Abstract
The T cell antigen receptor (TCR) expressed on thymocytes interacts with self peptide-major histocompatibility complex (pMHC) ligands to signal apoptosis or survival. Here we found that negative-selection ligands induced thymocytes to exert forces on the TCR and the coreceptor CD8 and formed cooperative TCR–pMHC–CD8 trimolecular ‘catch bonds’, whereas positive-selection ligands induced less sustained thymocyte forces on TCR and CD8 and formed shorter-lived, independent TCR–pMHC and pMHC–CD8 bimolecular ‘slip bonds’. Catch bonds were not intrinsic to either the TCR–pMHC or the pMHC–CD8 arm of the trans (cross-junctional) heterodimer but resulted from coupling of the extracellular pMHC–CD8 interaction to the intracellular interaction of CD8 to TCR-CD3 via associated kinases to form a cis (lateral) heterodimer capable of inside-out signaling. We suggest that the coupled trans-cis heterodimeric interactions form a mechanotransduction loop that reinforces negative-selection signaling that is distinct from positive-selection signaling in the thymus.
T-cell antigen recognition occurs upon interactions between T-cell receptors (TCRs) and antigen peptides bound to the major histocompatibility complexes (pMHCs). The invariant coreceptors CD4 or CD8 also bind pMHC class II or I molecules, respectively. This recognition is highly discriminative, as small differences in the peptide sequence can induce distinctive phenotypic outcomes in T-cells. Extensive studies have been devoted to these molecular interactions1. Still, our understanding of how the TCR and/or CD4/CD8 interact with pMHC in situ, what measures of these interactions provide the most informative features to predict T-cell behavior, how such interactions transmit (and are regulated by) extracellular and intracellular signals, and how they drive T-cell function and developmental fate is incomplete.
Existing models of thymocyte selection are based on measurements of TCR kinetics and affinity, e.g., the TCR–pMHC dwell-time determines the selection outcome2, 3. The differences in TCR dwell-times with positive- and negative-selection ligands measured using purified proteins in solution (three-dimensional or 3D measurements) are very small; however, signaling initiated by these ligands results in the survival or death of thymocytes2, 4. Whereas making in situ measurements with TCRs and coreceptors on live cells (i.e., 2D measurements) represents an important advance in kinetic analysis5–7, these measurements are made at zero-force, which assesses bond quantity only and may miss key information required to predict all T-cell behaviors8. Additional information is found by measuring TCR–pMHC dissociation under forces, which assesses bond quality and reveals TCR dynamic bonds, including ‘catch bonds’ with agonist pMHCs in which force prolongs bond lifetime, and ‘slip bonds’ with antagonist pMHCs in which force shortens bond lifetimes, which amplifies the kinetic differences of TCR bonds with different pMHCs9–11. Furthermore, T-cells exert endogenous forces on the TCR and CD8 that depends on the ligand and the protein tyrosine kinase Lck, supporting the physiological relevance of kinetic measurement under force12.
To find better predictors for thymocyte selection outcomes, we made in situ measurements of affinities, molecular stiffness13 and force-dependent lifetimes of the bonds of two panels of pMHCs with two TCRs and/or CD8, and the thymocyte forces exerted on these bonds. Our data suggest that mechanotransduction amplifies the ligand discriminative power of the TCR to determine the outcome of thymocyte selection.
RESULTS
TCR dynamic bonds distinguish negative- and positive-selection ligands
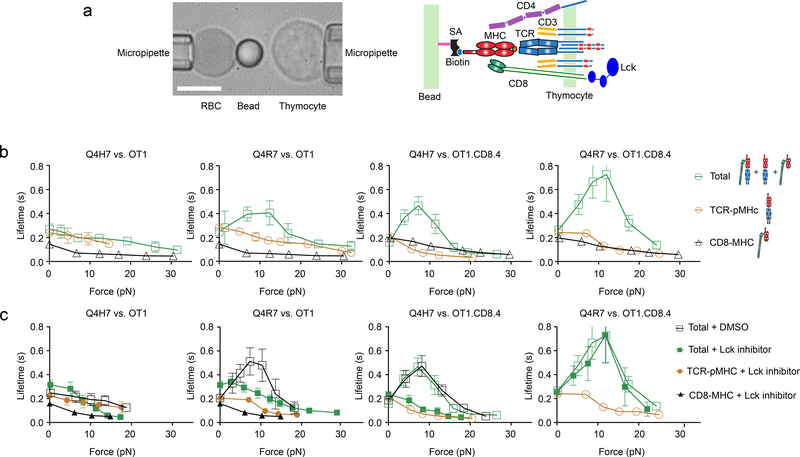
We used the biomembrane force probe (BFP)9 to measure the force-dependent lifetimes of single pMHC bonds on the surface of CD4+CD8+ double-positive (DP) thymocytes from OT1 TCR transgenic mice. The probe was coated with H2-Kb or H2-Kbα3A2 (in which the mouse α3 domain was replaced with that of human HLA-A2 to abolish CD8 binding9) to present Q4H7, the strongest positive-selection peptide or Q4R7, the weakest negative-selection peptide in a panel of peptides mutated from OVA (SIINFEKL) previously identified using fetal thymic organ culture (FTOC)4 (Fig. 1a). The OT1 TCR interactions with Q4H7 and Q4R7 presented by H2-Kbα3A2 (mQ4R7 and mQ4H7 hereafter) showed comparable slip bonds, in which the mean bond lifetime decreased monotonically with increasing force (Fig. 1b). CD8 interaction with VSV (RGYVYQGL) presented by H2-Kb (wVSV hereafter), a noncognate ligand of the OT1 TCR, showed short-lived slip bonds (Fig. 1b). Presenting cognate peptides by H2-Kb to allow interaction with TCR, CD8 and both (total interaction hereafter), wQ4R7 formed a catch bond that prolonged lifetime at low forces, then transitioned to a slip bond at >13 pN forces, whereas wQ4H7 formed a short-lived slip bond nearly identical to that of mQ4H7 (Fig. 1b). These data suggest that whether a ligand forms a CD8-dependent catch bond with the TCR at low forces determines whether it is a negative-selection ligand.
Figure 1 |. Lck-dependent Dynamic Bonds of TCR and/or CD8 with Q4H7 and Q4R7.
a, BFP setup (left) and a schematic showing the interacting molecules (right). The BFP uses a pressurized RBC as a force transducer with an attached glass bead to present pMHC. A thymocyte is aspirated by an apposing micropipette and driven to contact the bead. Scale bar = 5 μm. pMHC on the BFP bead is coupled via biotin–streptavidin (SA) and interacts with TCR/CD3 complex as well as CD4 and CD8 coreceptors on the DP thymocyte. b, c, The top and bottom rows show untreated and Lck inhibitor (or DMSO) treated data, respectively. The left and right groups (of two columns each) show data from OT1 and OT1.CD8.4 thymocytes, respectively. The first and third columns show data for Q4H7 whereas the second and fourth columns show data for Q4R7, respectively. Colored symbols and legend letters indicate OT1 thymocyte negative selection outcomes: red for negative selection and blue for positive selection, which are used throughout this paper. Mean ± s.e.m. of lifetime vs. force plots of TCR bonds with Q4H7 or Q4R7 presented by H2-Kbα3A2 (brown circle) or total bonds with either peptide presented by H2-Kb (green square), or CD8 bonds with VSV:H-2Kb (black triangle). Open and closed symbols denote untreated and Lck inhibitor-treated systems, respectively. Open black square denotes DMSO-treated controls. The numbers of lifetime measurements per curve for different ligands are summarized in Supplementary Table 1a. Results of statistical tests examining the trends of the bond lifetime vs. force curves and their differences are summarized in Supplementary Tables 2a and 3a, b.
A shift in the negative-selection threshold has been observed in thymocytes OT1.CD8.4 thymocytes2, in which the wild-type (WT) CD8 is replaced by a chimeric coreceptor consisting of the CD8 ectodomain fused with the CD4 cytoplasmic tail14. wQ4H7 interaction with OT1.CD8.4 thymocytes at low forces changed from the slip bond to a catch bond (Fig. 1b), correlating with the negative-selection threshold shift that changes Q4H7 from a positive-to a negative-selection ligand. wQ4R7 interaction with OT1.CD8.4 thymocytes showed a more pronounced longer-lived catch bond than OT1 thymocytes (Fig. 1b), corresponding to the Q4R7 change from the weakest negative-selection ligand for OT1 thymocytes to a stronger negative-selection ligand for OT1.CD8.4 thymocytes2. mQ4H7 and mQ4R7 interaction with the OT1.CD8.4 thymocytes formed slip bonds, similar to those formed with the OT1 thymocytes (Fig. 1b), indicating that the catch bonds of wQ4H7 and wQ4R7 with the OT1.CD8.4 thymocytes depended on the chimeric CD8.4 coreceptor.
The cytoplasmic tails of CD4 and CD8 associate with Lck, which phosphorylates the CD3 to initiate signaling. CD4 (and CD8.4) associates with Lck more (hence has a higher signaling capacity) than CD82. Our data suggest that enhancing the signaling capacity of the TCR stabilized its ligand binding. Because Lck activity regulates the formation of CD8-dependent TCR–pMHC bonds at zero-force7, we tested whether Lck activity would also regulate the prolongation of CD8-dependent TCR–pMHC bond lifetimes. We treated OT1 thymocytes with a Lck inhibitor15 and repeated the above measurements. In OT1 thymocytes, Lck inhibition had minimal effect on the wQ4H7 bond lifetime, but converted the catch bond of wQ4R7 to a slip bond (Fig. 1c). In OT1.CD8.4 thymocytes, Lck inhibition converted the catch bond of wQ4H7 to a slip bond, but was unable to do so for wQ4R7 (Fig. 1c). Together, these data indicate that Lck signaling capability and activity regulated the CD8-dependent conversion of TCR–pMHC slip bond to catch bond at the negative selection border.
