Significance
Structures of G protein-coupled receptors (GPCRs) in complex with ligands mainly provide frozen pictures with little information about the actual molecular mechanism of action of the ligand in the normally highly dynamic receptor. Through computer-based molecular dynamics simulations of a receptor for long-chain fatty acids, free fatty acid receptor 1 (FFAR1), we discover that an unoccupied, solvent-exposed pocket closes on removal of the lipid-like agonist; that is, during a major conformational change of the receptor. Importantly, a compound designed to prevent closure of this previously unrecognized, dynamic pocket was identified through structure-based virtual screening and shown to function as an allosteric agonist for the receptor. The study demonstrates that molecular dynamics simulations can be used in drug discovery to identify different modes of stabilizing specific receptor states.
Keywords: free fatty acid receptor 1, virtual screening, GPCR, molecular dynamics, allosteric modulator
Abstract
The long-chain fatty acid receptor FFAR1/GPR40 binds agonists in both an interhelical site between the extracellular segments of transmembrane helix (TM)-III and TM-IV and a lipid-exposed groove between the intracellular segments of these helices. Molecular dynamics simulations of FFAR1 with agonist removed demonstrated a major rearrangement of the polar and charged anchor point residues for the carboxylic acid moiety of the agonist in the interhelical site, which was associated with closure of a neighboring, solvent-exposed pocket between the extracellular poles of TM-I, TM-II, and TM-VII. A synthetic compound designed to bind in this pocket, and thereby prevent its closure, was identified through structure-based virtual screening and shown to function both as an agonist and as an allosteric modulator of receptor activation. This discovery of an allosteric agonist for a previously unexploited, dynamic pocket in FFAR1 demonstrates both the power of including molecular dynamics in the drug discovery process and that this specific, clinically proven, but difficult, antidiabetes target can be addressed by chemotypes different from existing ligands.
The Gq-coupled free fatty acid receptor 1 (FFAR1/GPR40) is a clinically validated antidiabetes G protein-coupled receptor (GPCR) target that binds medium- to long-chain fatty acids (C8–C22) (1, 2) to stimulate gut hormone and insulin secretion (2–5). In addition to the relatively broad group of endogenous lipid ligands, many synthetic agonists have been developed, of which several have entered clinical trials for the treatment of type 2 diabetes, as reviewed by Li et al. (6). Although most of these ligands appear to share chemical properties, characterized by an elongated topology and a terminal carboxylic acid group, molecular pharmacology studies have indicated the presence of several mutually interacting binding sites in FFAR1 (7, 8). Certain synthetic agonists are, similar to the endogenous lipid agonists, only able to induce Gq-like signaling, whereas other agonists induce both Gq and Gs-like signaling, resulting in more robust hormone responses. Radioligand binding experiments have demonstrated that these two different types of FFAR1 ligands act as positive allosteric modulators, as they do not compete for, but instead increase binding of, each other, indicating that they act on distinct receptor sites (7).
Srivastava et al. (9) solved the X-ray crystal structure of FFAR1 at 2.3 Å resolution in complex with the synthetic agonist TAK-875 (PDB 4PHU), a previous clinical phase 3 drug candidate. This structure revealed a unique binding mode of TAK-875 compared with agonists in other GPCR structures. The elongated hydrophobic part of the agonist was partly bound in a hydrophobic, tunnel-like interhelical site between transmembrane helix (TM)-III and TM-IV, and partly extruded into the outer layer of the lipid bilayer, suggesting entry from there. Importantly, the carboxylic acid moiety of TAK-875 interacted directly with two arginines (R183 and R258), as well as two tyrosines (Y91 and Y240), which are part of an anchoring polar network at the center of the receptor exposed to the extracellular space (Fig. 1). Recently, Lu et al. (10) published two X-ray crystal structures of FFAR1, one of them in complex with two agonists; that is, the Gq-only synthetic agonist MK-8666 bound in the interhelical site in a very similar manner as TAK-875 and the ago-allosteric modulator AP8, bound in an open extrahelical, groove-like site located between TM-III, TM-IV, and TM-V, facing the inner leaflet of the lipid bilayer. Interestingly, in the previously published FFAR1 structure in complex with TAK-875 (9), and in several other GPCR structures (11–13), this extrahelical site is occupied by either a detergent or lipid molecule.
