Significance
Crucihimalaya himalaica is a close relative of Arabidopsis with typical Qinghai–Tibet Plateau (QTP) distribution. Here, by combining short- and long-read sequencing technologies, we provide a de novo genome sequence of C. himalaica. Our results suggest that the quick uplifting of the QTP coincided with the expansion of repeat elements. Gene families showing dramatic contractions and expansions, as well as genes showing clear signs of natural selection, were likely responsible for C. himalaica’s specific adaptation to the harsh environment of the QTP. We also show that the transition to self-pollination of C. himalaica might have enabled its occupation of the QTP. This study provides insights into how plants might adapt to extreme environmental conditions.
Keywords: adaptive evolution, natural selection, extreme environment, Qinghai–Tibet Plateau, S-locus
Abstract
Crucihimalaya himalaica, a close relative of Arabidopsis and Capsella, grows on the Qinghai–Tibet Plateau (QTP) about 4,000 m above sea level and represents an attractive model system for studying speciation and ecological adaptation in extreme environments. We assembled a draft genome sequence of 234.72 Mb encoding 27,019 genes and investigated its origin and adaptive evolutionary mechanisms. Phylogenomic analyses based on 4,586 single-copy genes revealed that C. himalaica is most closely related to Capsella (estimated divergence 8.8 to 12.2 Mya), whereas both species form a sister clade to Arabidopsis thaliana and Arabidopsis lyrata, from which they diverged between 12.7 and 17.2 Mya. LTR retrotransposons in C. himalaica proliferated shortly after the dramatic uplift and climatic change of the Himalayas from the Late Pliocene to Pleistocene. Compared with closely related species, C. himalaica showed significant contraction and pseudogenization in gene families associated with disease resistance and also significant expansion in gene families associated with ubiquitin-mediated proteolysis and DNA repair. We identified hundreds of genes involved in DNA repair, ubiquitin-mediated proteolysis, and reproductive processes with signs of positive selection. Gene families showing dramatic changes in size and genes showing signs of positive selection are likely candidates for C. himalaica’s adaptation to intense radiation, low temperature, and pathogen-depauperate environments in the QTP. Loss of function at the S-locus, the reason for the transition to self-fertilization of C. himalaica, might have enabled its QTP occupation. Overall, the genome sequence of C. himalaica provides insights into the mechanisms of plant adaptation to extreme environments.
The Qinghai–Tibet Plateau (QTP), also known as Himalayan Plateau, is the highest (average elevation above 4,000 m) and largest (ca. 2.5 million km2) plateau in the world. The QTP mountains reached their current elevations mainly between the late Miocene and late Pliocene, according to recent syntheses of different studies (1). The uplift of the QTP resulted in profound climatic and environmental changes in both the plateau region and Asia at large (2). Conditions on the QTP are now characterized by low temperature, low oxygen, reduced pathogen incidence, and strong UV radiation, which together provide a unique environment to study adaptive evolution (3, 4). Previous genome-wide studies on adaptive evolution on the QTP have focused mainly on humans and vertebrates (reviewed in ref. 5). These studies have revealed that genes involved in hypoxia responses, energy metabolism, and skeletal development are under positive selection and rapid evolution. In contrast, hitherto, no study of adaptive evolution based on whole-genome analysis has been conducted on wild plants (excluding the domesticated Tibetan highland barley) (6) in this region.
Arabidopsis thaliana (Cruciferae/Brassicaceae) is the best-known model organism in plant biology. Relatives of A. thaliana are important for studies on evolutionary ecology and comparative genomics of plants (7–9), especially those found in specific and challenging environments. Crucihimalaya himalaica (Edgew.) Al-Shehbaz, O’Kane & R.A.Price, a self-fertilizing, diploid (2n = 16) relative of A. thaliana, was previously recognized as A. himalaica (Edgew.) O.E.Schulz, and the names Arabis rupestris Edgew. and Arabis brevicaulis Jafri are also accepted as synonyms (10). C. himalaica mainly grows on rocky hillsides, sandy slopes, alpine meadows, and screes in the Himalayas and Hengduan Mountains. Previous estimates suggested that Crucihimalaya diverged from the closely related Pachycladon ∼10.94 Mya (11), while the species C. himalaica originated about 3.56 Mya (12). Given the postulated timing of the most recent rapid uplift of the QTP during the period of late Miocene to late Pliocene (1, 13), many of the genetic changes in C. himalaica might reflect its adaptation to the extreme environment of QTP (4).
In a previous study (4), we identified 487 positively selected genes (PSGs) in the transcriptome of C. himalaica. Predicted functions of these PSGs indicate that they potentially contribute to miscellaneous traits of adaptive importance, such as response to UV radiation, DNA repair, and membrane stabilization, which presumably are important for the adaptation of C. himalaica to the specialized environment on the QTP. Therefore, we believe that this species represents a model system for the study of speciation and ecological adaptation in extreme environments. In the present study, we generated de novo whole-genome sequences of C. himalaica and applied comparative and evolutionary genomics approaches to clarify the origin of this species and to investigate signals of adaptation. Our goal is to gain a deeper understanding into the mechanisms by which C. himalaica has adapted to the complex extreme conditions on the QTP at the whole-genome level.
Results and Discussion
Genome Assembly and Annotation.
The genome size of C. himalaica was estimated to be 265.23 Mb with a heterozygosity of 0.70% on the basis of k-mer statistics (14) (SI Appendix, Table S1). In total, we generated 50.68 Gb of paired-end (PE) and mate-pair (MP) reads with different insert sizes using an Illumina HiSeq platform combined with PacBio sequencing technology (SI Appendix, Table S2). A total assembly of 234.72Mb, consisting of 583 scaffolds (scaffold N50 length, 2.09 Mb; longest scaffold, 8.34 Mb), was achieved by combination of the ALLPATHS–LG (15) and PBJelly2 (16) assembly strategies (Table 1). To assess assembly accuracy, we remapped raw sequencing reads of a small fragment library to the assembled genome. With a 96.69% mapping rate and 224.57× average sequence depth, the reads covered 96.43% of the genome, which implies that the current assembly covered almost all unique genomic regions. About 94.05% of the assembly was covered by at least 20× reads, which guaranteed a highly accurate assembly at the single-nucleotide level (SI Appendix, Table S3).
Table 1.
Genome assembly of C. himalaica
| Genome features | Contigs | Scaffolds |
| Total length, bp | 230,905,116 | 234,722,603 |
| Total no. | 3,983 | 583 |
| Longest length, bp | 1,756,581 | 8,343,586 |
| No. of length ≥2,000 | 3,586 | 429 |
| Length of N50, bp | 136,392 | 2,088,603 |
| No. of N50 | 406 | 34 |
| Length of N90, bp | 32,421 | 470,087 |
| No. of N90 | 1,711 | 129 |
| GC content, % | 36.38 | — |
| No. of genes | — | 27,019 |
A total of 27,019 protein-coding genes were predicted (Table 1 and SI Appendix, Table S4). Among these, 26,806 genes (99.21%) were functionally annotated (SI Appendix, Table S5). In addition to protein-coding genes, various noncoding RNA sequences were identified and annotated (SI Appendix, Table S6), including 448 microRNAs, 577 transfer RNAs, 153 ribosomal RNAs, and 974 small nuclear RNAs. Gene region completeness was evaluated by RNA sequencing data (SI Appendix, Table S7). Of the 29,420 transcripts assembled by Trinity, 99.74% were mapped to the genome assembly, and 92.37% were complete. The completeness of gene regions was further assessed using BUSCO (Benchmarking Universal Single Copy Orthologs) (17), which showed that 96% of the plant single-copy orthologs were complete (SI Appendix, Table S8). Compared with other close relatives, the C. himalaica genome contains a similar number of transcription factors (1,711) and transcription regulators (413) (SI Appendix, Table S9). The C. himalaica genome showed strong homology with the genome of A. thaliana, Arabidopsis lyrata, and Capsella rubella, except in centromeres and certain high-repeat regions (Fig. 1 and SI Appendix, Figs. S1–S3). These results implied that the assembly was of high quality, thus ensuring the reliability of subsequent comparative genomic analyses in this study.
