (H2N2), 1968 (H3N2) [5], and after emergence of a swine-origin virus in 2009 (a novel H1N1 with a classical swine-derived H1 hemagglutinin [HA] genetically and antigenically similar to, and directly descended from, the H1 HA gene of the original human 1918 virus [4]); several intrasubtypic reassortant seasonal viruses that have spread pandemically [5, 6]; and all annual/seasonal IAVs that have circulated endemically and epidemically on an ongoing basis for the last 100 years [4] (Figure 2).
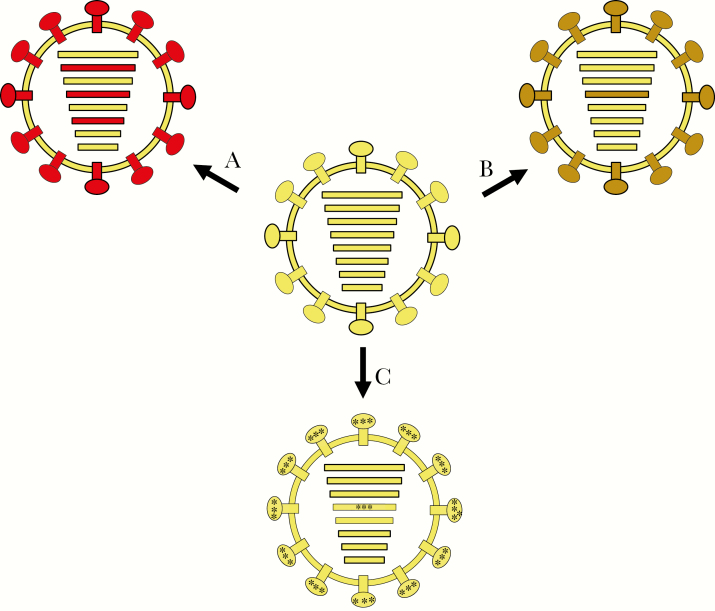
Figure 2.
Influenza A viruses and mechanisms of antigenic change. Hemagglutinin (HA) and neuraminidase (NA) are the major surface glycoproteins that elicit protective humoral immunity. Antigenic change can occur via several different mechanisms [7], as diagrammed here. A, Antigenic shift, or gene segment reassortment with another influenza A virus (following mixed infection), can lead to viruses with novel gene segment combinations. In the example shown, reassortment of the pre-1957 human H1N1 virus with one or more unknown avian H2N2 influenza A viruses led to the emergence of the 1957 pandemic virus containing 3 novel avian influenza–derived gene segments, PB1, HA, and NA. B, Intrasubtypic reassortment in which 2 co-circulating human influenza viruses of the same HA subtype can undergo reassortment to create a novel genotype, as occurred in both the postpandemic H1N1 and H3N2 viruses [5]. In the example shown, 2 clades of H3N2 viruses reassorted and led to the antigenically variant 2003 Fujian-like epidemic [6]. C, Antigenic drift, where coding mutations in the antigenic regions of HA and NA lead to continual antigenic alteration of circulating influenza viruses [8] (see Figure 3).
Such seasonal viruses mutate continually via “antigenic drift” that creates new, and may remove old, HA and neuraminidase (NA) antigenic epitopes, and which may also add N-linked glycosylation sites, all to escape human population immunity elicited by the circulation of recent IAV ancestors (Figure 2). As deadly as the 1918 pandemic was, US mortality data, adjusted for population growth, suggest that over the past century about 3 times as many deaths have been caused by descendants of the 1918 pandemic virus than by the pandemic virus itself.
The targets of a universal influenza vaccine are therefore many, variable, and constantly changing. The past century has revealed the extraordinary ability—unmatched in communicable disease history—of a single introduced infectious agent to successfully overcome, on a continuing basis, the serial adaptations of population immunity that the virus itself elicits by constant antigenic change. It is a useful, if challenging, thought experiment to realize that even at a minimum, a truly universal influenza vaccine would have to reliably protect against all of the past century’s worth of influenza viruses. Not only that, but the ideal universal vaccine would also need to protect against the other influenza types (especially influenza B viruses), not further discussed here [9].
But to complicate matters even further, out of the global reservoir of wild-bird IAVs that apparently gave rise to the 1918 pandemic virus have also emerged IAVs that infect other animals and have subsequently gone on to infect humans, often causing severe diseases in individuals, even when the viruses are not transmitted between humans and when they are not necessarily proceeding toward pandemicity. We have little understanding of the risks posed by such IAVs.
