Significance
Mutations in the master quorum-sensing signal receptor LasR are common in Pseudomonas aeruginosa isolated from chronically infected lungs of cystic fibrosis patients. In many of these LasR mutant isolates, a quorum-sensing system involving the transcription factor RhlR has escaped dependence on LasR. We have modeled the escape of RhlR activity from LasR regulation in laboratory evolution experiments. We gain insights into the genetic basis of the escape and show that an escape variant has a fitness advantage over a RhlR-null derivative. We also propose a hypothesis for the course of P. aeruginosa quorum-sensing evolution in chronic cystic fibrosis lung infections. Our work points to RhlR as a potential target for therapeutic development.
Keywords: acyl-homoserine lactone, cystic fibrosis, LasR, RhlR, sociomicrobiology
Abstract
The bacterial pathogen Pseudomonas aeruginosa activates expression of many virulence genes in a cell density-dependent manner by using an intricate quorum-sensing (QS) network. QS in P. aeruginosa involves two acyl-homoserine-lactone circuits, LasI-LasR and RhlI-RhlR. LasI-LasR is required to activate many genes including those coding for RhlI-RhlR. P. aeruginosa causes chronic infections in the lungs of people with cystic fibrosis (CF). In these infections, LasR mutants are common, but rhlR-rhlI expression has escaped LasR regulation in many CF isolates. To better understand the evolutionary trajectory of P. aeruginosa QS in chronic infections, we grew LasR mutants of the well-studied P. aeruginosa strain, PAO1, in conditions that recapitulate an environment where QS signal synthesis by other bacteria might still occur. When QS is required for growth, addition of the RhlI product butyryl-homoserine lactone (C4-HSL), or bacteria that produce C4-HSL, to LasR mutants results in the rapid emergence of a population with a LasR-independent RhlI-RhlR QS system. These evolved populations exhibit subsequent growth without added C4-HSL. The variants that emerge have mutations in mexT, which codes for a transcription factor that controls expression of multiple genes. LasR-MexT mutants have a competitive advantage over both the parent LasR mutant and a LasR-MexT-RhlR mutant. Our findings suggest a plausible evolutionary trajectory for QS in P. aeruginosa CF infections where LasR mutants arise during infection, but because these mutants are surrounded by C4-HSL–producing P. aeruginosa, variants rewired to have a LasR-independent RhlIR system quickly emerge.
The opportunistic pathogen Pseudomonas aeruginosa is the cause of a variety of human infections, including chronic infections of the airways of people with cystic fibrosis (CF). Expression of many P. aeruginosa virulence factors and other genes is regulated by two acyl-homoserine lactone (AHL) quorum-sensing (QS) systems, LasI-LasR (LasIR) and RhlI-RhlR (RhlIR). LasI catalyzes production of the diffusible QS signal N-3-oxo-dodecanoyl homoserine lactone (3OC12-HSL). 3OC12-HSL binds to its cognate receptor, the transcription factor LasR, which can then activate expression of multiple genes, including lasI, rhlI, and rhlR. In an analogous manner, RhlI catalyzes production of the diffusible signal butyryl-HSL (C4-HSL), which binds to the transcription factor RhlR. Signal-bound RhlR then activates expression of rhlI and other genes, some of which overlap with the LasR regulon (for reviews, see refs. 1–3). In commonly studied P. aeruginosa strains such as PAO1, the RhlIR QS system requires induction by LasR. As a result, deletion of either lasR or lasI in strain PAO1 diminishes expression of QS-activated genes. LasR-null mutants of strain PAO1 also show impaired virulence in acute infection animal models (for example, see refs. 4 and 5).
These two AHL QS systems interact with a third system, which is mediated by the signal 2-heptyl-3-hydroxy-4-quinolone [Pseudomonas quinolone signal (PQS)] and its biosynthetic precursor 2-heptyl-4-quinolone (HHQ) (6–9). The pqs operon pqsABCDE, together with pqsH, codes for HHQ and PQS production. PQS and HHQ bind to the transcriptional regulator PqsR (also called MvfR), which then activates expression of the PQS synthesis genes. The PQS system activates multiple genes, many of which are also regulated by LasR or RhlR (8–10). LasR activates expression of the PQS genes, and in turn, a product of pqsE increases expression of RhlR-activated genes (9–11).
A majority of QS-activated genes code for secreted factors or the production of secreted factors. Such factors can serve as public goods if they benefit the entire population of cells. Public goods production carries a metabolic cost to individual cells, and this in turn provides an incentive for the emergence of cheating cells that do not produce those goods. When P. aeruginosa PAO1 growth requires QS-regulated protease production, LasR mutant cheaters emerge, and these mutants have a fitness advantage (12–16). In some circumstances, the frequency of cheaters can become high enough to cause collapse of the population (14, 15). One mechanism whereby P. aeruginosa restricts infiltration by cheaters is policing mediated by the RhlR-induced production of cyanide (CN−) (15). RhlR mutants are more sensitive to CN− than the wild type, and both CN−-synthesis mutants and RhlR mutants are particularly vulnerable to cheating (15).
LasR mutations are common among P. aeruginosa isolated from the lungs of chronically infected CF patients (17–22). However, unlike strain PAO1, many LasR mutants from chronic infections have an active RhlIR QS system and display QS-regulated phenotypes when grown under laboratory conditions (18, 22). It is likely that the apparent differences in QS regulatory networks of clinical isolates in comparison with PAO1 reflect adaptation of P. aeruginosa during the long-term infections. We are interested in understanding why some clinical isolates from CF lungs maintain an active QS system in a LasR-null background, and we want to identify laboratory conditions that would give rise to such a phenotype. We discovered that the presence of C4-HSL or C4-HSL–producing bacteria in cultures of a LasR-null mutant led to a rapid emergence of LasR mutant variants capable of growth on casein. These variants had characteristics similar to those of many CF clinical isolates. During preparation of our manuscript, Oshri et al. (23) reported that LasR-null mutants with an active RhlIR system emerged in casein broth without added C4-HSL but only after a prolonged incubation of 4 to 6 wk. We propose a course of evolution for P. aeruginosa QS during CF lung infections, whereby once LasR mutants emerge among populations capable of C4-HSL production, they can rapidly evolve an active LasR-independent RhlIR QS system. We also present experiments leading to a plausible explanation for why RhlR mutants are rare in the context of CF lung infections.