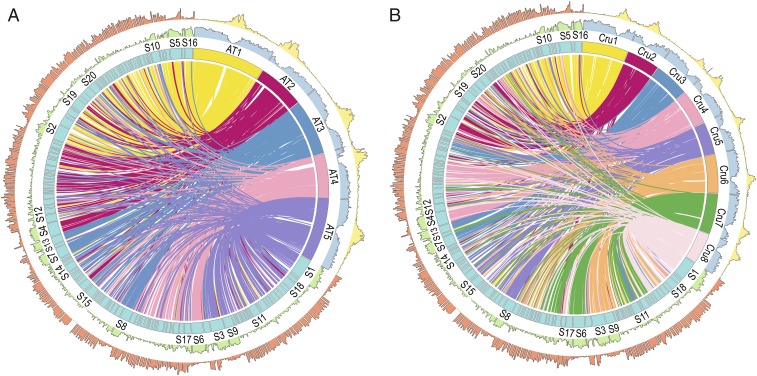
Fig. 1.
Comparative analyses of genomic features of C. himalaica vs. A. thaliana (A) and C. himalaica vs. C. rubella (B). Tracks from inside to outside are collinearity between both genomes, number of chromosomes/scaffolds, gene density, and TE density.
Repeat Elements.
Compared with the genome size of closely related species, such as C. rubella (which has an assembled genome size of 134.8 Mb, whereas the genome size estimated from flow cytometry was 219 Mb) (18) and A. thaliana [125 Mb (19) and the estimated genome size of the reference accession Col-0 from flow cytometry is 166 Mb (20)], the C. himalaica genome is considerably larger. It has been well documented that polyploidization (whole-genome duplication, WGD) events and transposable element (TE) amplification are major causes of genome expansion (21, 22). Analyses of age distributions built from transversion substitutions at fourfold degenerate sites (4DTv) indicated that, except for the α (4DTv distance = ∼0.3) and β (4DTv distance = ∼0.6) polyploidy events shared among members of the Brassicaceae (23), C. himalaica has not undergone an additional species-specific WGD event (Fig. 2A). Next, we investigated the content and evolutionary history of TEs in C. himalaica. Using de novo prediction of TEs (Materials and Methods), we identified and marked 46.91% of the assembly as repeat regions, among which TEs occupied 45.78% of the genome assembly length (SI Appendix, Table S10), which is higher than that reported for A. lyrata and A. thaliana (29.7% and 23.7%, respectively) (24). This was also apparent in a CIRCOS genomic comparison plot, which showed that the density of TEs in C. himalaica was higher than that in A. thaliana or C. rubella, whereas the density of genes in C. himalaica was lower than that in A. thaliana or C. rubella (Fig. 1).
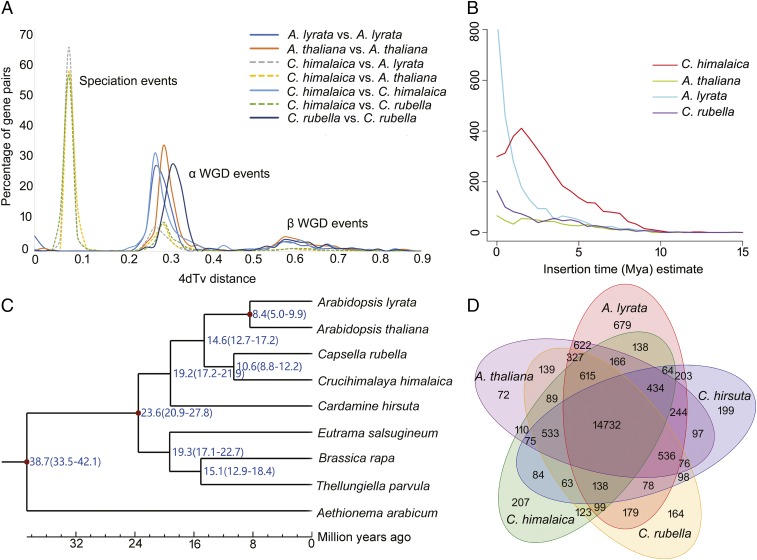
Fig. 2.
Evolutionary analyses of the C. himalaica genome. (A) Age distribution of 4DTv distance values between orthologs of C. himalaica and A. thaliana, C. himalaica and A. lyrata, and C. himalaica and C. rubella. (B) Insertion time distribution of LTR retrotransposons. (C) Estimation of divergence times of nine species in the Brassicaceae. (D) Venn diagram showing unique and shared gene families between genomes of C. himalaica and four close relatives.
In particular, a high proportion of TEs in C. himalaica were LTR retrotransposons (30.37%), whereas other retrotransposons (short and long interspersed nuclear elements) only constituted 3.31% collectively (SI Appendix, Table S10). The retrotransposon proliferation might be responsible for the genome-size expansion in C. himalaica. To investigate the evolutionary dynamics of LTR retrotransposons, we estimated their insertion dates in four closely related species (Materials and Methods). Compared with A. thaliana, which shows a large number of microdeletions in noncoding DNA and transposons (24), A. lyrata has a comparatively high proportion of recent insertions (18) (Fig. 2B), possibly contributing to its larger genome size (207 Mb). The proliferation of LTR retrotransposons in C. himalaica (Fig. 2B and SI Appendix, Fig. S4 and Table S11) peaks at ∼2.0 Mya, shortly after the dramatic elevational and climatic changes of the QTP between the late Miocene and late Pliocene (1, 13). The activity of TEs (including LTR retrotransposons) can lead to diverse genetic changes (e.g., chromosomal rearrangements and gene duplication, creation, and disruption) that may drive lineage-specific diversification and adaptation (25, 26). We therefore hypothesize that the proliferation of LTR retrotransposons likely contributed to the diversification, speciation, and adaptation of C. himalaica, similar to the proliferation of BARE-1 retrotransposons in wild barley (27) and DIRS1 retrotransposons in Antarctic teleost (28), which might have facilitated the adaptation of plants to drier slopes and subzero temperatures, respectively. Another example from apples involves a major burst of retrotransposons ∼21.0 Mya, which coincided with the uplift of the Tianshan mountains, the postulated center of origin of apples (29).
Phylogenetic Tree Construction and Estimation of Divergence Times.
Previous studies suggest that Crucihimalaya forms an endemic genus in the Himalayas and is most closely related to Arabidopsis (30, 31). However, our earlier phylogenetic analyses of transcriptome sequences found that C. himalaica formed a clade sister to C. rubella (4). Applying OrthoMCL (32) to nine published whole-genome sequences from Brassicaceae (SI Appendix, Table S12), we identified a total of 22,670 orthogroups. Among these orthogroups, 4,586 contained putative single-copy gene families. To verify the phylogenetic position of C. himalaica, we generated a maximum likelihood phylogenetic tree with a trimmed and concatenated protein sequence alignment from 4,586 single-copy genes in nine species. The resulting phylogeny indicated that C. himalaica was most closely related to C. rubella, and that these two species in turn formed a clade with A. lyrata and A. thaliana (Fig. 2C), confirming our previous results (4). The above-mentioned four genera (which are classified in the tribe Camelineae), together with the allied Cardamine hirsuta, were often recognized as Lineage I or Clade A in previous phylogenetic studies (33, 34).