Molecular stiffness distinguishes trimolecular from bimolecular bonds
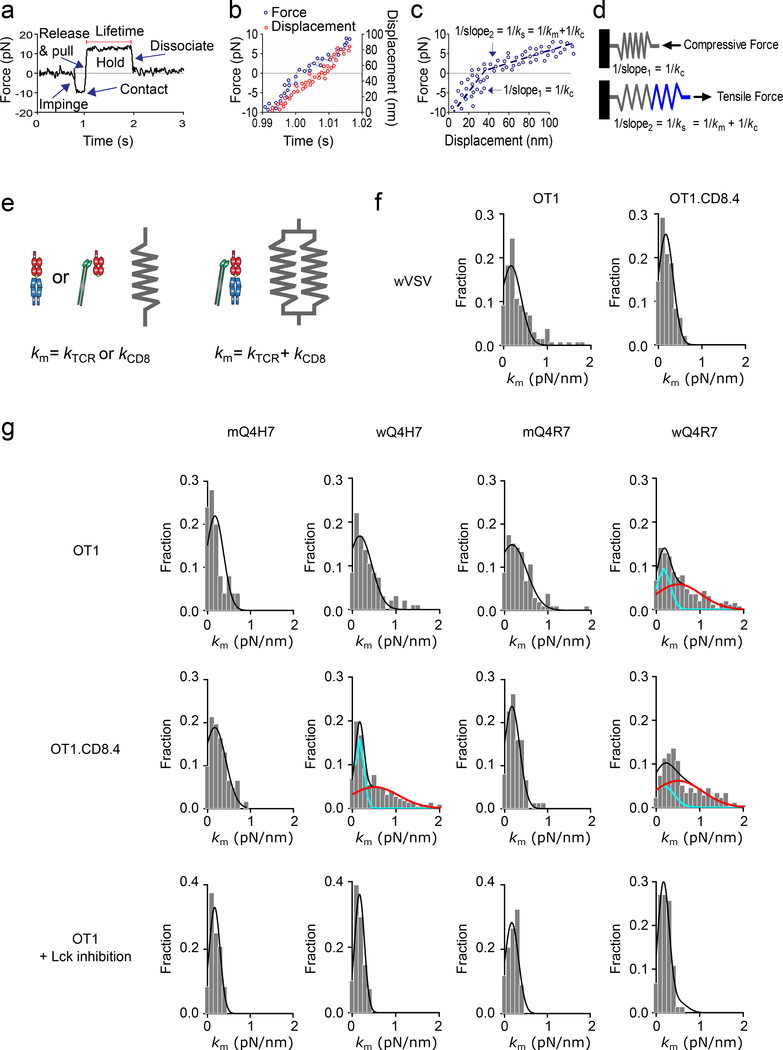
The detection of catch bond of wQ4H7 with the OT1.CD8.4 thymocytes and of wQ4R7 with both the OT1 and OT1.CD8.4 thymocytes indicates the presence of TCR–pMHC–CD8 trimolecular interaction in addition to TCR–pMHC and pMHC–CD8 bimolecular interactions, because any linear superposition of two slip bonds cannot produce a catch bond, no matter how their relative contributions are adjusted16. The finding that catch (hence trimolecular) bonds were formed only with negative, but not positive, selection ligands, even though both contained CD8 binding sites, prompted us to measure the molecular stiffness of TCR bonds from the same BFP experiment from which their lifetimes were measured. The stiffness was measured from increasing force segment, and the lifetime from the constant force segment, of the force vs. time trace (Fig. 2a-d)17. Because the TCR–pMHC–CD8 bond could be viewed as a heterodimeric bond and modeled as two springs in parallel with an equivalent spring constant approximating to the sum of the spring constants of its two monomeric members (Fig. 2e), the TCR–pMHC–CD8 bond was predicted to have a greater spring constant than the TCR–pMHC or MHC–CD8 bond13.
Figure 2 |. Molecular Stiffness Analysis Distinguishes Heterodimeric and Monomeric Bonds.
a, A representative force vs. time trace of a BFP force-clamp assay cycle. Indicated are thymocyte impingement to and contact with the BFP bead, retraction to release compression and to pull with an increasing tension, and holding on a clamp force for lifetime measurement until bond dissociation. b, Force (left ordinate, blue) and displacement (right ordinate, red) vs. time data corresponding to the increasing force portion in (a). c, Force vs. displacement data obtained by combining the two sets of data from (b) to eliminate time. Two line segments (blue dashed lines) were fit to data to evaluate the compliances 1/kc and 1/ks (= reciprocal slopes of the two lines). d, The molecular spring constant km is solved from 1/ks = 1/kc + 1/km because for two springs in series, the system compliance equals the sum of compliances of the two springs. e, Mechanical model for the TCR–pMHC and MHC–CD8 monomeric complexes (one spring) and for the TCR–pMHC–CD8 heterodimeric complex (two springs in parallel). f, Molecular stiffness histograms of bimolecular bonds of wVSV with CD8 on OT1 (left) or OT1.CD8.4 (right) thymocytes. g, The first and second rows show data from untreated OT1 and OT1.CD8.4 thymocytes, respectively. The third row shows data from Lck inhibitor-treated OT1 thymocytes. The left two columns show Q4H7 data whereas the right two columns show Q4R7 data. The peptides are labeled by “m” or “w” to indicate whether they were presented by the MT or WT MHC. Molecular stiffness histograms of monomeric bonds of TCR with mQ4H7 (1st column) or mQ4R7 (3rd column), or total bonds with wQ4H7 (2nd column) or wQ4R7 (4th column). Data (bar) were fitted by a single (black curve) or double (black curve = cyan curve + red curve) Gaussian. The fitting parameters and statistics for their comparisons are summarized in Supplementary Tables 4 and 5a.
When we measured wVSV interaction with the OT1 or OT1.CD8.4 thymocytes, which yields MHC–CD8 slip bonds, we obtained single-mode molecular spring constant (km) histograms (Fig. 2f). When we measured mQ4H7 or mQ4R7 interaction with the OT1 thymocytes, OT1.CD8.4 thymocytes or OT1 thymocytes treated with the Lck inhibitor, which yields TCR–pMHC slip bonds, we also obtained mono-modal km histograms (Fig. 2g). These histograms had statistically indistinguishable means and were well-fitted to Gaussian distributions (Fig. 2f, g), suggesting that they comprised bimolecular bonds. When we measured wQ4R7 interaction with the OT1 thymocytes and wQ4H7 or wQ4R7 interaction with the OT1.CD8.4 thymocytes, which yields catch bonds and includes TCR–pMHC, MHC–CD8 and TCR–pMHC–CD8 three bond species, we obtained bimodal km histograms (Fig. 2g). These histograms were statistically better fitted to double Gaussian distributions than to single distributions, with the smaller means indistinguishable to, and the larger means approximately twice as much the means of the mono-modal histograms of the TCR–pMHC or MHC–CD8 bonds (Fig. 2g), suggesting that they included both bimolecular and trimolecular bonds. These data demonstrate that molecular stiffness can distinguish trimolecular from bimolecular interactions.
When we measured wQ4H7 interaction with the OT1 thymocytes and wQ4H7 or wQ4R7 interaction with the OT1 thymocytes treated with the Lck inhibitor, which yields slip bonds, we obtained mono-modal km histograms that were not statistically better fitted to double Gaussian distributions than to single distributions (Fig. 2g). This indicates that these interactions only yielded biomolecular bonds, despite that the peptides were presented by H2-Kb that could have formed TCR–pMHC–CD8 trimolecular bonds. Thus, the presence or absence of a stiffer subpopulation in the molecular spring constant histogram correlated with the presence or absence of catch bond.
In control experiments, mono-model histograms were observed for wQ4H7 interaction with DMSO-treated OT1 thymocytes and Lck inhibitor-treated OT1.CD8.4 thymocytes (Supplementary Fig. 1), which form slip bonds. By comparison, bimodal histograms were observed for the interaction of wQ4R7 with DMSO-treated OT1 thymocytes or Lck inhibitor-treated OT1.CD8.4 thymocytes and of wQ4H7 with DMSO-treated OT1.CD8.4 thymocytes (Supplementary Fig. 1), all of which form catch bonds. Together, these data indicate that catch bonds resulted from TCR–pMHC–CD8 trimolecular interactions. The finding suggests that whether a ligand forms a trimolecular bond with DP thymocytes determines whether it is a negative-selection ligand.
Thymocyte force distinguishes negative from positive selection ligands
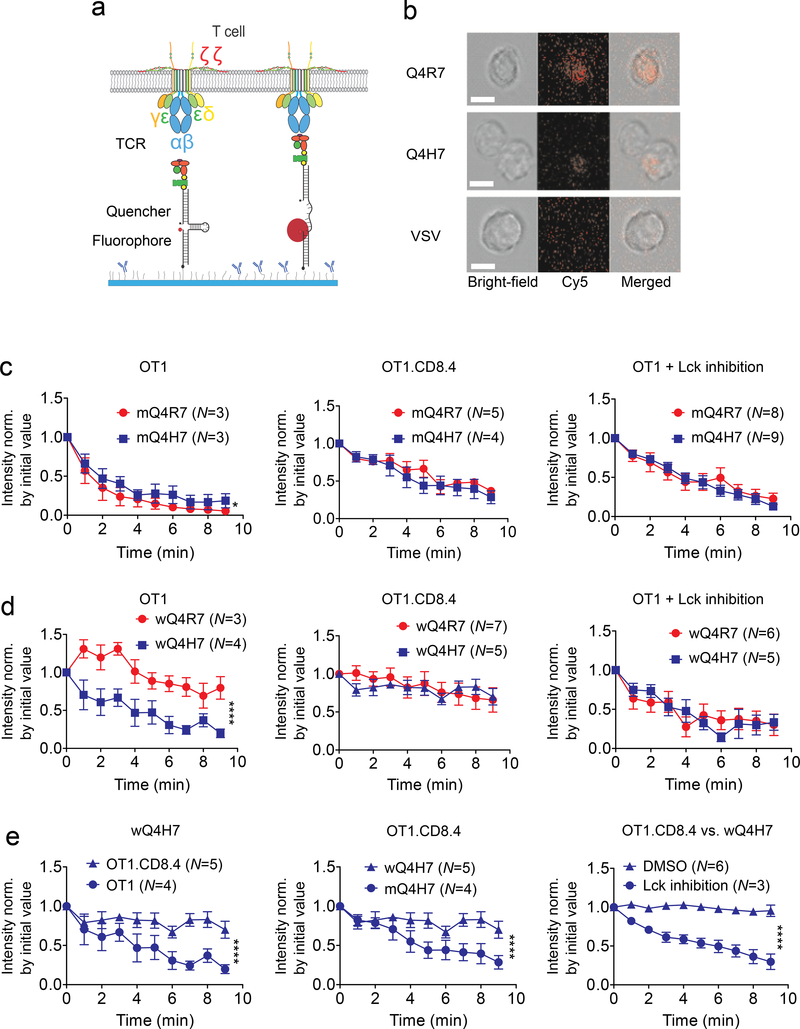
Dynamic bond and molecular stiffness analyses require applying external force on the TCR and/or coreceptor. T-cells can also exert endogenous forces on the TCR and/or coreceptor via engaged pMHC or anti-CD3 antibodies12, 18. To determine whether endogenous force could produce readouts of thymocyte fates, we placed DP thymocytes on glass surfaces bearing pMHC tagged with a molecular tension probe (MTP), which contains a DNA hairpin designed to unfold at a threshold tension to de-quench a Cy5 fluorophore, thereby reporting a force above this threshold (Fig. 3a)12, 19. This is exemplified by the fluorescence images (shown together with the bright-field and merged images) of OT1 thymocytes on surfaces coated with wQ4R7, wQ4H7 or wVSV tagged by MTP of 13.1-pN threshold tension (Fig. 3b).
Figure 3 |. Thymocytes Exert Different Forces on Q4R7 and Q4H7 via TCR and/or CD8.
a, Schematic of pMHC-tagged single hairpin DNA force probe before (left) and after (right) being engaged with and pulled by TCR complex. b, Representative images of thymocytes placed on wQ4R7, wQ4H7 and wVSV tagged with a 13.1 pN DNA force probe viewed in the bright-field (left), fluorescence (middle), and merged (right) channels. Scale bar = 5 μm. c, d, Comparison of forces on either MT or WT Q4H7 (blue) and Q4R7 (red) pulled by untreated OT1 thymocytes (left), OT1.CD8.4 thymocytes (middle), and Lck inhibitor-treated OT1 thymocytes (right). e, Replot of the wQ4H7 data; replot of the OT1.CD8.4 data; and comparison of forces on wQ4H7 pulled by DMSO-treated and Lck inhibitor-treated OT1.CD8.4 thymocytes. Data are presented as mean ± s.e.m. of indicated number of cells, fluorescence intensities were normalized by the corresponding values at the initial time (0 min). * and **** denote significance with p < 0.05 and 0.0001, respectively, by two-way ANOVA.
