Significance
Nonreceptor tyrosine kinases are major players in cell signaling. Among them, focal adhesion kinase (FAK) is the key integrator of signals from growth factors and cell adhesion. In cancer, FAK is frequently overexpressed, and by promoting adhesion to the tumor stroma and ECM, FAK provides important signals for tumor invasion and metastasis. Although autoinhibitory mechanisms have previously been described, FAK activation in response to force generated by ECM attachment is currently not understood. Here, we use experimental and computational approaches to demonstrate how mechanical forces can induce conformational changes in FAK that result in activation. This mechanistic insight enables the design of alternative strategies for the discovery of potential anticancer drugs that inhibit both catalytic and scaffolding functions of FAK.
Keywords: atomic force microscopy, mechanobiology, focal adhesion signaling, protein kinase regulation, single-molecule force spectroscopy
Abstract
Focal adhesion kinase (FAK) is a key signaling molecule regulating cell adhesion, migration, and survival. FAK localizes into focal adhesion complexes formed at the cytoplasmic side of cell attachment to the ECM and is activated after force generation via actomyosin fibers attached to this complex. The mechanism of translating mechanical force into a biochemical signal is not understood, and it is not clear whether FAK is activated directly by force or downstream to the force signal. We use experimental and computational single-molecule force spectroscopy to probe the mechanical properties of FAK and examine whether force can trigger activation by inducing conformational changes in FAK. By comparison with an open and active mutant of FAK, we are able to assign mechanoactivation to an initial rupture event in the low-force range. This activation event occurs before FAK unfolding at forces within the native range in focal adhesions. We are also able to assign all subsequent peaks in the force landscape to partial unfolding of FAK modules. We show that binding of ATP stabilizes the kinase domain, thereby altering the unfolding hierarchy. Using all-atom molecular dynamics simulations, we identify intermediates along the unfolding pathway, which provide buffering to allow extension of FAK in focal adhesions without compromising functionality. Our findings strongly support that forces in focal adhesions applied to FAK via known interactions can induce conformational changes, which in turn, trigger focal adhesion signaling.
Focal adhesions (FAs) are dense molecular assemblies that anchor cells via integrin receptors to the ECM and intracellularly connect to actin stress fibers (1). FAs not only form a structural link between the cell and its surroundings but also, are important for exchanging mechanical force cues and regulatory signals (2, 3). A key regulator in FAs is the nonreceptor tyrosine kinase focal adhesion kinase (FAK) that triggers FA signals on cell adhesion to the ECM. Apart from its function as a signaling kinase, it acts as a scaffolding hub for diverse interaction partners in FAs. Via its interactions and embedding in the FA complex, FAK is exposed to forces arising from inside or outside the cell. Cell-based studies show that increased forces exerted on FAs result in activation of FAK (4–6). Moreover, FAK seems to have an important force sensing role, since FAK is required for cells to respond to externally applied forces or to migrate toward stiffer substrates, which allows generation of higher forces in FAs (7). However, current studies lack a clear hint of whether FAK represents a first responder to force or is indirectly force activated by downstream signaling. An activation mechanism based on the direct application of mechanical force on an enzyme was previously described for the mammalian titin kinase, the related twitchin kinase in nematode (8, 9), and the smooth muscle myosin light-chain kinase (10), which are located in the load-bearing environment of the muscle sarcomeres. However, no nonmuscle enzyme was shown to be directly activated by mechanical force yet.
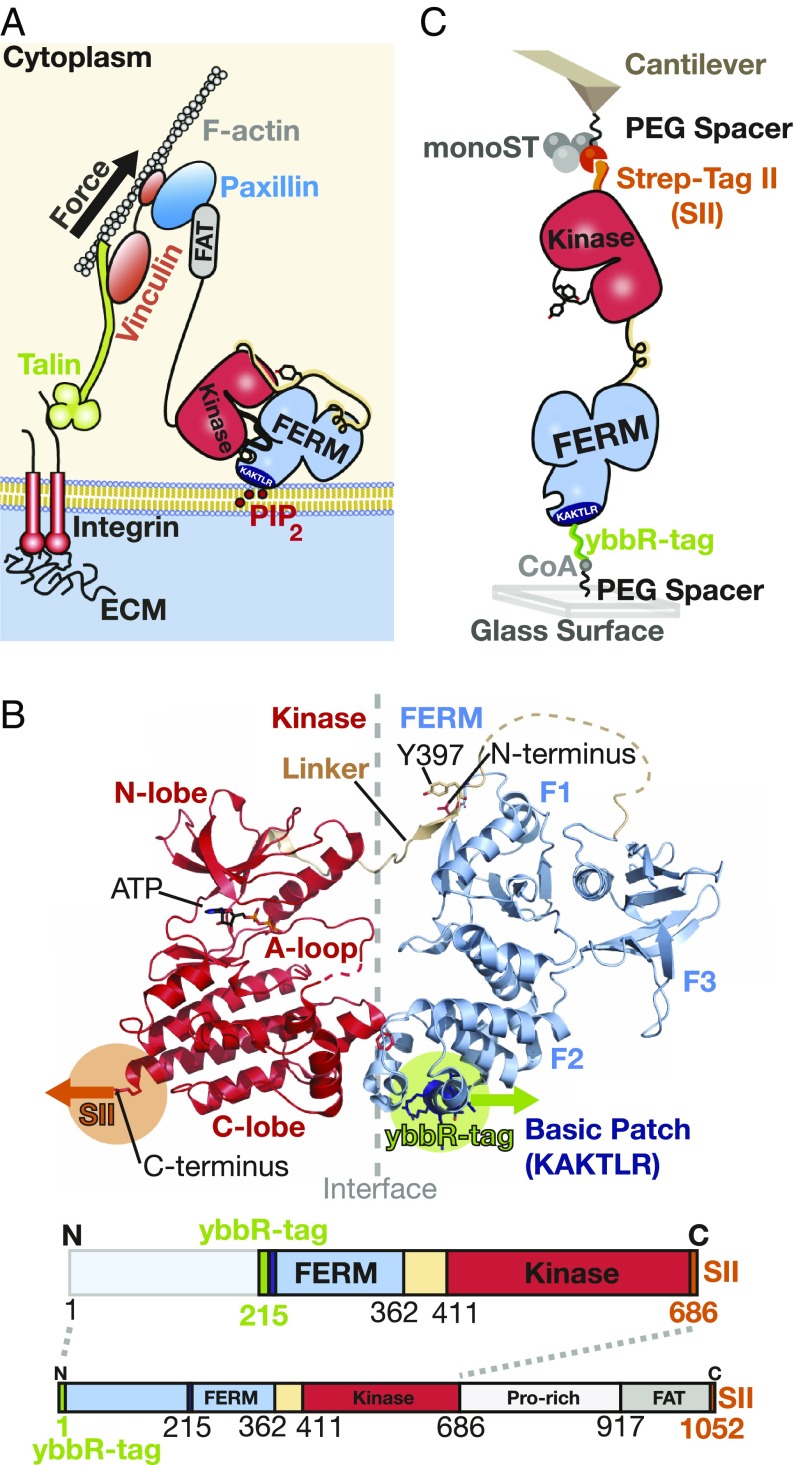
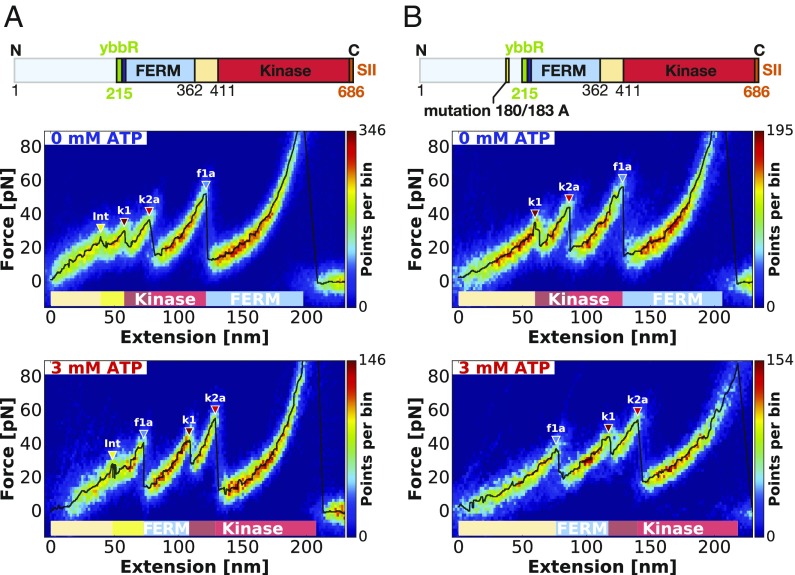
FAK is a multidomain protein that is subdivided into three major domains. The central catalytic kinase domain is flanked by an N-terminal 4.1 protein, Ezrin, Radixin, Moesin homology domain (FERM) and a C-terminal focal adhesion targeting (FAT) domain (Fig. 1A). In the basal state, the FERM and kinase domains interact to form a closed and autoinhibited conformation, where the active site and several regulatory phosphorylation sites are sequestered (Fig. 1B) (11). On integrin-mediated cell adhesion, the FAT domain targets FAK into FAs. Super-resolution optical microscopy has localized FAK to an integrin signaling layer in FAs in close proximity to the plasma membrane (12). Accordingly, the FERM domain contains a stretch of basic residues forming a basic patch that interacts with phosphatidylinositol 4,5-bisphosphate (PIP2) (13, 14), which is clustered in the cell membrane at FA sites (15, 16). Activation of FAK can be initiated by an orchestrated process starting with PIP2 binding to the basic patch, resulting in exposure of a linker region containing the autophosphorylation site (14). After autophosphorylation, this site becomes a docking site for the Src kinase, which in turn, phosphorylates tyrosine residues in the activation loop of the FAK kinase. This results in full activation of FAK. The last step of Src phosphorylation is strongly promoted by prior separation of FERM and kinase domains (11). Here, we test the hypothesis that force is a key stimulus driving domain separation for FAK activation. With the N-terminal domain docked to the cell membrane and the C-terminal FAT domain tethered via paxillin and structural FA components to the actin cytoskeleton, FAK is likely exposed to stretching forces in FAs. Force-induced domain opening and activation were indeed already proposed and supported by molecular dynamics (MD) simulations, showing that forces applied to FAK result in rupture of autoinhibitory interactions and exposure of Src phosphorylation sites in the FAK activation loop (17). Furthermore and consistent with this hypothesis, FAK is known to undergo domain opening when localized to FAs in cells (13). Additionally, mutational disruption of autoinhibitory interactions and resulting FAK opening has experimentally been shown to strongly promote phosphorylation by Src (11).
