The bacterium Pseudomonas aeruginosa is an important opportunistic pathogen causing life-threatening acute infections in individuals with compromised immune systems. It is also the most common cause of chronic respiratory infections and the leading cause of morbidity and mortality in patients with the genetic disease cystic fibrosis (CF). In the CF lung, the P. aeruginosa population expands, and a chronic infection ensues. Chronic phenotypes and genotypes emerge during these infections due to ecological adaptation to the CF lung milieu as well as intraspecies competition between P. aeruginosa strains. A perplexing observation is that P. aeruginosa from chronic CF infections appears to frequently lose the ability to perform quorum sensing (QS), a mechanism of chemical communication that allows bacteria to detect the density of a population within a given space. This finding is surprising because animal models of infection implicate P. aeruginosa QS as essential for productive infections. In PNAS, two papers (1, 2) use experimental evolution to address this discrepancy and identify mutations that reengineer the P. aeruginosa QS regulatory pathway, suggesting that QS is not lost during infection but rather rewired.
The QS system of P. aeruginosa is highly complex and controlled by two acyl-homoserine lactone (AHL) systems, LasI-LasR and RhlI-RhlR (RhlIR). LasI catalyzes the production of the autoinducer (AI) signal N-3-oxo-dodecanoyl-l-homoserine lactone, which binds to its cognate receptor LasR (3, 4), whereas RhlI catalyzes the production of AI N-butanoyl-l-homoserine lactone (C4-HSL), which binds to the transcription factor RhlR (5, 6). In addition, P. aeruginosa has a third, non-AHL QS system, the Pseudomonas quinolone signal (PQS), which is mediated through several quinolone AI signaling molecules (7).
In P. aeruginosa, QS was originally recognized as a regulatory mechanism controlling the transcription of genes associated with virulence in the laboratory strain PAO1 (3, 4). In this setting, a hierarchy was described, with LasR as the “master regulator” controlling the expression of RhlR and PQS pathways (8, 9). Thus, LasR has been the focus of efforts to create QS antagonists that could inhibit P. aeruginosa infections. But as the field has progressed, the complexity and overlapping nature of these systems has been revealed to indicate that the situation is not that simple. Furthermore, the characterization of clinical isolates makes the picture even murkier. Of particular interest, a number of laboratories have noted that some isolates from chronic CF infections have null mutations in lasR (10–12). Based on the standing dogma in the field of the role of QS in virulence, these findings were originally seen as counterintuitive. Moreover, these findings suggested that LasR inhibitors may not work in the clinic because they would merely select for mutants that bypass LasR QS. It is therefore critical to understand the role of P. aeruginosa QS during CF infections to not only appreciate how this bacterium colonizes the lung, but also to direct drug development efforts at the appropriate targets.
The diversity of Pseudomonas clinical isolates and the complex environment in CF lungs has hampered our understanding of how and why lasR mutants evolve during infection and how these mutants are able to cause disease. Therefore, two papers in PNAS address this question using a reductionist approach by performing experimental evolution studies in minimal casein medium (1, 2). In this condition, growth of P. aeruginosa requires production of the extracellular protease elastase, encoded by lasB, which is dependent on both the Las and Rhl QS systems (13). LasB functions as a public good that can be exploited by nonproducing cells, and continuous culture of a QS-proficient strain in casein medium selects for QS-defective cheaters that utilize LasB produced by cooperators (14, 15). Because QS is required for growth in this medium, and amino acids are major nutrient sources in the CF lung, this growth condition replicates multiple aspects of growth in vivo and can thus be harnessed for experimental evolution studies.