Results
Reprogramming the QS Circuit: A LasR-Independent RhlIR System.
The observation that many P. aeruginosa LasR-null isolates obtained from chronically infected CF lungs have a functioning RhlIR system (18, 22) suggests that regaining or maintaining expression of at least a subset of the QS regulon is advantageous in the context of a long-term infection. Although LasR-null mutants are not expected to activate expression of QS-regulated genes, in the CF lung they would be exposed to the QS signals produced by neighboring cells with an active LasIR QS system. Production of biologically relevant levels of C4-HSL has been reported for clinical isolates from CF lungs (18, 22, 24). Given the key role of C4-HSL in RhlR gene regulation, we asked whether C4-HSL could promote emergence of a PAO1 LasR mutant with an activated RhlIR QS system, under conditions that require QS for growth. We conducted a laboratory evolution experiment, in which we grew a LasR mutant with or without C4-HSL in casein broth, a minimal medium which contains casein as the sole carbon and energy source. Growth in casein broth requires QS-regulated production of the extracellular protease elastase. Expression of the elastase gene lasB in PAO1 is activated primarily by LasR, with RhlR playing a lesser role (25). Wild-type bacteria readily grow in casein broth, but LasR mutants do not. However, when we added 30 µM C4-HSL to a monoculture of LasR mutants at the time of inoculation and again 4 d later, growth was evident in less than a week (SI Appendix, Fig. S1).
To test whether growth of the LasR mutant in casein broth was conditional or a fixed heritable trait, we transferred the growing cultures to fresh casein broth without added C4-HSL and continued to monitor growth. Growth was observed within a few days and continued with subsequent transfers and no additional C4-HSL. We saw a similar outcome with a PAO1 LasI mutant (SI Appendix, Fig. S1). Growth was not observed over a period of 4 wk for either the LasR or the LasI mutant when C4-HSL was not added to the casein broth (SI Appendix, Fig. S1). Our observations are consistent with a recent report that in casein broth without added C4-HSL, growth of LasR-null P. aeruginosa was evident after an incubation period of over 1 mo (23).
After several transfers, we used a skim milk agar plate assay to identify representative protease-secreting isolates from each of three biological replicates. We verified that these isolates were able to grow on casein as the sole carbon and energy source (SI Appendix, Fig. S2). It was possible that in contrast to wild-type PAO1, elastase production in the selected isolates did not require QS. To address this possibility, we added an AHL-degrading enzyme, AiiA lactonase (26), to casein broth cultures. Growth of the evolved variants was impaired by AiiA (SI Appendix, Fig. S2), indicating that protease production was regulated by an AHL-responsive factor.
Reprogrammed LasR Mutants Have a Functioning RhlIR QS System.
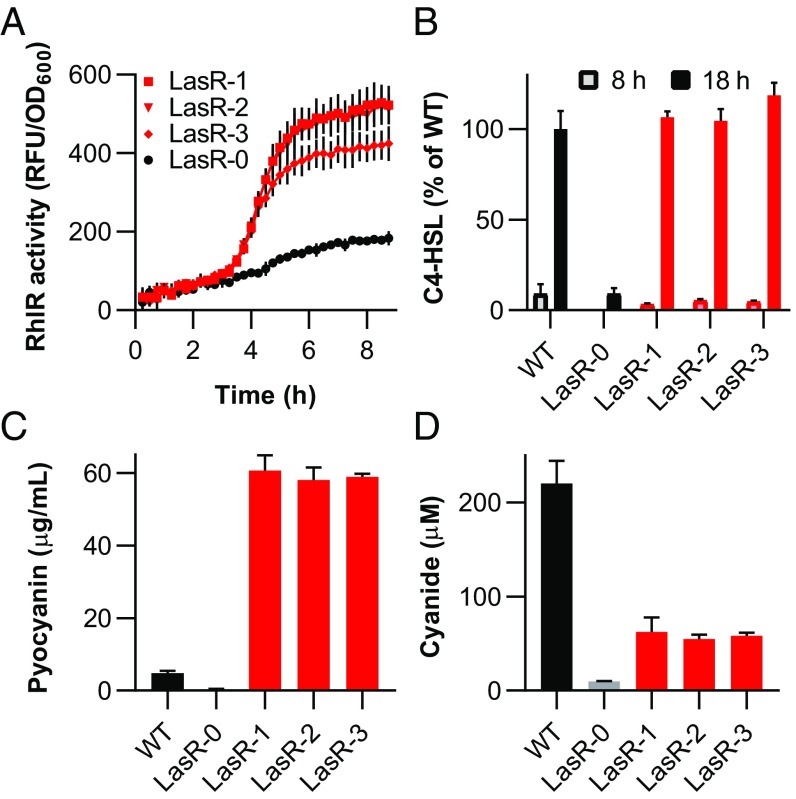
We hypothesized that the LasR mutant variants, which grow in casein broth, had a functioning RhlIR system. To test this hypothesis, we transformed several such variants with a reporter plasmid, which contains the RhlR-responsive rhlA promoter fused to gfp (18). After 8 h of growth in LB-Mops broth, these variants displayed GFP fluorescence well above that of the parent LasR mutant (Fig. 1A). This result indicates that the reprogrammed variants had an active RhlIR QS system despite having a LasR-null mutation. These variants also produced C4-HSL and CN−, unlike the parent LasR mutant strain. Furthermore, when grown under phosphate limitation, these isolates produced pyocyanin at levels more than 10-fold higher than those of the wild-type PAO1 (Fig. 1 B–D), which is likely due to the loss of repression by the LasR-dependent negative regulator RsaL (27). The evolved strains displayed characteristics similar to many of the LasR-null isolates from chronically infected lungs of CF patients described by Feltner et al. (18).