Fig. 1.
Mimicking the attachment of FAK in FAs in the AFM force spectroscopy assay. (A) Schematic depiction of an FA connecting the ECM with the actin cytoskeleton. FAK is anchored to PIP2 in the lipid membrane via its N-terminal basic patch (amino acid sequence: “KAKTLR”; dark blue). On the other end, FAK is attached with its C-terminal FAT (gray) to paxillin. (B) The structure of autoinhibited FAK (Protein Data Bank ID codes 2J0J and 2IJM) is shown with placement of the affinity tags for AFM. The FERM domain directly attaches to the kinase domain and blocks the active site and phosphorylation of the activation loop (A loop). On the bottom, the schematic sequence of FAK used in the experiments (FK-FAK, residues 1–686) is shown with the placement of the 11-aa ybbR-tag in green for site-specific immobilization to CoA. Additionally, the SII in orange and the lipid binding basic patch in dark blue are depicted. The amino acid sequence from residues 1 to 215 is not stressed directly by force during the experiment and is, therefore, colored in light blue. The full-length FAK (1–1,052) with the C-terminal region lacking in FK-FAK was used for control measurements (SI Appendix, Fig. S1) and is depicted here to show the complete FAK protein. (C) The scheme illustrates the measurement setup with immobilized FAK on a PEG functionalized glass surface and monoST on the cantilever. The ybbR-tag ensures site-specific covalent attachment to the surface via an Sfp-catalyzed reaction with CoA connected to a PEG spacer. At the C terminus, FK-FAK is equipped with an SII for reversible tethering to monoST on the cantilever.
In this study, we aim to obtain insight into the force response of FAK on a single-molecule level by means of atomic force microscopy (AFM)-based force spectroscopy in conjunction with force probe molecular dynamics (FPMD) simulations. We are able to measure highly reproducible force profiles recorded during FAK stretching and map-detected force events to structural features in FAK. By adapting experimental conditions, we are able to identify interface rupture of FERM and kinase domains as a discrete force peak. This is verified by control force profiles of FAK mutants lacking autoinhibitory FERM–kinase interactions and comparison with MD simulations. We conclude that the inhibitory interface ruptures at forces of around 25 pN for pulling speeds of 12,800 nm/s (or at smaller forces for slower pulling)—significantly before any domain unfolding—and that functionally important regions in FAK start to unfold at an extension of about 50 nm. Hence, we demonstrate that force activation of FAK can occur at physiological forces in FAs and that FAK maintains an active state when exposed to extensions expected to occur in FAs.
Results
Relative Mechanical Stabilities of FERM and Kinase Are ATP Dependent.
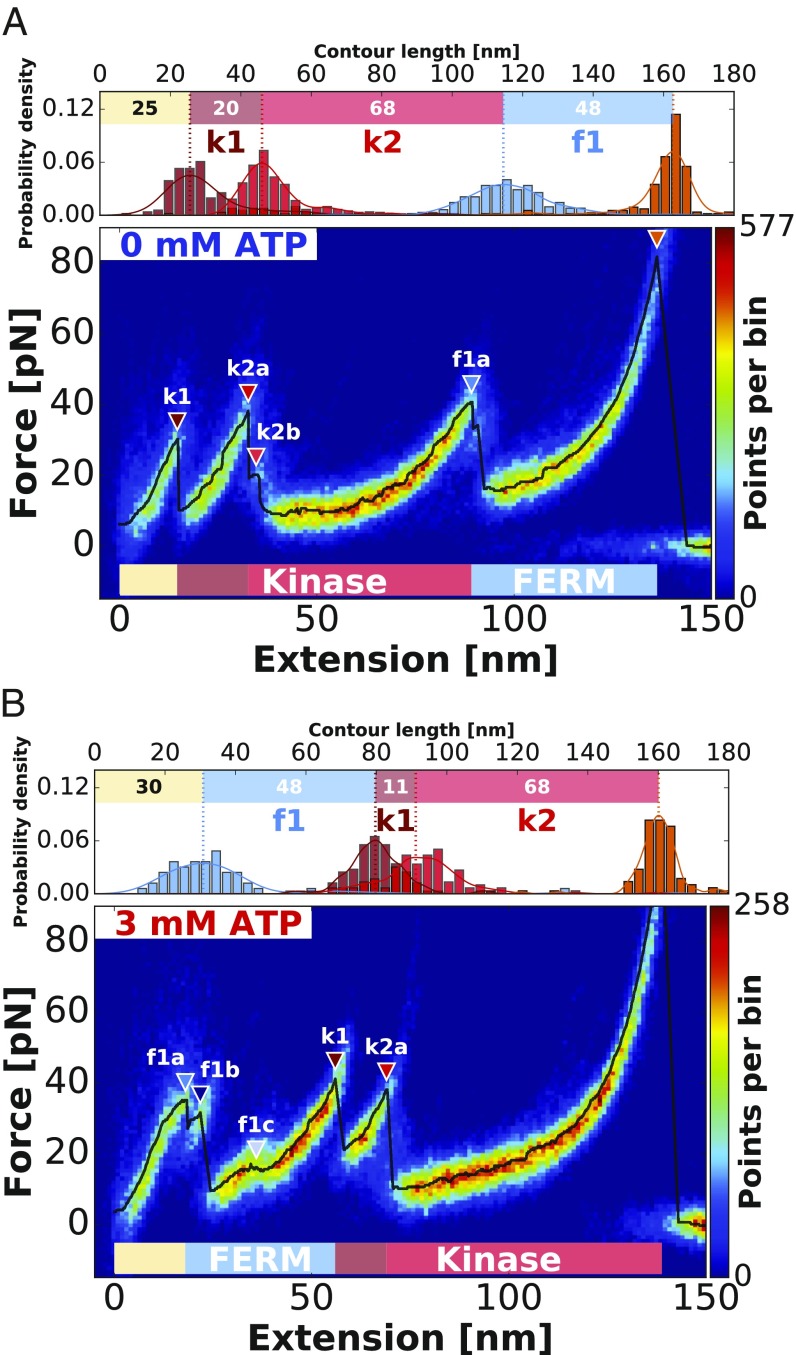
In this study, we record force–distance traces for FAK containing FERM and kinase domain (residues 1–686; referred to hereafter as FK-FAK) (Fig. 1C) attached via short PEG linkers (425.39 Da) and pulling speeds of 800 nm/s. The curves show force peaks between 20 and 50 pN, with reliably recurring unfolding features and a final Strep-Tag II (SII) rupture at around 70 pN (Fig. 2). Force application guides the protein through a sequence of conformational states obeying a strict hierarchy as suggested by the heatmap, and the most probable unfolding curve in black (Fig. 2). All detected peaks above the FWHM of the noise level are highlighted with a colored triangle (the process is described in Data Analysis) (Fig. 2). These unfolding peaks indicate transitions between these conformational states and mark the unfolding of stable structural modules within the protein structure leading to a release of previously hidden contour lengths. To determine contour length increments of the polypeptide chain unfolded for each peak, we fitted the data before each rupture event with the worm-like chain model (18, 19). We determine contour length increments of 20 nm (k1), 68 nm (k2), and 48 nm (f1) for FK-FAK (Fig. 2A and Table 1).
