Abstract
Among gastrointestinal stromal tumors (GISTs) of 10–15% are negative for KIT and PDGFRA, and most of these cases are SDH deficient. Recent studies have provided data on additional molecular alterations such as KRAS in KIT mutant GISTs. We aimed to assess the frequency and spectrum of somatic mutations in common oncogenes as well as copy number variations in GISTs negative for KIT and PDGFRA mutations. GISTs with wild type KIT/PDGFRA were tested via next generation sequencing for somatic mutations in 341 genes. SDHB immunohistochemistry to evaluate for SDH deficiency was also performed. Of 267 GISTs tested for KIT and PDGFRA mutations, 15 were wild type, of which eight cases had material available for further testing. All eight cases had loss of SDHB expression and had various molecular alterations involving ARID1A, TP53, and other genes. One case had a KRAS G12V (c.35G>T) mutation in both the primary gastric tumor and a post-imatinib recurrence. This tumor had anaplastic features and was resistant to multiple tyrosine kinase inhibitors, ultimately resulting in cancer-related mortality within 2 years of diagnosis. In conclusion, KRAS mutations occur in rare GISTs with wild type KITand PDGFRA. These tumors may display immunohistochemical positivity for KITand primary resistance to tyrosine kinase inhibitors.
INTRODUCTION
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal neoplasms of the gastrointestinal tract. These tumors are characteristically positivity for KIT immunohistochemistry as well as mutations within the KIT or PDGFRA receptor tyrosine kinase genes (Miettinen and Lasota, 2006). The majority of these tumors are sensitive to imatinib or second generation tyrosine kinase inhibitors such as sunitinib or sorafenib. However, 10–15% are KIT and PDGFRA wild type, some of which harbor mutations in NF1, BRAF (V600E), or succinate dehydrogenase SDH complex genes (Wang et al., 2011; Falchook et al., 2013).
KRAS mutations have recently been described in GISTs, and unlike mutually exclusive BRAF V600E mutations, have only been reported to date in the presence of either KIT or PDGFRA mutations (Miranda et al., 2012; Antonescu et al., 2013; Serrano et al., Epub ahead of print). Further, their presence and prevalence of KRAS mutation in GIST has been disputed (Lasota et al., 2013). While numerous recent studies have yielded a high number of KIT mutant GISTs tested for KRAS mutation, wild type GISTs have not been held to such rigorous study, likely due to relative rarity of these tumors. Accordingly, KRAS mutations have never been reported in KIT/PDGFRA wild type GISTs. In this study, we have surveyed GISTs wild type for KIT and PDGFRA for the presence of KRAS mutations. We report the clinicopathologic and molecular features of a case of a KRAS G12V mutant, malignant gastric GIST with immunohistochemical positivity for KIT and we review the literature on KRAS mutations in GIST.
MATERIALS AND METHODS
Patient Selection
After approval by our local institutional review board, molecular results for KIT and PDGFRA mutation testing performed in the Laboratory of Diagnostic Molecular Pathology were reviewed for all 267 GISTs clinically analyzed between January 2009 and December 2013. The diagnosis of GIST was based on morphology and positive immunohistochemical staining for either KIT or DOG1. Cases with wild type KIT and PDGFRA results (n = 15) underwent both SDHB immunohistochemistry and KRAS sequencing.
Mutation Analysis
Genomic DNA was extracted from formalinfixed paraffin-embedded tissue after macrodissection using the DNeasy Tissue KIT (Qiagen, Valencia, CA), following the manufacturer’s standard protocol. KIT and PDGFRA mutations were routinely detected by a sizing assay, Sanger sequencing, and next generation sequencing (NGS). KRAS mutation was detected via NGS and mass spectrometry.
Briefly, insertions and deletion of exons 9 and 11 of KIT were detected by length analysis of fluorescently labeled polymerase chain reaction products on a capillary electrophoresis instrument (ABI 3730) (Pan et al., 2005). Negative cases underwent Sanger sequencing of KIT exons 9, 11, 13, and 17 and PDGFRA exons 12, 14, and 18. The coding regions of these exons were amplified using Hot-Start Taq DNA polymerase and appropriate primers. The PCR products were purified using Spin Columns (Qiagen) and sequenced using BigDye Terminator v3.1 Cycle Sequencing KIT (Applied Biosystems) according to the manufacturer’s protocol on an ABI 3730 running ABI Prism DNA Sequence Analysis Software. All PCR products were sequenced with forward and reverse primers. Concordant results in duplicate in forward and reverse directions were required for mutation calling.