For example, the 2009 pandemic virus contained gene segments derived from the 1918 pandemic virus, which had been transmitted to pigs in 1918, and had gone on to circulate in pigs over the past century [4]. Are any, or all, swine influenza viruses potential threats to humans? What about IAVs that circulate in horses or dogs? A panzootic equine influenza virus has switched hosts to cause a canine panzootic [10]; are humans also at risk of future pandemics arising from similar mammalian host-switching events? And, what about poultry-adapted influenza viruses that occasionally infect humans, sometimes fatally, such as the H5N1 and H7N9 viruses [11–13], which have together killed approximately 1100 people since 1997, but which have not become adapted to human transmission? Do these viruses pose a pandemic risk; if so, would their emergence cause extremely high case fatality?
What are the implications of these many potential forms of IAV emergence for universal influenza vaccine prevention? Clearly, influenza has displayed an array of deadly mechanisms for host switch, evolution, and escape from population immunity. How many additional mechanisms are we completely unaware of, and what sorts of vaccines could be produced to prevent emergences in humans of novel IAVs introduced by any such mechanism? To make universal vaccines, we need to understand and synthesize an enormous amount of virologic, pathologic, immunologic, epidemiologic, and historical information about many, diverse, and ever-changing influenza viruses.
Threats Posed by Influenza Viruses Emerging From Their Natural Hosts
With so many different types of IAVs infecting so many different hosts, it is helpful to start by considering the global universe of IAVs, which includes genetically diverse and well-adapted viruses of wild waterfowl and shore birds, hosts which constitute the reservoir of all IAVs. These viruses circulate silently, and reassort continually, causing little or no harm to their billions of avian hosts, while retaining high antigenic stability because they are under little immune pressure within avian enteric tracts. Only rarely (apparently only once in the past 100 years) has one of these waterfowl viruses somehow host-switched, via unknown direct or indirect mechanisms, to cause a pandemic (in 1918). Of the 16 waterfowl HA subtypes and 9 waterfowl NA subtypes known to exist in nature, only 3 HAs (H1, H2, H3) and 2 NAs (N1, N2) have ever been documented to occur in a pandemic virus, and epidemiologic evidence supports the possibility that this may be true as far back as the earlier pandemics of 1831 and 1889.
However, we have no assurance that other HAs and NAs do not pose as great a threat (see Preventing Disease Severity Caused by Pathogenic Influenza Viruses section), requiring that truly universal influenza A vaccines protect against viruses possessing all 25 of these highly different surface glycoproteins (in 144 combinations, each of which may be associated with different viral properties), each expressing multiple and unique critical epitopes. This represents an extraordinarily steep challenge to vaccine development, but it is only a part of the total threat that universal vaccines must face.
The Challenge of Postemergence Viral Antigenic Change in Humans/Other Mammals
The challenges to preventive vaccines discussed above address only pandemic viruses that arise from wild waterfowl and may be of limited relevance to waterfowl viruses that adapt to humans or to other nonhuman hosts and then evolve in their new hosts. As a result of high-level transmission necessary for survival in these new adapted hosts, both human- and other mammalian-adapted and poultry-adapted influenza viruses drift continually and change antigenically over time. Unlike the genetically stable viruses within the waterfowl reservoir [14], human-adapted IAVs [8], as well as poultry- and mammalian-adapted viruses, are restless moving targets of ever-changing antigenicity. This can be visualized dramatically by examining antigenic drift in the 4 different pandemic/postpandemic viruses prevalent since 1955 [15] (Figure 3).
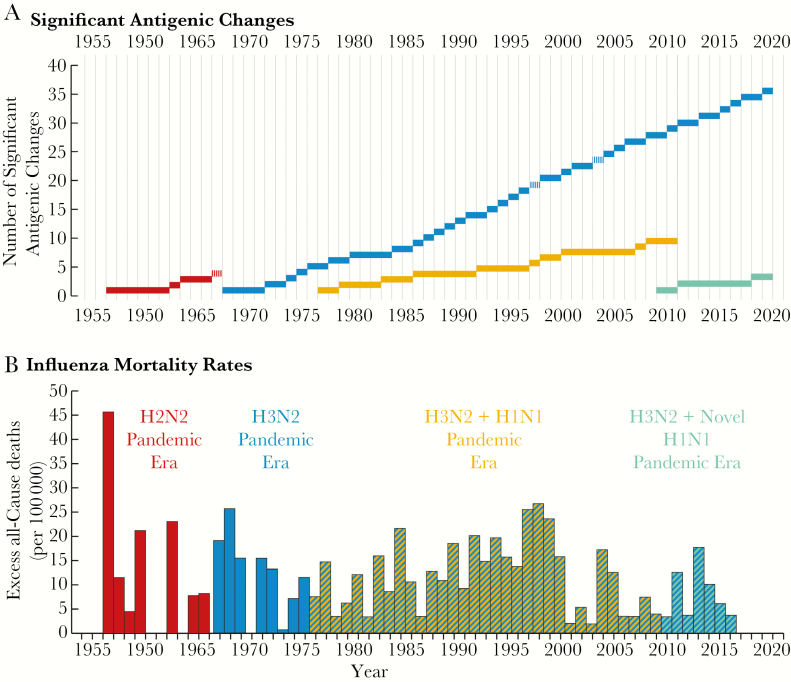
Figure 3.