C. himalaica and C. rubella were estimated to have diverged ∼10.6 (8.8 to 12.2) Mya in our analyses using MCMCtree (35) with three calibration points (Materials and Methods and Fig. 2C); the two species diverged from Arabidopsis ∼14.6 (12.7 to 17.2) Mya and from Brassica rapa ∼23.6 (20.9 to 27.8) Mya. These results are in agreement with previous estimates (11, 36). Given the absence of genomic information for the remaining species of Crucihimalaya, we are unable to date the exact origin of C. himalaica, which was estimated previously at ∼3.56 Mya (11, 12). However, C. himalaica must have evolved after the split with Capsella and almost certainly less than 10.6 Mya. Such estimation is in accordance with the timing of the most recent rapid uplift of the QTP from late Miocene to late Pliocene (1, 13). The genus Crucihimalaya is assigned to the tribe Crucihimalayeae, which includes two additional genera, Ladakiella and Transberingia (37, 38). The tribe Crucihimalayeae is mainly distributed in Central Asia, with one species (Transberingia bursifolia) extending into North America. Therefore, we speculate that the ancestor of C. himalaica dispersed from Central Asia into the Himalayas, and speciation of C. himalaica was likely triggered by the rapid uplift of the Himalayas, which resulted in the evolution of specialized phenotypic and physiological characters as adaptations to the extreme environment (discussed further below).
Gene Family Expansion and Contraction.
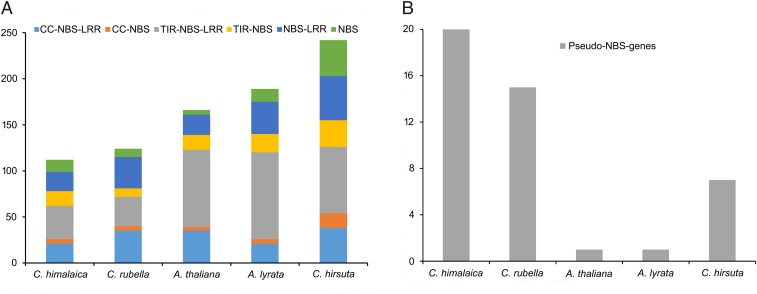
Significant expansion or contraction in the size of particular gene families is often associated with the adaptive divergence of closely related species (39, 40). Comparisons of the genomes of C. himalaica and four close relatives (Fig. 2D) identified a total of 151 gene families that are significantly (P < 0.01) expanded in C. himalaica and 89 gene families that are significantly contracted (SI Appendix, Table S13). Based on Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) annotations, expanded gene families were highly enriched in ubiquitin-conjugating enzyme (E2) activity (P = 7.17E-40) and DNA repair pathways (P = 1.45E-12) (Table 2). It is notable that the most significantly contracted gene families in the C. himalaica genome were found to be functionally related to disease and immune responses, such as the Toll-like receptor and NF-κB signaling pathway (Table 2). The majority of bacteria interact with Toll-like receptors on the surface of the host cell membrane, stimulating the NF-κB signaling pathway in the immune response (41). The Toll and interleukin-1 receptor (TIR) is an N-terminal component of the nucleotide-binding site (NBS) disease resistance protein family, which includes the TIR-NBS-LRR (TNL) and CC-NBS-LRR (CNL) subfamilies. In the C. himalaica genome, both TNL and CNL subfamilies underwent severe contractions compared with their close relatives (Fig. 3A). In particular, disease resistance RPP8-like proteins (belonging to CNL) have undergone the most significant contraction in the C. himalaica genome (6 vs. 16∼29 members, P = 1.26E-10). These proteins are major players in plant defense against pathogens by triggering hypersensitive responses (42). The rapid evolutionary expansion or contraction of the NBS gene family may be a fundamental strategy for plants to adapt to the rapidly changing species-specific pathogen spectrum (43, 44). As fewer microorganisms exist on the QTP owing to the harsh environments characterized by cold temperatures, aridity, and high UV radiation (45), it is reasonable to presume that the reduction in number of NBS genes in the C. himalaica genome is due to a lighter load of pathogens present in the environment and therefore the scarcity of pathogen infection on the QTP. We performed additional searches for NBS pseudogenes, which were assumed to have frameshift mutations and/or premature stop codons. Only one pseudogene could be found in A. thaliana and A. lyrata, but 20, 15, and 7 NBS pseudogenes could be identified in the C. himalaica, C. rubella, and C. hirsuta genomes, respectively (Fig. 3B). Therefore, pseudogenization of NBS disease resistance genes in C. himalaica is at least partly responsible for the contraction of this gene family. Similar observations were made for the genome of the ground tit (Parus humilis), native to the QTP, where MHC genes involved in the cellular immune defense against pathogens also show significant contractions (46). Although the mechanism of MHC gene evolution in ground tit remains unclear, it is worth noting that both ground tit and C. himalaica show a convergent evolution with respect to the contraction of gene families involved in defense against pathogens.
Table 2.
Functional annotation of the most significantly expansive and contract gene families in C. himalaica
| Gene families | KEGG terms | Input no. | Background no. | P value |
| Expanded gene | Ubiquitin-mediated proteolysis | 19 | 137 | 5.59E-29 |
| families | Mismatch repair | 10 | 80 | 9.44E-12 |
| Homologous recombination | 10 | 105 | 9.12E-11 | |
| DNA replication | 10 | 103 | 9.12E-11 | |
| Nucleotide excision repair | 10 | 137 | 9.31E-10 | |
| RNA polymerase | 4 | 32 | 1.59E-06 | |
| Pyrimidine metabolism | 4 | 105 | 9.53E-05 | |
| Purine metabolism | 4 | 176 | 5.66E-04 | |
| Contracted gene | NF-κB signaling pathway | 64 | 93 | 1.96E-106 |
| families | Toll-like receptor signaling pathway | 64 | 106 | 5.32E-104 |
| Retinol metabolism | 21 | 65 | 3.60E-29 | |
| Metabolism of xenobiotics by P450 | 18 | 73 | 2.89E-23 | |
| Tryptophan metabolism | 13 | 40 | 2.89E-18 | |
| Steroid hormone biosynthesis | 13 | 58 | 1.73E-16 | |
| Metabolic pathways | 31 | 1,243 | 1.03E-12 | |
| Arachidonic acid metabolism | 10 | 62 | 1.35E-11 | |
| Linoleic acid metabolism | 8 | 29 | 4.96E-11 | |
| Lysosome | 7 | 123 | 1.27E-05 | |
| Apoptosis | 4 | 33 | 9.75E-05 |
Fig. 3.
Size of the NBS gene family (A) and number of pseudo-NBS genes (B) in the C. himalaica genome compared with those of related species.
Positive Selection on Single-Copy Genes.
Orthologs that show signs of positive selection usually underwent adaptive divergence (47). Previously, we observed that the ratio of nonsynonymous to synonymous substitutions (dN/dS or ω) in C. himalaica is higher than those of closely related species, suggestive of accelerated evolution in C. himalaica after divergence from its ancestral lineage (4). In the present study, we conducted a positive selection analysis using the genomic sequences of C. himalaica and four close relatives. Among the 21,383 orthogroups, 11,085 contained single-copy orthologous genes. We used the branch-site model of the PAML 4 package (35) to identify genes with signs of positive selection. As a result, 844 genes possibly under positive selection were identified in the C. himalaica genome (ω > 1, P < 0.05). Of these genes, 610 showed highly significant (P < 0.01) positive selection (SI Appendix, Table S14). A KEGG functional classification of the 610 significant PSGs in the C. himalaica genome (SI Appendix, Table S15) showed that several categories associated with DNA repair, the ubiquitin system, as well as plant hormone biosynthesis and signal transduction were enriched. Genes involved in DNA repair were also identified as being under positive selection pressure in a previous transcriptomic study of C. himalaica (4). It is notable that significantly expanded gene families and PSGs were both enriched in DNA repair and protein ubiquitination pathways. Signal transduction-related CheY-like genes were also found to have undergone both significant positive selection and expansion events in our previous study on the adaptive evolution of the cyanobacterium Trichormus sp. NMC-1 on the QTP (3), again providing evidence that expansion/duplication and (subsequent) positive selection of genes are an important mechanism for plant adaptive evolution.