Fig. 1.
Characteristics of three protease-producing LasR mutant variants (LasR-1, LasR-2, and LasR-3) derived from the LasR mutant (LasR-0). (A) RhlR activity of each variant and their nonprotease producing parent LasR-0, measured as GFP fluorescence and reported as relative fluorescence units (RFU). All strains contained the PrhlA-gfp fusion plasmid pProbe-GT-PrhlAgfp. (B) Relative levels of C4-HSL in culture fluid after 8 or 18 h of cell growth. Values are relative to those produced by P. aeruginosa PAO1 (WT) at 18 h. (C) Pyocyanin production by P. aeruginosa PAO1, the parent LasR mutant, and LasR mutant variants. (D) Cyanide levels of cultures. Data in A are means of triplicate experiments, and data in B–D are means of duplicate experiments. In all panels, error bars represent the SEM.
Mutations in mexT Lead to an Active RhlIR QS System in the LasR Mutant.
To understand the genetic changes responsible for the reprogrammed QS phenotype, we sequenced the genomes of the parent LasR mutant, three protease-positive isolates, and three protease-negative isolates from the casein broth evolution experiments. We identified one gene, mexT (PA2492), which was mutated or deleted in all three protease-producing isolates but not in the non–protease-producing isolates or the parent strain (Table 1). Two of the protease-producing isolates had unique nonsynonymous point mutations in the mexT coding sequence, and the third had a 19.7-kb deletion, which included all of mexT along with the neighboring genes spanning PA2475 to PA2494. We also found mexT mutations in three protease-producing LasI mutant variants that we sequenced (Table 1) (28). mexT was also mutated in the slowly-evolved LasR− variants reported by Oshri et al. (23).
Table 1.
Genetic changes in protease-producing variants of PAO1 LasR and LasI mutants
| Variant* | mexT mutation | MexT change | Other affected genes |
| LasR mutants | |||
| LasR-1 | T353A | L118Q | None |
| LasR-2 | A122G | S41G | None |
| LasR-3 | Δ(2793006-2812975) | ΔmexT | PA2475, dsbG, PA2477-PA2490, mexS, mexE, mexF |
| LasR130-1 | Δ(2808103-2808997) | Truncation | NA† |
| LasR130-2 | Δ(A768) | Frameshift | NA† |
| LasR130-3 | A559C | T560P | NA† |
| LasR131-1 | A55C | T19P | NA† |
| LasR131-2 | G31C, C478G | V11L, L160V | NA† |
| LasR131-3 | NP‡ | NP‡ | NA† |
| LasI mutants | |||
| LasI-1 | A55C | T19P | psdR |
| LasI-2 | A55C | T19P | psdR |
| LasI-3 | A55C | T19P | psdR, fleQ |
Variants LasR-1–3 were isolated from casein broth inoculated with the LasR mutant only, LasR130-1-3 and LasR131-1-3 were isolated from casein broth coinoculated with the LasR mutant and clinical P. aeruginosa isolates E130 or E131, and LasI-1–3 were isolated from casein broth inoculated with the LasI mutant. Further details about the variants can be found in SI Appendix, Table S1.
NA, other genes were not analyzed in these isolates.
NP, no product. We could not amplify mexT for sequencing.
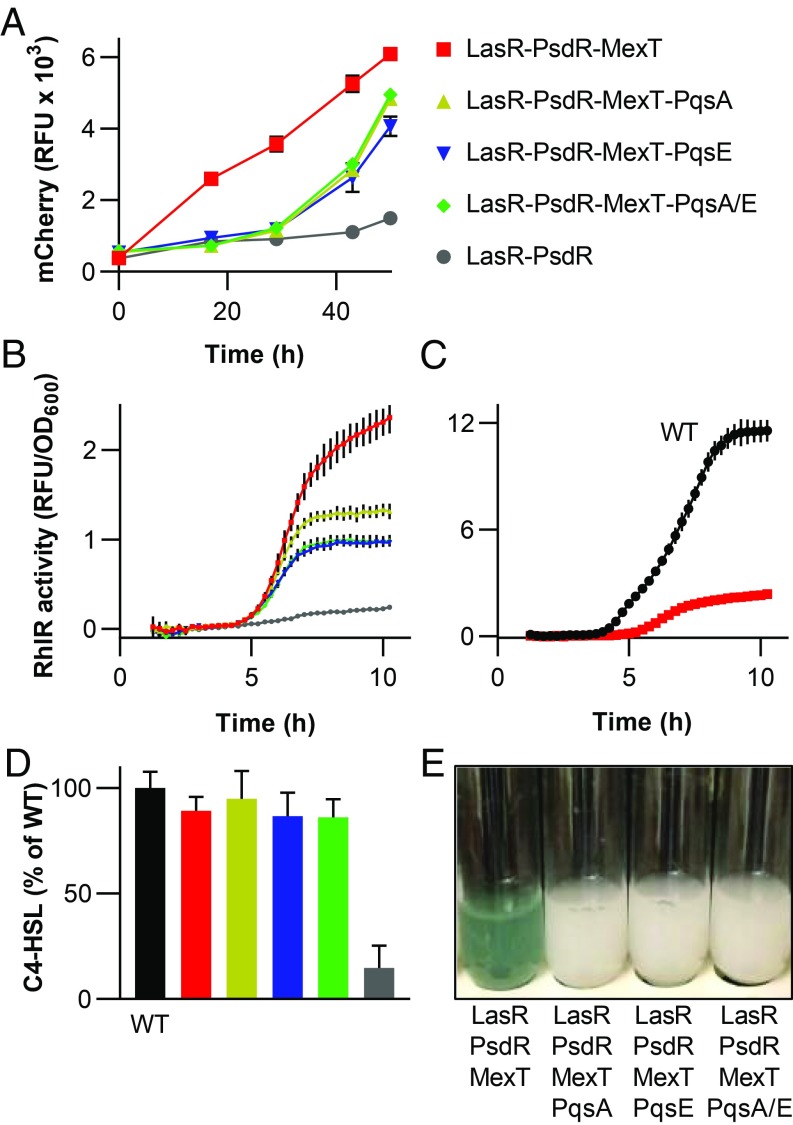
MexT is a transcription factor that affects expression of at least 40 genes (29), and its overexpression negatively affects known RhlR-regulated phenotypes, including C4-HSL, CN−, elastase, and pyocyanin production (30, 31). To confirm that the mexT mutations in the evolved isolates were involved in reprograming QS so that RhlIR was functioning in the absence of LasR, we deleted mexT from the PAO1 LasR-null mutant. We also deleted psdR because inactivation of this gene has been shown to improve growth on casein independently of QS and psdR mutations commonly arise early in evolution experiments on casein (16, 23). Surprisingly, mutations in psdR were present only in the analyzed evolved isolates of the LasI mutant but not the LasR mutant (Table 1). Consistent with the idea that MexT-inactivating mutations result in an altered QS hierarchy, the PAO1 LasR-PsdR-MexT triple mutant grew in casein broth, displayed RhlR gene activation measured with the PrhlA-gfp reporter, produced levels of C4-HSL comparable to wild-type PAO1, and produced high levels of pyocyanin (Fig. 2). Deletion of rhlR from the triple mutant abrogated these QS-dependent phenotypes (SI Appendix, Fig. S3). Thus, we concluded that inactivation of MexT results in reprogramming of the QS circuits such that RhlIR QS is independent of LasR. As noted above, this is in agreement with Oshri et al. (23).