NGS of KIT/PDGFRA wild type GISTs (n = 8) was performed with the clinically validated NGS assay, MSK-IMPACT (Memorial Sloan Kettering- Integrated Mutation Profiling of Actionable Cancer Targets). MSK-IMPACT is a custom hybrid capture-based deep sequencing assay that interrogates 341 cancer-associated genes (including all exons of KIT, PDGFRA, BRAF, and KRAS, see Supporting Information Table 1 for a full list of genes) for single nucleotide variants (SNVs), small indels, and somatic copy number alterations (Cheng et al., in press). Briefly, DNA isolated from FFPE samples were subjected to shearing, followed by Library preparation. Matched normal tissue was processed in the same manner where available and samples were pooled together for sequencing on an Illumina HiSeq 2500. 100bp paired end sequence reads were aligned to reference human genome (hg19) using Burrows-Wheeler Aligner BWA (Li and Durbin, 2010). SNVs were detected using MuTect (Cibulskis et al., 2013); small indels were identified using SomaticIndelDetector. Matched normal DNA was used to filter out germ line variants. In cases where matched normal tissue was not available (n = 3), a mixture of pooled FFPE normal DNA was used as unmatched normal for mutation calling. SNVs and indels required at least 5% alternate allele frequency for novel mutations and 2% for hotspot mutations. Minimum read coverage, at the position where the mutation is called, required was 20 for both classes of mutations. To detect copy number changes, an in-house developed algorithm was used. Briefly, Loess normalized coverage values for each exon sequenced were used to derive tumor/normal ratios, which were subsequently log-transformed. The log-ratio values were used in segmentation by circular binary segmentation (Olshen et al., 2004). Segmented values were used to derive a null distribution to estimate significant changes. Fold change >2.0 (gain) and <2.0 (loss) along with P <0.05 (FDR corrected) were used as criteria for calling copy number changes. All candidate mutations were manually reviewed using IntegratedG-enomicsViewer (Robinson et al., 2011). Technical details of this NGS assay are described elsewhere (Jelinic et al., 2014).
KRAS mutation testing on a separate sample of any positive GIST was performed with separate methodology for confirmation and assessment of heterogeneity. The separate methodology used was the MassARRAY system (Sequenom) with primers as previously described (Arcila et al., 2011; Chaft et al., 2012) at the following hotspots in duplicate with forward and reverse primers: c.34, 35 and forward only primers: 37, 38, 181, 182, 183, 351, and 437. Allele frequency was estimated by dividing mutant peak height by the sum of mutant and nonmutant peak height.
RESULTS
Prevalence of SDH Deficiency and KRAS Mutation
Of 267 GISTs analyzed for KIT and PDGFRA mutation, 15 were negative for mutations in these genes. Eight KIT/PDGFRA wild type GISTs had material available for KRAS testing, of which one case was positive for a KRAS G12V (c.35G>T) mutation. All eight cases wild type for KIT and PDGFRA by previously performed sizing assays and Sanger sequencing were also negative for KIT and PDGFRA mutations via NGS. The details of this case are presented below. All eight cases, including the KRAS mutant, were positive for KIT expression by immunohistochemistry and showed absence of immunohistochemical expression of SDHB. Two cases harbored frame-shift mutations in TP53, making TP53 the most commonly mutated tumor suppressor in the cohort. One case had an ARID1A mutation with loss of ARID1A immunohistochemical expression. Somatic alterations from all cases are summarized in Table 1.
TABLE 1.