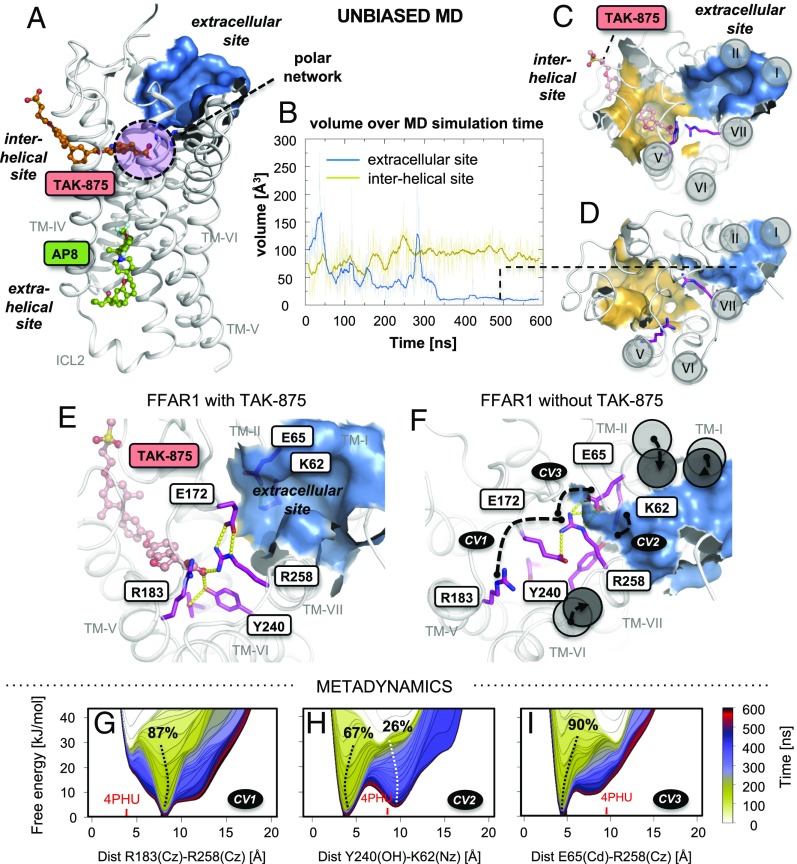
Fig. 1.
FFAR1 shows a third extracellular binding site that is stabilized by the presence of the partial agonist TAK-875. (A) FFAR1 in complex with TAK-875 and AP8 docked into the interhelical site (PDB 4PHU). (B) Pocket volume of the interhelical (yellow) and extracellular (blue) site over MD simulation time as 5 ns running averages (solid line) and for every 0.2 ns (faint line). (C and D) Comparison between representative structures, obtained by 1 Å rmsd geometric clustering from a 600 ns unbiased MD simulation in the presence (snapshot at 171.40 ns) and absence (snapshot at 493.24 ns) of TAK-875. (E and F) Rearrangement of the polar network stabilized by TAK-875, as shown by representative structures from a 600-ns unbiased MD simulation of FFAR1 in the presence and absence of TAK-875. Key amino acid side chains involved in the polar network are shown as purple sticks. (G–I) Free-energy profiles of selected atomic distances (CV1-3) obtained by a well-tempered metadynamics approach, using the same MD setup as for the unbiased MD simulation in absence of TAK-875. The free-energy profile was recorded every 10 ns, as indicated by the color scale. The population of conformational states are indicated as percentages. The corresponding atomic distances are displayed as black dotted lines in F.
In all three published FFAR1 crystal structures, a third, solvent-exposed pocket (termed site 3 in ref. 9) could be observed between the extracellular poles of TM-I, TM-II, and TM-VII; that is, on the opposite side of the polar network of residues coordinating the carboxylic acid moieties of TAK-875 and MK-8666 (Fig. 1A). This extracellular site is the main focus of the present study, in which we investigated the structural dynamics of FFAR1 in both the presence and absence of TAK-875 in the interhelical site by use of all-atom classical molecular dynamics (MD) simulations combined with metadynamics simulations. Using experimental pharmacological testing of compounds, as well as mutational analyses, we describe an ago-allosteric modulator and highlight the extracellular site as an avenue for the structure-based design of FFAR1 ligands.