Fig. 2.
The influence of mexT, pqsA, and pqsE mutations on RhlR activity in the P. aeruginosa LasR-PsdR mutant. (A) Growth of P. aeruginosa mutants in casein broth. Bacteria were tagged with mCherry, and fluorescence (RFU) was used as a proxy for growth, as described in Materials and Methods. (B) RhlR activity in the LasR-PsdR mutant and mutants derived from this strain, measured as GFP fluorescence (RFU). All strains contained the PrhlA-gfp fusion plasmid pProbe-GT-PrhlAgfp. (C) RhlR activity in strain PAO1 compared with RhlR activity in the LasR-PsdR-MexT mutant, measured as in B. (D) Relative concentrations of C4-HSL produced by the WT and mutants after 18 h. (E) Image of culture tubes after 2 d of growth of the indicated strains in casein broth. The LasR-PsdR-MexT mutant culture is blue green as a result of pyocyanin production. Data in A and D are means of duplicate experiments, and data in B and C are means of triplicate experiments. In all panels, error bars represent the SEM.
A Functional RhlIR QS System Provides a Competitive Advantage to Cells in Coculture.
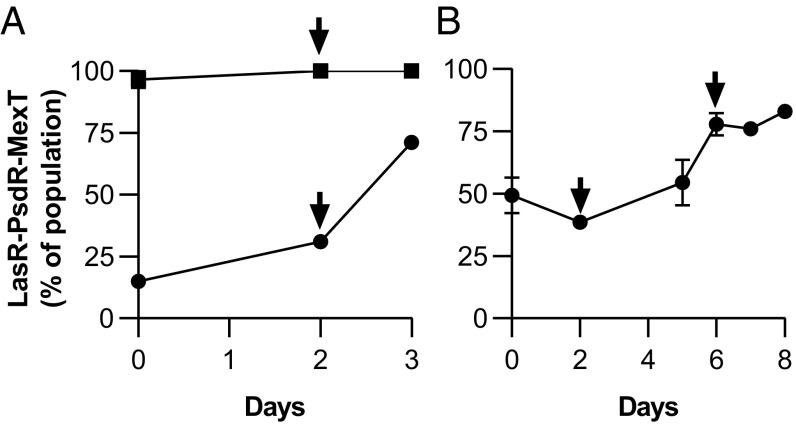
When grown on casein as the sole carbon and energy source, LasR mutants, which do not incur the metabolic cost of QS, have a negative frequency-dependent fitness advantage over wild-type strain PAO1 in coculture (12, 13). For the same reason, a similar advantage could be expected for a LasR mutant in competition with an isogenic LasR-MexT mutant, which bears the cost of protease production for the coculture. To the contrary, we found that LasR-PsdR-MexT mutants outcompeted LasR-PsdR mutants when inoculated either at low or high initial frequency (Fig. 3A). The fitness advantage of the LasR-PsdR-MexT mutant could be due to either the mexT mutation itself or the activated RhlIR system in the MexT mutant background. The latter possibility is attractive because previous work showed that the RhlR-dependent production of CN− by wild-type strain PAO1 provides a fitness advantage over LasR or RhlR mutants (15). To distinguish between the two possible explanations, we performed competition experiments with a LasR-PsdR-MexT mutant and a LasR-PsdR-MexT-RhlR mutant. Although the LasR-PsdR-MexT mutant bears the costs associated with an active QS system, it still outcompeted the QS-null LasR-PsdR-MexT-RhlR mutant (Fig. 3B). This result is consistent with the idea that RhlR induces production of a self-produced stressor or toxin and corresponding resistance factors.
Fig. 3.
Competition between the LasR-PsdR-MexT mutant and QS mutants that cannot grow in casein broth by themselves. (A) Competition experiments with the LasR-PsdR parent. The competitions were initiated with either a low (circles) or high (squares) frequency of LasR-PsdR-MexT. (B) Competition with the LasR-PsdR-MexT-RhlR mutant. Arrows indicate a transfer to fresh casein broth (inoculum 2% vol/vol). Data are the means of duplicate experiments, and error bars represent the SEM.
To test whether policing observed in the competition experiments was mediated by the RhlR-regulated production of CN− (15), we generated a CN−-deficient mutant, LasR-PsdR-MexT-HcnC, and competed it against either the LasR-PsdR or LasR-PsdR-MexT-RhlR mutants (SI Appendix, Fig. S4). In both competitions, the LasR-PsdR-MexT-HcnC mutant outcompeted the QS-null strain in a similar fashion as in the above experiments, indicating that the fitness advantage of the LasR-PsdR-MexT mutant cannot be explained by cyanide-mediated policing.