The evolution of, and annual mortality associated with, 4 pandemic influenza viral descendants of the 1918 pandemic virus that arose by antigenic shift, 1955–2016. A, Antigenic changes in postpandemic viruses. The colored bars represent prevalence of the 1957 H2N2 pandemic virus (red); the 1968 H3N2 pandemic virus (blue); the unexpected return of a 1950s-era descendant of the 1918 pandemic virus, presumably released accidentally from viral storage (amber); and the 2009 H1N1 pandemic virus (green). Antigenic drift changes of sufficient magnitude to require reformulation of the annual vaccine for use in the Northern Hemisphere are represented on the y-axis. Notably, the 1968 H3N2 has been drifting at a greater rate (an average 0.70 significant genetic changes per year) than the other 3 pandemic viruses (an average of 0.27 genetic changes per year for the 3 combined). Antigenic changes in postpandemic influenza viruses have been associated with antigenic drift, which introduces new epitopes or new glycosylation sites, and by intrasubtypic reassortment of an antigenically different HA of the same subtype, represented by vertical hash marks. B, Annual excess mortality rates attributed to influenza. Data are missing for some early years. The figures are obtained from or extrapolated from Centers for Disease Control and Prevention data to reflect excess all-cause mortality, the most common calculation method available for all of the years, although arguably represent overestimations of mortality. Figure updated and modified from Morens et al [15].
Defining significant antigenic change as drift or intrasubtypic reassortant that requires a new vaccine formulation (a rough marker of a virus’s neutralization escape), it is clear (Figure 3) that all human IAVs have been changing antigenically after every few years of human circulation, with H3N2 viruses in particular showing an extraordinary degree of antigenic drift associated with alarming mortality. It is noteworthy that when this H3N2 virus first appeared in pandemic form in 1968, introducing a novel waterfowl-origin H3 onto the backbone of a human-adapted virus, it was of very low pathogenicity. In the 50 years since then, its further evolution in humans has led to increased pathogenicity and increased mortality.
Analogous viral antigenic changes associated with viral circulation in other mammals and in domestic poultry have also been documented [16, 17]. Evidence from all of these secondarily infected host species leads to the conclusion that adaptation of wild waterfowl IAVs to nonnatural hosts begins a process of continuing viral hyperevolution to escape host population immunity that it keeps on creating, a process that may have no definite end unless host populations can outlive the mutational repertoire of the IAVs.
Preventing Disease Severity Caused by Pathogenic Influenza Viruses
The above considerations about preventing IAV infection draw our attention to historical observations that influenza pandemics are of variable severity [18], from the milder pandemics of 1510 and 2009, to the deadly pandemics of 1557 and 1918. In 1918, for example, the pandemic virus appeared to produce at least 4-fold higher mortality across the entire age spectrum than did the pandemic virus of 1889. In animal studies using chimeric viruses that express different modern waterfowl HAs on the same isogenic 7-gene viral backbone, HAs were shown to be independent virulence factors, with some avian HA subtypes being inherently and significantly more pathogenic in mammals than other subtypes, including H1, H6, H7, H10, and H15 [19, 20].
This is an alarming observation, since these pathogenic HAs exist in nature around the globe in the wild waterfowl reservoir and will remain in existence for the foreseeable future, constituting a threat that must be prevented because it cannot be eliminated. It also raises an issue for universal vaccine development: Are some influenza virus subtypes more important than others with respect to their ability to cause fatality? If so, must candidate vaccines demonstrate an even higher degree of protection against them? In this regard, are the pathogenetic mechanisms of severe influenza disease different from those of mild disease, and will it be possible to make universal vaccines that not only prevent infection but also prevent disease severity if infection should occur? Much remains unknown, but it is abundantly clear that whether emerging directly from the wild waterfowl reservoir, or from antigenic change associated with circulation in intermediate hosts, extreme viral pathogenicity is an important aspect of the threat of pandemic emergence and of postpandemic/postpanzootic viral circulation, and must be effectively countered by any truly universal vaccine.