Results
MD Simulations Reveal a Major Rearrangement of Polar Anchor-Point Residues.
Initially, we performed 600 ns of unbiased MD simulations and compared representative frames that were obtained by geometric clustering. Although FFAR1 is crystalized in complex with agonists, the overall structure of the intracellular part of the helical bundle of FFAR1 is rather similar to the structure of various published inactive receptor conformations, conceivably because of thermostabilizing mutations, which allow for high-affinity agonist binding, but also prevent the receptor from signaling in solution (9). In analogy to previous MD simulation studies of GPCRs in their inactive form (14, 15), we did not observe any substantial change of the overall helical configuration at the intracellular side, which remained similar to the inactive-like state of, for example, the β2-adrenergic receptor (16) (SI Appendix, Fig. S2A). However, during the course of the MD simulation of FFAR1 in the presence of TAK-875, the intracellular segment of TM-VI moved slightly (∼4 Å) toward TM-V, which likely is related to the insertion of ICL3 in the FFAR1 model, which in the X-ray crystal structure is replaced with a fusion protein (PDB 4PHU).
At the extracellular solvent-exposed face of the FFAR1 receptor, we observed a major rearrangement of residues of the polar network during the MD simulations in the absence of TAK-875, which was not observed in the presence of the stabilizing carboxylic acid group of the agonist (Fig. 1 C and D). This disruption lead to a substantial conformational change of not only the side chains of the polar network but also a shifting movement of the extracellular part of TM-I, TM-II, and TM-VII, ultimately resulting in total closure (<10 Å3) of the solvent-exposed extracellular site after ∼340 ns of simulation time (Fig. 1 B and D). The movement of TM-VII was facilitated by the structural flexibility induced by the proline of the conserved, central NPxxY motif. In contrast to these changes observed in the extracellular site, the hydrophobic tunnel-like part of the interhelical TAK-875 site remained surprisingly stable after the ligand had been removed (Fig. 1 B and D). We compared the structural heterogeneity of all the MD trajectory structures by geometric clustering of residues forming both the interhelical and the extracellular site. Only 35 clusters were found for the trajectory in the presence of TAK-875, and 64% of all trajectory conformations belonged to a single large cluster based on a 1 Å rmsd cutoff. In comparison with this, more than a 100 different clusters were identified for the trajectory in the absence of TAK-875. This further illustrates the structural dynamics that occur in the absence of TAK-875 (SI Appendix, Fig. S3).
Metadynamics Simulations of the Rearrangement of the Polar Network.
The MD simulations described here provide an unbiased view of the dynamics of FFAR1. We supplemented these by metadynamics simulations that allow us to map out the conformational free-energy landscape (17, 18). Thus, we estimated the free-energy landscape for three selected collective variables (CVs), chosen to represent the rearrangement of the polar network as distances between key sidechains. Together, these CVs describe the major conformational changes and exchange of interaction partners we observed in the MD simulation in the absence of TAK-875 (Fig. 1F): CV1 represents the separation of the two positively charged guanidine groups of R183(Cζ) and R258(Cζ) (from 3.5 Å in the X-ray crystal structure to 10.0 Å in the representative MD snapshot), CV2 describes the rotation of Y240(OH) toward K62(Nζ) (from 8.67 Å to 4.5 Å), and CV3 indicates the formation of a salt bridge between R258(Cζ) and E65(Cδ) (from 9.9 Å to 3.9 Å). During the metadynamics simulation, a time-dependent bias potential was introduced to discourage previously visited conformations. Thermodynamic reweighting provides access to the unbiased free energy as a function of the simulation time to examine whether the observed conformation represents energetic “trapping” resulting from insufficient sampling or corresponds to a true global free-energy minimum (17). In general, the global energy minima for the three atomic distances that were chosen as CVs were in good agreement with the atomic distances observed in the representative structures for the unbiased simulation absence of TAK-875 (CV1: 8.1 vs. 10.0 Å; CV2: 3.9 vs. 4.5 Å; CV3: 4.6 vs. 3.9 Å; Fig. 1 G–I). For CV2, a second, less occupied local minimum is present at 9.5 Å, probably indicating a second stable rotamer of Y240. However, when plotting the 2D free-energy plots for each of the combinations of the three collective variables (SI Appendix, Fig. S3), it is apparent that on rotation of Y240 toward K62, shown by a shift to a lower value of CV2 at 3.9 Å, the free-energy landscape for CV1 and 3 converges into single minima at 8.1 Å and 4.6 Å, respectively. This suggests that the rotation of Y240 stabilizes the formation of the salt bridge between R258 and E65 and keeps R183 and R258 separated. Overall, these results point to a distribution of conformational states in which the rearranged polar network (Fig. 1F) leading to the closure of the extracellular site is in fact the dominant conformation in absence of TAK-875.