The Relationship Between PQS and RhlIR Independence from LasR.
Overexpression of MexT has been shown to strongly decrease transcript levels of the pqsABCDE operon (29), which is involved in biosynthesis of PQS. The PQS system induces many RhlR-regulated phenotypes (9, 10). Thus, mexT mutations might lead to a functioning RhlIR QS in LasR mutants by activation of pqsABCDE transcription. To test this hypothesis, we deleted, independently and together, pqsA and pqsE from the LasR-MexT-PsdR triple mutant. PqsA is an essential component of the PQS synthesis operon, and its deletion results in loss of PQS production (6). PqsE is an effector protein that modulates the activity of the PQS system (9), and it has been shown to function even in the absence of PQS or PqsR (11). Recently, it was shown to be involved in the synthesis of a putative alternative ligand for RhlR (32). As previously reported, deletion of either gene resulted in a loss of pyocyanin production (Fig. 2E) (7, 11, 32). However, growth on casein and RhlR gene activation, measured with the PrhlA-gfp reporter, were only partially reduced, with no additive effect by inactivation of both pqsA and pqsE (Fig. 2 A and B). C4-HSL levels were not affected by either mutation (Fig. 2D). These results indicate that genes in the pqsABCDE operon contribute to but do not fully account for the observed RhlR-independent phenotype.
LasR Mutants Can Evolve the Ability to Grow on Casein in Coculture with C4-HSL-Producing Clinical Isolates.
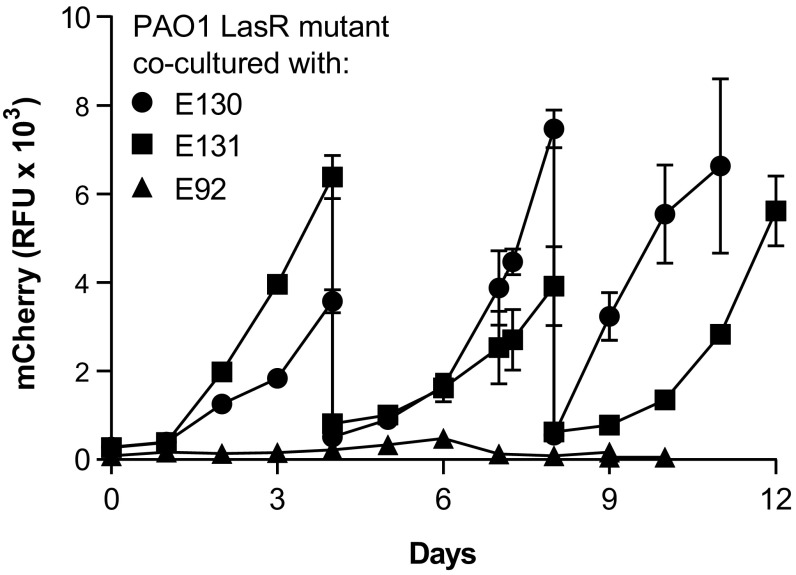
In chronic CF lung infections, the emergence of LasR mutants with an active RhlIR system may be encouraged by other bacteria that produce C4-HSL. Although it cannot provide proof, one test of this hypothesis is to coincubate a LasR mutant with a C4-HSL–producing clinical isolate and ask whether reprogrammed LasR mutants emerge. We therefore selected three clinical P. aeruginosa isolates, E92, E130, and E131, which secrete little elastase but produce more C4-HSL than does our wild-type strain PAO1 (18), and coinoculated casein broth with each of the isolates along with our mCherry-marked PAO1 LasR mutant (Fig. 4). Reprogrammed PAO1 LasR mutants emerged in cocultures with E131 and 132 but not E92. We did not investigate why such variants did not arise during coincubation of the LasR mutant with E92. After two transfers of the cocultures with E130 and E131, we identified protease-positive isolates on skim milk agar plates (SI Appendix, Fig. S5) and verified that these were from the PAO1 LasR mutant lineage by measuring mCherry fluorescence of the isolates. Sequencing of mexT revealed nonsynonymous mutations and deletions in five out of six selected isolates (Table 1). We were unable to amplify mexT in the sixth isolate, which suggests that there was a large deletion encompassing mexT. The results of these coincubation experiments indicate that evolution of an active RhlIR QS system in a LasR-null P. aeruginosa strain could be driven by cooccurrence with some but not all C4-HSL producing bacteria.
Fig. 4.
Emergence of PAO1 LasR mutant variants from coinoculation of PAO1 LasR mutant with clinical P. aeruginosa isolates in casein broth. PAO1 LasR mutant was tagged with mCherry and its growth was measured by monitoring mCherry fluorescence (RFU), as described in Materials and Methods. The clinical isolates were E92 (triangles), E130 (circles), and E131 (squares). Cocultures with E130 and E131 were transferred on days 4 and 8; those with E92 were transferred on days 6 and 9 (inoculum 2% vol/vol). Data are means of duplicate experiments, and error bars represent the SEM.
Discussion
P. aeruginosa has a complicated QS network, which likely plays a significant role in its ability to inhabit many environments (3). QS appears to be important for P. aeruginosa fitness. For example, QS-null mutants are attenuated for virulence in acute animal infection models (4, 5). It is therefore notable that LasR mutants commonly arise in chronic CF lung infections (17–21). The presence of LasR-null, RhlR-active isolates in chronically infected CF lungs (18, 22) suggests that this environment exerts pressure on LasR mutant cells to restore at least part of their QS regulon through the activation of the RhlIR circuit. Here we demonstrate that when a simple selective pressure is applied to a LasR-null mutant of strain PAO1, it can rapidly evolve a LasR-independent, active RhlIR QS system (Fig. 4 and SI Appendix, Fig. S1). Our results suggest that in a mixed culture, production of C4-HSL by a QS-proficient strain can facilitate the alteration of the QS hierarchy. It is not hard to imagine that similar interactions could take place in the context of a chronic P. aeruginosa infection. The reprogrammed LasR-deficient, RhlR-active variant outcompeted the parental LasR mutant as well as the RhlR mutant of rewired variants (Fig. 3). The competitive advantage of the LasR-PsdR-MexT strain over the QS-null strains could not be explained by cyanide-mediated policing (SI Appendix, Fig. S4), which suggests that other policing mechanisms may contribute to the stabilization of QS-regulated cooperation in P. aeruginosa.