We measured the time course of single-cell fluorescence intensity normalized to the initial value, which reports the changing number of pMHCs pulled by that thymocyte with >13.1 pN forces over time (Supplementary Movie 1), and compared data pairs obtained under different conditions to correlate with the TCR dynamic bond type. On mQ4H7 and mQ4R7, which form slip bonds with thymocytes regardless of whether the OT1 or OT1.CD8.4 cells were used and whether they were treated by Lck inhibitor, we observed similar rapidly decreased force signals (Fig. 3c). On wQ4H7 and wQ4R7, which form catch bonds with OT1.CD8.4 thymocytes but slip bonds with Lck inhibitor-treated OT1 thymocytes, we observed similar force signals from the same cells but the signals decreased less rapidly from the OT1.CD8.4 thymocytes than the Lck inhibitor-treated OT1 thymocytes (Fig. 3d). When OT1 thymocytes were put on wQ4R7, which forms catch bonds, we observed significantly less rapidly decreased forces than when placed on wQ4H7, which forms slip bonds (Fig. 3d and Supplementary Movie 1). Thus, the more sustained force correlates with the catch bond and the less sustained force correlates with the slip bond.
On wQ4H7 we observed more sustained pulling by OT1.CD8.4 thymocytes, which forms catch bond, than OT1 thymocytes, which forms slip bond (Fig. 3e). The force signals from the OT1.CD8.4 thymocytes were more sustained on wQ4H7, which forms catch bond, than mQ4H7, which forms slip bond (Fig. 3e). On wQ4H7, we observed more sustained pulling by DMSO-treated than Lck inhibitor-treated OT1.CD8.4 thymocytes (Fig. 3e), correlating with the catch bond of the former interaction and of slip bond of the latter interaction.
Together, these data indicate that thymocytes exert more sustained endogenous forces on negative- than positive-selection ligands, which correlates with the ability of thymocytes to form longer-lasting trimolecular catch bonds with negative-selection ligands and shorter-lasting bimolecular slip bonds with positive-selection ligands.
Force distinguishes negative-selection ligands in other TCR-pMHC systems
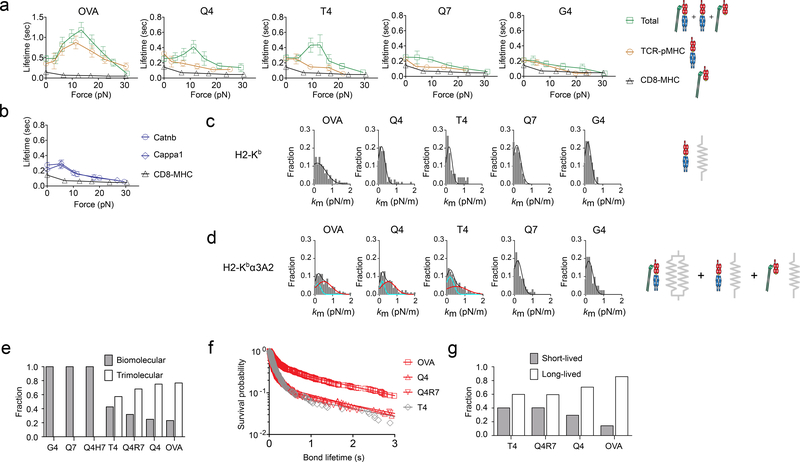
Next we tested additional peptides to cover a broader span of functional avidity on both sides of the negative-selection threshold for the OT1 thymocytes2, 4. When presented by H2-Kbα3A2 to prevent trimolecular bond formation, we observed slip bonds for all peptides except for mOVA, which is known to form catch bond with the TCR on naïve OT1 T-cells9 (Fig. 4a). When presented by H2-Kb to analyze total interactions, we observed catch bonds for negative-selection ligands (OVA, Q4) and a threshold ligand (T4), and slip bonds for positive-selection ligands (Q7, G4) (Fig. 4a). We also observed slip bonds for two endogenous positive-selection ligands (Catnb, Cappa1)20, 21 in their interactions with the TCR, CD8 and both (Fig. 4b). We tested another TCR (2C) and found that negative-selection ligands (SIYR, dEV8:H2-Kbm3) formed catch bonds, whereas positive-selection ligands (dEV8, EVSV, p2Ca) formed slip bonds with 2C thymocytes (Supplementary Fig. 2a). These experiments confirmed that whether a ligand forms a CD8-dependent catch bond with the TCR at low forces determines whether it is a negative-selection ligand.
Figure 4 |. The Same Criterion Distinguishes Other Positive and Negative Selecting Ligands.
a, b, Mean ± s.e.m. of lifetime vs. force plots of bonds of OT1 thymocytes with the indicated peptides presented by H2-Kbα3A2 (brown circle) or H2-Kb (green square, blue circle, and blue diamond), or with VSV:H-2Kb (black triangle). The numbers of lifetime measurements per curve for different ligands, the results of statistical tests examining the trends of the curves and their differences are summarized in Supplementary Table 1b, 2b and 3a, b, respectively. c, d, Molecular stiffness histograms of TCR bonds with indicated peptides presented by H2-Kbα3A2 (top) or total bonds of these peptide presented by H2-Kb (bottom). Data (bar) were fitted by a single (black curve) or double (black curve = cyan curve + red curve) Gaussian. The fitting parameters and statistics for their comparisons are summarized in Supplementary Tables 4 and 5b. e, Fractions of bi- and tri-molecular complexes returned from the Gaussian fits. f, Survival probability vs. bond lifetime data (point) and their two-state model fits (curve) of OT1 thymocyte bonds with the indicated peptides bound to H2-Kb measured at 10–15 pN force (N≥100 per curve) by the force-clamp assay. g, Short- and long-lived fractions returned from the dual exponential fits. Data are presented and statistics were analyzed as in Fig. 3.
Furthermore, molecular stiffness measurements showed mono-modal histograms for all peptides presented by H2-Kbα3A2 (Fig. 4c), and bimodal histograms for negative-selection ligands but mono-modal histograms for positive-selection ligands when presented by H2-Kb (Fig. 4d). We calculated the fractions of bimolecular (ϕb) and trimolecular (ϕt) bonds from the areas under the first and the second peaks of the bimodal histograms and found that ϕt increased with the ligand biological activity at the expense of ϕb (to satisfy ϕb + ϕt = 1; Fig. 4e), suggesting that the trimolecular bonding is peptide-dependent and hence TCR-induced. Similar results were obtained using the 2C TCR system (Supplementary Fig. 2b).
To examine the dissociation characteristics of the stiff and soft bonds, we fitted the survival probability of total bonds to a two-state model22 (Fig. 4f), which assumes that the total bonds consist of two subpopulations, one short-lived (fraction ω1) with a fast off-rate (koff1) and one long-lived (fraction ω2 = 1 - ω1) with a slow off-rate (koff2). The best-fit ω2 increased with the ligand biological activity at the expense of ω1 (Fig. 4g), similar to ϕt increasing with the ligand biological activity at the expense of ϕb (Fig. 4e). These results confirmed that whether a ligand forms a stiffer longer-lived trimolecular bond determines whether it is a negative-selection ligand. Note that this conclusion would not be affected by ligand densities, as they were adjusted to keep the adhesion frequencies uniformly low (<20%), a necessary condition for the lifetimes and spring constants to be measured predominately (>89%) from single-bond events.
Moreover, MTP experiments showed only background fluorescence intensity when OT1 thymocytes were put on wVSV surfaces, higher Cy5 intensity on wCatnb and highest Cy5 intensity on wOVA surfaces (Supplementary Fig. 2c, d), indicating that ligand recognition by the TCR is required for these force signals. When OT1 thymocytes were put on wOVA, which form trimolecular bonds, we observed significantly less rapidly decreased forces than when put on mOVA (Supplementary Fig. 2e), which form bimolecular bonds. We also confirmed these findings using MTP of a force threshold of 4.7 pN instead of 13.1 pN (Supplementary Fig. 2f-h). Compared to the DMSO control, the Cy5 signals were diminished over time when thymocytes were treated with latrunculin A to disrupt actin filaments or ROCK inhibitor Y-27632 to prevent myosin contraction (Supplementary Fig. 2i, j), indicating that pMHCs were pulled by actomyosin-based forces generated by the thymocytes, consistent with prior reports23. These data indicate that whether a ligand induces more sustained thymocyte forces determines whether it is a negative-selection ligand.
Force amplifies the discriminative power of thymocyte negative selection
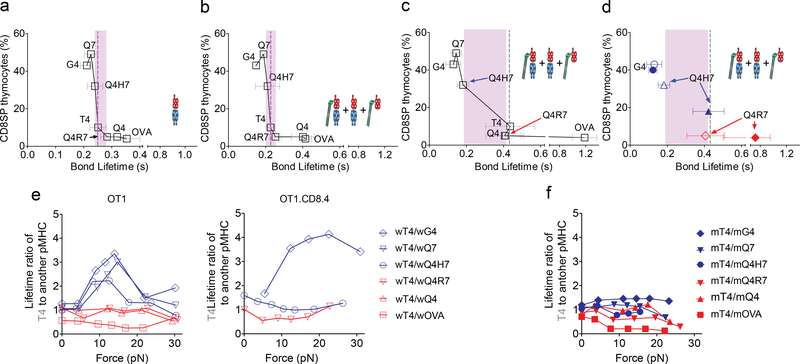
Force amplifies the discriminative power of naïve T cells by eliciting TCR catch bonds with agonist pMHCs and slip bonds with antagonist pMHCs9,11. Therefore, we asked whether force played a similar amplifying role in thymocyte selection and whether such role depended on CD8. When we plotted the survival frequency of CD8+ single-positive (SP) thymocytes previously reported in FTOC assays4 against the bond lifetime measured at zero force using the thermal fluctuation assay24, we found that, for the same panel of peptides presented by H2-Kbα3A2 or H2-Kb, the rank order of the functional avidities matched that of the lifetimes of bimolecular bonds and total bonds (Fig. 5a, b). These plots showed a sharp separation at the border between the positive- and negative-selection ligands with very small differences in their mean bond lifetimes (Fig. 5a, b). Because there were extensive overlaps among the exponentially-distributed individual bond lifetimes of different ligands (Supplementary Fig. 3a), it remained unclear how thymocytes make a ‘live or die’ decision based on the small mean bond lifetime differences, a problem encountered by previous reports using 3D measurements to predict thymocyte selection2–4.
Figure 5 |. Force Amplifies the Discriminative Power of Thymocyte Negative Selection.
a-d, Published percent of CD8 SP thymocytes from FTOC assay4 was plotted vs. average bond lifetimes of OT1 thymocytes with indicated peptides presented by H2-Kbα3A2 (a) or H2-Kb (b-d) measured at 0 pN (a, b) or 10–15 pN (c, d) forces. Data for three ligands (G4, Q4H7, and Q4R7) measured using the OT1 system (open symbol) are replotted in (d) to compared corresponding data measured using the OT1.CD8.4 system (filled symbol). Grey dash line indicates the threshold ligand T4. Pink area indicates gap between the strongest positive selecting ligand and the weakest negative selecting ligand. e, f, Plots of force-dependent mean lifetime ratios of T4 to indicated peptides presented by H2-Kb (e for OT1 and OT1.CD8.4 systems) or H2-Kbα3A2 (f for OT1).
When we plotted the SP thymocyte survival frequency vs. total bond lifetime measured at 10–15 pN, the differential mean bond lifetime across the selection threshold was increased by 4.9-fold from the zero-force measurement, as shown by the widened gap between the weakest negative-selection ligand and the strongest positive-selection ligand (Fig. 5c). The gap was widened because force induced two distinct dynamic bond types, depending on the positive or negative selection-inducing potential of the ligands. wOVA, wQ4, wQ4R7 and wT4 interacted with both TCR and CD8 to form long-lived catch bonds, while wQ4H7, wQ7 and wG4 interacted with only TCR to form short-lived slip bonds (Fig. 5c).