The conformational dynamics and apparent connectivity between the different FFAR1 binding sites prompted us to investigate whether the closure of the extracellular site is reversible. The addition of TAK-875 after 600 ns of unbiased MD simulation in fact causes a reopening of this site, causing the pocket volume to rise from <10 Å3 to ∼100–250 Å3 (SI Appendix, Fig. S1). Likewise, the associated relocation of TM-I, TM-II, and TM-VII to their initial position (SI Appendix, Fig. S1B) leads to a Cα-rmsd decrease from 2.8 Å (representative structure from the MD simulation in absence of TAK-875) to 1.8 Å for the MD simulation in which TAK-875 was added after 600 ns, compared with the X-ray crystal structure of FFAR1 (PDB 4PHU). As a consequence, the observed structural connectivity appears to be based on a concerted dynamic allosteric mechanism in which the presence of an agonist in the interhelical site is associated with formation of a relatively large well-defined pocket; that is, the extracellular site on the opposite side of the polar network (Fig. 1E). The observed structural dynamics of this part of the receptor leads to the hypothesis that prevention of the closure of the extracellular site through binding of a ligand specifically designed for this site would stabilize the active conformation by preventing the receptor from moving into the presumably inactive conformation, and consequently would function as an agonist.
Structure-Based Design of an Ago-Allosteric Modulator.
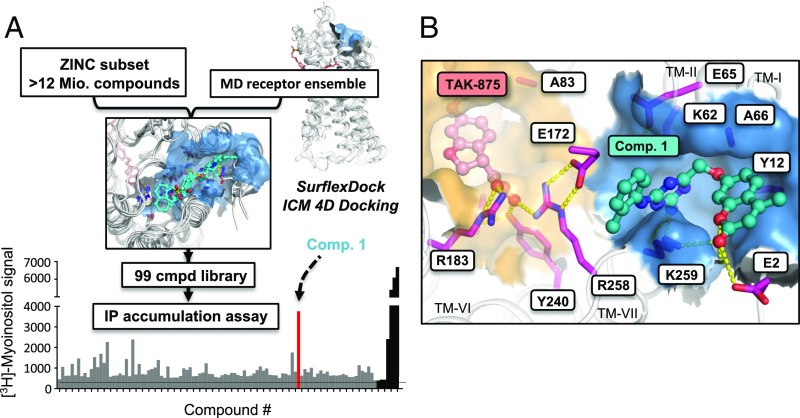
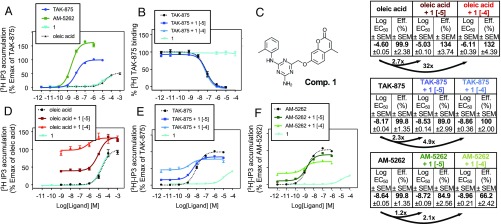
Accordingly, we probed the extracellular site by conducting a structure-based virtual screening of >12 million commercially available small synthetic compounds (Fig. 2). These compounds were each docked in two steps into an ensemble of four representative structures from the unbiased MD simulation in the presence of TAK-875, and filtered on the basis of general chemical criteria and docking scores (19). A library of the 99 best-scored compounds was acquired and tested for their ability to activate FFAR1-mediated signal transduction in transfected HEK293 cells. Compound 1 (ZINC9634346/Z18477911), a derivative of 1,3,5-triazine-2-amine (Fig. 3C), was the most efficacious, with similar potency and efficacy as the endogenous ligand, oleic acid (Fig. 3A). Radiolabeled competition binding experiments showed that Compound 1, in contrast to unlabeled TAK-875, was unable to displace [3H]TAK-875 from the receptor, indicating that this agonist can bind simultaneously with an agonist binding to the interhelical site, in agreement with its expected binding in the extracellular site (Fig. 3B). We then tested the ability of Compound 1 to modulate activation of FFAR1 by oleic acid and the two different types of synthetic agonists, TAK-875 and AM-5262 (Fig. 3 D–F). Compound 1 acted not only as an agonist but also as an allosteric modulator by increasing the potency of oleic acid ∼32-fold and increasing its efficacy (Emax) with 34% at 100 µM of Compound 1 (Fig. 3D). However, although increasing the potency of TAK-875 and AM-5262 by fivefold and twofold, respectively, Compound 1 surprisingly had a negative effect on the efficacy of these two agonists; that is, reducing their Emax by 22% and 35%, respectively (Fig. 3 E and F). Thus, Compound 1 acts as a positive allosteric modulator on the endogenous lipid agonist, with respect to both potency and efficacy, but increases the potency of the two types of synthetic agonist types only slightly, and even decreases their maximal efficacy. In this context, it is probably important to note that the maximal efficacy of Compound 1 itself is less than that of TAK-875 and AM-5262 (Fig. 3A). Therefore, it seems possible that Compound 1, by stabilizing the extracellular site, is shifting the dynamic equilibrium to an “active-like” conformation that can be described as an intermediate between the conformations induced by oleic acid, TAK-875, and AM-5262.
Fig. 2.
Structure-based discovery of Compound 1. (A) Ligand screening approach used in this study. The best-scored 100.000 compounds from a virtual screening of the In Stock subset of the ZINC database were docked into an ensemble of representative FFAR1 structures retrieved from a 600-ns MD trajectory. After applying further filtering criteria, a library of 99 compounds was purchased and experimentally tested in an IP accumulation assay. (B) Docking pose of Compound 1 in FFAR1. Polar receptor-ligand interactions are indicated with yellow spheres.
Fig. 3.
Pharmacologic profile of Compound 1. (A) Dose–response curves of Compound 1 in comparison with known synthetic and endogenous FFAR1 agonists. (B) Radiolabeled ligand binding of [3H]TAK-875 in combination with Compound 1. (C) Chemical structure of Compound 1 (D–F) Dose–response curves of Compound 1 alone and in combination (10 and 100 µM fixed concentration) with oleic acid, TAK-875, and AM-5262. Values for log EC50, efficacy, and potency fold-changes are indicated for each compound in the table (Right).
During follow-up experiments, we acquired 96 commercially available analogs of Compound 1, which were subsequently experimentally screened in an inositol phosphate (IP) accumulation assay. We initially identified two compounds, the γ-lactam ZINC7064600/Z107326816 and the ester-linked 3-methyl-7-fluorobenzofurane ZINC45546728/Z154646582, with similar pharmacological activity as Compound 1 (SI Appendix, Fig. S7). In general, the available analogs were chemically rather diverse, especially around the coumarin motif and the attached ether linker, which was only present in three of 96 compounds. We performed a more systematic exploration of the structure–activity relationship, based on custom-made synthesis (SI Appendix, Fig. S8). We found several active compounds with improved potency and efficacy, among which the meta- substituted Compounds 4 and 5 showed the highest EC50 values of ∼1 µM. It was also possible to replace the coumarin motif in Compound 1 with an ester-linked 3-methyl-7-fluorobenzofuran (Compound 6) moiety to achieve a modest potency gain at the cost of some loss of efficacy.
To rule out colloid aggregation-based activation, we tested the ability of Compound 1 to inhibit AmpC β-lactamase (SI Appendix, Fig. S9). At 31.6 µM concentration, the compound showed 0.4% enzyme inhibition, comparable to TAK-875 and well below the level of known aggregators, such as tetra-iodophenolphthalein (16.66%) (20). Further, Compound 1 did not show any unspecific activity on mock-transfected cells (SI Appendix, Fig. S10).