The rewiring of QS circuits such that RhlIR functions in a LasR-null background is caused by inactivation of the LysR-type transcriptional regulator MexT (Fig. 2 and ref. 23). Mutations in mexT have been reported among isolates obtained from chronically infected CF lungs (17) and from other P. aeruginosa infections (33, 34). Among the genes activated by MexT are those in the operon that encodes the efflux pump MexEF-OprN (29). Overexpression of MexT and MexEF-OprN leads to the attenuation of known RhlR-regulated phenotypes that include the production of C4-HSL, elastase, CN−, and pyocyanin (30, 31). Interestingly, although overexpression of MexEF-OprN results in lower levels of PQS (29, 30, 35), our results indicate that activation of the RhlIR QS system by inactivating mutations in mexT is only partially mediated through the PQS system (Fig. 2). This observation suggests that one or more other genes in the MexT regulon influence RhlIR QS. It is unlikely that MexT directly affects the expression of rhlR or rhlI, because neither of these genes contains the conserved MexT binding motif in the promoter region (29). Furthermore, inactivation of MexEF-OprN in a LasR-null background results in many of the same phenotypes that are observed for the inactivation of MexT (SI Appendix, Fig. S6, ref. 23). Finally, in our competition experiments (Fig. 3), the decrease in frequency of the RhlR-null mutant (Fig. 3B) was slower than that of the LasR-null mutant (Fig. 3A). Because the RhlR mutant also contains a mexT deletion, it is possible that the enhanced resilience of this mutant in competition is attributable to a QS-independent benefit to cells resulting from inactivation of MexT, as proposed by Oshri et al. (23). This effect could be either direct or indirect because the MexT regulon includes several genes coding for proteins of unknown function (29).
Our experiments and those of Oshri et al. (23) indicate that RhlIR QS system can be activated in the absence of an active LasR, and this requires only a mutation in mexT. Based on earlier studies (12, 13, 21), we propose that in the CF lung, and in similar environments, LasR mutants arise because they are either social cheaters on QS-intact bacteria, have a nutritional advantage over the wild type, or both. However, because some QS-regulated products are important in CF lung infections, a variant that uses RhlR as the primary AHL QS regulator emerges from the QS-null subpopulation. Evolution of such strains is accelerated by C4-HSL, which can be provided by neighboring cells. Our competition experiments provide a plausible explanation for why RhlR mutants are rare among CF isolates. That is, cells with an active RhlR have fitness advantage over RhlR mutants. Our data indicate that even though LasR is often considered to be at the apex of the P. aeruginosa hierarchy, this hierarchy can be easily reprogrammed such that the PQS and RhlIR systems are independent of LasR through a surprisingly simple genetic change. Although many questions remain, our work and other recent publications [accompanying article Chen et al. (36) and refs. 23, 32, and 37] point toward RhlIR QS as a potential anti-QS therapeutic target.
Materials and Methods
Bacteria and Growth Conditions.
Bacteria used are described in SI Appendix, Table S1. P. aeruginosa was grown in Luria-Bertani broth buffered with 50 mM 3-(N-morpholino) propanesulfonic acid, pH 6.8 (LB-Mops broth), casein broth (38) containing 1% sodium caseinate as the sole source of carbon and energy, casamino acid broth (38) with 1% casamino acids as the sole source of carbon and energy, or Pseudomonas P broth (20 g/L pancreatic digest of gelatin, 1.4 g/L magnesium chloride, 10 g/L potassium sulfate) (39), as indicated. Escherichia coli was grown in LB broth. For colony growth we used LB agar (1.5% agar) or milk agar plates (13), as indicated. Liquid cultures were grown at 37 °C with shaking. Where indicated, gentamicin was included in media at a concentration of 100 μg per mL (Gm100).
Construction of P. aeruginosa Mutants.
In all experiments, we used strain P. aeruginosa PAO1-UW (40). However, our strain differs from the originally sequenced strain by ∼20 SNPs and lacks an inactivating 8-bp insertion (41) at the mexT locus that is present in the sequenced strain. Mutants with deletions of lasR, lasI, mexT, mexEF, psdR, rhlR, pqsA, and pqsE were derived from PAO1-UW by a homologous recombination-based two-step allelic exchange approach (42). Briefly, 300–500 bp of DNA flanking the gene of interest were PCR-amplified and cloned in pEXG2 by using E. coli-mediated DNA assembly (43) or standard restriction enzyme-based techniques. The primers used to generate the pEXG2 knockout plasmids are listed in SI Appendix, Table S2. E. coli S17-1 was used to deliver knockout plasmids to P. aeruginosa via conjugation. Merodiploids were selected by plating on Pseudomonas Isolation agar containing Gm100, and deletion mutants were then selected on LB agar containing 10–15% sucrose and no sodium chloride. All deletion mutations were confirmed by PCR of genomic DNA.
Because growth in casein broth cannot be measured using optical density, we used mCherry fluorescence of the LasR and LasI mutant strains as a proxy for growth. We integrated a constitutively expressed mCherry at the att site using the mini-Tn7 insertion system (44). Briefly, P. aeruginosa mutants were transformed with mCherry-containing pUC18-mini-Tn7-Gm and pTNS2 plasmids. Gentamicin-resistant transformants were selected on LB agar containing Gm100. Integration of the mCherry-containing cassette was confirmed by PCR of genomic DNA. To excise the gentamicin resistance marker, we transformed strains with pFLP2 and selected transformants on LB agar containing 5% sucrose and no sodium chloride. Marker excision was confirmed by PCR. mCherry was present in all PAO1-derived strains. Expression of mCherry does not have a measurable metabolic cost when strains with and without mCherry are grown in LB-Mops.