Clinicopathologic Characteristics and Somatic Mutations in SDH-Deficient Wild Type Gastric GISTs
| Age | Sex | Morphology | Size (cm) | Mitotic rate (50 HPF) | AJCC stage, 7th ed. | Mean sample coverage | Somatic alterations (Allele frequency) |
|---|---|---|---|---|---|---|---|
| 67 | M | Epithelioid | 4.7 | 99 | II | 440 |
KRAS p. G12V (26%) ATRX p. G75X (74%) ATRX p. EI509_1510delinsX (65%) DICER1 p. L805Q (70%) MSH2 p. T568fs (28%) NF1 p.Q4234fs (8%) PMS1 p. WI98L (27%) IGF1 amplification (3.3 fold change) MDM2 amplification (3.2 fold change) |
| 35 | M | Epithelioid | 8 | 71 | IV, liver metastasis | 343 |
ARID1A p. S499fs (43%) with loss of ARID1A immunohistochemical expression |
| 32 | F | Spindle | 2 | 12 | II | 379 | none |
| 43 | F | Epithelioid | 3.5 | 1 | IV, nodal metastases | 501 |
SDHA p. R352X (26%) DAXX p. T5681 (5%) BMPR1A splice site (c. 6751+1G>T) (27%) |
| 59 | F | Mixed | 4 | 4 | IA | 999 |
aBRD4 p.1113del (43%) aCREBBP p.686Q (52%) aFLT1 p. L927S (49%) aHNF1A p. N62S (46%) aSDHB p. D204fs (63%) aTP53 p. A138V (6%) aTP53 p. R306fs (6%) aTP53 p. 191_192del (29%) |
| 28 | F | Epithelioid | 4 | >5 | IV, nodal metastases | 395 |
TP53 p. R342fs (29%) TP53 p. 191-192del (28%) |
| 61 | F | Epithelioid | 7.5 | 1 | IV, nodal metastases | 1034 |
aSDHA p. R210X (31%) aRAD52 p. G118D (46%) aBRCA2 p. T598A (46%) aBRCA2 p. T2097M (49%) aAXL p. T328M (44%) |
| 32 | F | Epithelioid | 10 | 2 | IV, liver metastasis | 776 |
aNOTCH2 p. R2105W (46%) aMSH2 p. M813W (48%) aKDR p. N205S (49%) aSDHA p. C311F (41%) |
Sequenced against pooled normal due to unavailability of matched normal DNA.
HPF= High-powered field, AJCC= American Joint Committee on Cancer, M= Male, F= Female
KRAS Mutant GIST Clinical History
The patient displaying a KRAS mutation was a 67-year-old male with a 25-pack year history of smoking, a negative family history, and a remote history of prostate cancer, presented with abdominal pain. A computed tomography scan revealed massive lymphadenopathy involving the chest, abdomen, and pelvis as well as a 4.7 cm gastric mass. Biopsies revealed stage IV follicular lymphoma and synchronous GIST. The patient received chemotherapy for his lymphoma with positron emission tomography negative response.
KRAS Mutant GIST Pathology Findings
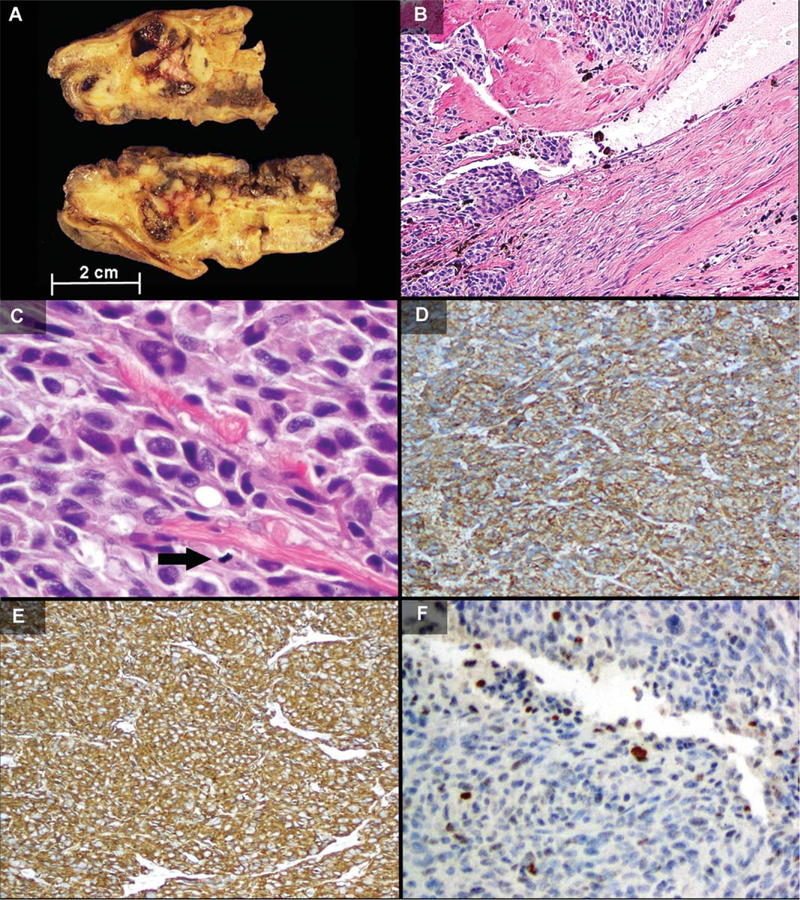