Fig. 2.
Assignment of force peaks to structural modules of FERM and kinase domain (residues 1–686; FK-FAK) using AFM unfolding patterns. (A) The heatmap obtained from an overlay of 224 curves shows the typical unfolding pattern of FK-FAK in the absence of ATP, revealing low-force unfolding below 50 pN and final SII rupture at around 70 pN. In Upper, the increments between identified peaks are depicted, allowing assignment of the rupture events to structural modules in the crystal structure. (B) Depiction of the FK-FAK unfolding pattern by an overlay of 115 curves in the presence of 3 mM ATP. Both plots are created from one dataset recorded with the same cantilever, and therefore, they are directly comparable in absolute force. Although the unfolding pattern in B looks different from the one in A, the increments stay conserved. Comparison of the two conditions shows that the increment of f1 is swapping its position with k1 and k2 on ATP binding. This can be attributed to stabilization of the kinase domain on addition of ATP, shifting peak k1 to higher forces and thereby, changing the force hierarchy between FERM and kinase. (A and B) The black lines show the most probable unfolding patterns with all detected peaks above the FWHM of the noise level (described in Data Analysis) highlighted with colored triangles according to their assigned domain.
Table 1.
All measured contour length increments of the various FAK constructs used in this study
| Protein segment | f1 | k1 | k2 |
| FK-FAK −ATP | 48 | 20 | 68 |
| FK-FAK +ATP | 48 | 11 | 68 |
| FERM −ATP | 46 | ||
| f1a/f1b/f1c | 7/25/14 | ||
| FERM +ATP | 46 | ||
| f1a/f1b/f1c | 7/21/18 | ||
| Kinase −ATP | 15 | 66 | |
| Kinase +ATP | 14 | 66 |
The contour length increment values are given in nanometers and were determined as most probable values from a KDE.
Additionally, FK-FAK was stretched in presence of 3 mM ATP to probe the effect of ATP binding to the kinase domain. Although the presence of ATP substantially changes the unfolding pattern of FK-FAK (as observed in Fig. 2), the identified increments remain remarkably conserved [48 nm (f1ATP), 11 nm (k1ATP), and 68 nm (k2ATP)] (Fig. 2B and Table 1). This suggests that the unfolding hierarchy changes on ATP addition as increment f1 moves from the back to the beginning of the curve (Fig. 2). The most probable rupture force of peak k1 (Fig. 2) significantly increases on ATP binding, corroborating its association with the kinase domain (20, 21). This is probably due to a stabilization of the part of the kinase domain involved in ATP binding, thereby yielding higher forces for peak k1 and consequently, changing the force hierarchy between FERM and kinase domain. The kinase domain maintains the internal sequence of unfolding (k1 then k2a) but unfolds after the FERM domain in the presence of ATP. Their sums of all contour lengths (88 and 79 nm, respectively) agree with expected values for the FAK kinase domain [91 nm for residues 422–686 (19, 22), subtracting 5 nm to account for the initial distance between residues 422 and 686 in the folded domains according to the crystal structure]; hence, we assign k1 and k2 to the kinase domain. The increment f1 is assigned to FERM unfolding, and a contour length of 48 nm reproduces an expected length of 50 nm for folded FERM from residues 216–362 plus an additional 12 residues for the inserted ybbR-tag, subtracting an initial folded distance of 6 nm. We, therefore, conclude that stabilizing the kinase domain by ATP binding results in a reversed unfolding sequence, with the kinase unfolding first in absence of ATP but as the last event in the presence of ATP. All determined contour length increments are in good agreement with defined structural features in the crystal structure of FK-FAK in Fig. 1B (11).
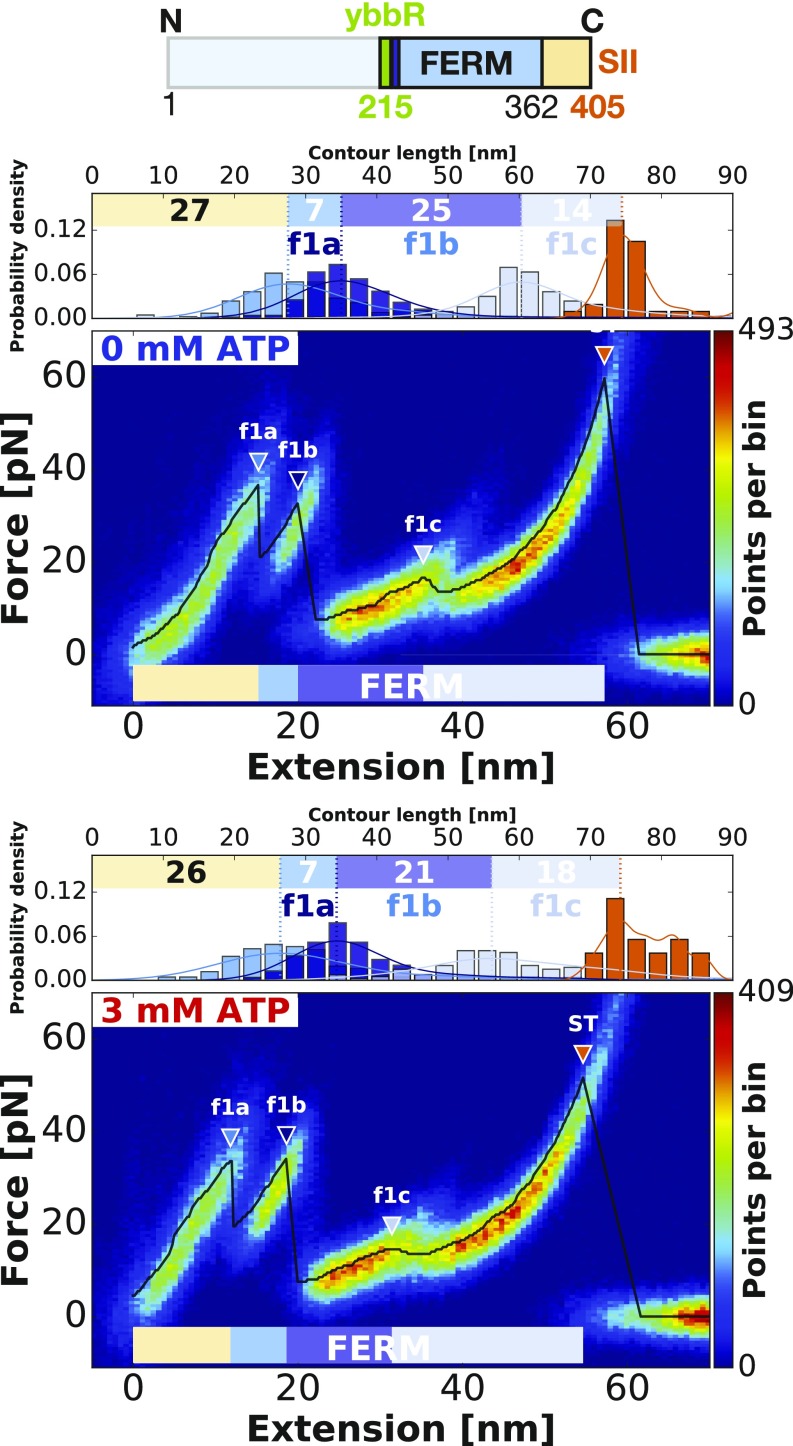
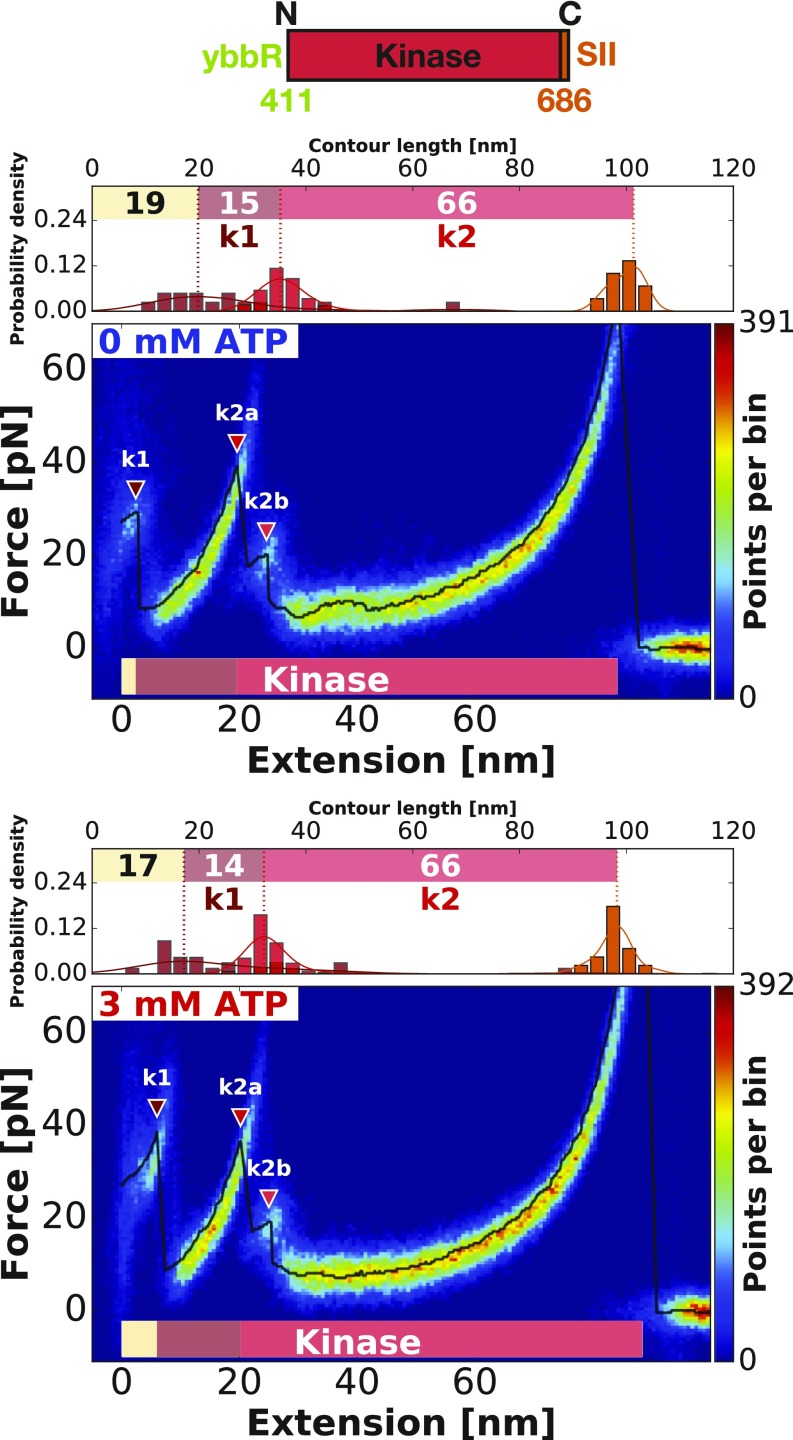
To further validate our assignment of unfolding increments, we used single-domain constructs with only the isolated FERM or kinase domains. The curves were analyzed as previously described (Figs. 3 and 4). The data confirm our assignment of the structural modules, with determined contour length increments matching well with the ones assigned in FK-FAK plots (Table 1). In agreement with our assignment in FK-FAK, the isolated kinase likewise unfolds in two main modules (k1 and k2a). Furthermore, we identify additional unfolding intermediates that were previously hidden in the noise. For the FERM domain, we now resolve three peaks with contour lengths 7 nm (f1a), 25 nm (f1b), and 14 nm (f1c). For the kinase domain, k2a is followed by another peak k2b, but it is too short to determine a proper contour length increment. Close inspection reveals that these intermediate states (k2b, f1b, f1c) are in fact also present in FK-FAK (Fig. 2). They can be detected in the beginning of the unfolding curve where the total free length is still short enough, leading to higher loading rates (as property of the worm-like chain model) and consequently, to higher forces, thereby allowing us to resolve more subtle peaks. These subtle peaks are, however, too small (and their extension is too short) to determine their contour length accurately. All determined contour length increments are summarized in Table 1.