The extremely intense UV radiation on the QTP may influence plant growth and developmental processes such as photoperiodism and flowering, or cause DNA, RNA, and protein damage (48). Our results showed that 10 PSGs in the C. himalaica genome are involved in the DNA repair pathway (SI Appendix, Table S15). Notably, several genes in the nucleotide excision repair pathway showed signatures of positive selection. One of them encodes the DNA excision repair protein ERCC-1 (or UV hypersensitive 7), which was also identified previously (4) and is reported to function as a component of a structure-specific endonuclease that cleaves on the 5ʹ side of UV photoproducts in DNA (49). In addition to ERCC-1, we identified PSGs that encode the AP endonuclease 2 protein (APEX2), DNA polymerase delta subunit 2 (POLD2), DNA mismatch repair protein MLH1, and DNA repair and recombination protein RAD54 (SI Appendix, Table S14). These PSGs are involved in base excision repair, mismatch excision repair, and homologous recombination, suggesting that C. himalaica has evolved an integrated DNA-repair mechanism to adapt to the harsh habitats caused by the uplift of the QTP. Moreover, high UV-B radiation is a common stress that both animals and plants on the QTP must cope with. The DNA repair and radiation responses pathways similarly have played crucial roles in the highland adaptation of the Tibetan highland barley (6), Tibetan antelope (50), Tibetan chicken (51), and Tibetan hot-spring snake (52).
Our analyses also indicated that ubiquitin system-related gene families underwent significant expansion and natural selection. Ubiquitin-mediated proteolysis impacts almost every aspect of plant growth and development, including plant hormone signal transduction, photomorphogenesis, reproduction, and abiotic stress responses (53). Such a multifunctional biological process is of even greater importance for plants in the complex harsh environment on the QTP. Among the seven PSGs in the ubiquitin system based on the KEGG annotation, for instance, one encodes an ortholog of the COP10 protein in A. thaliana, which can enhance the activity of ubiquitin-conjugating enzymes (E2s) (54) and participates in light signal transduction and photomorphogenesis (55). An additional PSG encodes the ortholog of the auxin transport protein BIG (E3 ubiquitin-protein ligase UBR4) in A. thaliana, which not only influences multiple auxin-mediated developmental processes (e.g., lateral root production and inflorescence architecture) but also plays a critical role in a multitude of light and phytohormone pathways (56). These PSGs enriched in the ubiquitin system might be important for Crucihimalaya to better survive the harsh environment in the QTP. Similarly, in three mangrove species, the ubiquitin-mediated proteolysis pathway includes 12 genes that have experienced convergent evolution at conservative sites, presumably important for adaptation of mangroves to the harsh coastal habitat (57).
Moreover, many of the identified PSGs were associated with reproduction pathways (92 PSGs, P = 5.40E-06) based on GO annotation (SI Appendix, Table S16). One of the PSGs encodes the phytochrome and flowering time regulatory protein 1 (PFT1), which represses the PhyB-mediated light signaling and regulates the expression of FLOWERING LOCUS T (FT) and CONSTANS (CO) (58, 59). In a low-red to far-red ratio environment, plants require CO and FT to fully accelerate flowering in long days (58, 59). PFT1 acts as a hub to integrate a variety of interdependent environmental stimuli, including light quality and jasmonic acid-dependent defense responses. An additional gene encodes TERMINAL FLOWER 1 (TFL1), which controls inflorescence meristem identity and regulates flowering time in the long-day flowering pathway. As the QTP experiences a long sunshine duration and a short vegetation growing season, flowering time is particularly critical and affects both the life cycles and reproductive success of many alpine plants (60). In the wild, C. himalaica initiates flowering relatively early (April). However, this species rarely blossoms in low-altitude areas outside the QTP. These observations suggest that C. himalaica has evolved specific reproductive strategies on photoperiodism and flowering-related pathways as an adaptation to the long sunshine duration and short growing season on the QTP.
S-Locus Structure and Self-Fertilization of C. himalaica.
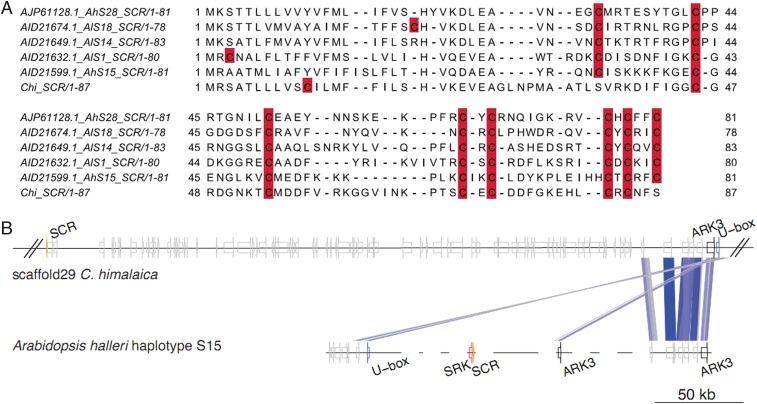
Self-incompatibility in Brassicaceae is controlled by the S-locus recognition system, which prevents self-fertilization (61, 62). Whereas self-fertilization often leads to decreased fitness of homozygous offspring, it also ensures reproduction in absence of pollinators or suitable mates, and therefore can be advantageous for plants to expand their distribution edges and occupy new niches (63, 64). The QTP is characterized by harsh conditions including a short growing season and low pollinator activities. Therefore, it is not surprising that C. himalaica is a selfing plant (9). The Arabidopsis/Capsella-like S-locus contains the female specificity gene (S-receptor kinase, SRK) and the male specificity gene (S-locus cysteine-rich protein, SCR) (65). Self-compatibility is achieved when the SRK receptor fails to recognize the SCR ligand, usually through a loss of SCR function (66–69). We manually annotated the S-locus flanking genes (AKR3 and U-box) and the SCR gene on scaffold 29 of the C. himalaica genome assembly. Alignment of Crucihimalaya SCR coding sequences with homologs from miscellaneous Arabidopsis halleri, A. lyrata, A. thaliana, Capsella grandiflora, C. rubella, and C. hirsuta S-locus haplotypes showed that the closest haplotype to C. himalaica was the one from A. halleri S15 (SI Appendix, Fig. S5) (66, 68, 70, 71). Note that, due to strong balancing selection on the S-locus, highly diverged S-alleles are shared across species and SCR gene phylogeny does not follow species phylogeny (SI Appendix, Fig. S5) (72, 73). SCR protein sequence alignment showed that C. himalaica appears to have lost two out of eight conserved cysteines (Fig. 4A) that are essential for structural and functional integrity of SCR ligand (66, 74). This alone may explain the transition to self-fertilization in C. himalaica. Interestingly, neither SRK from the S15 haplogroup nor any other SRK had a BLAST hit to the C. himalaica genome (Fig. 4B). To investigate whether SRK is indeed lost from the genome, we mapped short reads (450-bp insert size library) of C. himalaica to the combination of the C. himalaica genome assembly and A. halleri S15 haplogroup of S-locus. The SRK gene region did not have any reads mapped, and therefore we conclude that SRK is not present in the C. himalaica genome and likely has been lost. Taken together, we reason that the transition to self-compatibility of C. himalaica was accomplished by both (i) loss of function in the male recognition gene SCR and (ii) loss of the female recognition gene SRK. It is often suggested that self-fertilization rates of alpine plants increase at higher altitudes (75), due to the reduction in pollinator abundance and seed production in alpine plants (76, 77). This suggestion is consistent with the fact that self-fertilizing populations also exist in Arabis alpina, a close relative of C. himalaica that grows at high altitudes (78, 79). Moreover, previous studies also found that the rate of self-fertilizing hermaphroditic plants is increasing on the QTP and autonomous self-fertilization provides substantial reproductive assurance in the pollinator scarcity condition of the QTP (80, 81). Therefore, the transition to a self-compatibility mating system in C. himalaica likely facilitated its occupation of the QTP.