Because the ability for a ligand to form a TCR–pMHC–CD8 trimolecular catch bond was regulated by Lck, we examined the plots of published functional avidity readouts2 vs. bond lifetimes measured here using OT1.CD8.4 thymocytes, which have increased association between Lck and the coreceptor. Overlaying this data with those from the OT1 thymocytes showed that the negative-selection threshold remained unshifted and the differential bond lifetime across the threshold also remained the same (Fig. 5d). Only the Q4H7 data point shifted from the left to the right side of the negative-selection threshold (Fig. 5d), because its bond lifetime was lengthened as its dynamic bond type changed from slip to catch bond.
We plotted the mean bond lifetime ratios of the threshold ligand T4 to other ligands vs. force and observed that the positive-selection ligand plots displayed a high peak around 10–15 pN, whereas the negative-selection ligand plots were insensitivity to force (Fig. 5e). The reason is that the T4 curve was similar to the catch bond curves of the negative-selection ligands, but distinct from the slip bond curves of the positive-selection ligands. As such, the bond lifetimes of negative- and positive-selection ligands were discriminated by a 2–4 fold separation at 10–15 pN. Q4H7 changed from being a positive-selection ligand for the OT1 thymocytes to being a negative-selection ligand for the OT1.CD8.4 thymocytes, and its bond lifetime ratio curve changed accordingly (Fig. 5e). However, all bond lifetime ratio plots became insensitive to force when we used H2-Kbα3A2 to present the peptides instead of H2-Kb (Fig. 5f), highlighting CD8’s role in facilitating the force amplification of TCR ligand discrimination.
We also used the adhesion frequency assay25 to test whether 2D binding measurements at zero-force could discriminate positive- from negative-selection ligands (Supplementary Fig. 3b-e). When we plotted the 2D affinity for bimolecular interactions side-by-side with published peptide:H2-Kbα3A2 tetramer data4, we observed that, except for OVA, which had the highest affinity, the measurements were comparable for all positive- and negative-selection ligands (Supplementary Fig. 3f), despite their >100-fold variations of functional avidities4. When we converted the adhesion frequency Pa of total interactions to average adhesion bonds <n> = – ln(1 – Pa) and normalized it by pMHC density mpMHC, we observed the collapse of all binding curves except for those of wOVA and wVSV (Supplementary Fig. 3e). When we plotted the plateau level of the normalized total adhesion bonds (<ntotal>/mpMHC) side-by-side with published 3D data measured by peptide:H2-Kb tetramer4, we observed that, similar to the 2D affinity, the normalized total adhesion bonds showed minimal differences between negative- and positive-selection ligands across the selection threshold (Supplementary Fig. 3g). These negative results obtained at zero-force contrast sharply to the positive results obtained at 10–15 pN, indicating the importance of force in thymocyte selection.
Force enhances the accuracy of thymocyte selection
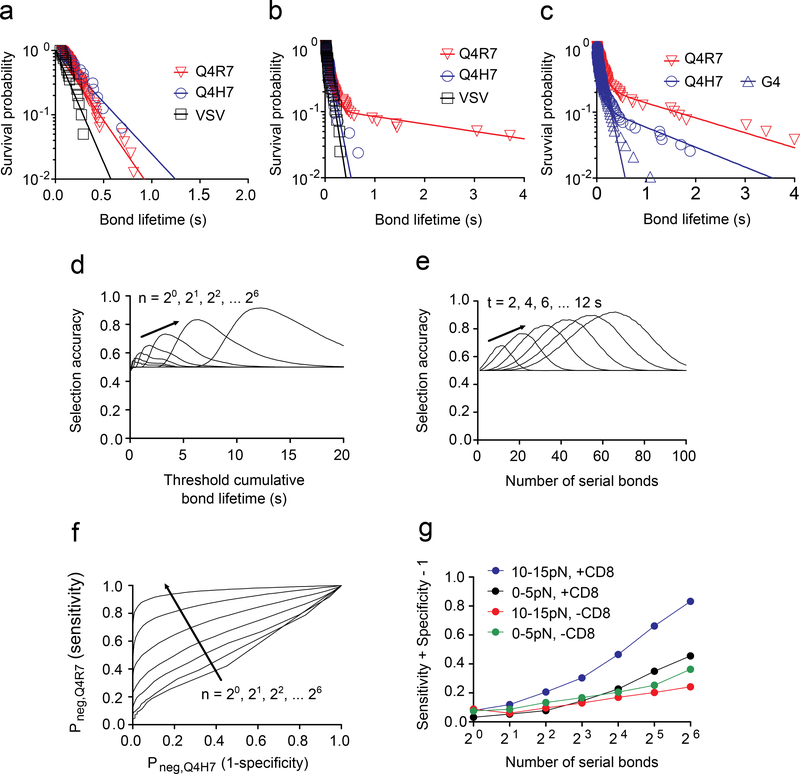
Thymocyte signaling was triggered by individual TCR bonds whose lifetimes were distributed multi-exponentially (Supplementary Fig. 4). This observation predicted that a small difference in the mean lifetime would translate into a large difference in the fractions of bond survival over time, which has been hypothesized as a basis for cell fate decision3. However, the difference in dwell times between Q4R7 and Q4H7 measured from tetramer4 and Quantum-dot2 dissociation from TCR and CD8 is too small to produce sufficiently large difference in their survival probabilities. Similarly, the survival probability difference between wQ4R7 and wQ4H7 was too small based on their minute bond lifetime difference at zero-force (Fig. 6a). As 10–15 pN of force, the survival probability curves of wQ4R7 and wQ4H7 became separated because a subpopulation of the wQ4R7 catch bond dissociated much more slowly than the wQ4H7 slip bond (Fig. 6b and Supplementary Fig. 4a). In addition, the Q4H7 survival probability curve changed from fast decay as seen in the OT1 thymocytes (Fig. 6b) to slow decay in the OT1.CD8.4 thymocytes (Fig. 6c), yielding a much greater fraction of bonds lasting >1 s and explaining the conversion of Q4H7 from a positive-selection ligand for the OT1 thymocytes to a negative-selection ligand for the OT1.CD8.4 thymocytes.
Figure 6 |. Force Enhances Thymocyte Selection Accuracy.
a-c, Survival probability vs. lifetime of total TCR and/or CD8 bonds with H2-Kb bound to the indicated peptides measured at 0 pN (N≥20 per curve) (a) and at 10–15 pN force (N≥100 per curve) using either OT1 (b) or OT1.CD8.4 (c) thymocytes. Each survival probability data set (points) was fitted with a one-state (a) or two-state (b, c) model (curve). d, e, Selection accuracy plots. The combined successful rate SA of a thymocyte to have a 50–50 chance to form n bonds in a row either with wQ4H7 and be positively selected or with wQ4R7 and be negatively selected, calculated using the survival probabilities in b, is plotted vs. tth for a range of n (d) or vs. n for a range of tth (e). The respective criteria for positive and negative selections are whether the single-bond lifetimes accumulated from the n serial bonds are shorter and longer, respectively, than the threshold cumulative bond lifetime tth required to trigger negative selection. f, ROC curves. The thymocyte’s probability to be negatively selected by wQ4R7 (true positive rate or sensitivity) is plotted vs. its probability to be negatively selected by wQ4H7 (false positive rate or 1 -selectivity) for a range of n. g, Youden’s J statistic (sensitivity + selectivity – 1) is plotted vs. the number n of serial bonds over which bond lifetimes are accumulated for the indicated cases using the ROC curves in f and in Supplementary Fig. 5c.
To understand how catch bonds impact fate decisions in thymocytes, we defined a metric for ligand discrimination using the lifetime distributions of Q4R7 and Q4H7 bonds. We assumed that a thymocyte has a 50–50 chance to encounter an APC that expresses either ligand and that the two cells would form n single bonds in a row during this encounter. We calculated the combined successful rate for the cumulative bond lifetimes to exceed (Q4R7) and not exceed (Q4H7) a threshold cumulative bond lifetime, tth, required to induce negative selection. This parameter, named ‘selection accuracy’ (SA) hereafter, represents the likelihood of an appropriate selection, i.e. the thymocyte would be negatively or positively selected if the APC expressed Q4R7 or Q4H7 respectively. As such, an SA of 0.5 would indicate equal likelihood of inducing negative selection or not, regardless of the ligands, i.e., no discrimination, whereas an SA approaching 1 would indicate a 100% probability for inducing negative selection by Q4R7 and not by Q4H7, i.e., perfect discrimination. Plotting SA vs. tth for a range of n showed that, for a given n, the SA vs. tth curve was biphasic rather than increasing monotonically (Fig. 6d). The reason is that, for short tth, both ligands had similarly high probabilities to cumulate >tth bond lifetimes, which yielded small SA, and for long tth, both ligands had similarly low probabilities to cumulate >tth bond lifetimes, which also yielded small SA. Only in mid-range tth did Q4R7 have a high probability, and Q4H7 a low probability, to cumulate >tth bond lifetimes to differentially induce (or not induce) negative-selection in the thymocyte, yielding a peak in the SA vs. tth curve (Fig. 6d). The peak SA increased with increasing n, reaching >90% at n = 64 (Fig. 6d), indicating the more information the thymocyte gathers by forming more TCR bonds with pMHC over time, the more accurate a selection decision it can make. This increasing trend in the SA vs. tth plots was greatly suppressed when they were calculated from the data from experiments when CD8 binding was prevented or when force was reduced (Supplementary Fig. 5a), indicating the important role of force and CD8. In addition, as n increased, the SA vs. tth curve shifted rightward towards a longer tth (Fig. 6d), because the greater the number of bond formation, the longer the expected cumulative bond lifetime. Of note, should one choose the probability of n bonds to have at least one lifetime (instead of the cumulative lifetime of n bonds) to (Q4R7) or not to (Q4H7) exceed tth as the respective criteria for negative- or positive-selection, then the SA vs. tth curves would have the same properties, except for the right-shift. Finally, for a fixed tth, the SA vs. n curve was biphasic, which had an optimal n value where SA reached maximum (Fig. 6e and Supplementary Fig. 5b). Because a certain amount of time is required to form a given number of bonds, an optimal n corresponds to an optimal time window. This result is consistent with previous reports that T cell triggering depends on two factors: a cumulative bond lifetime and the time window within which the bond lifetimes are accumulated9.
The biphasic shape of the SA vs. tth curves reflected a well-recognized difficulty of the existing models of antigen discrimination: the inability to achieve high sensitivity while retaining high specificity26. To better understand this we used the bond lifetime distributions to calculate and plot the probability that a thymocyte to be negatively-selected by Q4R7 (true positive rate, or sensitivity) vs. the probability to be negatively-selected by Q4H7 (false positive rate, or 1 - selectivity) for varying tth and a range of n (Fig. 6f and Supplementary Fig. 5c). These receiver operating characteristic (ROC) curves visually depict the discriminatory capacity of SA: a suboptimal SA could be due to a low probability of negative-selection by Q4R7 (low sensitivity) or a high probability of negative-selection by Q4H7 (low selectivity). The up- and left-shift of the ROC curve with increasing n showed the benefit of forming more thymocyte–APC bonds (Fig. 6f), which was suppressed by reducing the force to 0–5 pN or by preventing CD8 binding (Supplementary Fig. 5c). As a single-value quantification of a ROC curve, we plotted the Youden’s J statistic (= sensitivity + selectivity − 1) vs. the number n of serial bonds. In the presence of 10–15 pN force and allowing CD8 binding, J increased with n to achieve >80% at n = 64 (Fig. 6g). J increased much less with n when force was reduced to 0–5 pN or CD8 binding was prevented (Fig. 6g). Observations similar to the above made in the OT1 TCR thymocytes were also obtained using the 2C TCR thymocytes (Supplementary Fig. 4c, d). Together, these data suggest the existence of a CD8-dependent, force-regulated checkpoint, which amplifies the differential bond lifetimes to decide thymocyte survival or deletion.