Viral–Bacterial Copathogenicity
Further regarding viral pathogenicity, in the 1918 pandemic the great majority of deaths were caused not by the virus itself but by secondary bacterial pneumonias associated with various pneumopathogens, especially Streptococcus pneumoniae, Streptococcus pyogenes, and Staphylococcus aureus [21]. Copathogenic phenomena seem to be responsible for most influenza deaths today as well, although, fortunately, age-specific incidence rates of secondary bacterial pneumonia in persons with influenza are significantly lower in 2018 than they were in 1918. A growing body of research has examined numerous variables potentially associated with postinfluenza development of bacterial pneumonia [22]. However, there remains much to be learned about the natural history and pathogenesis of influenza, its elicitation of inflammatory and immune responses, and its ability to act as a cofactor in the initiation and development of a secondary bacterial pneumonia (see Influenza Infection and the Human Host section).
Influenza Infection and the Human Host
The 1918 virus is more pathogenic than most other IAVs in experimental animals, causing greater cytopathicity and eliciting more robust and potentially host-destructive inflammatory responses [23]. Even so, the vast majority of infected people in 1918, probably around 98% in the United States, had an asymptomatic or typical self-limited illness that was no different in type or severity than influenza today. (The other 2% had one or more complications, including pneumonia, empyema, and sepsis. About half of those persons died, resulting in an overall influenza case-fatality ratio of around 1% or less). Moreover, ironically, autopsies of fatal 1918 cases show prompt repair of viral damage in areas without bacterial destruction [24–26], as remains true for influenza pneumonia today [27], indicating that even severe influenza with pneumonia is a “recoverable” disease [28], and that a universal influenza vaccine needs to be able to prevent not only infection, but also extreme viral pathogenesis should infection occur.
Are there unidentified genetic, host, or environmental susceptibility factors that drive influenza disease severity and copathogenicity? It is important to answer this question in designing universal vaccines, not the least because the most important goal of such vaccines is not to prevent influenza, but to prevent influenza deaths. Another challenging problem is recent controversial evidence from population studies in Canada and elsewhere suggesting that sequential influenza immunization can, in some situations, cause an increased risk of subsequent wild virus infection [29]. In attempting to understand these data, scientists have begun to examine complex immunological phenomenon such as “original antigenic sin” [30] and have postulated a new pathogenetic mechanism referred to by some as immunologic “imprinting” [31].
It is also noteworthy that during the 1918 pandemic, numerous clinical trials of what were then called influenza vaccines (actually crude preparations of inactivated bacteria) demonstrated efficacy in preventing influenza deaths [32]. In the modern era, we have vaccines for only one of the major bacterial copathogens that routinely caused fatal influenza in 1918, S. pneumoniae, in the form of 2 vaccines that each contain immunogens of common but different circulating bacterial types. We have argued elsewhere that, hand in hand with improved influenza vaccine prevention, we need to develop better means of preventing deaths from secondary bacterial pneumonia [2]. The highest priorities are efficacious vaccines against the most important secondary pneumopathogens, such as S. aureus and S. pyogenes, and identification of early reliable biomarkers of impending bacterial pneumonia in people with influenza illnesses.
What Should a Truly Universal Influenza Vaccine Be Able to Do?
Considering the protean evolutionary strategies that influenza has demonstrated, it may be useful to suggest benchmark criteria for an “ideal” universal vaccine, so that we can identify targets at which to aim [33, 34] (Table 1), and so that we can examine knowledge gaps that must be bridged (see KNOWLEDGE GAPS IN DESIGNING “UNIVERSAL” INFLUENZA VACCINES section). It should be emphasized that these “ideals,” representing vaccine “perfection,” may not all be realistic given the many complexities discussed herein. In fact, a truly “universal” vaccine that meets all desirable qualities noted—for individual protection, prepandemic protection, and public health control during times of endemic/epidemic drift—will probably remain beyond the reach of science for the foreseeable future. Nevertheless, as discussed in many of the publications in this supplement, greatly improved influenza vaccines, capable of inducing broader and more durable immunity, are very definitely on the horizon.
Table 1.
Ideal Properties of a Universal Influenza Vaccine
| Necessary | Desirable |
|---|---|
| Vaccine properties that relate to protection of vaccinated individuals against infection, disease, and death | |
| Prevents clinical disease | Is highly efficacious in 1 dose |
| Prevents infection | Induces robust lifelong immunity |
| Prevents all types of influenza (A, B, C, D) | |
| Prevents all subtypes of IAV | Induces robust lifelong mucosal immunity |
| Prevents infection by viral drift variants | Immunity is boosted by wild virus exposure |
| Immune response is rapid and robust | Does not alter respiratory microbiome |
| Vaccine “take” is not prevented by preexisting immunity | Is affordable |
| Induces immunity to multiple viral components | |
| Is generally safe | |
| Is safe for pregnant women | |
| Does not induce ADE upon subsequent wild virus exposures | |
| Is used in persons of all ages | |
| Is efficacious in immunosuppressed persons | |
| Is cross-protective against related viruses | |
| Vaccine properties that relate to public health utility | |
| Covers all wild waterfowl HAs and NAs | Prevents transmission |
| Covers all poultry-adapted viruses | Reduces/shortens viral shedding |
| Covers all mammalian-adapted viruses | Creates durable herd immunity |
| Can be used for pandemic prevention | Does not elicit neutralization escape mutants |
| Based on platform that is easily upgraded with new antigens | Is stable in storage |
| Sequential vaccinations boost immune protection |
These ideal properties are not meant to be confused with various operational definitions/criteria for universal vaccines (eg, those of the National Institutes of Health or the World Health Organization [33, 34]), but rather to encourage thought and discussion about the medical and public health implications of improved influenza vaccines.