Fig. 4.
Transitioning to self-fertilization in C. himalaica. (A) Protein sequence alignment of S-locus SCR genes from C. himalaica, A. halleri, and A. lyrata. Cysteine residues are highlighted in red, showing eight conserved sites important for structural and functional integrity of the protein (66, 74). The C. himalaica SCR protein appears to have lost two out of eight conserved cysteines and therefore is probably nonfunctional. (B) Structure of the S-locus part of scaffold 29 in C. himalaica genome assembly compared with A. halleri S15 haplogroup (70). Colors of the lines between S-loci correspond to BLAST scores from highest (blue) to lowest (gray).
Conclusion
Organisms that live on the QTP face a variety of abiotic stresses under the harsh environmental conditions and presumably have been subjected to a series of adaptive evolutionary changes. In the present study, we de novo-sequenced the genome of C. himalaica, a species exclusively found in the Himalayan region. Based on phylogenetic reconstruction and estimated divergence times, we propose that the speciation of C. himalaica was triggered by the uplift of the QTP, mediated by genomic evolution to adapt to the altered environment. The proliferation of LTR retrotransposons may at least partly be responsible for the increased genome size of C. himalaica. The significant contraction and pseudogenization of NBS gene families reflect the strong reduction in pathogen incidence on the QTP. Gene families that underwent significant expansion and genes that show signs of positive selection are enriched in DNA repair and protein ubiquitination pathways, which probably reflect to the adaptation of C. himalaica to high radiation, low temperature, and pathogen-depauperate environments on the QTP. Occupation of the QTP by C. himalaica was likely facilitated by self-compatibility, which, as we have shown here, involved both male and female components of the recognition system. Both similarities (e.g., DNA repair and disease-resistance pathways) and differences (e.g., reproduction and plant hormone-related pathways) in adaptive mechanisms have been identified among plants and animals that grow at high altitudes. Although further experimental verification is needed, our results provide insights into how plants adapt to harsh and extreme environments.
Materials and Methods
Plant Material, Genome Sequencing, and Assembly.
Seedlings of C. himalaica were sampled from Batang County (altitude 4,010 m, N 30.313°, E 99.358°) of QTP. Seedlings from the same individual were cultivated in the greenhouse at Kunming Institute of Botany. High-quality genomic DNA was extracted using the Qiagen DNeasy Plant Mini Kit. The C. himalaica genome assembly was performed using sequence data obtained from a combination of sequencing technologies: Illumina PE reads, Illumina MP reads, and PacBio RS II reads (SI Appendix, Table S2). The whole step of library construction and sequencing was performed at Novogene Bioinformatics Technology Co., Ltd. First, six PE libraries were prepared to sequence the C. himalaica genome. These included two PE libraries with insert sizes of 250 and 450 bp and four MP libraries with insert sizes of 2, 5, 10 and 15 kb. Whole genomic sequence (44.49 Gb) data were generated solely using Illumina platforms (HiSEq. 2500) and assembled using ALLPATHS -LG (15). Next, the PacBio reads (6.19 Gb) were used to fill gaps using the PBJelly2 tool (16), followed by scaffold assembly with SSPACE (82) using long-insert-size PE reads. The accuracy and completeness of the assemblies were assessed by aligning the reads from short-insert-size libraries back to the scaffolds using BWA-mem (v. 0.7.17) (83).
Gene Prediction and Annotation.
Gene prediction was performed using a combination of homology, de novo, and transcriptome-based approaches. Gene models were integrated by EvidenceModeler (evidencemodeler.github.io/). Gene models were further updated by PASA (84) to generate UTRs and provide information on alternative splicing variants. The predicted genes were analyzed for functional domains and homologs using InterProScan and BLAST against the NCBI nonredundant protein sequence database, TrEMBL and SwissProt with an E-value cutoff 1E-15 and Blast2GO with default parameters. Completeness of the genome was also assessed by performing core gene annotation using the BUSCO (17) methods. Transcription factors were identified and classified into different families using the iTAK pipeline (bioinfo.bti.cornell.edu/cgi-bin/itak/index.cgi) (85).
Repetitive Elements Identification and Dynamic Analysis.
A combined strategy based on homologous sequence alignment and de novo searches was used to identify repeat elements in the C. himalaica genome. We de novo predicted TEs using RepeatModeler (www.repeatmasker.org/RepeatModeler), RepeatScout (www.repeatmasker.org), Piler (86) (www.drive5.com/piler/), and LTR-Finder (87) (tlife.fudan.edu.cn/tlife/ltr_finder) with default parameters. For alignment of homologous sequences to identify repeats in the assembled genome, we used RepeatProteinMask and RepeatMasker (www.repeatmasker.org) with the repbase library (88). TEs overlapping with the same type of repeats were integrated, while those with low scores were removed if they overlapped over 80% of their lengths and belonged to different types.
Intact LTR retrotransposons were identified by searching the genomes of C. himalaica with LTRharvest (89) (-motif tgca -motifmis 1) and LTR_Finder (LTR length 100 to 5,000 nt; length between two LTRs: 1,000 to 2,0000 nt). We combined results from both analyses after integrating the overlap sites. Candidate sequences were filtered by a two-step procedure to reduce false positives. First, LTRdigest (90) was used to identify the primer binding site (PBS) motif, and only elements containing PBS were retained; then, protein domains (pol, gag, and env) in candidate LTR retrotransposons were identified by searching against HMM profiles considered as intact. Second, clustering of candidate LTR retrotransposons was performed using the SiLiX software package (lbbe.univ-lyon1.fr/SiLiX). Using a substitution rate (r) of 7*10−9 substitutions per site per year (91, 92), the insertion date (T) can be calculated for each LTR retrotransposons (T = K/2r, K: genetic distance). Tandemly repeated gene arrays were identified using BLASTP with a threshold E value < 10−8; fragmental alignments were conjoined for each gene pair. The tandem repeat gene clusters were identified using a cutoff of sequence identity >70% and distance <150 kb. Each cluster with more than two genes was retained.
Whole-Genome Alignment and Duplication Analysis.
We aligned the C. himalaica genome to those of A. thaliana, A. lyrata, and C. rubella using LASTZ (93) with the following parameter values: M = 254 K = 4500 L = 3000 Y = 15000 -seed = match 12 -step = 20 -identity = 85. To avoid the interference caused by repetitive sequences for sequence alignment, RepeatMasker and RepBase library were used to mask repetitive sequences of the above four genomes. The raw alignments were combined into larger blocks using the ChainNet algorithm. We identified orthologous genes among the C. himalaica, A. thaliana, A. lyrata, and C. rubella genomes and paralogous genes within C. himalaica and other relatives using BLASTP (E value < 1E-7). MCscanx (94) was used to identify syntenic blocks within the genome. For each gene pair in a syntenic block, the 4DTv distance was calculated; values of all gene pairs were plotted to identify putative whole-genome duplication events and divergence in two species.