Fig. 3.
Verification of the assignments of structural modules by probing single FERM (residues 1–405) domains. The heatmaps show single-FERM unfolding events without (Upper) and in the presence of 3 mM ATP (Lower). The increments and forces do not change in the presence of ATP as expected. The same peaks detected in detail here (f1b and f1c) can also be found in the FERM domain in Fig. 2B. Due to longer linkers at the end of the unfolding curve and therefore, lower loading rates leading to lower forces, they cannot be detected in Fig. 2A.
Fig. 4.
Verification of the assignments of structural modules by probing single-kinase (residues 411–686) domains. The heatmaps show single-kinase unfolding events without (Upper) and in the presence of 3 mM ATP (Lower). The first peak k1 increases in force by addition of ATP, suggesting a binding event of ATP to the kinase domain. The same peak detected in detail here (k2b) can also be found in the kinase domain in Fig. 2A. Due to longer linkers at the end of the unfolding curve and therefore, lower loading rates leading to lower forces, they cannot be detected in Fig. 2B.
Rupture of FERM–Kinase Interface Precedes Domain Unfolding.
The single-domain constructs demonstrated that subtle peaks can be hidden within prominent peaks. The interface opening between kinase and FERM likely represents such a small peak that is expected to release a contour length of only 20 nm. Assuming physiological activation under stress in vivo, the opening has to occur at relatively small forces and before any other unfolding, since it is shielding the remaining structure. However, since in this region of the force profiles, we do not detect an additional distinct force peak in FK-FAK profiles with a pulling speed of 800 nm/s, we conclude that domain separation may occur anywhere before the first unfolding event (i.e., below 40 pN in this loading rate regime) but likely is hidden in the measurement noise.
Aiming to resolve domain separation, we used higher pulling speeds of 12,800 nm/s to increase force responses and used long PEG (5,000 Da) for FAK attachment to facilitate analysis early in the force profile. This indeed enabled us to resolve an additional force peak at the beginning of the unfolding pattern (yellow triangles in Fig. 5A), where we expect domain separation to occur. To verify that this peak originates from the domain opening, we probed a mutant of FK-FAK (FK-FAKmut) with point mutations at the domain interface (Y180 A, M183 A). These mutations cause FERM and kinase domains to be permanently dissociated (14). As shown in Fig. 5B, this mutant lacks the low-force rupture event, which we identified as the interface opening in the FK-FAK wild-type profile (yellow triangles and Int label in Fig. 5A), but still retains all other unfolding features. As expected, domain dissociation in the FK-FAK wild type precedes any unfolding events. In conclusion, these data indicate that forced domain dissociation and thereby, conformational activation happens before any other subdomain unfolding at a force around 25 pN at pulling speeds of 12,800 nm/s.
Fig. 5.
Resolving the FERM–kinase domain interface rupture by comparing FK-FAK with a permanently open FK-FAKmut—recorded with long PEG (5,000-Da) linkers at a pulling speed of 12,800 nm/s. (A) The heatmap shows the same construct as measured in Fig. 2 but with improved force resolution in the investigated region. This way, an additional subtle peak in the beginning of the curve can be observed highlighted by yellow triangles. This peak is not detected in the permanently open FK-FAKmut construct measured in B. We conclude that this rupture event corresponds to the interface opening between FERM and kinase domain. With measurements in the presence of 3 mM ATP (Bottom), the same behavior is observed, hence showing no association of the rupture event with a protein domain and validating its assignment to interface opening. SI Appendix, Fig. S2 provides additional contour length histograms for the peaks detected in this graph.
MD Simulations Confirm Conformational Activation of FAK.