Initial wedge gastrectomy revealed a 6.5 × 6.5 × 4.5 cm encapsulated, lobulated, and partially cystic fleshy tan yellow mass within the wall of the stomach. The patient’s second, post-imatinib, gastric resection specimen on the recurrent GIST revealed multiple nodules of similar appearance (Fig. 1A) Surgical pathology revealed mostly solid architecture with lymphovascular invasion (Fig. 1B), epithelioid to rhabdoid morphology with areas of anaplasia, multinucleation, intracytoplasmic vacuolization, and an increased mitotic rate of 99 mitoses per 50 high powered fields (Fig. 1C). Multinodular or plexiform architecture was not present. The tumor demonstrated immunohisto-chemical reactivity for KIT (Fig. 1D), CD34 (Fig. 1E), desmin, smooth, muscle actin, and MDM2 (Fig.1F) and was negative for S100 and lost expression of SDHB. Histologic examination and immunohistochemistry for CD20, CD10, and BCL2 revealed the resected GIST to be negative for microscopic involvement by the patient’s known lymphoma.
Figure 1.
Surgical pathology findings. A) Gross pathology of the recurrent post-imatinib gastric tumor showed multiple tan fleshy nodules with cystic hemorrhage within the gastric wall. B–F) The original resection demonstrated areas of lymphovascular invasion (B, H&E, 103), anaplasia with large tumor cells with multinucleation, intracytoplasmic vacuoles, and mitotic figure (arrow) (C, H&E, 40×), diffuse immunohistochemical positivity for KIT and CD34 (D and E, 10×), and patchy nuclear expression of MDM2 (F, 20×). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
KRAS Mutant GIST Molecular Findings
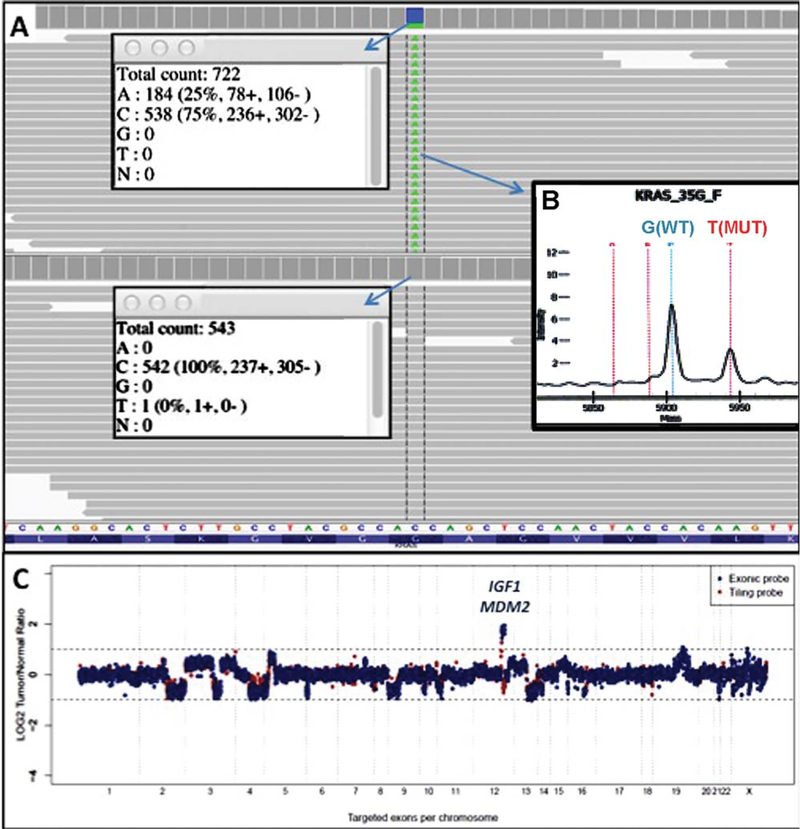
The original resection was analyzed first. Sizing assay and Sanger sequencing were negative for mutations in both KIT and PDGFRA. NGS (MSK-IMPACT assay) supported these results and also revealed the missense mutation KRAS G12V(c.35G>T) at an allele frequency of 25% (Fig. 2A). Mass spectrometry analysis of the post-imatinib gastric recurrence revealed the same KRAS mutation that was found in the original resection at similar estimated allele frequency of 29% (Fig. 2B). Amplification of IGF1 and MDM2 was also identified within the primary tumor by MSK-IMPACT (Fig. 2C). The patient progressed on imatinib at 400 mg and sunitinib at 37.5 mg, with widespread disease in the stomach, colon, and liver at 12 months after diagnosis. After secondary resection, the patient failed multiple other therapies including sorafenib plus temozolomide, pazopanib, and gemcitabine. The patient died of complications related to metastatic GIST 24 months after initial diagnosis.
Figure 2.