Abbreviations: ADE, antibody-dependent enhancement; HA, hemagglutinin; IAV, influenza A virus; NA, neuraminidase.
KNOWLEDGE GAPS IN DESIGNING “UNIVERSAL” INFLUENZA VACCINES
Understanding Natural History and Pathogenesis
We have very little precise information about the natural history and pathogenesis of human influenza infection (ie, the sequence of biological events that occur during the course of influenza infection) and the mechanisms by which infections produce disease. Influenza viruses infect the ciliated and goblet respiratory epithelial cells of the upper respiratory tract, the nasopharynx, and sinuses. Most patients have no clinical evidence of infection below the nasopharynx. A minority of patients develop laryngitis, tracheitis, bronchitis/bronchiolitis, or pneumonia, but it is unclear whether the virus extends down the respiratory tract in persons who do not manifest lower respiratory tract symptoms, nor is it clear what cell types might be infected or how cell tropism may play a role in infection and disease.
Because of this lack of knowledge, we know little about the specific characteristics of the protective immune response a vaccine needs to elicit. With regard to prevention of upper respiratory infection, it is likely that elicitation of local immunity in the mucosa-associated lymphoid tissue (MALT) is of great significance [35], with a complementary role for systemic immunity related to humoral antibody and circulating immune cells [36]. With respect to limiting influenza progression and its associated complications, the immune response to infection of the bronchial tree resides in the inducible bronchial-associated lymphoid tissue (iBALT) [37], and the immune response to infection of the lung periphery resides in the network of pulmonary alveolar macrophages (PAMs), recruited macrophages, and local niches of B and T cells induced by prior influenza infection [38]. These 4 separate immune compartments, at the various level of the bronchopulmonary system, may act independently.
Such issues are of importance, as an ideal vaccine must stimulate influenza immunity in all anatomical compartments in which influenza viruses can cause infections and must be able to prevent severe disease if infection occurs. Although it is not clear that lower respiratory tract cells are infected in uncomplicated influenza, autopsy studies of persons with viral or viral/bacterial pneumonia suggest that respiratory epithelial cells can, at least in some severe cases, be infected all the way down the respiratory tree to the alveoli, and that alveolar epithelial cells and alveolar macrophages may also be infected by virus [24, 27]. Few studies addressing these aspects of natural history have been conducted in the past 50 years.
With respect to vaccine induction of immunity in the anatomical compartments, there are insufficient data on the impact of route of viral vaccine administration in eliciting important aspects of the protective immune response. That the current live attenuated influenza vaccine (given intranasally) is only moderately effective in eliciting broad protective immunity could indicate that stimulation of mucosal immunity is inherently insufficient for protection, or might simply reflect overattenuation of the vaccine, or other phenotypic vaccine properties that are underappreciated (eg, viral interference) [39], arguing strongly for a need to study mucosal vaccination immunity in greater depth.
We also need to better understand the mechanisms that lead to viral and to secondary bacterial pneumonia. Secondary pneumonias are typically caused by commensal bacteria that are resident in the nasopharynx, but how they get down into the lungs is not fully understood. Evidence from autopsies is consistent with direct extension of bacterial growth following initial viral cytopathic destruction of epithelial, ciliated, and goblet cells [21, 24], but although this pathophysiologic mechanism has to a limited extent been studied in experimental animals [40], it has not been well studied in humans. Moreover, other proposed mechanisms of viral–bacterial copathogenesis have been demonstrated in experimental animals, including exposure of bacterial receptors and enhanced inflammatory responses [22, 23, 41].
Is viral pneumonia (as opposed to viral bronchitis/bronchiolitis) a prerequisite for bacterial bronchopneumonia, or can bacterial bronchopneumonia develop in the absence of alveolar viral infection? If enhanced viral–bacterial copathogenicity is a result of properties of viral pathogenicity, what are they: cytopathicity, viral growth, cell tropism, or elicitation of an aberrant inflammatory responses? Of particular interest is the role of PAMs, which, among many other roles, moderate the balance between controlling viral infection and limiting immune damage, and which may also enhance bacterial damage via interferon release [42].