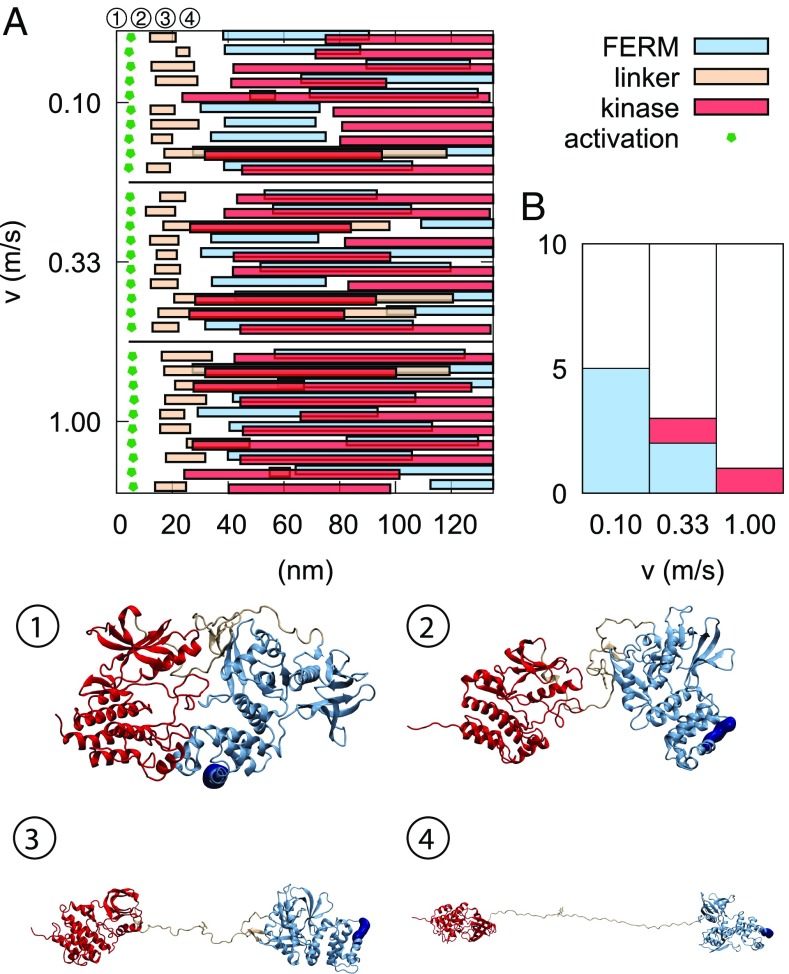
Fig. 6 summarizes our unfolding simulations (detailed unfolding data are given in SI Appendix, Figs. S3–S11). Consistent with previously simulated results (17), the first event is invariably dissociation of the FERM–kinase interface. After this, there is considerable heterogeneity in our data, with most of the simulations featuring both domains being in the process of unfolding. However, it should be considered that pulling speeds in simulations are significantly faster than in AFM experiments, and Fig. 6B shows a clear trend of slower simulations featuring the experimentally observed hierarchy: the FERM domain unfolds before the kinase. We, therefore, restrict the following analysis to these cases (five at 0.1 m/s and two at 0.33 m/s).
Fig. 6.
Domain dissociation and linker detachment precede domain unfolding. (A) Rupture events across the 30 simulations are depicted. The green pentagons show the moment of activation (i.e., FERM–kinase dissociation; measured by a sudden increase in V215–K630 distance), and the three different types of bars show when unfolding of these elements begins and ends (monitored by end-to-end distance changes). (B) Preferred unfolding mechanism as a function of pulling velocity: FERM unfolds first before the kinase (blue), kinase unfolds first before the kinase (red), or a “mixed” unfolding pathway (white). The bars show proportions of the 10 simulations performed at that pulling velocity. Snapshots 1–4 show the process of conformational activation and linker release observed in MD simulations as illustrated by a sample trajectory at 0.1 m/s. In the native state (snapshot 1), the FERM domain is in contact with the kinase. After an initial kinase C-terminal helix unwinding (SI Appendix, Fig. S6), these two domains lose contact (snapshot 2). The interdomain linker gradually detaches and elongates, leading to a release of Tyr397 (snapshot 3). After an additional extension of 12 nm, the linker is fully stretched (snapshot 4). The approximate positions in time of these snapshots are shown in A.
The first force-induced conformational change is the loss of the FERM–kinase interface, giving 10 nm of extension (Fig. 6 and SI Appendix, Table S1). This is followed by another 12-nm extension due to the linker losing contact with the F1 lobe in the FERM domain (Fig. 6B). In agreement with simulations, domain separation of FAK was also experimentally observed as the first event (Fig. 5), and FERM–linker separation, which simulations show to require less force (SI Appendix, Fig. S11) and to occur shortly after domain separation, is not detected in experiments.
The FERM domain unfolding was observed experimentally to occur in three steps: one at 7–8 nm, a second one around 21–25 nm, and a third one around 14–19 nm. In our MD simulations, the FERM domain unfolding happens in two stages: first, lobe F3 unfolding is observed, amounting to 30 nm, and second, lobe F2 stretching is observed, corresponding to an increase of 9 nm. Since the F2 stretching never happens before F3 unfolding in our trajectories, we assign the first two events observed experimentally to F3 unfolding. The F2 stretching contributes to an increase in end-to-end distance of the protein by about 9 nm, which in experiments, amounts to around 13 nm considering the additional loop contained in the handle of the experimental FK-FAK construct. Kinase domain unfolding leads to FAK deactivation independent of the detailed sequence of events. We, therefore, discuss kinase unfolding events of the kinase subdomains observed in MD pulling simulations vs. AFM in SI Appendix. Importantly, the same unfolding sequences of various lobes of the FERM and kinase domains were observed in simulations performed on the subdomains only, further validating the experiments on individual domains. SI Appendix has details.
Discussion
Here, we report a detailed mechanical characterization of FAK by using an AFM setup to apply stretching forces on single FAK molecules and record force–extension profiles with high sensitivity. Combining our measurements with structural information of the FERM–kinase region of FAK (11) and FPMD simulations has allowed us to assign measured force peaks to unfolding of defined structural features in FAK. Importantly, increasing the force loading rate by applying high pulling speeds has enabled the detection of a low-force event corresponding to rupture of the autoinhibitory FERM–kinase interaction. We show that domain separation occurs at a low-force regime around 25 pN for pulling speeds of 12,800 nm/s. The fact that forces required for domain separation are much lower than those required for domain unfolding supports the hypothesis that tensile forces in FAs applied to N- and C-terminal regions in FAK can trigger activation via domain separation. Mutational dissociation of FERM and kinase domains has previously been shown to activate FAK (11).
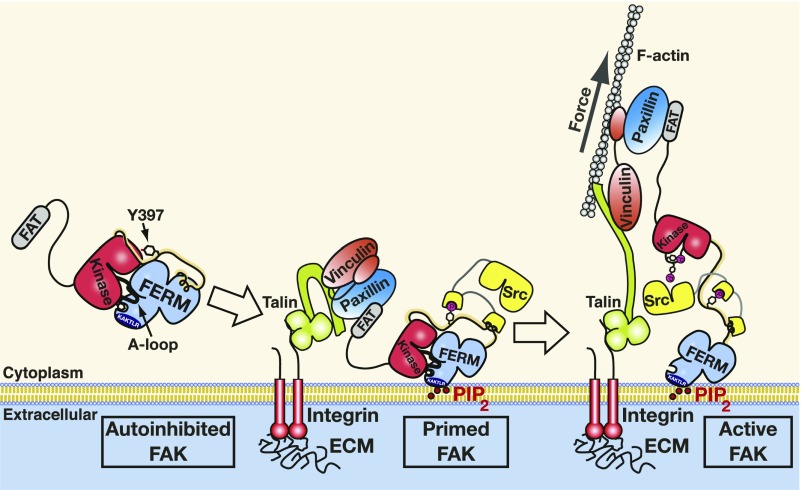
We propose that tension forces in FAK are built up between the membrane-bound N-terminal FERM domain and the C-terminal FAT domain engaged via paxillin to vinculin and the actin cytoskeleton (Fig. 7). This scenario was also suggested by our previous MD simulations of the first conformational activation step (17), where FERM and kinase domains detached from one another via these attachment sites. These previous simulations explicitly included a PIP2-containing bilayer and indicated that, for membranes enriched in PIP2, such as is the case at FA sites (15, 16), the membrane–FERM interaction is mechanically significantly more stable than the autoinhibitory FERM–kinase interaction. Force applied at the FAK C terminus in the opposite direction of the membrane resulted in FERM–kinase separation independent of the pulling angle or loading rate (17). Consistent with AFM measurements and the MD simulations presented here, the previous membrane-containing simulations also suggest that domain separation occurs before domain unfolding. In the previous simulations as well as in the full unfolding simulations shown here, the force required for separating FERM and kinase domains is 150 pN; however, the much higher pulling speed in simulations (6 mm/s or higher) is known to cause overestimation of rupture forces. Our combined AFM experiments and MD simulations suggest that the FAK domain organization protects against unfolding of functional domains of FAK: that is, the kinase domain and the PIP2 binding site in the F2 lobe of FERM. First, FERM–kinase domain dissociation yielding around 10-nm extension is followed by around 12-nm-long extension (SI Appendix, Table S1, Linker-F1) due to the stretching of the interdomain linker including the Tyr397 phosphorylation site. This would indicate that the length of the linker further protects the kinase domain from forced unfolding after the dissociation of the FERM and kinase domains. Second, F3 unfolding involves an increase in extension of about 30 nm (SI Appendix, Table S1, F3 unfold) and happens when the F2 lobe is still capable of binding PIP2. All in all, this would mean that FAK has a “safety margin” up to a total length of 50 nm (10-nm domain dissociation, 12 nm of linker stretching plus at least 28 nm for F3 unfolding) where it is still catalytically active and also, capable of binding PIP2. Consistent with our model where force to the C terminus of FAK is applied via paxillin and vinculin (Fig. 7), vinculin is found to transition from a signaling layer close to the membrane in FAs (which also contains FAK) to a force transduction layer closer to actin (23). Both layers have an approximate thickness of 30 nm measured vertical to the membrane; therefore, the average movement of vinculin approximates 30 nm toward the force transduction layer. This suggests that the 50-nm safety margin appears sufficient to protect the average engaged FAK molecules from force-induced deactivation. The fraction of FAK molecules exceeding this margin would expect to unfold their F2 lobe, hence losing contact with the membrane but retaining an active kinase. At an average of 30-nm extension, our data suggest that FAK molecules, after they are extended, no longer experience significant stretching forces other than what is required to keep FAK in an extended conformation. Likely, forces generated in FAs are mainly carried by structural components, such as talin and vinculin. In contrast for FAK, force seems to act as an activation catalyst by operating a digital distance switch, which is “on” when forces stretch FAK into an open conformation.