Molecular pathology findings. A) IntegrativeGenomics-Viewer of KRAS sequencing by MSK-IMPACT assay from original tumor shows 25% of bidirectional reads show a KRAS c.35G>T (p.G12V) mutation in reverse direction in tumor (top, arrow pointing to total and nucleotide-specific reads) but not matched normal (below, arrow pointing to total and mutant specific reads). B) Confirmatory Sequenom Mass Spectrometry (arrow) results show the wild type (WT) KRAS peak, as well as the same mutant (MUT) peak (c.35G>T) in the GIST recurrence. C) Log ratio of copy number alterations graph show MDM2 and IGF1 amplification relative to normal tissue, with fold changes of 3.3 and 3.2, respectively. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
DISCUSSION
In summary, we describe a lethal gastric GIST with KRAS mutation that did not respond to multiple lines of therapy. The identification of KRAS mutations in GIST is still relatively new, and recent previously reported cases of KIT-mutated, KRAS-mutated GISTs are summarized in Table 2.
TABLE 2.
Previously Reported Cases of KRAS-Mutant GISTs
| Case | KRAS mutation |
KIT/PDGFRA mutation |
Organ | Risk | KIT IHC | Response to Imatinib |
|---|---|---|---|---|---|---|
| Miranda et al. (2012) | G12D | KIT Δ570–576 | Stomach | High | + | Unknown |
| Miranda et al. (2012) | G12A/ G13D | KIT Δ 579 | Small bowel | Intermediate | + | Unknown |
| Miranda et al. (2012) | G13D | PDGFRA D842V | Stomach | Low | + | Unknown |
| Antonescu et al. (2013) | G12V | KIT Δ 557–558 | Small bowel | High | − | None |
| Serrano et al. (2014) | G12R | KIT Δ 554–559 | Stomach | High | + | Limited to KRAS wild type nodule |
| Current | G12V | None | Stomach | High | + | None |
The molecular profile of the tumor is summarized in Table 1. Several other mutations in tumor suppressors including ATRX, NF1, MSH2, PMS1, and DICER1 were also detected in this case. The contribution of several of these mutations to GIST development, progression, and imatinib resistance has not been well established. Interestingly, amplification of the proto-oncogenes MDM2 and IGF1 was identified in the GIST. The MDM2 gene, perhaps most well known in association with well-differentiated liposarcoma and rarely reported in GISTs, encodes an E3 ubiquitin protein ligase, a negative regulator of TP53. In vitro studies have suggested that MDM2 cooperates with RAS in tumor formation (Alarcon-Vargas and Ronai, 2002). The IGF1 gene encodes the ligand, insulin-like growth factor. The IGF system has been indicated to be activated in GISTs, and recent data have shown that increased IGF1 correlates with poor prognosis in GISTs (Braconi et al., 2008). Interaction between this ligand and its receptor has been shown to activate the PI3K and MAPK pathways, leading to cellular proliferation (Yu and Rohan, 2000). Like MDM2, recent data have shown that the IGF system cooperates with KRAS mutation in lung cancer (Molina-Arcas et al., 2013).