What role is played by infected pneumocytes and infected macrophages? What is the nature of the local inflammatory and immune responses, and how can they best be modified by vaccination? Can a protective immune response in the lower respiratory tract be elicited by vaccination and, if so, how? These are questions of critical importance that cannot yet be answered.
Understanding Immune Correlates of Protection
It has become accepted wisdom that serum hemagglutination inhibition (HI) titers ≥1:40 or, even better, ≥1:80, are correlates of protection against natural infection. Support for this idea comes from epidemiologic studies and from study of passive immunotherapy with immune serums in humans and experimental animals. Yet it is far from clear that HI antibody alone reliably prevents asymptomatic or symptomatic infection in humans. In experimental human challenge studies, HI antibody is not a reliable determinant of protection [43], and some persons with significant levels of HI antibody can be repeatedly infected by the same virus to which they have preexisting antibody (Memoli and Taubenberger, unpublished data). It seems increasingly likely that HI titers are by themselves only modest indicators of protection, but that, following natural infection, they may be correlated with other aspects of immunity such as NA, HA stalk, and cell-mediated immunity, suggesting that universal vaccines must elicit comprehensive immune responses. In this regard, we draw attention to a large but poorly remembered body of research on NA immunity. Human observational and experimental studies, supported by studies in experimental animals, indicate an important role for NA immunity in preventing and controlling infection, reducing viral growth and tissue pathology, and limiting transmission [44]. Current influenza vaccines have unstandardized and generally low NA activity. Within the immediate future, regulating NA content and immunogenicity in current vaccines may represent the lowest of the “low-hanging fruit” in terms of preventing uncomplicated and severe influenza and in reducing disease severity and transmission [44], and thus may represent a very important first step in universal vaccine development.
Understanding Host Susceptibility Factors and Disease Severity
As noted, even in the deadly 1918 pandemic, the majority of infected persons (probably ≥98% in the United States) had self-limited, mild, or asymptomatic infection and recovered completely. Although people in the various high-risk groups (eg, young children, the elderly, pregnant women, and unique to 1918, persons aged 20 to 40 years [2, 18]) had higher incidences of severe and fatal complications in 1918, there is little evidence to suggest that uncomplicated infections in these groups were any more severe, or any different in character, signs, or symptoms, than influenza infections seen today. What was different about those persons who, in 1918, went on to develop severe and fatal disease? The high-risk groups for severe influenza in 1918, including both expected and unexpected groups, is a powerful clue that both viral pathogenicity and host variables are of critical importance.
What variables may be associated with increased disease severity and higher fatality? Are they immunologic? If so, can they be overcome by higher vaccine doses or more intensive regimens? In addition, epidemiologic data from study of human poultry-adapted H5N1 cases strongly suggest that rare individual host susceptibilities may play a major role in severe disease caused by these IAVs, which are otherwise poorly adapted to humans and unable to productively infect most persons [45]. Is there one, or more likely multiple, genetic host susceptibility factors (eg, the IFITM3 rs12252 polymorphism [45, 46] and/or others not yet identified) that predispose to severe influenza disease? If so, such persons might be identified and targeted for vaccination. Would such individuals respond normally to an influenza vaccine—and if not, what alternative vaccination strategies are needed?
Experimental Study in Universal Vaccine Development
There are no ideal animal models to provide needed information on influenza immunity sufficient to bring a universal vaccine to licensure. Human challenge studies are, and will continue to be, of paramount importance. Expanding challenge study capacity is a key priority. Viral challenge studies are required not only to understand natural history and pathogenesis, but also to characterize the protective immune responses in the various immune compartments, to conduct proof-of-principle passive immunotherapy studies, and to evaluate safety and efficacy of candidate vaccines themselves. It is noteworthy that to evaluate the “universality” of a putative universal vaccine, we must challenge humans with live or live chimeric viruses that are not among those that currently circulate in humans, including mammalian and avian IAVs with subtypes other than H1N1 and H3N2 (out of the 144 possible HA/NA subtype combinations found in nature).
Without such research, a vaccine’s universality remains unproven even for the viruses with which humans continually come into contact, and its utility as a stockpiled prepandemic or pandemic control vaccine would therefore be doubtful. It will be also crucially important to develop and validate standardized, efficient assays to evaluate antibodies against all HA and NA subtypes from serum and nasopharyngeal samples. Also of importance will be development of better primate challenge models and primate challenge studies, since human challenge studies are inevitably conducted in persons already immune to influenza viruses. Understanding vaccine responses and protective vaccine responses in the immunologically naive, as well as the role of sequential influenza infection in shaping the immune response, including “original antigenic sin” and examination of the role of vaccination/infection sequences in the controversial risk data noted above, will also be necessary for optimal vaccine development. These sorts of studies cannot be done optimally in humans because human subjects already have had influenza exposure, and their study exposures cannot be controlled for, or easily characterized going forward, while subjects live in open populations and are naturally exposed to different viruses at different times.