Fig. 7.
Model of force-induced FAK activation. FAK is recruited into FAs via C-terminal FAT interactions with paxillin and talin. The N-terminal FERM domain docks via PIP2 to the lipid membrane to promote a primed FAK state where Tyr397 in the linker between FERM and kinase is autophosphorylated. Forces generated via the actin cytoskeleton pull FAK’s C terminus away from the membrane, resulting in kinase release from the FERM domain and membrane. Src is recruited to autophosphorylated FAK and phosphorylates the exposed FAK activation loop to trigger full FAK activity.
In contrast to force activation of FA signaling, force-induced changes on structural FA components, such as talin or vinculin, have been characterized in detail. Mean forces experienced by talin and vinculin in FAs in cells have been determined to be in the range of 7–10 pN and around 2.5 pN, respectively (24, 25). However, force estimates from bulk measurements can be deceiving, since the load could be carried by only a fraction of molecules, whereas many others might not be engaged. Indeed, for integrins, average forces have initially been estimated at 1–2 pN (10); however, DNA-based tensions sensors that are irreversibly ruptured above a threshold force indicate peak forces of up to 40 pN for single integrin molecules (26). In our experiments, FERM–kinase separation occurs at around 25 pN at a pulling speed of 12,800 nm/s. Considering that average cellular force application in FAs is likely slower or even constant over certain time periods, it is highly likely that engaged FA molecules build up sufficient force (at a maximum of 40 pN for integrins) to separate FERM and kinase domains in FAK. Intriguingly, we find that unfolding of the FAK kinase domain in the more physiological ATP-loaded state occurs at around 50 pN; therefore, cellular forces in FAs seem well suited to allow FERM–kinase separation but not kinase unfolding, which are both prerequisites for FAK activation.
Force-induced separation of FERM and kinase domains will expose both the autophosphorylation site in the linker and the Src phosphorylation sites in the kinase activation loop. Mechanical extension of the Tyr397-containing linker might enhance Tyr397 exposure and autophosphorylation. This would likely only be the case in transphosphorylation mode, as force would act against folding back of Tyr397 into the active site of the same FAK molecule. However, we showed previously that membrane binding and resulting FAK oligomerization are sufficient to promote highly efficient FAK autophosphorylation also in the absence of force (14). This study also indicated significant membrane-induced conformational changes that expose the autophosphorylation site but apparently not the kinase active site, since contrary to domain separation, membrane binding did not catalytically activate FAK (14). Together, the two studies, therefore, support a model where initial membrane binding promotes a primed state of FAK by exposing the linker for efficient autophosphorylation, but subsequent buildup of tensile forces in FAK exposes the active site for efficient phosphorylation of the activation loop by Src (Fig. 7). It is the latter event that promotes full catalytic activity of FAK. It was recently shown that the kinase domain of FAK also contributes to binding to PIP2 membranes (27); therefore, force might be responsible for removing the kinase from both the FERM domain and the membrane. MD simulations indeed support such a scenario and found that the pulling angle can dictate which occurs first (17).
In conclusion, our mechanical analysis of FAK supports a model where physiological stretching forces in FAs can cause conformational changes in FAK, promoting its catalytic activation and thereby, triggering of FA signals. Multiple cellular studies have previously shown that FAK is activated in response to various mechanical stimuli (4–6), and our analysis on single FAK molecules demonstrates the feasibility of direct force activation of FAK. Force-induced activation of FA signals is highly relevant in disease. In tumors, stiffening of the stroma that allows increased force generation triggers strong adhesion signals that promote tumor invasion (28). Understanding the direct relation between tumor stiffness, force-induced adhesion signaling, and tumor invasion can, therefore, provide the basis for the development of specific agents targeting this mechanism.
Materials and Methods
AFM Setup for Characterizing FAK.
To mimic physiological FAK stretching as occurring in FAs and to identify force-induced structural changes in FAK under stress, we developed an AFM-based single-molecule force spectroscopy assay. This allows for the detection of subtle force-induced events for FAK with high sensitivity during its guided stretching. We engineered FAK proteins to harbor affinity pulling handles for attachment to sample surface and cantilever. We introduced a ybbR-tag (29) for covalent linkage to the glass surface and an SII (30) for reversible tethering to an AFM cantilever tip functionalized with a monovalent Strep-Tactin (monoST) (26). Both attachments are formed via heterobifunctional PEG linkers (Fig. 1C has a schematic of FAK attachment to AFM). Before the experiment, FAK proteins are covalently immobilized to the glass surface. Typically, several thousand single-molecule AFM measurements are then performed, and force extension profiles are recorded by repeatedly approaching and retracting the functionalized AFM cantilever at constant speed. The measured curves are aligned and overlaid to generate heatmaps highlighting recurring features in the plots. These recurring unfolding events were identified by creating a most probable unfolding curve as described in Data Analysis.
In a previous study, the autoregulatory region of FAK was defined as FERM interacting with the kinase domain (14). Initial AFM experiments were conducted with full-length FAK (residues 1–1,052 in Fig. 1B, Bottom) and a construct containing only the FERM and kinase domains (residues 1–686, FK-FAK in Fig. 1B, Bottom), both equipped with affinity tags at their N and C termini. These experiments indicated that the FAT domain does not contribute to the force profile of the autoregulatory region (SI Appendix, Fig. S1); hence, subsequent experiments were only performed with FK-FAK. To mimic the physiological force path through the molecule, we introduced the N-terminal tag close to the lipid binding site, which in vivo attaches to the cell membrane. To prevent perturbation in protein folding, we inserted the 11-residue ybbR-tag into an unstructured loop immediately before the K216AKTLRK PIP2 binding site in the FERM domain. We confirm that these insertion mutants retain basal activity of wild-type FAK and that the FERM domain still maintains the ability to autoinhibit the catalytic activity of FAK (SI Appendix, Fig. S13). Previous MD simulations confirmed the PIP2–FERM linkage to be significantly more mechanically robust than the FERM–kinase interaction at relevant PIP2 concentrations (17), rationalizing the choice of a covalent handle to mimic of the FERM–membrane interaction.
FAK Expression.
Chicken FAK constructs were engineered to contain the 11-aa ybbR-tag after V215, just before the K216-AKTLRK basic patch sequence (29), and the 8-aa SII tag (30) at the C terminus. FAK constructs containing full-length, FERM and kinase, or kinase-only regions were expressed by transient transfection of HEK293GnT1 cells using polyethyleneimine as a transfection agent (31). FERM-only constructs were expressed in Escherichia coli BL21 (DE3) as in ref. 32. All proteins were expressed with an N-terminal 6xHis tag. Initial purification was performed by Ni-chelate affinity purification (GE Healthcare) followed by protease cleavage to remove the 6xHis tag. Proteins containing an SII tag were further purified by Strep-Tactin (GE Healthcare) affinity and size exclusion (Superdex 200; GE Healthcare) chromatography. Proteins without SII tag were further purified by anion exchange (Source 15Q; GE Healthcare) and size exclusion chromatography.
Sample Preparation for Surface and Cantilevers.
The preparation of the experiment includes specifically immobilizing (29, 33) the FAK construct on the glass surface and functionalizing the cantilever with an monoST. This ensures a well-defined pulling geometry for minimizing multiple interactions. All FAK constructs harbored a ybbR-tag for covalent immobilization on a glass surface and an SII for binding to the monoST-functionalized cantilever (Fig. 1C). All measured constructs were derived from chicken FAK and expressed in HEK cells (compare with FAK Expression).
Both cantilevers and glass surfaces were passivated by short 425.39-Da [SM(PEG)2; PEGylated SMCC cross-linker; Thermo Scientific Pierce] or long 5,000-Da (molecular mass 5,000; Rapp Polymere) heterobifunctional PEG spacers to avoid unspecific interactions between the cantilever and the glass surface. The PEG spacers offer an N-hydroxy succinimide group on one side for attachment to the amino silanized surface of the cantilever. The other end provides a Maleimide (Mal) group for attachment of the thiol group found in the Cysteine of the monoST.