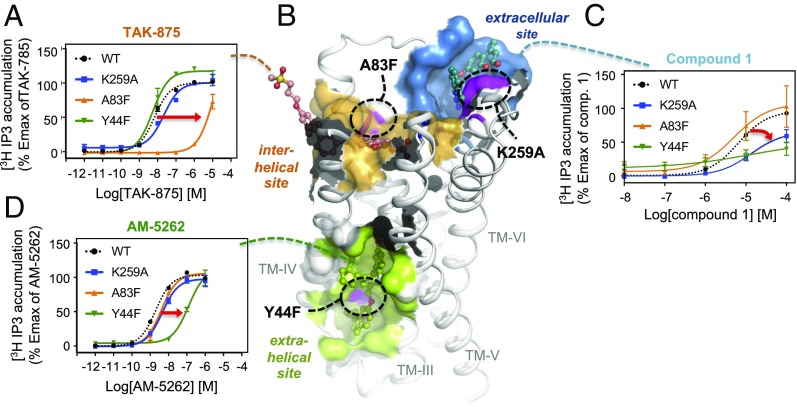
To verify the binding sites and mode of action of the three different types of synthetic agonists, we introduced a series of, respectively, four, two, and seven mutations in the suspected extrahelical, intrahelical, and extracellular binding sites and tested the ability of the agonists to activate FFAR1, as listed in SI Appendix, Table S1 and shown for key selected mutations in Fig. 4. A very clear result was obtained by steric hindrance mutation, A83F, in the hydrophobic tunnel of the interhelical site, which, as expected, impaired the potency of TAK-875 considerably (i.e., 1,740-fold; Fig. 4A), without affecting the function of AM-5262 or Compound 1 (Fig. 4 C and D). Conversely, mutations in the lipid-exposed extrahelical site impaired the potency of AM-5262 as anticipated, without affecting TAK-875, as shown for Y44F (Fig. 4 A and D). However, two of the mutations in the extrahelical site (Y114F and S123A) unexpectedly decreased the potency and efficacy of Compound 1 (Fig. 4C and SI Appendix, Table S1), although this less lipophilic compound is expected to act through binding to the extracellular site distant from the mutations in the lipid-exposed extrahelical site. Although the possibility of Compound 1 binding to the extrahelical site cannot be excluded, the other two mutations in this site (Y44F and G95F) actually improved potency of Compound 1 while dramatically decreasing potency of AM-5262. Specifically, the bulky phenylalanine, which is introduced for G95, is located in a central position in the extrahelical site and would be expected to occupy a large section of this pocket. This steric hindrance mutation has a major effect on the pharmacologic activity of AM-5262 (67-fold potency decrease), as well as the positive allosteric modulator AP8 (10), but remarkably, it has an unexpected positive effect on the activity of Compound 1, roughly doubling both its potency and efficacy. This observation might be related to an effect on a potential allosteric network that controls the dynamic structural connectivity between these two sites. A similar finding has been described in a mutational mapping study involving the M1 and M4 muscarinic receptors, in which mutation of a large number of residues involved in an allosteric network, but not in direct ligand contact, led to reduction of the activity of the positive allosteric modulator LY2033298 (21, 22).
Fig. 4.
Mutational analysis of the interplay between FFAR1’s three binding sites. (A, C, and D) Dose–response curves of the receptor mutants K259A, A83F, and Y44F are shown for TAK-875, AM-5262, and Compound 1, respectively. (B) Surface/cartoon representation of FFAR1 (PDB 5TZY) with the three binding sites indicated. Docking poses for TAK-875 (orange), AM-5262 (green), and Compound 1 (blue) are shown in stick representation. The location of key mutations tested in this study is marked in purple.