Reducing CD3 ITAMs weakens TCR–pMHC–CD8 trimolecular interaction
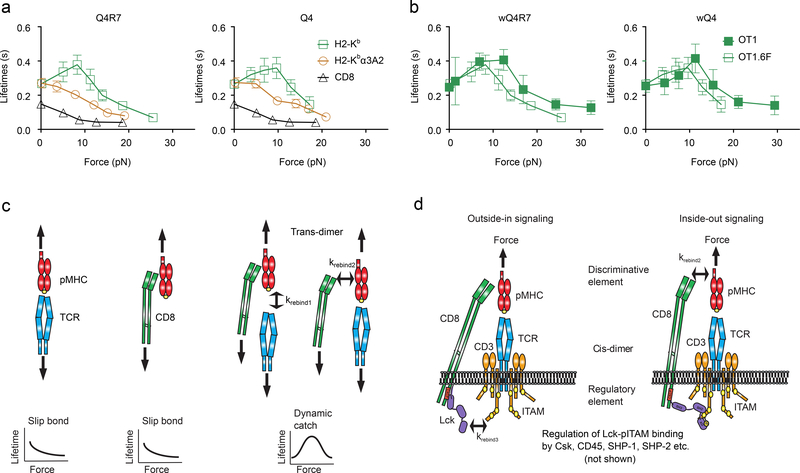
Because Lck phosphorylates ITAMs on the cytoplasmic tails of CD3, we tested whether mutating CD3 ITAMs would decrease Lck signaling and reduce TCR–pMHC–CD8 bond lifetime. We used thymocytes from OT1 transgenic mice homozygous for the 6F knock-in mutation (OT1.6F), in which the tyrosines in the six CD3ζ ITAMs were mutated to phenylalanine, resulting in a predicted 60% reduction in the signaling capacity of the TCR complex27, 28. OT1.6F thymocytes formed catch bonds with wQ4R7 or wQ4 and slip bonds with wVSV, mQ4R7 or mQ4, similar to OT1 thymocytes (Fig. 7a). At >10 pN forces, however, wQ4R7 and wQ4 formed shorter-lived bonds with OT1.6F thymocytes than OT1 thymocytes, resulting in a left- and down-shift of the OT1.6F curves relative to the OT1 curves (Fig. 7b), indicating that decreasing the number of signaling-competent ITAMs reduces the TCR–pMHC–CD8 trimolecular bond lifetime. These results imply that the dynamic bonds of pMHC with TCR and/or CD8 can be regulated by the kinase activity of Lck and/or by the docking sites available for other signaling molecules, such as Csk, SHP-1/2, ZAP-70 (Fig. 7c, d)29–31.
Figure 7 |. Outside-in/Inside-out Loop of the TCR Mechanotransduction apparatus.
a, Mean ± s.e.m. of lifetime vs. force plots of OT1.6F TCR bonds with mQ4R7 and mQ4 (brown circle), CD8 bond with wVSV (black triangle), and total TCR and/or CD8 bonds with wQ4R7 and wQ4 (green square). The numbers of lifetime measurements per curve, the results of statistical tests examining the trends of the curves and their differences are summarized in Supplementary Table 1c, 2d and 3b, respectively. b, Mean ± s.e.m. of lifetime vs. force plots of total bonds of OT1 (green square, reports from Figs. 1b and 4a) and OT1.6F (black square) thymocytes with wQ4R7 and wQ4. The results of statistical tests examining their differences are summarized in Supplementary Table 3c. c, d, Model of the outside-in/inside-out loop of the TCR mechanotransduction apparatus. The loop consists of two orthogonal heterodimeric interactions: An extracellular trans-interaction of TCR and CD8 with pMHC to form a cooperative trimolecular catch bond, despite that in the absence of synergy, the two TCR–pMHC and CD8–pMHC bimolecular interactions only form slip bond (c). This is coupled to an intracellular cis-interaction of CD8-associated Lck with TCR/CD3 ITAMs, which allows signaling differentially triggered from outside-in by the discriminative element (TCR recognition) to modulated by the regulatory element (Lck and related molecules), resulting in inside-out signaling (d).
DISCUSSION
Dynamic bond formation occurs at the ligand binding sites of the TCR and CD8 ectodomains. Our data indicate that increasing Lck association with CD8, inhibiting Lck kinase activity and reducing the number of CD3ζ ITAMs, which represent an Lck substrate resulted in enhanced, suppressed, and reduced ability for TCR and CD8 to form a catch bond with pMHC, respectively. The intracellular regulation of the extracellular binding of a receptor has been observed in integrins, and is termed ‘inside-out’ signaling32. The ability of Lck to regulate ligand binding of TCR and CD8 could be interpreted as inside-out signaling. However, distinct from integrin inside-out signaling, TCR may not associate with Lck before binding the pMHC and outside-in signaling by the TCR does not require priming of the TCR to bind pMHC. Integrin inside-out signaling involves conformational changes. While the mechanism for such a TCR-CD8 inside-out signaling remains unknown, our data support a model involving cis- and trans-heterodimeric interactions. Cross-junctional interactions between the T-cell and the APC are trans-interactions and include both TCR–pMHC and pMHC–CD8 bimolecular interactions and TCR–pMHC–CD8 trimolecular interaction. Lateral interactions on the T-cell surface are cis-interactions and include the intracellular interactions of Lck with CD8 and CD3 and the extracellular CD8 interaction with the TCR mediated by a common ligand pMHC. As a kinase, Lck transduces TCR signals by phosphorylating CD3 ITAMs. Our data suggest that Lck can also connect the TCR and CD8 intracellularly and enhance their binding of pMHC extracellularly, i.e., serving as an adaptor. The dependence of the TCR–pMHC–CD8 catch bonds on the kinase activity of Lck and its docking sites on CD8 and CD3 indicates the interdependence of Lck’s dual role.
The Lck inhibitor effect depended on the ligand’s biological activity, indicating that the efficiency of TCR inside-out signaling depends on its outside-in signaling. Upon becoming a stronger agonist for the OT1.CD8.4 than OT1 thymocytes, Q4R7 retained its ability to form catch bond in the trans-interactions, despite the presence of the Lck inhibitor, implying that Lck inhibition could not suppress the cis-interaction. This can be explained by mass action, because the propensity of Lck binding to CD8 and CD3 is determined not only by their affinities but also by the amount of Lck associated to CD8. Thus, the trans-cis-interaction loop operates in a progressive rather than an all-or-none fashion. Ligands at the negative selection border may be most sensitive to Lck modulation. The sensitivity may decrease when the weakest negative selecting ligand become stronger, as in the case of Q4R7 in the OT1.CD8.4 system. Lck inhibition may decrease the affinities of Lck for CD8 and/or CD3, which may be compensated by the increased amount of CD8-associated Lck in the OT1.CD8.4 thymocytes. This implies that the dynamic bonds of pMHC with TCR and/or CD8 can be regulated by the kinase activity of Lck and/or by the docking sites available for other signaling molecules, such as Csk, SHP-1/2, ZAP-7029–31.
Our data support a positive feedback loop to amplify differential ligand binding. Differential TCR–pMHC binding would transduce differential signals, which may induce differential Lck-mediated interactions of TCR and CD8, which may bring them to different degrees of close proximity to allow or prevent TCR–pMHC–CD8 interaction, thereby amplifying the differential TCR engagement times with different pMHCs. The proposed trans-cis-interaction loop also suggests a physical mechanism for amplification. When either the TCR–pMHC or pMHC–CD8 interaction in the trimeric complex dissociate, it may readily rebind due to the close proximity of the dissociated TCR or CD8 to the pMHC in the remaining bimolecular complex. This more favorable configuration may greatly enhance the rebinding rate to a level much higher than its original on-rate, thereby increasing the avidity and prolonging the overall lifetime of the trimeric bond until both TCR and CD8 simultaneously dissociate. Alternatively, the formation of the trimolecular bond may induce conformational changes to better fit the interfaces of the TCR–pMHC, CD8–pMHC or both arms to strengthen their stability.
Because cooperativity in biophysical interactions may result from the cooperativity in biochemical signaling, TCR signaling can also be amplified by the prolonged TCR engagement and increased accessibility to active Lck29. Prolongation of the time that the TCR feels the antigen by trans- and cis-interactions may translate into signaling enhancement. Thus, the trans-cis-interaction loop and TCR mechanotransduction signaling loop may be self-reinforcing within each other, as well as cross-reinforcing between the two loops. This may provide two levels of positive feedback and a synergistic mechanism for amplifying the discrimination of different pMHCs by the TCR.
Thus, our model consists of two coupled trans- and cis- heterodimeric interactions that employ Lck to selectively prolong TCR engagement time. The trans-interaction is the demonstrated cross-intercellular junction binding of TCR and CD8 to pMHC, which forms a vertical heterodimer. The cis-interactions include the hypothetical Lck bridging between CD8 and TCR-CD3 to comprise the intracellular arm of the horizontal heterodimer. Its extracellular arm is the demonstrated CD8 binding to the TCR–pMHC complex. This shared element provides the coupling between the horizontal and vertical heterodimers.
The intracellular cis-interaction has been supported previously, studies indicating the association of CD8 with TCR-CD333–35, a process that could be dynamically regulated by TCR-CD3 stimulation33. Co-immunoprecipitation studies in Jurkat cells suggested a molecular model that involves CD8-associated Lck binding to CD3-associated tyrosine-phosphorylated ZAP-70 using its SH2 domain30, 36. It is also possible that the SH2 domain of Lck binds to phosphorylated CD3 ITAMs directly37. CD3–ZAP-70–Lck–LAT interactions were shown to facilitate LAT phosphorylation38. Considering the abundance of CD3 ITAMs, the intracellular cis-interaction may include both direct and indirect binding of Lck to CD3, manifesting the dependence of the TCR–CD8–pMHC bond on Lck kinase activity, Lck-CD8 association and CD3 ITAM availability.
We suggest that the trans-cis-interaction or signaling loops may function to set a criterion to distinguish ligands that induce distinct cell fates in thymocyte selection. This assertion is supported by the concurrent shifts in the selection threshold and the dynamic bond type of the threshold ligands in the OT1.CD8.4 system. The assertion is also consistent with the observation that weaker negative selecting ligands changed to positive selecting ligands in the absence of CD839. This shift in the thymocyte negative selection threshold agrees with our data on the importance of CD8, as all ligands, except for OVA, formed slip bonds with the TCR when CD8 binding was prevented. Of note, OVA behaves differently from self-ligands because it is the specific antigen of (and hence interacts strongly with) the OT1 TCR. Similarly, the super agonist SIYR is also a special case in the 2C TCR system.
We propose that this binary criterion represents a force-regulated checkpoint for bond quality and includes three aspects: catch bonds, trimolecular interaction and sustained pulling. The mechanism of signaling-regulated catch bond formation is distinct from previously studied mechanisms that only involve force-induced atomistic contacts at the interface of the two binding partners40. The phenomenon of force-induced formation of a trimolecular catch bond, despite the fact that the two arms of the heterodimeric interactions behave as slip bonds, is termed dynamic catch, and was observed in the integrin α5β1–Thy-1–syndecan-4 trimolecular interaction16.