For silanization, the cantilevers were first oxidized in a UV ozone cleaner (UVOH 150 LAB; FHR Anlagenbau GmbH) and subsequently silanized for 2 min in (3-aminopropyl)dimethylethoxysilane [ABCR; 50% (vol/vol) in ethanol]. For rinsing, the cantilevers were stirred in 2-Propanol (IPA) in MilliQ and afterward, dried at 80 °C for 30 min. After that, the cantilevers were incubated in a solution of 25 mM heterobifunctional PEG spacer and 50 mM Hepes for 30 min (for short PEG first solved in half DMSO and then filled to 50 mM Hepes). Finally, the monoST was bound to the cantilevers for 1 h at room temperature followed by a washing step in 1 PBS. The functionalized cantilevers were stored in measurement buffer (40 mM Hepes, pH 7.4, 10 mM MgCl2, 200 mM NaCl, 1 mM DTT) until use.
The preparation of the glass surfaces is in a lot of steps similar to the functionalization of the cantilevers as seen in Fig. 1C. The glass surfaces are amino silanized followed by a passivation with PEG linkers. The Mal of PEG offers a binding site for the thiol group of CoA. Via an Sfp-catalyzed reaction, the CoA can bind the ybbR-tag harbored by the FAK protein construct. This way, the protein gets attached and tethered in an uncompromisingly specific way.
Before silanization, the glass surfaces have to be cleaned by sonification in 50% (vol/vol) IPA in MilliQ for 15 min. For oxidation, the glass surfaces are soaked for 30 min in a solution of 50% (vol/vol) hydrogen peroxide (30%) and sulfuric acid. Afterward, they have to be thoroughly washed in MilliQ and then blown dry in an N2 stream. Then, the glass surfaces get silanized by incubating them in ABCR [1.8% (vol/vol) in ethanol]. Thereafter, they were washed again in IPA and MilliQ and then dried at 80 °C for 40 min. Then, the PEG is applied as described for the cantilevers. Subsequent to rinsing, the surfaces were incubated in 20 mM CoA (Calbiochem) dissolved in coupling buffer (sodium phosphate, pH 7.2) to react with Mal. After washing the glass surfaces, 8 L of the FAK construct (20 M) was mixed with 1 L Sfp-synthase (132 M) and 1 L of 10 reaction buffer (100 mM Tris, pH 7.5, 100 mM MgCl2); then, it was pipetted on the surfaces and incubated for 2 h at room temperature. Finally, the surfaces were rinsed thoroughly in measurement buffer (40 mM Hepes, pH 7.4, 10 mM MgCl2, 200 mM NaCl, 1 mM DTT).
Force Spectroscopy Experiments.
The AFM measurements were conducted on an Asylum research controller (Asylum Research) providing analog-to-digital converter and digital-to-analog converter channels as well as a digital signal processor board for setting up feedback loops. The controller operated either a custom-built AFM head (34) or an xyz-movable piezo-driven sample stage. Data were recorded automatically by cycling through the following steps: (i) approach of the functionalized AFM tip to the surface to allow coupling to the SII of surface-immobilized FAK; (ii) retraction of the AFM cantilever with nanometer precision at a constant speed and simultaneous recording of the mechanical force response with pN precision; and (iii) after monoST:SII separation, the piezo stage of the AFM is moved to probe a new spot on the sample surface in the next cycle. This process was operated by using an IgorPro6 (Wavemetrics) program controlling the z piezo in the AFM head (or sample stage) and the xy piezos. The surface is sampled in steps of 100-nm distance in a snail trace to avoid probing a spot multiple times. The BioLever Mini (BLAC40TS) cantilevers (Olympus; 10-nm nominal tip radius, sharpened probe) were indented with 180 pN, applying no additional dwell time. Cantilevers were chemically modified (compare with Sample Preparation for Surface and Cantilevers) and were calibrated after the measurement using the equipartition theorem method (28). The datasets were recorded in the course of a few hours and contained around 50,000–90,000 curves saved in hdf5 files for additional data analysis.
Previous studies on smooth muscle myosin light-chain kinase (10) were conducted with 5,000-Da PEG linkers at a pulling speed of 800 nm/s. For this study, these parameters did not provide sufficient force resolution for clearly identifying contour length increments of the subtle peaks measured. By using shorter PEG linkers (35) (in this case, 425 Da) and thereby, increasing the loading rate as seen by the molecule, rupture forces of the detected events could be increased (SI Appendix, Fig. S13). These higher forces enable reliable worm-like chain fits for accurate analysis of the contour length increments.
Data Analysis.
To show the characteristic unfolding patterns of the probed FAK construct, heatmaps were assembled with all curves that showed the correct total contour length (indicating correct site-specific attachment) as well as the presence of the characteristic unfolding peaks. Denoised (based on Savitzky–Golay, length 35, two polynomial for 800 nm/s and length 21, two polynomial for 12,800 nm/s) force spectroscopy data were aligned manually in force–distance space (only by translating along the distance axis to account for length differences in PEG); they were binned from −15 to 150 nm in distance and from −15 to 150 pN in force for measurements with short PEG linkers (425.39 Da) and binned from −15 to 250 nm in distance and from −15 to 250 pN in force for measurements with long PEG linkers (5,000 Da) to create a heatmap. The number of bins (equal for both distance and force axis) is dependent on the curves contained in the heatmap (Fig. 2, 150 bins; Fig. 3, 250 bins; Fig. 4, 250 bins; and Fig. 5, 150 bins).
The denoised data points (Savitzky–Golay) in force–distance space were binned on the distance axis into 2.5-nm- (for Fig. 2), 3-nm- (for Figs. 3 and 4), and 3.5-nm-sized slices (for Fig. 5) (moving the slice window by 0.2 nm each step), and their densities on the force axis (y axis) were estimated by a kernel density estimate (KDE) with a bandwidth of 0.2 pN (compare with SI Appendix, Fig. S14). The resulting most probable values are then assembled to form the most probable unfolding curve (shown as a black line in SI Appendix, Fig. S14) and analyzed to find the most probable unfolding peaks. The FWHMs of the distance slices were then taken as selection criteria for the unfolding peaks. If slices contain a rupture event, the drop in force results in broad distributions, thereby clearly deviating from the noise level. The peaks were first detected by a simple peak detection based on taking the first order difference and then validated by the FWHM of the distance slices. To be accepted as a peak, the FWHM of the distance slices has to be above the FWHM of the KDE of the accumulated FWHMs of the distance slices of the curve, which gives a good representation of the noise level of the curve (compare with SI Appendix, Fig. S14). The procedure of assembling the most probable curve does not necessarily reproduce absolute rupture forces but yields a good result for the most probable and most representative pathway (36).
For additional analysis of the contour length increments, each stretch preceding an unfolding event is fitted with the worm-like chain model. This is done for every single curve contained in the heatmap. The most probable contour length for each peak is determined using a KDE. The increments between these most probable contour lengths were used to compare them with structural elements of the crystal structure.
Data analysis was completely carried out in Python 2.7 and is available online together with all used datasets (https://gitlab.physik.uni-muenchen.de/Magnus.Bauer/fak_analysis).
MD Simulations.
We use the FK-FAK construct developed previously (17) and solvate the protein in a 150 × 10 × 10-nm box. The total system contains ∼1.5 million atoms, including 908 Na+ and 903 Cl− ions, corresponding to a ionic strength of 0.1 M. We use GROMACS (15), version 2016 for all of our simulations. As force field, we use Amber99SB-ILDN* force field (16) with Joung ions (27) and a transferable intermolecular potential with 3 points (TIP3P) water model (37). We use a time step of 2 fs and freeze all bonds in our simulations through a linear constraint solver (LINCS) procedure (BP Hess) of fourth order. Two Nosé–Hoover thermostats, one for protein and one for nonprotein atoms, were used with a time constant of 0.6 ps to keep the temperature at 300 K. An isotropic Parrinello–Rahman barostat with a time constant of 2 ps and a compressibility of 4.5 10−5 with a reference pressure of 1 atm was used for pressure coupling. Verlet neighbor lists with a cutoff of 1.0 nm were used with an initial frequency of 0.03 ps. These parameters were automatically updated during the simulations by GROMACS for optimal performance. For long-range electrostatics, we use a fourth-order particle mesh Ewald method (38) with a grid spacing of 0.16.
We perform a total of 30 pulling simulations, each with a spring constant of 830 pN/nm: 10 simulations each at the velocities of 1, 1/3, and 1/10 nm/ns. The simulations were performed in the presence of an ATP molecule and an Mg2+ ion. To obtain single-domain pulling simulations, we start from the coordinates of the full FK-FAK construct and keep only the residues in the relevant domains. We relax the structures in 100-ns equilibrium simulations and solvate the FERM domain and the kinase domain in 67 × 9 × 9 and 100 × 8.5 × 8.5 nm, respectively. These correspond to 534,000 (FERM) and 706,000 (kinase) atoms. In both cases, we remove the ATP molecule and the Mg2+ ion from the simulation. We pull only using the fastest pulling velocity (1.0 m/s) and otherwise, keep all parameters unchanged.