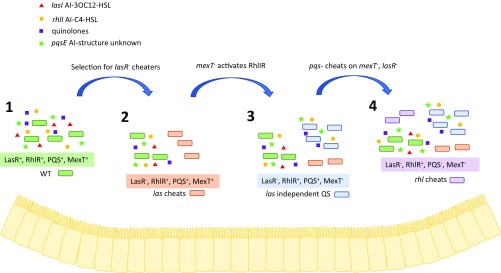
The study by Kostylev et al. (1) examines the evolution of PAO1 lasR mutants in casein broth. In concurrence with previous results, inoculation of lasR mutants into casein broth did not generate any growth for up to 4 wk. However, the authors made a key insight when they realized that evolution of a lasR mutant in vivo would occur within a population of QS-active P. aeruginosa that is still producing AIs, including C4-HSL, the molecule sensed by RhlR. They then repeated the lasR evolution experiment in casein broth with 30 μM C4-HSL and observed outgrowth of mutants in less than a week. These mutants exhibited heritable activity of RhlR activation, including synthesis of C4-HSL and production of LasB. Whole-genome sequencing identified mutations in the transcription factor MexT, a global regulator of gene expression that has been linked to both induction of the MexEF-OprN efflux pump and repression of PQS biosynthesis (16) (Fig. 1). This target of selection in a lasR mutant was also recently identified by Oshri et al. (17), although the absence of C4-HSL dramatically increases the time in which the mexT mutants evolve. Both Kostylev et al. (1) and Oshri et al. (17), therefore, suggest that loss of MexT is the primary mechanism by which the RhlIR pathway can become active when the LasR pathway is nonfunctional. Kostylev et al. (1) also demonstrate that mutation of the PQS pathway in the LasR-MexT mutants leads to a partial loss of RhlIR activity, which could potentially be explained by recent work of the Bassler laboratory suggesting that the PQS system synthesizes an unidentified AI that activates RhlR (18).
Fig. 1.
Speculative and simplified model of P. aeruginosa evolution in the CF lung (no. 1). The CF lung is initially colonized by P. aeruginosa that has wild-type (WT) Las, Rhl, and PQS QS systems, leading to high concentrations of all AIs (no. 2). Over time, LasR− strains evolve, possibly due to the selection for cheats that utilize the public good of the WT strain. As the LasR− cells increase in frequency, concentrations of AIs decrease (no. 3). LasR− cheats can regain functionality of the Rhl QS system by mutation of mexT. The LasR− and MexT− strains are able to produce proteases, C4-HSL, and PQS, but not N-3OC12-HSL (no. 4). The Rhl-activated strains can themselves be cheated on by null mutations to the PQS system. All of these strain phenotypes have been observed in clinical isolates, although the mutations underpinning them are not always understood. We also expect that each of these morphotypes can coexist in an infected lung due to spatial heterogeneity. RhlR− strains do not emerge as cheats because they have a fitness defect when growing in the presence of the reprogrammed LasR mutants with functioning Rhl systems. This also represents one evolutionary pathway, and we expect that there are other targets of selection that can lead to similar P. aeruginosa morphotypes.
Once lasR-inactivated mutants have evolved to have an activated RhlIR system, can these new mutants lead to the evolution of new cheats? Chen et al. (2) explore this question by studying the evolution of the CF isolate P. aeruginosa E80. This isolate has a 4-bp deletion in lasR but displays enhanced protease production and growth in casein broth similar to those evolved by Kostylev et al. (1), suggesting it has an active RhlIR system, although whether this is due to a mutation in mexT was not determined. Nevertheless, passage of E80 in casein broth led to the evolution of protease-deficient freeloaders in ∼1 to 2 wk, producing a tragedy of the commons and collapse of the entire population. The simplest explanation for such freeloaders would be a loss-of-function mutation in rhlR. However, whole-genome sequencing did not detect mutations in the rhlI-rhlR genomic region, but instead identified mutations in pqsR, the master regulator of the PQS system that disrupted protease production (Fig. 1). Indeed, E80 PqsR mutants exhibited elevated fitness when competed with the parent E80 in a frequency-dependent manner. On the other hand, E80 RhlR mutants were less fit than the parent strain, suggesting that even though these mutants no longer had to invest resources to synthesize protease that could be exploited, mutation of rhlR was not a viable path for the emergence of cheats. Chen et al. (2) explain this finding by demonstrating that the loss of rhlR leads to sensitivity to cyanide, which is a policing mechanism produced in an RhlIR-dependent fashion. The work of Chen et al. adds yet another potential layer to this evolutionary trajectory by suggesting that the LasR− Rhl+ QS strains are themselves targets of exploitation. Importantly, these studies show that even during experimental evolution, RhlR function remains.