Our force-based thymocyte negative selection model extends existing models3. Due to stochastic dissociation of TCR–pMHC–CD8 complexes, the probability of a complex to remain associated decreases exponentially, with its half-life or dwell-time as the reciprocal decay rate. However, previous measurements at zero force by SPR41, tetramer technology4 or Quantum dot-pMHC monomer2, yielded very small differential 3D half-lives across the selection border. This predicts that the vast majority of TCR molecules would have overlapping engagement times with pMHCs across the negative-selection border, making discrimination difficult. Based on the suggestion of a computational study that the coreceptor binding to MHC primarily acts to enhance Lck delivery with a minor effect on stabilizing TCR–pMHC interaction42, it was proposed that a TCR–pMHC bond with long dwell-time would allow it to scan a pool of CD8 molecules, each binding and unbinding rapidly, to search for one that associates with an active Lck2. Our data indicate that Lck-mediated cis-interaction enhances the trans-interaction of CD8 to synergize with that of TCR. This may allow rebinding to the same pMHC by the same TCR and CD8 molecules to lengthen their total bond lifetime under force for negative selecting ligands, enabling initiation, prolongation and accumulation of signaling events. By contrast, positive selecting ligands do not induce Lck-mediated TCR-CD8 cooperation and force shortens TCR–pMHC bond lifetime to restrict signaling to occur, sustain or accumulate, resulting in a distinct thymocyte fate.
Differential lifetimes of TCR dynamic bonds also induced differential endogenous forces on TCR and CD8 bonds, providing yet another level of positive feedback and self-reinforcement. Note that the MTP used here reported an active response of the cell induced by TCR and CD8 upon engaging pMHC. Bond formation per se, as governed by affinity and kinetics, does not produce force. Only when the cell signals to generate actomyosin-based force and exert such force on the TCR and CD8 to pull on pMHC, will the DNA hairpin unfold to de-quench fluorescence. This is different from the BFP assay, in which force is applied externally on the TCR and/or CD8 to trigger signaling. The TCR bond lifetimes were much shorter than the decay timescale for the thymocyte force, so the force signals report pulling on intermittent bonds that form, dissociate and reform over time, mimicking serial engagement to sustain signaling. The slower the decay in the force signal, the more sustained the TCR signaling, which should directly correlate with the level of thymocyte activation, translating into clonal deletion (negative selection) or survival (positive selection). Higher and more efficient calcium signaling can be generated by moving the pMHC-coated microsphere in a tangential direction to its contact surface with the T-cell than the normal direction43. Although we used BFP to exert forces normal to the T-cell surface, the purpose was to analyze TCR dynamic bonds and molecular stiffness to allow us to correlate (rather than trigger) thymocyte selection outcomes in vivo. Besides, the DNA force probe reports tensile (not exclusively shear) force on the pMHC transmitted by the TCR and/or CD8 despite its unknown (possibly variable) directions in vivo.
Our findings regarding Lck and CD8 may be closely related to previous reports of ligand-dependent TCR-CD8 interaction33, Lck-dependent T-cell response to lower-affinity pMHC31 and coreceptor-dependent MHC restriction44, antigen recognition33 and ligand-dependent thymocyte migration45. Future studies should further elucidate the mechanism of how slip bonds and catch bonds drive functional and developmental outcomes and the possible parallel role of CD4. Whether and how catch bonds contribute to the development of other subset of T-cells such as γδ T-cells and regulatory T-cells are some of the interesting follow-up questions.
METHODS
Cells and protein
OT1 and 2C TCR transgenic mice were housed at the Emory University Department of Animal Resources facility and followed protocols approved by the Institutional Animal Care and Use Committee of Emory University. OT1 CD8.4 Rag−/− MHCII−/− (OT1.CD8.4) and OT1 Rag−/− MHCII−/− transgenic mice were housed at the NCI and OT1 transgenic mice homozygous for the CD3ζζ 6F knock-in mutation (OT1.6F) were housed in NICHD. OT1.CD8.4 and OT1.6F mice were shipped from the NIH to the Zhu lab on the day of the experiment. Pre-selected DP thymocytes46 were purified from a mouse thymus with CD53− CD4+CD8+ thymocyte enrichment by magnetic bead immunoaffinity cell sorting (MACS) according to the manufacturer’s instructions (Miltenyi Biotec, San Diego, CA). Since CD53 expression and positive selection strongly correlate, we used CD53 as a purification marker47. Recombinant pMHC monomers were from the NIH Tetramer Core Facility at Emory University. For analysis of OT1 DP thymocytes, the following peptides were synthesized and presented by C-terminally biotinylated mouse MHC class I H2-Kb or its mutant H2-Kbα3A2 (replacing the mouse α3 domain by that of human HLA-A2): chicken ovalbumin-derived peptide OVA257–264 (SIINFEKL, agonist and negative selecting ligand) and its altered peptides4 Q4 (SIIQFEKL, weak agonist and negative selecting ligand), Q4R7 (SIIQFERL, weak agonist and negative selecting ligand), T4 (SIITFEKL, weak agonist and negative selecting ligand), Q4H7 (SIIQFEHL, weak agonist and positive selecting ligand), Q7 (SIINFEQL, weak agonist and positive selecting ligand), and G4 (SIIGFEKL, weak agonist/antagonist and positive selecting ligand) as well as endogenous peptides F-actin capping protein A-derived Cappa192–99 (ISFKFDHL, positive selecting ligand)20,21 and β-catenin-derived Catnb329–336 (RTYRYEKL, positive selecting ligand)21. In addition, vesicular stomatitis virus-derived nucleoprotein VSV52–59 (RGYVYQGL) bound to H2-Kb was prepared in the same way as noncognate ligand to test CD8 binding. For the 2C TCR, SIYR (SIYRYYGL, super agonist and negative selecting ligand), dEV8 (EQYKFYSV, agonist and positive selecting ligand), EVSV (RGYVYQEL, antagonist and positive selecting ligand), and p2Ca (LSPFPFDL, weak agonist and endogenous positive selecting ligand) peptides were bound to H2-Kb or H2-Kbm3 (two mutations in α1 domain, Asp77Ser and Lys89Ala48)49, 50. All pMHC monomers were engineered to have a biotin tag on the α chain C-terminus.
Coating pMHC on RBC/bead
These procedures have been described5, 51. Briefly, human RBCs were isolated from the whole blood of healthy volunteers according to a protocol approved by the Institutional Review Board of the Georgia Institute of Technology. pMHCs were coupled onto RBCs by first biotinylating cells with Biotin-XNHS (Calbiochem, San Diego, CA) at pH 7.2 for 30 min at room temperature, next incubating with 0.5 mg/ml streptavidin (Pierce, Rockford, IL) for 30 min at 4 °C, finally incubating with pMHCs. To coat proteins on glass beads for BFP experiments, borosilicate glass beads (Duke Scientific, Palo Alto, CA) were first covalently coupled with mercapto-propyltrimethoxy silane (United Chemical Technologies, Bristol, PA), followed by covalently linking to tetravalent streptavidin-maleimide (Sigma-Aldrich, St. Louis, MO) in phosphate buffer saline (PBS) (pH 6.8) by overnight incubation at room temperature. Streptavindylated beads were then incubated with biotinylated pMHCs for 2 h at room temperature and finally resuspended in HEPES buffer saline (HBS) plus 0.5% bovine serum albumin (BSA).
Site density measurement
Site densities of TCR, CD8, and pMHC were measured by flow cytometry5 using PE-conjugated antibodies: anti-mouse Vα2 TCR monoclonal antibody (mAb) (B20.1, BD Pharmingen, San Jose, CA), anti-mouse CD4 (RM-45, eBioscience, San Diego, CA), anti-mouse CD8 (53–6.7, BD Pharmingen), anti-mouse OVA257–264 bound H2-Kb (25-D1.16, eBioscience), anti-mouse H2-Kb (AF6–88.5, BD Pharmingen), and β2 microglobulin (S19.8, Santa Cruz Biotechnology, Dallas, TX). PE-conjugated rat IgG2a κ (eBioscience), mouse IgG2a (Santa Cruz Biotechnology), and hamster IgG3 λ1 (BD Pharmingen) were used as isotype controls. Cells and beads were incubated with appropriate antibodies at 10 μg/ml in 100 μl of FACS buffer (PBS without calcium and magnesium, 5 mM EDTA, 1% BSA, 25mM HEPES, 0.02% sodium azide) at 4 °C for 30 min; measured the fluorescent intensity by the BD LSR II flow cytometer (BD Biosciences, San Jose, CA); and calibrated by the BD QuantiBRITE PE standard beads (BD Biosciences) to determine the site densities using the cell or bead surface area (154 μm2 for a thymocyte; 140 μm2 for a RBC; and 12.6 μm2 for a bead)5.
Adhesion frequency assay
Force-free 2D kinetics of pMHC interactions with TCR and/or CD8 were measured by the micropipette adhesion frequency assay as described previously25. In brief, a thymocyte and an apposing pMHC-coated RBC were manipulated to move in repeated approach-retract cycles for detection of adhesion events by the deflection of the RBC membrane (Supplementary Fig. 3b). For bimolecular interactions, the contact time (tc) dependent adhesion frequency (Pa) was measured and fitted with a probabilistic kinetic equation25, Pa =1 − exp{−mr ml Ac Ka [1 − exp(−koff tc )]}, to derive an effective 2D affinity AcKa and 2D off-rate koff with separately measured molecular densities (mr and ml). The curve fit uses the least mean squares method (Excel Solver) and the parameters were evaluated from repeated experiments (N≥3) with different TCR and pMHC densities. For blocking of OT1 TCR or CD8, DP thymocytes were incubated with 50 μg/ml anti-mouse TCR Vα2 mAb B20.1 and anti-CD8 CT-CD8a, respectively, for 30 min in 4°C prior to the experiment and the mAbs were continuously present in the experiment chamber at the same concentration7.
Thermal fluctuation assay
Thermal fluctuation assay was used to obtain more actuate measurements of zero-force off-rate koff as described previously24. In brief, a pMHC-coated bead was attached to the apex of a RBC and an apposing thymocyte was driven to near the bead, allowing intermittent contacts by thermal fluctuation to prompt bond formation between the two (Fig. 1a). With high-speed edge detection analysis, bond formation and dissociation were identified by the reduction and resumption of the thermal fluctuations of the BFP bead, respectively24. Bond lifetimes (tb), i.e., from the instant of bond association to the instant of bond dissociation, were obtained from the displacement and standard deviation analysis of the bead movement. Modeling the kinetic process as a single-step first-order dissociation of a single monomeric bond (one-state model, see below), the probability Pb of a bond formed at time 0 to survive at time tb is Pb = exp(−koff tb). Taking a natural log to linearize the exponential function, the ln(# of events with a lifetime ≥ tb) vs. tb plot was fitted by a straight line. The off-rate koff was estimated from the negative slope of the fitted line or from the reciprocal of average bond lifetime, koff = 1/<tb>52. The average bond lifetime <tb> is the same as dwell time, and is related to half-life of a bond by t1/2 = <tb>ln2.