This notion of an allosteric connectivity between the three FFAR1 binding sites was further supported by the mutational analysis of the extracellular site (Fig. 4 and SI Appendix, Table S1), in which all seven mutations had a profound effect on the activity of Compound 1, ranging from 34.6% reduction of Emax (K259A) to a complete loss of activity (K259W, A66W, and Y12W). However, we also observed that several of these mutations impaired the potency of, in particular, TAK-875 (400–500-fold decrease), and in one case (K259W) also decreased the potency of AM-5262 (20-fold). A single mutation (K259A) could be identified that selectively affected the efficacy of Compound 1 without affecting the two other compounds (blue curves in Fig. 4 A, C, and D). Because of their size and lipophilicity, it is not conceivable that TAK-875 or AM-5262 bind to the solvent-exposed extracellular site, also with respect to the crystallographically confirmed binding mode of TAK-875 in the tunnel-like interhelical site. It is more likely that the close proximity between these two sites (L262 is, e.g., a close neighbor to R258, a residue in direct contact with TAK-875) is the underlying cause for the observed effects, and that inserting a mutation into the extracellular site affects affect the dynamic coordination of the neighboring polar network required for anchoring the carboxylic acid moiety of the TAK-875 ligand.
Discussion
The main discovery of the present study is that an extracellularly facing, solvent-exposed empty pocket, present in all three agonist-bound X-ray crystal structures of FFAR1, closes during MD simulations on removal of the prototype agonist TAK-875 from its interhelical site, and that a ligand designed to occupy the extracellular pocket functions as an agonist and allosteric modulator of the receptor. Compound 1 has been designed to bind to the extracellular site, and by presumably preventing the closure of this pocket, it might stabilize an active, signaling conformation of the receptor. In this case, Compound 1 was identified on the basis of its favorable docking score (−28.04) in a representative receptor conformation originating from the largest cluster from the 600-ns MD trajectory in the presence of TAK-875. Docking of Compound 1 into the unrefined X-ray crystal structure (PDB 4PHU), in contrast, resulted only in a score of −18.37, well above the chosen cutoff (−25.00) that was applied in the virtual screening protocol. The MD simulations were thus necessary for the identification of Compound 1, which might underline the importance of accounting for receptor dynamics in structure-based drug discovery efforts and the potential of using MD simulations to study the dynamics of unexploited pockets, which deliberately can be targeted.
Although rational drug design efforts for GPCRs have traditionally focused on pockets between the extracellular segment of the transmembrane helices and loops (23), a number of recent X-ray crystal structures has revealed that ligands in fact can act as agonists or antagonists through binding to multiple alternative sites in the receptors. This includes allosteric sites located more superficially between the extracellular loops (24); extrahelical sites in the transmembrane region (25), similar to the binding site of AP8 in FFAR1 (10); or even sites at the intracellular face of the receptor, involving interactions with the intracellular poles of the TMs as well as ICL1 and helix 8 (26). This is in accordance with hypotheses dating more than 2 decades ago, in which it was proposed that GPCRs would have multiple different agonist binding sites, based on the notion that an agonist did not have to interact with a specific active site, but only needed to stabilize an active conformation of the dynamic receptor structure (27). FFAR1 is a prime example of this, as it appears to possess at least three very distinct binding sites for agonists: the interhelical site for the hydrophobic para-substituted O-phenyl-alkyl acids, such as TAK-875 and MK-8666; an extrahelical lipid-exposed site for meta-substituted O-phenyl-alkyl acids, such as AM-5262 and AP8; and a solvent-exposed extracellular site with more polar properties (Fig. 4). It is, however, still unclear to which sites the endogenous long-chain fatty acid (LCFA) agonists, including the newly identified 20-HETE (28, 29), bind, as they readily can be docked into both the interhelical and the extrahelical site (SI Appendix, Fig. S4) (7). From a signal transduction point of view, endogenous LCFAs function as Gq-only agonists (i.e., similar to TAK-875 and MK-8666), which would indicate that the interhelical binding site is the orthosteric site. Further, it is possible, and even likely, that two LCFA molecules may in fact bind to FFAR1 at the same time, and access both the inter- and extrahelical sites via the lipid bilayer. This might also be the case for some of the synthetic agonists, such as AM-5262, as they dock very well into both sites (7).
In contrast, the extracellular solvent-exposed site is more solvent-exposed and of a rather polar character, as well as a different shape, in relation to both of the two other agonist binding sites, and it is unlikely that ligands binding to either the extrahelical or the interhelical site are able to act on the extracellular site as well. Compared with other GPCRs with crystallographically resolved allosteric binding sites, such as the muscarinic M2 receptor with its modulator binding site in the extracellular vestibule (24), FFAR1’s extracellular site is located in a partially overlapping location, but slightly smaller in size (30) and of more polar character. The M2 allosteric binding site is largely preformed in the presence of an agonist, similar to the stabilizing effect that we observed for the extracellular allosteric pocket in FFAR1 during the MD simulations in the presence of TAK-875.