These in vitro studies are important because they provide a framework and set of predictions that can now be further explored in clinical isolates (Fig. 1). For example, it is expected that in addition to lasR mutations, chronic CF isolates would be expected to also have mutations in mexT, and such mutations have been observed (19–21). Furthermore, PQS mutants should be present as cheaters of the Rhl QS strains, and indeed LasR− PQS− clinical isolates have been discovered (22, 23). Four recent papers (1, 2, 17, 18) demonstrate connections between RhlIR and the PQS pathway, and this connection is likely through the PqsE-generated AI. Thus, one key question to be addressed is the nature of this AI, which would allow a better understanding of its role in the evolution of these QS circuits. Furthermore, RhlR-regulated genes can be C4-HSL dependent, PqsE AI dependent, or dependent on either one. The adaptive benefits of the genes regulated by these signals in both casein medium and the CF lung remain to be defined. Spatial structuring of P. aeruginosa within the CF lung is also a key consideration when considering the evolution of QS variants. It has been recently suggested that P. aeruginosa forms highly heterogeneous micropopulations within a CF lung and that cells in small aggregates are only able to communicate within the aggregate itself (24, 25). How these local dynamics and small population sizes impact the evolution and rewiring of P. aeruginosa QS during CF infection remains a key avenue of future research. Most importantly, all of these studies suggest that RhlR is playing a central role to mediate in vivo QS. Efforts to generate P. aeruginosa QS inhibitors to prevent infection and treat CF have been underway for a few decades, but no inhibitor has yet made it to the clinic. While these works suggest that shifting the target to RhlR could produce the magic bullet needed to inhibit in vivo QS of P. aeruginosa during CF infections, caution should be taken with this approach; the ability of this pathogen to evolve and adapt to such an assault should not be underestimated.
Acknowledgments
C.M.W. acknowledges support from NIH Grants GM109259, GM110444, and AI130554.
Footnotes
References
- 1.Kostylev M, et al. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc Natl Acad Sci USA. 2019;116:7027–7032. doi: 10.1073/pnas.1819796116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen R, Déziel E, Groleau M-C, Schaefer AL, Greenberg EP. Social cheating in a Pseudomonas aeruginosa quorum-sensing variant. Proc Natl Acad Sci USA. 2019;116:7021–7026. doi: 10.1073/pnas.1819801116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Passador L, Cook JM, Gambello MJ, Rust L, Iglewski BH. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 4.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pesci EC, et al. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 9.Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman LR, et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros. 2009;8:66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjarnsholt T, et al. Scandinavian Cystic Fibrosis Study Consortium Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS One. 2010;5:e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Høiby N. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: Strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology. 2010;156:1108–1119. doi: 10.1099/mic.0.033993-0. [DOI] [PubMed] [Google Scholar]
- 13.Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 15.Sandoz KM, Mitzimberg SM, Schuster M. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci USA. 2007;104:15876–15881. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olivares J, et al. Overproduction of the multidrug efflux pump MexEF-OprN does not impair Pseudomonas aeruginosa fitness in competition tests, but produces specific changes in bacterial regulatory networks. Environ Microbiol. 2012;14:1968–1981. doi: 10.1111/j.1462-2920.2012.02727.x. [DOI] [PubMed] [Google Scholar]
- 17.Oshri RD, Zrihen KS, Shner I, Omer Bendori S, Eldar A. Selection for increased quorum-sensing cooperation in Pseudomonas aeruginosa through the shut-down of a drug resistance pump. ISME J. 2018;12:2458–2469. doi: 10.1038/s41396-018-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S, et al. The PqsE and RhlR proteins are an autoinducer synthase-receptor pair that control virulence and biofilm development in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2018;115:E9411–E9418. doi: 10.1073/pnas.1814023115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherrard LJ, et al. Within-host whole genome analysis of an antibiotic resistant Pseudomonas aeruginosa strain sub-type in cystic fibrosis. PLoS One. 2017;12:e0172179. doi: 10.1371/journal.pone.0172179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith EE, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Mansfeld R, et al. Within-host evolution of the Dutch high-prevalent Pseudomonas aeruginosa Clone ST406 during chronic colonization of a patient with cystic fibrosis. PLoS One. 2016;11:e0158106. doi: 10.1371/journal.pone.0158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feltner JB, et al. LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. MBio. 2016;7:e01513-16. doi: 10.1128/mBio.01513-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung JC, et al. Genomic variation among contemporary Pseudomonas aeruginosa isolates from chronically infected cystic fibrosis patients. J Bacteriol. 2012;194:4857–4866. doi: 10.1128/JB.01050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorth P, et al. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe. 2015;18:307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darch SE, et al. Spatial determinants of quorum signaling in a Pseudomonas aeruginosa infection model. Proc Natl Acad Sci USA. 2018;115:4779–4784. doi: 10.1073/pnas.1719317115. [DOI] [PMC free article] [PubMed] [Google Scholar]