Force-clamp assay
Force-dependent kinetics of pMHC dissociation from TCR and/or CD8 was derived from forcec-lamp assay of bond lifetimes using BFP as described previously9. Briefly, after a short contact between a thymocyte and a pMHC-coated bead attached to a RBC, the thymocyte retracted to and was held at a desired force to wait for bond dissociation (Fig. 1a). Adhesion, if occurred, was detected by a tensile force caused by a bead displacement of that stretched the RBC. The bond lifetime was measured from the instant when the force reached the desired level to the instant of bond dissociation. After dissociation, the program returned the thymocyte to the original position for the next cycle. For each ligand tested, bond lifetimes were collected in a range of forces and presented in 5–7 bins as mean ± s.e.m (each bin with N≥20, see Supplementary Table 1 for the number of measurements for each curve). In order to affirm the condition for >89% single bonds, the pMHC densities on the beads were controlled to keep the adhesion frequency <20%25.
When the kinetic process did not follow single-step first-order dissociation of a single monomeric bond, the bond lifetime distributions were analyzed by the survival probability (fraction of events with a lifetime ≥ tb) for each force bin, which was plotted vs. tb and fitted with a one-state model for bimolecular interactions and a two-state model22 for total interactions that might include both bimolecular and trimolecular interactions. Instead of the one-state model used to analyze the thermal fluctuation data, a two-state model was used to analyze survival probability under force, Pb = ω1 exp(−koff1tb) + ω2 exp(−koff2tb). The model contains two off-rates, koff1 and koff2, and two associated fractions, ω1 and ω2 (ω1 + ω2 = 1) for the fast (subscripted 1) and slow (subscripted 2) dissociating states.
Selection accuracy, receiver operating characteristic curve and Youden’s J statistic
From the survival probabilities of the two bordering peptides, Q4R7 and Q4H7, we can calculate the combined successful rate or selection accuracy (SA), defined as the probability for a thymocyte in n binding events to accumulate enough Q4R7 bond lifetimes to exceed a threshold cumulative bond lifetime, tth, required to induce negative selection and not to accumulate enough Q4H7 bond lifetimes to exceed tth. The families of SA vs. tth or n plots in Fig. 6d, e and in Supplementary Fig. 5a, b were calculated for 0 pN and 10–15 pN as well as for wild-type and mutant pMHCs.
A receiver operating characteristic (ROC) curve was generated by plotting the probability for a thymocyte to be negatively-selected by Q4R7 (true positive rate, or sensitivity) vs. the probability to be negatively-selected by Q4H7 for a range of tth and a fixed n. By varying the n value we generated a family of ROC curves. Youden’s J statistic was calculated as sensitivity + selectivity − 1.
Molecular stiffness analysis
The stiffness of molecular bonds was measured from the ramping phase of BFP test cycles16, 17. The force vs. time data from the bead tracking system (blue points, Fig. 2b) and the displacement vs. time data from the piezoelectric actuator capacitance sensor feedback system (red points, Fig. 2b) were combined to obtain force vs. displacement data (Fig. 2c). Two straight lines were fit to the piece-wise linear data, one for compressive forces (<0) and the other for tensile forces (>0), to obtain two slopes that represent, respectively, the compressive stiffness of the thymocyte (slope1 = kc, assuming that the molecules could not support compression) and the tensile stiffness of a serially connected system (slope2 = ks) consisting of the thymocyte and the molecular bond (Fig. 2d). The Hooke’s law for springs in series states that 1/ks = 1/km + 1/kc where km and kc are, respectively, the molecular stiffness and the cellular stiffness. Assuming the cellular stiffness in tension equals to that in compression16, 17, km = 1/(1/ks − 1/kc) was calculated from each set of force vs. displacement data (Fig. 2c). An ensemble of km values were analyzed by histograms and fitted by single or double Gaussian distributions (Fig. 2f, g, 4c, d, Supplementary Fig. 1 and 2b).
DNA tension probe experiment
The DNA-based molecular tension probes (MTPs) were synthesized as previously described19. These MTPs were functionalized on to glass surfaces by NHS-PEG-Maleimide linker (JenKem Technology, Plano, TX) then linked to pMHC using biotin-streptavidin coupling. Anti-mouse CD11a (LFA-1) antibodies were incubated together with the MTPs in a 1:10 ratio to facilitate anchoring of thymocytes on the glass surface. These anti-LFA-1 antibodies were not conjugated to MTPs and did not contribute to fluorescence signal. OT1 thymocytes were injected onto the coverslips and allowed to settle for 5 min before imaging and imaged with Zeiss LSM 710 confocal microscope at room temperature for maximum of 30 min. During 30 min we performed either single snapshot of multiple spots containing thymocytes interacting with the functionalized surface or time lapse imaging of single cells for 9 min. For Lck inhibition treatment, cells were allowed to settle onto the coverslip containing 4 μM of Lck inhibitor for 5 min before being imaged in the presence of the inhibitor. For other drug treatments, 20 min time-lapse imaging experiments were performed with 5 min intervals on the multi-position imaging module.
Consistent with a previous report53, small numbers of thymocytes spread on glass surfaces, resulting in relatively low frequencies of Cy5 positive signaling (<10%). Nevertheless, thymocytes that did spread formed multifocal immunological synapses54, allowing us to image single cell. Upon unfolding of the DNA hairpin, an increase in Cy5 emission intensity was detected due to the separation between the fluorophore and the quencher. Normalized Cy5 intensity was calculated from single cell-sized ROI co-localized with a thymocyte in brightfield with high Cy5 intensity at the cell-glass interface (Supplementary Movie 2). Image analyses were performed using ImageJ. Mean Cy5 intensity of this ROI was normalized to that of the same size ROI without any thymocyte in the background. Any mean Cy5 intensities above the VSV condition were considered as the positive signal. For time lapse images, normalized Cy5 intensity was calculated from identical subcellular-sized ROI based on the accumulation of Cy5 signal (Supplementary Movies 1 and Supplementary Fig. 2c). For quantitative comparison in the time lapse images, normalized Cy5 intensities at different time points were normalized to the initial time point (0 min). The frequency of the Cy5 positive cells were calculated by dividing the number of all positive cells from all image frames by the total number of cells observed in all image frames. The threshold forces of the two DNA force probes were 4.7 and 13.1 pN, as calibrated by BFP19. Surface density of the DNA force probe was measured to be 119/μm2 from Rhodamine-DHPE calibration.
Selection accuracy, receiver operating characteristic curve and Youden’s J statistic
From the acquired lifetime data of the two bordering peptides, Q4R7 and Q4H7, we can calculate the combined successful rate or selection accuracy (SA), defined as the probability for a thymocyte in n binding events to accumulate enough Q4R7 bond lifetimes to exceed a threshold cumulative bond lifetime, tth, required to induce negative selection and not to accumulate enough Q4H7 bond lifetimes to exceed tth, by Monte Carlo selection of lifetimes. 10,000 cumulative lifetimes were simulated for each th and peptide within each force range. The families of SA vs. tth or n plots in Fig. 6d, e and in Supplementary Fig. 5a, b were calculated for force ranges of 0–5 pN and 10–15 pN as well as for wild-type and mutant pMHCs.
A receiver operating characteristic (ROC) curve was generated by plotting the probability for a thymocyte to be negatively-selected by Q4R7 (true positive rate, or sensitivity) vs. the probability to be negatively-selected by Q4H7 (false positive rate, or 1-specificity) for a range of tth and a fixed n. By varying the n value we generated a family of ROC curves. Youden’s J statistic was calculated as sensitivity + specificity − 1.
Statistical Analysis
Statistical analyses were done on bond lifetime, molecular stiffness and thymocyte pulling data. For the bond lifetime data, we addressed two types of questions using different statistical tests. The first question was determination of dynamic bond types, i.e., whether a given bond lifetime vs. force curve behaved as catch-slip bond or slip-only bond. Curves suspected to behave as catch-slip bond were divided into two segments of monotonically ascending and descending trends by visual inspection. If the ascending segment contained more than two force bins, it was analyzed by linear regression weighted with the reciprocal standard-error of each bin. The significance of the ascending trend was assessed by comparing the slope of the regression line to zero slope using Student’s t-test. Significant positive slope indicates catch bond. Curves suspected to behave as slip-only bond were directly analyzed by linear regression weighted by the reciprocal standard-error of each bin. The significance of the descending trend was assessed by comparing the slope of the regression line to zero slope using Student’s t-test. Significant negative slope indicate slip bond. When only the smallest force bin (measured by force-clamped assay) had a longer mean bond lifetime than that of the zero-force bin (measured by thermal fluctuation assay), as in the cases of Figs. 1c and 4b, unpaired Student’s t-test was used to compare the scatter lifetime data between these two force bins to assess the significance of the apparent catch bond. The results are summarized in Supplementary Table 2.
The second question was whether two force-dependent bond lifetime curves had similar or different trends, and if so, what the significant differences were. If two curves both showed slip bond behavior, two-way ANOVA (analyzed with GraphPad Prism) was used to assess the significance of their difference. The results are summarized in Supplementary Table 3a.
If one curve behaved as catch-slip bond but the other behaved as slip-only bond, or both curves had catch bonds but showed relative shifts, ANOVA could not be used because it is essentially the same as linear regression. To analyze cases where at least one of the two curves was biphasic, we employed an accelerated failure time model to compare the mean lifetimes between the two curves over a range of forces. We binned bond lifetime vs. force scatter data points in equal space of force levels and fitted the lifetime distributions in each force bin by Weibull distributions, which are an extended form of and more flexible than exponential distributions. Since the functional form of association between the force level and bond lifetime appears non-linear, we adopted a penalized spline smoothing function to fit the mean bond lifetime vs. force curve. The difference in non-linear patterns between groups were then compared based on the significance of group and force level interaction terms (including spline terms). The analysis was done using R (version: 3.4.2, with survival package)55. The results are summarized in Supplementary Table 3b.
The data in Fig. 7b were further analyzed to determine the two catch-slip bond curves obtained using OT1 or OT1.6F thymocytes in each panel exhibited significant relative shift. First, as a post-hoc analysis, the mean forces where mean bond lifetime peaked in the two curves were compared using two-sample t-test to examine whether the two peak locations (of force) are different. Second, the slip phases of the curves were compared for the relative vertical shift. Each curve was fitted with quadratic regression, weighted by the reciprocal of standard-error of each bin. Significance level of the shift was evaluated by Student’s t-test. The results are summarized in Supplementary Table 3c.
For the molecular stiffness data, the question was to determine whether the single or double Gaussian model better fits the molecular stiffness histograms of total bonds. The goodness-of-fit of these models were assessed by the extra sum-of-squares F test (analyzed with GraphPad Prism). The results are summarized in Supplementary Table 5.
For the pulling force data, statistical comparisons between of snapshots of initial forces pulled by thymocytes under two conditions were made using the Student’s t-test, whereas between two time courses were made using two-way ANOVA (analyzed with GraphPad Prism). The results are indicated in Fig. 3 and Supplementary Fig. 2d, e, g, h, j.
Data and code availability statement
The data and software code that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Lawrence for maintaining the mouse colonies; L. Doudy for thymocyte purification, W. Chen and Z. Li for helping with experiments and discussions, and the NIH Tetramer Core Facility at Emory University for providing the pMHC monomers. This work was supported by NIH grants CA214354 and AI124680 (to C.Z.), and NS071518 and AI096879 (to B.E.).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing interests.