CONCLUSIONS
Study of the 1918 influenza virus, including its numerous descendants, some of which are still circulating in humans, provides a backdrop for describing the desired and necessary properties of universal influenza vaccines. That there are so many different IAV subtypes and strains that circulate in humans and many other species, and that they are continually mutating antigenically, suggests that truly universal influenza vaccines—as strictly defined (Table 1)—could be beyond the reach of current technology. And even with optimal technologies, much more needs to be done, on an urgent basis, to begin to understand the natural history and pathogenesis of uncomplicated and severe influenza infection, including the bases of immune protection and bacterial copathogenesis.
Nevertheless, a rapidly growing knowledge base suggests new approaches to developing better influenza vaccines that can provide broader and more durable immunity than that offered by current vaccines. Fruitful lines of research include identifying critical epitopes shared by different influenza viruses (eg, stalk epitopes) and using these as immunogens; optimizing NA immunity; and eliciting robust local immunity within the MALT, iBALT, and lung immune compartments. The best future influenza vaccines will ideally elicit robust and persistent protective immune responses to multiple epitopes on multiple influenza proteins expressed in multiple anatomical compartments, and immune responses capable of being productively recalled upon wild virus exposure. The challenges to vaccinology are truly daunting, but the effort is necessary, and the rewards to human well-being and to the advancement of scientific understanding will be great. A fitting legacy for the 1918 influenza pandemic would be advances in basic understanding and vaccine development to prevent catastrophes of the same magnitude in the future.
Notes
Financial support. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Supplement sponsorship. This work is part of a supplement sponsored by the National Institute of Allergy and Infectious Diseases.
Potential conflicts of interest. Both authors: No reported conflicts of interest. Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Taubenberger JK, Morens DM. 1918 influenza: the mother of all pandemics. Emerg Infect Dis 2006; 12:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Morens DM, Taubenberger JK. The mother of all pandemics is 100 years old (and going strong)! Am J Public Health 2018; 108:1449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull Hist Med 2002; 76:105–15. [DOI] [PubMed] [Google Scholar]
- 4. Morens DM, Taubenberger JK, Fauci AS. The persistent legacy of the 1918 influenza virus. N Engl J Med 2009; 361:225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nelson MI, Viboud C, Simonsen L, et al. . Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog 2008; 4:e1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holmes EC, Ghedin E, Miller N, et al. . Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol 2005; 3:e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taubenberger JK, Kash JC. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 2010; 7:440–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fitch WM, Leiter JM, Li XQ, Palese P. Positive Darwinian evolution in human influenza A viruses. Proc Natl Acad Sci U S A 1991; 88:4270–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan J, Asthagiri Arunkumar G, Krammer F. Universal influenza virus vaccines and therapeutics: where do we stand with influenza B virus? Curr Opin Immunol 2018; 53:45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Collins PJ, Vachieri SG, Haire LF, et al. . Recent evolution of equine influenza and the origin of canine influenza. Proc Natl Acad Sci U S A 2014; 111:11175–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morens DM, Taubenberger JK, Fauci AS. Pandemic influenza viruses—hoping for the road not taken. N Engl J Med 2013; 368:2345–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morens DM, Taubenberger JK, Fauci AS. H7N9 avian influenza A virus and the perpetual challenge of potential human pandemicity. MBio 2013; 4. doi: 10.1128/mBio.00445-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morens DM, Subbarao K, Taubenberger JK. Engineering H5N1 avian influenza viruses to study human adaptation. Nature 2012; 486:335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dugan VG, Chen R, Spiro DJ, et al. . The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog 2008; 4:e1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morens DM, Taubenberger JK, Fauci AS. The 2009 H1N1 pandemic influenza virus: what next? MBio 2010; 1. doi: 10.1128/mBio.