Evolution Experiments and Growth in Casein Broth.
To initiate evolution experiments, we inoculated 3 mL of casein broth in 18-mm tubes with either the LasR mutant or the LasI mutant. Inocula were 150 µL of an overnight culture grown in LB-Mops broth, which corresponded to about 6–9 × 108 colony forming units (CFU). We added 30 µM C4-HSL (Cayman Chemical) to three of six tubes at the start of the experiment and again at day 4. When an increase in mCherry fluorescence was observed, we transferred 300 µL to 3 mL fresh casein broth. After several transfers, we incrementally decreased transfer volumes to 150, 100, 50, and 30 µL. Coculture evolution experiments were performed in a similar fashion. We added 60 µL of an overnight culture of each strain to 3 mL casein broth, and mCherry fluorescence was used to monitor growth of the LasR mutant. For the first and second passages, we transferred 300 and 150 µL of the culture to 3 mL casein broth, respectively.
To identify protease-producing variants, we isolated colonies on LB agar plates and then patched the colonies on skim milk agar plates (13). After about 18 h, the presence of a clear halo around a colony indicated protease production. To grow selected isolates on casein, we added 60 µL of an overnight LB-Mops broth culture to 3 mL casein broth. For experiments with Bacillus thuringiensis lactonase AiiA, we added either AiiA (100 µg lactonase per mL culture) or only buffer to the casein broth at the time of inoculation. AiiA was purified as described elsewhere (45). To monitor mCherry fluorescence, we transferred 150 µL to a 96-well plate and measured fluorescence intensity by using a BioTek Synergy H1 microplate reader.
Competition Experiments.
Overnight LB-Mops broth cultures were used to inoculate 3 mL of casein broth. A 1% inoculum (vol/vol) of the LasR-PsdR-MexT mutant was used, and competing bacteria were inoculated at 0.1, 1, or 10% (vol/vol). We performed transfers of 2% (vol/vol), as indicated. To determine cell yields, we obtained CFU on LB agar and patched 100–120 colonies on milk agar plates. Colonies surrounded by a clear halo were scored as LasR-PsdR-MexT mutants.
Assays for RhlR Activity, C4-HSL, CN−, and Pyocyanin.
We measured RhlR activity in cells grown in LB-Mops broth as previously described, by using a reporter plasmid containing a PrhlA-gfp fusion (18). We used a previously described bioassay to measure C4-HSL (46). We measured CN− concentration as follows. Cells were grown with shaking (200 rpm) in 45 mL of LB-Mops broth for 17 h. Cells were removed from the culture fluid by centrifugation, and 450 µL of 10 N sodium hydroxide was added to the fluid. The CN− in the base-treated fluid was then measured with a CN− electrode (Cole-Parmer). We prepared a CN− standard curve by using freshly prepared potassium cyanide in LB-Mops containing 1% (vol/vol) of 0.1 N sodium hydroxide. For pyocyanin measurements, we grew cells in Pseudomonas P broth for 20 h. Inocula were from overnight LB-Mops cultures (0.1% vol/vol). We added 2 mL chloroform to the cultures and vortexed for 30 s. After removal of the aqueous phase we added 800 µL of 0.2 N hydrochloric acid and then measured absorbance of the aqueous fraction at 520 nm and multiplied the value by 17.072, as described elsewhere (47).
Genome Variant Analysis.
DNA was isolated from bacteria grown in LB-Mops broth by using a Gentra Puregen kit (Qiagen). We used Illumina MiSeq to complete whole-genome sequencing with paired 150-bp reads and StrandNGS version 3.1 (Strand Life Sciences) to align the reads to the published sequence of P. aeruginosa PAO1 (accession no. NC_002516) and to identify genetic polymorphisms including point mutations, deletions, insertions, and inversions (48). Genomic sequences have been deposited at NCBI (BioProject PRJNA514363).
Supplementary Material
Acknowledgments
This work was supported by US Public Health Service (USPHS) Grants GM125714 (to A.A.D.) and GM059026 (to E.P.G.). A.A.D. was also supported by the Doris Duke Charitable Foundation (Grant 2017073) and the Burroughs–Wellcome Fund (Grant 1012253). M.K. was supported by Postdoctoral Fellowships from the Cystic Fibrosis Foundation. We acknowledge core support from USPHS Grant P30DK089507 and the Cystic Fibrosis Foundation (Grants SINGH15R0 and R565 CR11).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. PRJNA514363).
See Commentary on page 6525.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819796116/-/DCSupplemental.