Phylogenetic Tree Construction and Estimation of Species Divergence Times.
We selected genomes of C. himalaica and eight other species (A. thaliana, A. lyrata, C. rubella, C. hirsuta, Eutrema salsugineum, B. rapa, Schrenkiella parvula, and Aethionema arabicum) to identify orthologs. To remove redundancy caused by alternative splicing variations, we retained only gene models at each gene locus that encoded the longest protein sequence. To exclude putative fragmented genes, genes encoding protein sequences shorter than 50 aa were filtered out. All filtered protein sequences of these plants were compared with each other using BLASTP (E value < 1E-7) and clustered into orthologous groups using OrthoMCL (inflation parameter, 1.5) (33). Protein sequences from 4,586 single-copy gene families were used for phylogenetic tree construction. MUSCLE (95) was used to generate multiple sequence alignment for protein sequences in each single-copy family with default parameters. The alignments of each family were concatenated to a super alignment matrix, which was then used for phylogenetic tree reconstruction through the PROTCATJTT model in RAxML software.
Divergence time between nine species was estimated using MCMCtree in PAML (35) with the options “independent rates” and “GTR” model. A Markov chain Monte Carlo analysis was run for 10,000 generations, using a burn-in of 1,000 iterations. Three calibration points were applied based on a recent study using 53 plastomes of Brassicales: A. arabicum and other crucifers divergence time (29.0 to 41.8 Mya), core Brassicaceae origination time (21.3 to 29.8 Mya), and core Arabidopsis origination time (4.8 to 9.7 Mya) (36).
Gene Family Expansion and Contraction.
We selected four close relatives (A. thaliana, A. lyrata, C. rubella, and C. hirsuta) of C. himalaica to identify orthogroups (gene families). Expansions and contractions of orthologous gene families were determined using CAFÉ v. 4.1 (96). The program uses a birth and death process to model gene gain and loss over a phylogeny. For each significantly expanded and contracted gene family in C. himalaica, functional information was inferred based on its ortholog in A. thaliana. GO enrichment analyses of genes were conducted using web-based agriGO (systemsbiology.cau.edu.cn/agriGOv2) (97) with the singular enrichment analysis method and TAIR10 database. The KOBAS and BlastKOALA software (98) was also used to test the statistical enrichment of genes in KEGG pathways (99).
Identification and Classification of Putative NBS Resistance Genes.
A hidden Markov model search (hmmer.janelia.org) was used to identify NBS-encoding R genes in the C. himalaica genome and four close relatives (A. thaliana, A. lyrata, C. rubella, and C. hirsuta). The sequences were screened using HMMs to search for the Pfam NBS (NB-ARC) family PF00931 domain (pfam.xfam.org/). Pfam HMM searches were performed using a TIR model (PF01582) and several LRR models (PF00560, PF07723, PF07725, PF12799, PF13306, PF13516, PF13504, PF13855, and PF08263) to detect TIR domains and LRR motifs in the NBS-encoding amino acid sequences. CC motifs were detected using the COILS prediction program 2.2 (embnet.vital-it.ch/software/COILS_form.html) with a p-score cutoff of 0.9.
Gene Family-Based Positive Selection Tests.
For positive selection analyses we also selected the four most closely related species with assembled genomes (A. thaliana, A. lyrata, C. rubella, and C. hirsuta) and C. himalaica to identify orthologs based on our phylogenetic tree. The analysis procedure is similar to the phylogenetic tree analysis. First, we used OrthoMCL (32) to identify homologous gene clusters (orthogroups) among the five genomes. OrthoMCL was run with an E-value cutoff of 1E-15 and an inflation parameter of 1.5 due to the close genetic relationship between five relatives. Orthogroups with single-copy genes shared by all five genomes were retained for further analyses. Multiple sequence alignment was performed for each orthogroup using MUSCLE v. 3.8.31 (97) with default parameters. Poorly aligned regions were further trimmed using the trimAl v. 1.4 software (100) with the parameter “-gt 0.8 -st 0.001”. Maximum likelihood trees were generated using RAxML v7.0.4 (101) with the PROTCATJTT model. To calculate the nonsynonymous (Ka) and synonymous (Ks) substitution rates between pairs of orthogroups, we reverse-translated amino acid alignments to the corresponding codon-based nucleotide alignments using PAL2NAL (102). To increase the power of tests for positive selection, we applied the branch-site model (103) implemented in codeml program (35) to estimate the dN/dS substitution rates (ω value). We also deleted all gaps (clean data = 1) from the alignments to lower the effect of ambiguous bases on the inference of positive selection. A foreground branch was specified as the clade of C. himalaica. A likelihood ratio test was conducted to determine whether positive selection is operating in the foreground branch. In this study, PSGs were inferred if the P value was less than 0.01. The functional annotation of PSGs in C. himalaica was also conducted using the same approach and the same P-value cutoff with the gene family expansion and contraction analysis.
S-Locus Structure and Self-Fertilization of C. himalaica.
Known S-haplogroups from Arabidopsis and Capsella were used as query to search the genome assembly of C. himalaica (70, 104) using BLASTP. We manually annotated both flanking genes of the S-locus on scaffold 29: the ARK3 gene and U-box gene. However, no homologs were detected in our search for the SRK gene. Using the standard BWA-mem (v. 0.7.17) (83) and Samtools (v. 1.6) (105) pipeline, we mapped short reads (450-bp insert size library) of C. himalaica to the combined reference of C. himalaica genome and A. halleri S15 haplogroup of S-locus (70) and then manually explored the coverage of the SRK region of the S15 haplogroup using IGV (v. 2.4.13) (106). The coding sequence of the predicted SCR gene (ID = AT4G22105.1_Ath-D2 on scaffold29:1034290-1034749), together with SCR sequences from A. thaliana, C. rubella, C. grandiflora, and C. hirsuta, was added to the curated alignment of the publicly available SCR genes kindly provided by Vincent Castric (Unité Evo-Eco-Paléo, CNRS/Université de Lille 1, Villeneuve d’Ascq, France) from A. halleri and A. lyrata using MUSCLE (v. 3.8.31) (97), and a maximum likelihood tree with 500 bootstrap replicates was constructed using MEGAX (107) and visualized using Jalview2 (108). A comparative structure plot of C. himalaica and A. halleri S15 S-loci (Fig. 4B) was constructed using the R library genoPlotR (109).
Supplementary Material
Acknowledgments
We dearly cherish the memory of our respected mentor and friend Prof. Yang Zhong at Fudan University for his full support to this project. We thank Takashi Tsuchimatsu and Vincent Castric for their feedback on the S-locus part. This work was supported by National Natural Science Foundation of China Grants 31770408, 31590823, 31760082, 31760127, U1802232, and 91131901 (to T.Z., H.S., Q.Q., and L.Q.), National Key R & D Program of China Grant 2017YF0505200 (to H.S.), the Strategic Priority Research Program of Chinese Academy of Sciences Grant XDA 20050203 (to H.S.), National High Technology Research and Development Program of China Grant 2014AA020528 (to T.Z.), the Chinese Academy of Sciences “Light of West China” Program (J.H.), and European Union Seventh Framework Programme Grant FP7/2007-2013 under European Research Council Advanced Grant Agreement 322739 – DOUBLEUP (to Y.V.d.P.). P.Y.N. is a postdoctoral fellow of the Research Foundation–Flanders (12S9618N).
Footnotes
The authors declare no conflict of interest.