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lewis NS, Daly JM, Russell CA, et al. . Antigenic and genetic evolution of equine influenza A (H3N8) virus from 1968 to 2007. J Virol 2011; 85:12742–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nelson MI, Lemey P, Tan Y, et al. . Spatial dynamics of human-origin H1 influenza A virus in North American swine. PLoS Pathog 2011; 7:e1002077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morens DM, Taubenberger JK. Pandemic influenza: certain uncertainties. Rev Med Virol 2011; 21:262–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qi L, Pujanauski LM, Davis AS, et al. . Contemporary avian influenza A virus subtype H1, H6, H7, H10, and H15 hemagglutinin genes encode a mammalian virulence factor similar to the 1918 pandemic virus H1 hemagglutinin. MBio 2014; 5:e02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Qi L, Davis AS, Jagger BW, et al. . Analysis by single-gene reassortment demonstrates that the 1918 influenza virus is functionally compatible with a low-pathogenicity avian influenza virus in mice. J Virol 2012; 86:9211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis 2008; 198:962–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith AM, McCullers JA. Secondary bacterial infections in influenza virus infection pathogenesis. Curr Top Microbiol Immunol 2014; 385:327–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kash JC, Taubenberger JK. The role of viral, host, and secondary bacterial factors in influenza pathogenesis. Am J Pathol 2015; 185:1528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sheng ZM, Chertow DS, Ambroggio X, et al. . Autopsy series of 68 cases dying before and during the 1918 influenza pandemic peak. Proc Natl Acad Sci U S A 2011; 108:16416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol 2008; 3:499–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuiken T, Taubenberger JK. Pathology of human influenza revisited. Vaccine 2008; 26(Suppl 4):D59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gill JR, Sheng ZM, Ely SF, et al. . Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med 2010; 134:235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kash JC, Xiao Y, Davis AS, et al. . Treatment with the reactive oxygen species scavenger EUK-207 reduces lung damage and increases survival during 1918 influenza virus infection in mice. Free Radic Biol Med 2014; 67:235–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Belongia EA, Skowronski DM, McLean HQ, Chambers C, Sundaram ME, De Serres G. Repeated annual influenza vaccination and vaccine effectiveness: review of evidence. Expert Rev Vaccines 2017; 16:1–14. [DOI] [PubMed] [Google Scholar]
- 30. Monto AS, Malosh RE, Petrie JG, Martin ET. The doctrine of original antigenic sin: separating good from evil. J Infect Dis 2017; 215:1782–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Topham DJ, Nguyen P, Sangster MY. Pandemic influenza vaccines: what they have taught us about B cell immunology. Curr Opin Immunol 2018; 53:203–8. [DOI] [PubMed] [Google Scholar]
- 32. Chien YW, Klugman KP, Morens DM. Efficacy of whole-cell killed bacterial vaccines in preventing pneumonia and death during the 1918 influenza pandemic. J Infect Dis 2010; 202:1639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Erbelding EJ, Post DJ, Stemmy EJ, et al. . A universal influenza vaccine: the strategic plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis 2018; 218:347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ortiz JR, Hickling J, Jones R, et al. . Report on eighth WHO meeting on development of influenza vaccines that induce broadly protective and long-lasting immune responses: Chicago, USA, 23-24 August 2016. Vaccine 2018; 36:932–8. [DOI] [PubMed] [Google Scholar]
- 35. Braciale TJ, Sun J, Kim TS. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol 2012; 12:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seong Y, Lazarus NH, Sutherland L, et al. . Trafficking receptor signatures define blood plasmablasts responding to tissue-specific immune challenge. JCI Insight 2017; 2:e90233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hwang JY, Randall TD, Silva-Sanchez A. Inducible bronchus-associated lymphoid tissue: taming inflammation in the lung. Front Immunol 2016; 7:258. doi: 10.3389/fimmu.2016.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Turner DL, Bickham KL, Thome JJ, et al. . Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol 2014; 7:501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Singanayagam A, Zambon M, Lalvani A, Barclay W. Urgent challenges in implementing live attenuated influenza vaccine. Lancet Infect Dis 2018; 18:e25–32. [DOI] [PubMed] [Google Scholar]
- 40. Kash JC, Walters KA, Davis AS, et al. . Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. MBio 2011; 2. doi: 10.1128/mBio.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walters KA, D’Agnillo F, Sheng ZM, et al. . 1918 pandemic influenza virus and Streptococcus pneumoniae co-infection results in activation of coagulation and widespread pulmonary thrombosis in mice and humans. J Pathol 2016; 238:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Divangahi M, King IL, Pernet E. Alveolar macrophages and type I IFN in airway homeostasis and immunity. Trends Immunol 2015; 36:307–14. [DOI] [PubMed] [Google Scholar]
- 43. Memoli MJ, Shaw PA, Han A, et al. . Evaluation of antihemagglutinin and antineuraminidase antibodies as correlates of protection in an influenza A/H1N1 virus healthy human challenge model. MBio 2016; 7:e00417–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eichelberger MC, Morens DM, Taubenberger JK. Neuraminidase as an influenza vaccine antigen: a low hanging fruit, ready for picking to improve vaccine effectiveness. Curr Opin Immunol 2018; 53:38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morens DM, Taubenberger JK. How low is the risk of influenza A(H5N1) infection? J Infect Dis 2015; 211:1364–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prabhu SS, Chakraborty TT, Kumar N, Banerjee I. Association between IFITM3 rs12252 polymorphism and influenza susceptibility and severity: a meta-analysis. Gene 2018; 674:70–9. [DOI] [PubMed] [Google Scholar]