We quantify domain unfolding by measuring distances between residues as follows: for FERM, we used residues 216–362; for the linker, we used residues 362–418, and for the kinase, we used residues 418–686. We define the beginning and end of unfolding events as the times that the distances reach 10 and 45 nm for FERM, 7 and 15 nm for the linker, and 20 and 75 nm for the kinase. For the initial conformational activation, we also use a simple distance criterion: namely, whenever the distance 216–640 exceeds 10 nm. Since force profiles obtained in MD simulations include several intermediate ruptures, we identify peaks through a two-step procedure: (i) a Gaussian smoothing of the force profiles with an SD consistent with an extension of 0.1 nm and (ii) finding local maxima of the smoothed force profile in a window consistent with an extension of ±10 nm.
Supplementary Material
Acknowledgments
We thank Ivan Acebrón for help with activity measurements. M.S.B. acknowledges Leonard C. Schendel and Steffen M. Sedlak for experimental assistance; Angelika Kardinal and Thomas Nicolaus for laboratory support; Iris Ruider, Katherine Erlich, Marco Grison, and Wolfgang Ott for helpful discussions; Sylvia Kreuzer for administration; and the Nanosystems Initiative Munich for support. C.D. and F.G. are grateful for support from the state of Baden-Württemberg through high performance computing in Baden-Württemberg (bwHPC) and Deutsche Forschungsgemeinschaft (DFG) Grant INST 35/1134-1 FUGG. H.E.G. acknowledges funding from DFG Grant Sonderforschungsbereich 1032. F.G. acknowledges funding from the DFG through the research group SHENC (Shear Flow Regulation of Hemostasis—Bridging the Gap Between Nanomechanics and Clinical Presentation) and from the Klaus Tschira Foundation. D.L. acknowledges support from Spanish Ministry of Economy, Industry and Competitiveness Retos Grant BFU2016-77665-R cofunded by the European Regional Development Fund and Volkswagen Foundation Grant Az: 86 416-1. D.L. is the recipient of Worldwide Cancer Research Award 15-1177.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Both the code and the datasets used in this study can be accessed at https://gitlab.physik.uni-muenchen.de/Magnus.Bauer/fak_analysis.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820567116/-/DCSupplemental.
References
- 1.Winograd-Katz SE, Fässler R, Geiger B, Legate KR. The integrin adhesome: From genes and proteins to human disease. Nat Rev Mol Cell Biol. 2014;15:273–288. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]
- 2.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Z, Guo SS, Fässler R. Integrin-mediated mechanotransduction. J Cell Biol. 2016;215:445–456. doi: 10.1083/jcb.201609037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seong J, et al. Distinct biophysical mechanisms of focal adhesion kinase mechanoactivation by different extracellular matrix proteins. Proc Natl Acad Sci USA. 2013;110:19372–19377. doi: 10.1073/pnas.1307405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torsoni AS, Constancio SS, Nadruz WJ, Hanks SK, Franchini KG. Focal adhesion kinase is activated and mediates the early hypertrophic response to stretch in cardiac myocytes. Circ Res. 2003;93:140–147. doi: 10.1161/01.RES.0000081595.25297.1B. [DOI] [PubMed] [Google Scholar]
- 6.Wong VW, et al. Focal adhesion kinase links mechanical force to skin fibrosis via inflammatory signaling. Nat Med. 2011;18:148–152. doi: 10.1038/nm.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang HB, Dembo M, Hanks SK, Wang Y. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc Natl Acad Sci USA. 2001;98:11295–11300. doi: 10.1073/pnas.201201198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puchner EM, et al. Mechanoenzymatics of titin kinase. Proc Natl Acad Sci USA. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.von Castelmur E, et al. Identification of an n-terminal inhibitory extension as the primary mechanosensory regulator of twitchin kinase. Proc Natl Acad Sci USA. 2012;109:13608–13613. doi: 10.1073/pnas.1200697109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumann F, et al. Increasing evidence of mechanical force as a functional regulator in smooth muscle myosin light chain kinase. eLife. 2017;6:e26473. doi: 10.7554/eLife.26473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lietha D, et al. Structural basis for the autoinhibition of focal adhesion kinase. Cell. 2007;129:1177–1187. doi: 10.1016/j.cell.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai X, et al. Spatial and temporal regulation of focal adhesion kinase activity in living cells. Mol Cell Biol. 2008;28:201–214. doi: 10.1128/MCB.01324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goni GM, et al. Phosphatidylinositol 4,5-bisphosphate triggers activation of focal adhesion kinase by inducing clustering and conformational changes. Proc Natl Acad Sci USA. 2014;111:E3177–E3186. doi: 10.1073/pnas.1317022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pronk S, et al. Gromacs 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the amber ff99sb protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, et al. Mechanism of focal adhesion kinase mechanosensing. PLoS Comput Biol. 2015;11:e1004593. doi: 10.1371/journal.pcbi.1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouchiat C, et al. Estimating the persistence length of a worm-like chain molecule from force-extension measurements. Biophys J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hugel T, Rief M, Seitz M, Gaub HE, Netz RR. Highly stretched single polymers: Atomic-force-microscope experiments versus ab-initiotheory. Phys Rev Lett. 2005;94:048301. doi: 10.1103/PhysRevLett.94.048301. [DOI] [PubMed] [Google Scholar]
- 20.Hu X, Li H. Force spectroscopy studies on protein-ligand interactions: A single protein mechanics perspective. FEBS Lett. 2014;588:3613–3620. doi: 10.1016/j.febslet.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Verdorfer T, Gaub HE. Ligand binding stabilizes cellulosomal cohesins as revealed by afm-based single-molecule force spectroscopy. Sci Rep. 2018;8:9634. doi: 10.1038/s41598-018-27085-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietz H, Rief M. Exploring the energy landscape of gfp by single-molecule mechanical experiments. Proc Natl Acad Sci USA. 2004;101:16192–16197. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Case LB, Waterman CM. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol. 2015;17:955–963. doi: 10.1038/ncb3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonard TA, Hurley JH. Two kinase family dramas. Cell. 2007;129:1037–1038. doi: 10.1016/j.cell.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 26.Baumann F, et al. Monovalent strep-tactin for strong and site-specific tethering in nanospectroscopy. Nat Nanotechnol. 2015;11:89–94. doi: 10.1038/nnano.2015.231. [DOI] [PubMed] [Google Scholar]
- 27.Joung IS, Cheatham TE. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutter JL, Bechhoefer J. Calibration of atomic-force microscope tips. Rev Sci Instr. 1993;64:1868–1873. [Google Scholar]
- 29.Yin J, Lin AJ, Golan DE, Walsh CT. Site-specific protein labeling by sfp phosphopantetheinyl transferase. Nat Protoc. 2006;1:280–285. doi: 10.1038/nprot.2006.43. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt TG, Skerra A. The strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc. 2007;2:1528–1535. doi: 10.1038/nprot.2007.209. [DOI] [PubMed] [Google Scholar]
- 31.Durocher Y, Perret S, Kamen A. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-ebna1 cells. Nucleic Acids Res. 2002;30:E9. doi: 10.1093/nar/30.2.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ceccarelli DF, Song HK, Poy F, Schaller MD, Eck MJ. Crystal structure of the ferm domain of focal adhesion kinase. J Biol Chem. 2006;281:252–259. doi: 10.1074/jbc.M509188200. [DOI] [PubMed] [Google Scholar]
- 33.Zimmermann JL, Nicolaus T, Neuert G, Blank K. Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nat Protoc. 2010;5:975–985. doi: 10.1038/nprot.2010.49. [DOI] [PubMed] [Google Scholar]
- 34.Gumpp H, Stahl SW, Strackharn M, Puchner EM, Gaub HE. Ultrastable combined atomic force and total internal fluorescence microscope. Rev Sci Instr. 2009;80:063704. doi: 10.1063/1.3148224. [DOI] [PubMed] [Google Scholar]
- 35.Walder R, et al. Rapid characterization of a mechanically labile alpha-helical protein enabled by efficient site-specific bioconjugation. J Am Chem Soc. 2017;139:9867–9875. doi: 10.1021/jacs.7b02958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ott W, et al. Elastin-like polypeptide linkers for single-molecule force spectroscopy. ACS Nano. 2017;11:6346–6354. doi: 10.1021/acsnano.7b02694. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1998;79:926–935. [Google Scholar]
- 38.Darden T, York D, Pedersen L. Particle mesh ewald: An n log(n) method for ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.