The other seven KIT/PDGFRA wild type GISTs harbored various other mutations in tumor suppressor genes were detected, including TP53 frameshift mutations (two cases), and an ARID1A frameshift mutation with resulting loss of ARID1A immunohistochemical expression. TP53 mutations in GIST occur in approximately 8% of GISTs and are associated with shortened progression free survival as well as rare cases of dedifferentiated GIST (Romeo et al., 2009, Antonescu et al., 2013). To our knowledge, this is the first report of ARID1A mutations in GISTs. Loss of ARID1A has recently been linked to increased phosphorylation of AKT and activation of the PI3K/AKT pathway, making it a potential target for targeted therapy (Samartzis et al., 2014).
The prevalence of KRAS mutation in GISTs ranges from <0.2% of all GISTs (Lasota et al., 2013) to, in our series of eight wild type GISTs, 11%. In 2012, Miranda et al. first described the occurrence of KRAS mutations in GISTs in three patients, two of which harbored KIT mutations and one of which harbored a PDGFRA mutation. The estimated incidence of KRAS mutation in GIST was 5% in their series (Miranda et al., 2012). A year later, Lasota et al. described a paucity of KRAS mutations in a cohort of 514 GISTs (Lasota et al., 2013), including 117 KIT/ PDGFRA wild type cases while Antonescu et al described another KIT-mutant, KRAS-mutant GIST that developed in a patient receiving imatinib for chronic myelogenous leukemia (Antonescu et al., 2013). In 2014, Serrano et al. described a KIT exon 11 mutant (554–559del) gastric GIST with a KRAS G12R mutation limited to 1 of 3 nodules of tumor tested. While the KRAS wild type, KIT mutant areas responded to imatinib, the KRAS mutated area did not (Serrano et al., Epub ahead of print). This case thus represents the sixth report of KRAS mutation in GIST and the first report of KRAS mutant GIST in the absence of KIT or PDGFRA mutation.
Factors contributing to the development of a KRAS-mutant GIST are currently unknown. This patient had a history of multiple other cancers, as do many patients with GIST. However, he did not have either a family history of GIST or of any known cancer syndrome. Of interest, the patient had a smoking history of 25-pack years. Smoking has been linked to multiple types of cancer other than pulmonary and oropharyngeal, including pancreatic and colorectal carcinomas (Liang et al., 2009; Edderkaoui and Thrower, 2013). Transversion mutations are frequently seen in smoking-related malignancies. The particular missense transversion mutation present within this GIST, KRAS G12V (c. 35G>T) is frequently seen in smoking- associated pulmonary adenocarcinoma (Dogan et al., 2012).
Aside from this case, the case by Serrano et al. was the only other GIST with multiple intrapatient samples tested for KRAS mutation. Interestingly, the KIT/KRAS co-mutant case presented by Serrano et al. demonstrated KRAS heterogeneity, with only 1 of 3 GIST nodules positive for KRAS mutation (Serrano et al., Epub ahead of print). Not unsurprisingly, the KRAS mutant nodule demonstrated imatinib resistance while the nodule with only KIT mutation was sensitive. In contrast, both the pre- and post-imatinib samples from this case, which was KRAS mutation positive without activating mutation in KIT or PDGFRA, showed KRAS mutation homogeneity, with the same mutation of similar allele frequency in both samples tested. The differences and similarities between these two cases illustrate the following points: (1) KRAS mutation in KIT/PDGFRA wild type GISTs may be present as a part of the driving intracellular signaling alterations shared between multiple intrapatient samples, or as a resistance clone in a polyclonal background of KIT mutant GIST with imatinib resistance; and (2) KRAS mutant GISTs do not respond to imatinib.