REFERENCE
- 1.Rudolph M, Stanfield R, Wilson I. How TCRs bind MHCs, peptides, and coreceptors. Immunology 2006, 24: 419–466. [DOI] [PubMed] [Google Scholar]
- 2.Stepanek O, Prabhakar AS, Osswald C, King CG, Bulek A, Naeher D, Beaufils-Hugot M, Abanto ML, Galati V, Hausmann B, Lang R, Cole DK, Huseby ES, Sewell AK, Chakraborty AK, Palmer E. Coreceptor scanning by the T cell receptor provides a mechanism for T cell tolerance. Cell 2014, 159(2): 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer E, Naeher D. Affinity threshold for thymic selection through a T-cell receptor-co-receptor zipper. Nat Rev Immunol 2009, 9(3): 207–213. [DOI] [PubMed] [Google Scholar]
- 4.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NR, Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 2006, 444(7120): 724–729. [DOI] [PubMed] [Google Scholar]
- 5.Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature 2010, 464(7290): 932–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huppa JB, Axmann M, Mortelmaier MA, Lillemeier BF, Newell EW, Brameshuber M, Klein LO, Schutz GJ, Davis MM. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 2010, 463(7283): 963–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang N, Huang J, Edwards LJ, Liu B, Zhang Y, Beal CD, Evavold BD, Zhu C. Two-Stage Cooperative T Cell Receptor-Peptide Major Histocompatibility Complex-CD8 Trimolecular Interactions Amplify Antigen Discrimination. Immunity 2011, 34(1): 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez RJ, Andargachew R, Martinez HA, Evavold BD. Low-affinity CD4+ T cells are major responders in the primary immune response. Nature communications 2016, 7: 13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B, Chen W, Evavold BD, Zhu C. Accumulation of Dynamic Catch Bonds between TCR and Agonist Peptide-MHC Triggers T Cell Signaling. Cell 2014, 157(2): 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong J, Persaud SP, Horvath S, Allen PM, Evavold BD, Zhu C. Force-Regulated In Situ TCR-Peptide-Bound MHC Class II Kinetics Determine Functions of CD4+ T Cells. J Immunol 2015, 195(8): 3557–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, Brady SK, Wang JH, Wagner G, Reinherz EL, Lang MJ. Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci U S A 2015, 112(5): 1517–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Blanchfield L, Ma VP, Andargachew R, Galior K, Liu Z, Evavold B, Salaita K. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc Natl Acad Sci U S A 2016, 113(20): 5610–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarangapani KK, Marshall BT, McEver RP, Zhu C. Molecular stiffness of selectins. J Biol Chem 2011, 286(11): 9567–9576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erman B, Alag AS, Dahle O, van Laethem F, Sarafova SD, Guinter TI, Sharrow SO, Grinberg A, Love PE, Singer A. Coreceptor signal strength regulates positive selection but does not determine CD4/CD8 lineage choice in a physiologic in vivo model. J Immunol 2006, 177(10): 6613–6625. [DOI] [PubMed] [Google Scholar]
- 15.Hui E, Vale RD. In vitro membrane reconstitution of the T-cell receptor proximal signaling network. Nat Struct Mol Biol 2014, 21(2): 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiore VF, Ju L, Chen Y, Zhu C, Barker TH. Dynamic catch of a Thy-1-alpha5beta1+syndecan-4 trimolecular complex. Nature communications 2014, 5: 4886. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Lou J, Evans EA, Zhu C. Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J Cell Biol 2012, 199(3): 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashour KT, Gondarenko A, Chen H, Shen K, Liu X, Huse M, Hone JC, Kam LC. CD28 and CD3 have complementary roles in T-cell traction forces. Proc Natl Acad Sci U S A 2014, 111(6): 2241–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Ge C, Zhu C, Salaita K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nature communications 2014, 5: 5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogquist KA, Tomlinson AJ, Kieper WC, McGargill MA, Hart MC, Naylor S, Jameson SC. Identification of a naturally occurring ligand for thymic positive selection. Immunity 1997, 6(4): 389–399. [DOI] [PubMed] [Google Scholar]
- 21.Santori FR, Kieper WC, Brown SM, Lu Y, Neubert TA, Johnson KL, Naylor S, Vukmanovic S, Hogquist KA, Jameson SC. Rare, structurally homologous self-peptides promote thymocyte positive selection. Immunity 2002, 17(2): 131–142. [DOI] [PubMed] [Google Scholar]
- 22.Ju L, Dong JF, Cruz MA, Zhu C. The N-terminal flanking region of the A1 domain regulates the force-dependent binding of von Willebrand factor to platelet glycoprotein Ibalpha. J Biol Chem 2013, 288(45): 32289–32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dustin ML. T-cell activation through immunological synapses and kinapses. Immunol Rev 2008, 221: 77–89. [DOI] [PubMed] [Google Scholar]
- 24.Chen W, Evans EA, McEver RP, Zhu C. Monitoring receptor-ligand interactions between surfaces by thermal fluctuations. Biophys J 2008, 94(2): 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chesla SE, Selvaraj P, Zhu C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J 1998, 75(3): 1553–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dushek O, van der Merwe PA. An induced rebinding model of antigen discrimination. Trends Immunol 2014, 35(4): 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang S, Palin AC, Li L, Song KD, Lee J, Herz J, Tubo N, Chu H, Pepper M, Lesourne R, Zvezdova E, Pinkhasov J, Jenkins MK, McGavern D, Love PE. TCR ITAM multiplicity is required for the generation of follicular helper T-cells. Nature communications 2015, 6: 6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang S, Song KD, Lesourne R, Lee J, Pinkhasov J, Li L, El-Khoury D, Love PE. Reduced TCR signaling potential impairs negative selection but does not result in autoimmune disease. J Exp Med 2012, 209(10): 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nika K, Soldani C, Salek M, Paster W, Gray A, Etzensperger R, Fugger L, Polzella P, Cerundolo V, Dushek O, Hofer T, Viola A, Acuto O. Constitutively active Lck kinase in T cells drives antigen receptor signal transduction. Immunity 2010, 32(6): 766–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thome M, Duplay P, Guttinger M, Acuto O. Syk and ZAP-70 mediate recruitment of p56lck/CD4 to the activated T cell receptor/CD3/zeta complex. J Exp Med 1995, 181(6): 1997–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manz BN, Tan YX, Courtney AH, Rutaganira F, Palmer E, Shokat KM, Weiss A. Small molecule inhibition of Csk alters affinity recognition by T cells. eLife 2015, 4: e08088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinashi T Intracellular signalling controlling integrin activation in lymphocytes. Nat Rev Immunol 2005, 5(7): 546–559. [DOI] [PubMed] [Google Scholar]
- 33.Yachi PP, Ampudia J, Zal T, Gascoigne NR. Altered peptide ligands induce delayed CD8-T cell receptor interaction--a role for CD8 in distinguishing antigen quality. Immunity 2006, 25(2): 203–211. [DOI] [PubMed] [Google Scholar]
- 34.Arcaro A, Gregoire C, Bakker TR, Baldi L, Jordan M, Goffin L, Boucheron N, Wurm F, van der Merwe PA, Malissen B, Luescher IF. CD8beta endows CD8 with efficient coreceptor function by coupling T cell receptor/CD3 to raft-associated CD8/p56(lck) complexes. J Exp Med 2001, 194(10): 1485–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doucey MA, Goffin L, Naeher D, Michielin O, Baumgartner P, Guillaume P, Palmer E, Luescher IF. CD3 delta establishes a functional link between the T cell receptor and CD8. J Biol Chem 2003, 278(5): 3257–3264. [DOI] [PubMed] [Google Scholar]
- 36.Thome M, Germain V, DiSanto JP, Acuto O. The p56lck SH2 domain mediates recruitment of CD8/p56lck to the activated T cell receptor/CD3/zeta complex. Eur J Immunol 1996, 26(9): 2093–2100. [DOI] [PubMed] [Google Scholar]
- 37.Straus DB, Chan AC, Patai B, Weiss A. SH2 domain function is essential for the role of the Lck tyrosine kinase in T cell receptor signal transduction. J Biol Chem 1996, 271(17): 9976–9981. [DOI] [PubMed] [Google Scholar]
- 38.Lo WL, Shah NH, Ahsan N, Horkova V, Stepanek O, Salomon AR, Kuriyan J, Weiss A. Lck promotes Zap70-dependent LAT phosphorylation by bridging Zap70 to LAT. Nat Immunol 2018, 19: 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldrath AW, Hogquist KA, Bevan MJ. CD8 lineage commitment in the absence of CD8. Immunity 1997, 6(5): 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lou J, Zhu C. A structure-based sliding-rebinding mechanism for catch bonds. Biophys J 2007, 92(5): 1471–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR. T-cell-receptor affinity and thymocyte positive selection. Nature 1996, 381(6583): 616–620. [DOI] [PubMed] [Google Scholar]
- 42.Artyomov MN, Lis M, Devadas S, Davis MM, Chakraborty AK. CD4 and CD8 binding to MHC molecules primarily acts to enhance Lck delivery. Proc Natl Acad Sci U S A 2010, 107(39): 16916–16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng Y, Brazin KN, Kobayashi E, Mallis RJ, Reinherz EL, Lang MJ. Mechanosensing drives acuity of alphabeta T-cell recognition. Proc Natl Acad Sci U S A 2017, 114(39): E8204–E8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Laethem F, Tikhonova AN, Singer A. MHC restriction is imposed on a diverse T cell receptor repertoire by CD4 and CD8 co-receptors during thymic selection. Trends Immunol 2012, 33(9): 437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Borgne M, Ladi E, Dzhagalov I, Herzmark P, Liao YF, Chakraborty AK, Robey EA. The impact of negative selection on thymocyte migration in the medulla. Nat Immunol 2009, 10(8): 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
METHOD-ONLY REFERENCE
- 46.Sauer K, Huang YH, Lin H, Sandberg M, Mayr GW. Phosphoinositide and inositol phosphate analysis in lymphocyte activation. Curr Protoc Immunol 2009, Chapter 11: Unit11 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Puls KL, Hogquist KA, Reilly N, Wright MD. CD53, a thymocyte selection marker whose induction requires a lower affinity TCR-MHC interaction than CD69, but is up-regulated with slower kinetics. Int Immunol 2002, 14(3): 249–258. [DOI] [PubMed] [Google Scholar]
- 48.Nathenson SG, Geliebter J, Pfaffenbach GM, Zeff RA. Murine major histocompatibility complex class-I mutants: molecular analysis and structure-function implications. Annu Rev Immunol 1986, 4: 471–502. [DOI] [PubMed] [Google Scholar]
- 49.Degano M, Garcia KC, Apostolopoulos V, Rudolph MG, Teyton L, Wilson IA. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity 2000, 12(3): 251–261. [DOI] [PubMed] [Google Scholar]
- 50.Tallquist MD, Yun TJ, Pease LR. A single T cell receptor recognizes structurally distinct MHC/peptide complexes with high specificity. J Exp Med 1996, 184(3): 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang J, Edwards LJ, Evavold BD, Zhu C. Kinetics of MHC-CD8 interaction at the T cell membrane. J Immunol 2007, 179(11): 7653–7662. [DOI] [PubMed] [Google Scholar]
- 52.Marshall BT, Long M, Piper JW, Yago T, McEver RP, Zhu C. Direct observation of catch bonds involving cell-adhesion molecules. Nature 2003, 423(6936): 190–193. [DOI] [PubMed] [Google Scholar]
- 53.Hailman E, Allen PM. Inefficient cell spreading and cytoskeletal polarization by CD4+CD8+ thymocytes: regulation by the thymic environment. J Immunol 2005, 175(8): 4847–4857. [DOI] [PubMed] [Google Scholar]
- 54.Hailman E, Burack WR, Shaw AS, Dustin ML, Allen PM. Immature CD4(+)CD8(+) thymocytes form a multifocal immunological synapse with sustained tyrosine phosphorylation. Immunity 2002, 16(6): 839–848. [DOI] [PubMed] [Google Scholar]
- 55.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: 2017, https://www.R-project.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and software code that support the findings of this study are available from the corresponding author upon reasonable request.