References
- 1.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 2.Williams P, Cámara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. Acyl-homoserine lactone quorum sensing: From evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 4.Rumbaugh KP, Griswold JA, Iglewski BH, Hamood AN. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun. 1999;67:5854–5862. doi: 10.1128/iai.67.11.5854-5862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang HB, et al. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Déziel E, et al. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci USA. 2004;101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pesci EC, et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diggle SP, Cornelis P, Williams P, Cámara M. 4-quinolone signalling in Pseudomonas aeruginosa: Old molecules, new perspectives. Int J Med Microbiol. 2006;296:83–91. doi: 10.1016/j.ijmm.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 10.Rampioni G, et al. Unravelling the genome-wide contributions of specific 2-Alkyl-4-quinolones and PqsE to quorum sensing in Pseudomonas aeruginosa. PLoS Pathog. 2016;12:e1006029. doi: 10.1371/journal.ppat.1006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrow JM, 3rd, et al. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol. 2008;190:7043–7051. doi: 10.1128/JB.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 13.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci USA. 2007;104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dandekar AA, Chugani S, Greenberg EP. Bacterial quorum sensing and metabolic incentives to cooperate. Science. 2012;338:264–266. doi: 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, Schaefer AL, Dandekar AA, Greenberg EP. Quorum sensing and policing of Pseudomonas aeruginosa social cheaters. Proc Natl Acad Sci USA. 2015;112:2187–2191. doi: 10.1073/pnas.1500704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asfahl KL, Walsh J, Gilbert K, Schuster M. Non-social adaptation defers a tragedy of the commons in Pseudomonas aeruginosa quorum sensing. ISME J. 2015;9:1734–1746. doi: 10.1038/ismej.2014.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feltner JB, et al. LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. MBio. 2016;7:e01513–e01516. doi: 10.1128/mBio.01513-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman LR, et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros. 2009;8:66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilder CN, Allada G, Schuster M. Instantaneous within-patient diversity of Pseudomonas aeruginosa quorum-sensing populations from cystic fibrosis lung infections. Infect Immun. 2009;77:5631–5639. doi: 10.1128/IAI.00755-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Argenio DA, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjarnsholt T, et al. Scandinavian Cystic Fibrosis Study Consortium Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One. 2010;5:e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshri RD, Zrihen KS, Shner I, Omer Bendori S, Eldar A. Selection for increased quorum-sensing cooperation in Pseudomonas aeruginosa through the shut-down of a drug resistance pump. ISME J. 2018;12:2458–2469. doi: 10.1038/s41396-018-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh PK, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 25.Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong YH, Xu JL, Li XZ, Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci USA. 2000;97:3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cabeen MT. Stationary phase-specific virulence factor overproduction by a lasR mutant of Pseudomonas aeruginosa. PLoS One. 2014;9:e88743. doi: 10.1371/journal.pone.0088743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kostylev M, et al. 2019 Evolution of the Pseudomonas aeruginosa quorum sensing hierarchy. BioProject. Available at https://www.ncbi.nlm.nih.gov/bioproject/514363. Deposited February 20, 2019.
- 29.Tian ZX, et al. Transcriptome profiling defines a novel regulon modulated by the LysR-type transcriptional regulator MexT in Pseudomonas aeruginosa. Nucleic Acids Res. 2009;37:7546–7559. doi: 10.1093/nar/gkp828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Köhler T, van Delden C, Curty LK, Hamzehpour MM, Pechere JC. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J Bacteriol. 2001;183:5213–5222. doi: 10.1128/JB.183.18.5213-5222.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian ZX, et al. MexT modulates virulence determinants in Pseudomonas aeruginosa independent of the MexEF-OprN efflux pump. Microb Pathog. 2009;47:237–241. doi: 10.1016/j.micpath.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Mukherjee S, et al. The PqsE and RhlR proteins are an autoinducer synthase-receptor pair that control virulence and biofilm development in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2018;115:E9411–E9418. doi: 10.1073/pnas.1814023115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olivas AD, et al. Intestinal tissues induce an SNP mutation in Pseudomonas aeruginosa that enhances its virulence: Possible role in anastomotic leak. PLoS One. 2012;7:e44326. doi: 10.1371/journal.pone.0044326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luong PM, et al. Emergence of the P2 phenotype in Pseudomonas aeruginosa PAO1 strains involves various mutations in mexT or mexF. J Bacteriol. 2014;196:504–513. doi: 10.1128/JB.01050-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamarche MG, Déziel E. MexEF-OprN efflux pump exports the Pseudomonas quinolone signal (PQS) precursor HHQ (4-hydroxy-2-heptylquinoline) PLoS One. 2011;6:e24310. doi: 10.1371/journal.pone.0024310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen R, Déziel E, Groleau M-C, Schaefer AL, Greenberg EP. Social cheating in a Pseudomonas aeruginosa quorum-sensing variant. Proc Natl Acad Sci USA. 2019;116:7021–7026. doi: 10.1073/pnas.1819801116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee S, Moustafa D, Smith CD, Goldberg JB, Bassler BL. The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLoS Pathog. 2017;13:e1006504. doi: 10.1371/journal.ppat.1006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim MK, Harwood CS. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol Lett. 1991;83:199–203. [Google Scholar]
- 39.Zimbro MJ, Power DA, Miller SM, Wilson GE, Johnson JA, editors. Difco & BBL Manual: Manual of Microbiological Culture Media. 2nd Ed Becton, Dickinson and Company; Sparks, MD: 2009. [Google Scholar]
- 40.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 41.Maseda H, Saito K, Nakajima A, Nakae T. Variation of the mexT gene, a regulator of the MexEF-oprN efflux pump expression in wild-type strains of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2000;192:107–112. doi: 10.1111/j.1574-6968.2000.tb09367.x. [DOI] [PubMed] [Google Scholar]
- 42.Hmelo LR, et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc. 2015;10:1820–1841. doi: 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kostylev M, Otwell AE, Richardson RE, Suzuki Y. Cloning should be simple: Escherichia coli DH5α-mediated assembly of multiple DNA fragments with short end homologies. PLoS One. 2015;10:e0137466. doi: 10.1371/journal.pone.0137466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi KH, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat Protoc. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 45.Thomas PW, Fast W. 2011. Heterologous overexpression, purification, and in vitro characterization of AHL lactonases. Quorum Sensing: Methods and Protocols, Methods in Molecular Biology (Methods and Protocols), ed Rumbaugh K (Humana Press, New York), Vol 692, pp 275–290.
- 46.Ding F, et al. The Pseudomonas aeruginosa orphan quorum sensing signal receptor qscR regulates global quorum sensing gene expression by activating a single linked operon. MBio. 2018;9:e01274-18. doi: 10.1128/mBio.01274-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurachi M. Studies on the biosynthesis of pyocyanine. II. Isolation and determination of pyocyanine. Bull Inst Chem Res Kyoto Univ. 1958;36:174–187. [Google Scholar]
- 48.Toussaint JP, et al. Gene duplication in Pseudomonas aeruginosa improves growth on adenosine. J Bacteriol. 2017;199:e00261-17. doi: 10.1128/JB.00261-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.