Shortfalls of our study include small sample size as well as potential selection bias. KIT/PDGFRA sequencing is ordered by request of clinician or surgical pathologist as opposed to reflex testing. Therefore, cases with clinicopathologic features of SDH deficiency may undergo immunohistochemical staining without molecular analysis of KIT/ PDGFRA, resulting in a lower number of KIT/ PDGFRA wild type GISTS being tested in our clinical molecular lab. Additionally, low risk GISTs may not be tested if adjuvant therapy is not being considered. These factors may account for the fact that only 5.6% of out cohort was KIT/ PDGFRA wild type.
Regarding the use of next-generation sequencing in GISTs, recent studies have demonstrated this technique’s potential applications. Whole exome sequencing of a BRAF mutant GIST treated with dabrafenib revealed possible mechanisms of resistance including a PIK3CA p. H1047R mutation and a CDKN2A aberration (Falchook et al., 2013), and whole exome sequencing of a select set of GISTs was performed to elucidate recurrently altered candidate genes discovered as regions of interest of microarray, leading to the discovery of potentially prognostically significant genes SYNE2, DIAPH1, and RAD54L2 (Schopp-mann et al., 2013).
In conclusion, SDH deficient GISTs often have mutations or loss of known oncogenes including TP53 and ARID1A. KRAS mutation can occur in KIT/PDGFRA wild type GISTs that immunohisto-chemically express the KIT protein and, not unexpectedly, such cases may not respond to receptor tyrosine kinase inhibition. Molecular screening for mutations downstream of KIT and PDGFRA including KRAS and BRAF may be useful to help elucidate factors contributing to imatinib primary resistance to imatinib in KIT/PDGFRA wild type GIST patients as well as provide better-suited targeted therapy options in the future, such as small molecular KRAS inhibitors (Zimmermann et al., 2013).
Supplementary Material
ACKNOWLEDGMENT
The authors thank Lisa Zhen for assistance with Sequenom assays.
Supported by: Anbinder Fund.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Alarcon-Vargas D, Ronai Z. 2002. p53-Mdm2--the affair that never ends. Carcinogenesis 23:541–547. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Romeo S, Zhang L, Nafa K, Hornick JL, Nielsen GP, Mino-Kenudson M, Huang HY, Mosquera JM, Dei Tos PA, Fletcher CD. 2013. Dedifferentiation in gastrointestinal stromal tumor to an anaplastic KIT-negative phenotype: A diagnostic pitfall: Morphologic and molecular characterization of 8 cases occurring either de novo or after imatinib therapy. Am J Surg Pathol 37:385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcila M, Lau C, Nafa K, Ladanyi M. 2011. Detection of KRAS and BRAF mutations in colorectal carcinoma roles for high-sensitivity locked nucleic acid-PCR sequencing and broad-spectrum mass spectrometry genotyping. J Mol Diagn 13:64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braconi C, Bracci R, Bearzi I, Bianchi F, Sabato S, Mandolesi A, Belvederesi L, Cascinu S, Valeri N, Cellerino R. 2008. Insulin-like growth factor (IGF) 1 and 2 help to predict disease outcome in GIST patients. Ann Oncol 19:1293–1298. [DOI] [PubMed] [Google Scholar]
- Chaft JE, Arcila ME, Paik PK, Lau C, Riely GJ, Pietanza MC, Zakowski MF, Rusch V, Sima CS, Ladanyi M, Kris MG. 2012. Coexistence of PIK3CA and other oncogene mutations in lung adenocarcinoma-rationale for comprehensive mutation profiling. Mol Cancer Ther 11:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DT, Mitchell T, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O’Reilly C, Sadowska J, Casanova J, Yannes A, Hechtman JF, Yao J, Song W, Ross D, Oultache A, Dogan S, Borsu L, Hameed M, Nafa K, Arcila ME, Ladanyi M, Berger MF. MSK-IMPACT: A hybridization capture-based next generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan S, Shen R, Ang DC, Johnson ML, D’Angelo SP, Paik PK, Brzostowski EB, Riely GJ, Kris MG, Zakowski MF, Ladanyi M. 2012. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: Higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 18:6169–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edderkaoui M, Thrower E. 2013. Smoking and Pancreatic Disease. J Cancer Ther 4:34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falchook GS, Trent JC, Heinrich MC, Beadling C, Patterson J, Bastida CC, Blackman SC, Kurzrock R. 2013. BRAF mutant gastrointestinal stromal tumor: First report of regression with BRAF inhibitor dabrafenib (GSK2118436) and whole exomic sequencing for analysis of acquired resistance. Oncotarget 4: 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, Mueller JJ, Olvera N, Dao F, Scott SN, Shah R, Gao J, Schultz N, Gonen M, Soslow RA, Berger MF, Levine DA. 2014. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet 46:424–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasota J, Xi L, Coates T, Dennis R, Evbuomwan MO, Wang ZF, Raffeld M, Miettinen M. 2013. No KRAS mutations found in gastrointestinal stromal tumors (GISTs): Molecular genetic study of 514 cases. Mod Pathol 26:1488–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang PS, Chen TY, Giovannucci E. 2009. Cigarette smoking and colorectal cancer incidence and mortality: Systematic review and meta-analysis. Int J Cancer 124:2406–2415. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Lasota J. 2006. Gastrointestinal stromal tumors: Review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch Pathol Lab Med 130:1466–1478. [DOI] [PubMed] [Google Scholar]
- Miranda C, Nucifora M, Molinari F, Conca E, Anania MC, Bordoni A, Saletti P, Mazzucchelli L, Pilotti S, Pierotti MA, Tamborini E, Greco A, Frattini M. 2012. KRAS and BRAF mutations predict primary resistance to imatinib in gastrointestinal stromal tumors. Clin Cancer Res 18:1769–1776. [DOI] [PubMed] [Google Scholar]
- Molina-Arcas M, Hancock DC, Sheridan C, Kumar MS, Downward J. 2013. Coordinate direct input of both KRAS and IGF1 receptor to activation of PI3 kinase in KRAS-mutant lung cancer. Cancer Discov 3:548–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. 2004. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5:557–572. [DOI] [PubMed] [Google Scholar]
- Pan Q, Pao W, Ladanyi M. 2005. Rapid polymerase chain reaction-based detection of epidermal growth factor receptor gene mutations in lung adenocarcinomas. J Mol Diagn 7:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo S, Debiec-Rychter M, Van Glabbeke M, Van Paassen H, Comite P, Van Eijk R, Oosting J, Verweij J, Terrier P, Schneider U, Sciot R, Blay JY, Hogendoorn PC; European Organization for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group. 2009. Cell cycle/apoptosis molecule expression correlates with imatinib response in patients with advanced gastrointestinal stromal tumors. Clin Cancer Res 15: 4191–4198. [DOI] [PubMed] [Google Scholar]
- Samartzis EP, Gutsche K, Dedes KJ, Fink D, Stucki M, Imesch P. 2014. Loss of ARID1A expression sensitizes cancer cells to PI3K- and AKT-inhibition. Oncotarget. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoppmann SF, Vinatzer U, Popitsch N, Mittlböock M, Liebmann-Reindl S, Jomrich G, Streubel B, Birner P. 2013. Novel clinically relevant genes in gastrointestinal stromal tumors identified by exome sequencing. Clin Cancer Res 19: 5329–5339. [DOI] [PubMed] [Google Scholar]
- Serrano C, Wang Y, Mariño-Enríquez A, Lee JC, Ravegnini G, Morgan JA, Bertagnolli MM, Beadling C, Demetri GD, Corless CL, Heinrich MC, Fletcher JA. KRAS and KIT Gatekeeper Mutations Confer Polyclonal Primary Imatinib Resistance in GI Stromal Tumors: Relevance of Concomitant Phosphatidylinositol 3-Kinase/AKT Dysregulation. J Clin Oncol. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Lasota J, Miettinen M. 2011. Succinate Dehydrogenase Subunit B (SDHB) is expressed in Neurofibromatosis 1-associated gastrointestinal stromal tumors(Gists): Implications for the SDHB expression based classification of GISTs. J Cancer 2:90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Rohan T. 2000. Role of the insulin-like growth factor family in cancer development and progression. J Natl Cancer Inst 92:1472–1489. [DOI] [PubMed] [Google Scholar]
- Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI, Waldmann H. 2013. Small molecule inhibition of the KRASPDEd interaction impairs oncogenic KRAS signalling. Nature 497:638–642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.