Data deposition: The raw genomic reads generated in this study have been deposited in the NCBI Sequence Read Archive (BioProject PRJNA521295).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817580116/-/DCSupplemental.
References
- 1.Xing Y, Ree RH. Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc Natl Acad Sci USA. 2017;114:E3444–E3451. doi: 10.1073/pnas.1616063114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu XD, Dong BW. Influence of the Tibetan Plateau uplift on the Asian monsoon-arid environment evolution. Chin Sci Bull. 2013;58:4277–4291. [Google Scholar]
- 3.Qiao Q, et al. The genome and transcriptome of Trichormus sp. NMC-1: Insights into adaptation to extreme environments on the Qinghai-Tibet Plateau. Sci Rep. 2016;6:29404. doi: 10.1038/srep29404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiao Q, et al. Transcriptome sequencing of Crucihimalaya himalaica (Brassicaceae) reveals how Arabidopsis close relative adapt to the Qinghai-Tibet Plateau. Sci Rep. 2016;6:21729. doi: 10.1038/srep21729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simonson TS. Altitude adaptation: A glimpse through various lenses. High Alt Med Biol. 2015;16:125–137. doi: 10.1089/ham.2015.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng X, et al. The draft genome of Tibetan hulless barley reveals adaptive patterns to the high stressful Tibetan Plateau. Proc Natl Acad Sci USA. 2015;112:1095–1100. doi: 10.1073/pnas.1423628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koenig D, Weigel D. Beyond the thale: Comparative genomics and genetics of Arabidopsis relatives. Nat Rev Genet. 2015;16:285–298. doi: 10.1038/nrg3883. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell-Olds T. Arabidopsis thaliana and its wild relatives: A model system for ecology and evolution. Trends Ecol Evol. 2001;16:693–700. [Google Scholar]
- 9.Hall AE, Fiebig A, Preuss D. Beyond the Arabidopsis genome: Opportunities for comparative genomics. Plant Physiol. 2002;129:1439–1447. doi: 10.1104/pp.004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Shehbaz IA, O’Kane SL, Price RA. Generic placement of species excluded from Arabidopsis (Brassicaceae) Novon. 1999;9:296–307. [Google Scholar]
- 11.Hohmann N, Wolf EM, Lysak MA, Koch MA. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell. 2015;27:2770–2784. doi: 10.1105/tpc.15.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107:18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Fang X. Uplift of the Tibetan Plateau and environmental changes. Chin Sci Bull. 1999;44:2117–2124. [Google Scholar]
- 14.Liu B, et al. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Quant Biol. 2013;35:62–67. [Google Scholar]
- 15.Butler J, et al. ALLPATHS: De novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18:810–820. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.English AC, et al. Mind the gap: Upgrading genomes with Pacific biosciences RS long-read sequencing technology. PLoS One. 2012;7:e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 18.Slotte T, et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet. 2013;45:831–835. doi: 10.1038/ng.2669. [DOI] [PubMed] [Google Scholar]
- 19.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 20.Long Q, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013;45:884–890. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 22.Bennetzen JL. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica. 2002;115:29–36. doi: 10.1023/a:1016015913350. [DOI] [PubMed] [Google Scholar]
- 23.Iorizzo M, et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat Genet. 2016;48:657–666. doi: 10.1038/ng.3565. [DOI] [PubMed] [Google Scholar]
- 24.Hu TT, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warren IA, et al. Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates. Chromosome Res. 2015;23:505–531. doi: 10.1007/s10577-015-9493-5. [DOI] [PubMed] [Google Scholar]
- 26.Oliver KR, McComb JA, Greene WK. Transposable elements: Powerful contributors to angiosperm evolution and diversity. Genome Biol Evol. 2013;5:1886–1901. doi: 10.1093/gbe/evt141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc Natl Acad Sci USA. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auvinet J, et al. Mobilization of retrotransposons as a cause of chromosomal diversification and rapid speciation: The case for the Antarctic teleost genus Trematomus. BMC Genomics. 2018;19:339. doi: 10.1186/s12864-018-4714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daccord N, et al. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet. 2017;49:1099–1106. doi: 10.1038/ng.3886. [DOI] [PubMed] [Google Scholar]
- 30.Wu Z, Lu A, Tang Y, Chen Z, Li D. The Families and Genera of Angiosperms in China. Science; Beijing: 2003. [Google Scholar]
- 31.Wu Z, Sun H, Zhou Z, Li D, Peng H. Floristics of Seed Plants from China. Science; Beijing: 2011. [Google Scholar]
- 32.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beilstein MA, Al-Shehbaz IA, Kellogg EA. Brassicaceae phylogeny and trichome evolution. Am J Bot. 2006;93:607–619. doi: 10.3732/ajb.93.4.607. [DOI] [PubMed] [Google Scholar]
- 34.Huang CH, et al. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol Biol Evol. 2016;33:394–412. doi: 10.1093/molbev/msv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 36.Guo X, et al. Plastome phylogeny and early diversification of Brassicaceae. BMC Genomics. 2017;18:176. doi: 10.1186/s12864-017-3555-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Al-Shehbaz IA, O’Kane SL. Transberingia, a new generic name replacing the Illegitimate Beringia (Brassicaceae) Novon. 2003;13:396. [Google Scholar]
- 38.German DA. Six new synonyms in the central Asian Cruciferae (Brassicaceae) Nord J Bot. 2008;26:38–40. [Google Scholar]
- 39.Dassanayake M, et al. The genome of the extremophile crucifer Thellungiella parvula. Nat Genet. 2011;43:913–918. doi: 10.1038/ng.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudmant PH, et al. 1000 Genomes Project Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allan R. Vaccinia tricks Toll. Genome Biol. 2000;1:reports0079. [Google Scholar]
- 42.McDowell JM, et al. Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, et al. Unique evolutionary pattern of numbers of gramineous NBS-LRR genes. Mol Genet Genomics. 2010;283:427–438. doi: 10.1007/s00438-010-0527-6. [DOI] [PubMed] [Google Scholar]
- 44.Zhang YM, et al. Uncovering the dynamic evolution of nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes in Brassicaceae. J Integr Plant Biol. 2016;58:165–177. doi: 10.1111/jipb.12365. [DOI] [PubMed] [Google Scholar]
- 45.Zhang XJ, Yao TD, Ma XJ, Wang NL. Microorganisms in a high altitude glacier ice in Tibet. Folia Microbiol (Praha) 2002;47:241–245. doi: 10.1007/BF02817645. [DOI] [PubMed] [Google Scholar]
- 46.Qu Y, et al. Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat Commun. 2013;4:2071. doi: 10.1038/ncomms3071. [DOI] [PubMed] [Google Scholar]
- 47.Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 48.Frohnmeyer H, Staiger D. Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 2003;133:1420–1428. doi: 10.1104/pp.103.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reed E, Larkins T, Chau C, Figg W. DNA repair: ERCC1, nucleotide excision repair, and platinum resistance. In: Rudek MA, Chau CH, Figg WD, McLeod HL, editors. Handbook of Anticancer Pharmacokinetics and Pharmacodynamics, Cancer Drug Discovery and Development. Springer; New York: 2014. pp. 333–349. [Google Scholar]
- 50.Ge RL, et al. Draft genome sequence of the Tibetan antelope. Nat Commun. 2013;4:1858. doi: 10.1038/ncomms2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Q, et al. Genome resequencing identifies unique adaptations of Tibetan chickens to hypoxia and high-dose ultraviolet radiation in high-altitude environments. Genome Biol Evol. 2016;8:765–776. doi: 10.1093/gbe/evw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J-T, et al. Comparative genomic investigation of high-elevation adaptation in ectothermic snakes. Proc Natl Acad Sci USA. 2018;115:8406–8411. doi: 10.1073/pnas.1805348115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyzenga WJ, Stone SL. Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot. 2012;63:599–616. doi: 10.1093/jxb/err310. [DOI] [PubMed] [Google Scholar]
- 54.Yanagawa Y, et al. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 2004;18:2172–2181. doi: 10.1101/gad.1229504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki G, Yanagawa Y, Kwok SF, Matsui M, Deng X-W. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 2002;16:554–559. doi: 10.1101/gad.964602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanyuka K, et al. Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant J. 2003;35:57–70. doi: 10.1046/j.1365-313x.2003.01779.x. [DOI] [PubMed] [Google Scholar]
- 57.Xu S, et al. Genome-wide convergence during evolution of mangroves from woody plants. Mol Biol Evol. 2017;34:1008–1015. doi: 10.1093/molbev/msw277. [DOI] [PubMed] [Google Scholar]
- 58.Wollenberg AC, Strasser B, Cerdán PD, Amasino RM. Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol. 2008;148:1681–1694. doi: 10.1104/pp.108.125468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iñigo S, Alvarez MJ, Strasser B, Califano A, Cerdán PD. PFT1, the MED25 subunit of the plant mediator complex, promotes flowering through CONSTANS dependent and independent mechanisms in Arabidopsis. Plant J. 2012;69:601–612. doi: 10.1111/j.1365-313X.2011.04815.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhang L, Turkington R, Tang Y. Flowering and fruiting phenology of 24 plant species on the north slope of Mt. Qomolangma (Mt. Everest) J Mt Sci. 2010;7:45–54. [Google Scholar]
- 61.Barrett SC. The evolution of plant sexual diversity. Nat Rev Genet. 2002;3:274–284. doi: 10.1038/nrg776. [DOI] [PubMed] [Google Scholar]
- 62.Xing S, Li M, Liu P. Evolution of S-domain receptor-like kinases in land plants and origination of S-locus receptor kinases in Brassicaceae. BMC Evol Biol. 2013;13:69. doi: 10.1186/1471-2148-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barringer BC. Polyploidy and self-fertilization in flowering plants. Am J Bot. 2007;94:1527–1533. doi: 10.3732/ajb.94.9.1527. [DOI] [PubMed] [Google Scholar]
- 64.Goodwillie C, Kalisz S, Eckert CG. The evolutionary enigma of mixed mating systems in plants: Occurrence, theoretical explanations, and empirical evidence. Annu Rev Ecol Evol Syst. 2005;36:47–79. [Google Scholar]
- 65.Takayama S, Isogai A. Self-incompatibility in plants. Annu Rev Plant Biol. 2005;56:467–489. doi: 10.1146/annurev.arplant.56.032604.144249. [DOI] [PubMed] [Google Scholar]
- 66.Tsuchimatsu T, et al. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature. 2010;464:1342–1346. doi: 10.1038/nature08927. [DOI] [PubMed] [Google Scholar]
- 67.Novikova PY, et al. Genome sequencing reveals the origin of the allotetraploid Arabidopsis suecica. Mol Biol Evol. 2017;34:957–968. doi: 10.1093/molbev/msw299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boggs NA, Nasrallah JB, Nasrallah ME. Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000426. doi: 10.1371/journal.pgen.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang C, et al. The evolution of selfing in Arabidopsis thaliana. Science. 2007;317:1070–1072. doi: 10.1126/science.1143153. [DOI] [PubMed] [Google Scholar]
- 70.Goubet PM, et al. Contrasted patterns of molecular evolution in dominant and recessive self-incompatibility haplotypes in Arabidopsis. PLoS Genet. 2012;8:e1002495. doi: 10.1371/journal.pgen.1002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo YL, Zhao X, Lanz C, Weigel D. Evolution of the S-locus region in Arabidopsis relatives. Plant Physiol. 2011;157:937–946. doi: 10.1104/pp.111.174912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Castric V, Vekemans X. Plant self-incompatibility in natural populations: A critical assessment of recent theoretical and empirical advances. Mol Ecol. 2004;13:2873–2889. doi: 10.1111/j.1365-294X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- 73.Llaurens V, et al. Does frequency-dependent selection with complex dominance interactions accurately predict allelic frequencies at the self-incompatibility locus in Arabidopsis halleri? Evolution. 2008;62:2545–2557. doi: 10.1111/j.1558-5646.2008.00469.x. [DOI] [PubMed] [Google Scholar]
- 74.Kusaba M, et al. Self-incompatibility in the genus Arabidopsis: Characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell. 2001;13:627–643. [PMC free article] [PubMed] [Google Scholar]
- 75.Garcíacamacho R, Totland Ø. Pollen limitation in the alpine: A meta-analysis. Arct Antarct Alp Res. 2009;41:103–111. [Google Scholar]
- 76.Bingham RA, Orthner AR. Efficient pollination of alpine plants. Nature. 1998;391:238–239. [Google Scholar]
- 77.Kalisz S, Vogler DW. Benefits of autonomous selfing under unpredictable pollinator environments. Ecology. 2003;84:2928–2942. [Google Scholar]
- 78.Laenen B, et al. Demography and mating system shape the genome-wide impact of purifying selection in Arabis alpina. Proc Natl Acad Sci USA. 2018;115:816–821. doi: 10.1073/pnas.1707492115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tedder A, Ansell SW, Lao X, Vogel JC, Mable BK. Sporophytic self-incompatibility genes and mating system variation in Arabis alpina. Ann Bot. 2011;108:699–713. doi: 10.1093/aob/mcr157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peng DL, Zhang ZQ, Xu B, Li ZM. Patterns of flower morphology and sexual systems in the subnival belt of the Hengduan Mountains, SW China. Alp Bot. 2012;122:65–73. [Google Scholar]
- 81.Zhang ZQ, Li QJ. Autonomous selfing provides reproductive assurance in an alpine ginger Roscoea schneideriana (Zingiberaceae) Ann Bot. 2008;102:531–538. doi: 10.1093/aob/mcn136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 83.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Haas BJ. Analysis of alternative splicing in plants with bioinformatics tools. Curr Top Microbiol Immunol. 2008;326:17–37. doi: 10.1007/978-3-540-76776-3_2. [DOI] [PubMed] [Google Scholar]
- 85.Guo S, et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet. 2013;45:51–58. doi: 10.1038/ng.2470. [DOI] [PubMed] [Google Scholar]
- 86.Edgar RC, Myers EW. PILER: Identification and classification of genomic repeats. Bioinformatics. 2005;21:i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- 87.Xu Z, Wang H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 89.Ellinghaus D, Kurtz S, Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics. 2008;9:18. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Steinbiss S, Willhoeft U, Gremme G, Kurtz S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 2009;37:7002–7013. doi: 10.1093/nar/gkp759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Exposito-Alonso M, et al. The rate and potential relevance of new mutations in a colonizing plant lineage. PLoS Genet. 2018;14:e1007155. doi: 10.1371/journal.pgen.1007155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harris RS. 2007. Improved pairwise alignment of genomic DNA. PhD dissertation (Pennsylvania State Univ, State College, PA)
- 94.Wang Y, et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 97.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Xie C, et al. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stamatakis A, Ludwig T, Meier H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 102.Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 104.Bachmann JA, Tedder A, Laenen B, Steige KA, Slotte T. Targeted long-read sequencing of a locus under long-term balancing selection in Capsella. G3 (Bethesda) 2018;8:1327–1333. doi: 10.1534/g3.117.300467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guy L, Kultima JR, Andersson SG. genoPlotR: Comparative gene and genome visualization in R. Bioinformatics. 2010;26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.