The MD simulations of FFAR1 suggested that the presence of the agonist TAK-875 in the interhelical site between the extracellular segments of TM-III and TM-IV induced a helical conformation of ICL2, connecting the intracellular poles of these two helices. Secondary structure analysis of ICL2, which initially started in a random coil conformation, revealed that it was more likely to adopt an α-helical conformation in the presence (32% of all ICL2 residues over all frames) than in the absence (6%) of TAK-875 (SI Appendix, Fig. S1 A and B). This long-range allosteric effect appeared to be related to the relatively tight binding of the octadecanoyl tail of a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine molecule from the artificial membrane in the extrahelical groove between the intracellular segments of TM-III and TM-IV over the entire simulation time in a similar binding mode as the crystallographic pose of AP8 in FFAR1 (PDB 5TZY). A helical conformation of ICL2 has been observed in many X-ray crystal structures of agonist-bound GPCRs, and appears to be associated with active-state receptor conformations and interactions with the G proteins (16). Therefore, the extrahelical site of FFAR1 might play a role in stabilizing the important helical formation of ICL2. It is likely that this extrahelical site could function as an allosteric binding site in many other GPCRs, as a similar binding groove is found in many structures of GPCRs and is often occupied with a lipid or detergent molecule (11, 31, 32).
Compound 1 represents an exciting starting point for a different type of FFAR1 ago-allosteric modulators, with respect to both chemotype and allosteric mechanism, which could translate into alternative therapeutic strategies for the treatment of type 2 diabetes (29, 33). The modulator properties of Compound 1 might further enable a more fine-tuned effect on physiological signaling while maintaining the spatial and temporal attributes of endogenous free fatty acid ligands (34).
Materials and Methods
MD Simulations.
All MD simulations were performed in GROMACS 4.6.5 (35), using the CHARMM36 force field (36) and the TIP3P water model. The X-ray crystal structure of FFAR1 (PDB 4PHU) was embedded into a 50-ns preequilibrated palmitoyl-2-oleoylsn-glycero-3-phosphatidylcholine membrane, using g_membed (37). After solvation in a cuboid box, Na+ and Cl− ions were added to neutralize the overall electric charge and to model an ion concentration of 0.15 M. A steepest-descent energy minimization was conducted and the system containing TAK-875 was equilibrated in the NPT ensemble for 40 ns, using decreasing position restraints. Representative structures for the extracellular and interhelical sites were selected by performing a geometric clustering based on the 48 residues that form the extracellular site and the interhelical binding site for TAK-875 and MK-8666, using a Cα-rmsd cutoff value of 1 Å to determine cluster membership (SI Appendix, Fig. S3). Well-tempered metadynamics simulations were performed in GROMACS 5.1.2/PLUMED 2.2.1 (18) using the MD setup described above. We obtained the free-energy profiles of three selected atomic distances: CV1 = E65(Cδ)-R258(Cζ), CV2 = Y240(OH)-K62(Nζ), and CV3 = E65(Cδ)-R258(Cζ) (atom naming according to PDB standard). Additional details are included in the SI Appendix.
Molecular Docking and Virtual Ligand Screening.
We docked the In Stock subset of the ZINC database (38), using SurflexDock (version 2.5) (39), into FFAR1’s extracellular site. The 100,000 best-scored compounds (prefiltered for crash score >−2, polar score >2) were selected for a second docking protocol, using ICM 4D docking and an ensemble of four representative receptor structures (version 3.8–4a; Molsoft L.L.C.). A set of chemical filtering criteria was applied (MW ≤ 500, logP ≤ 5, nrotb ≤ 12, VLS score ≤−25, drug likeness score ≥0.2, internal ligand conformation energy ≤10) to yield 315 compounds from which a small library of 99 compounds was cherry-picked. Additional details are included in the SI Appendix.
Experimental Functional and Binding Studies.
FFAR1 was cloned into the expression vector pCMV-Tag 2B encoding a N-terminal FLAG tag epitope (Stratagene), using the QuikChange method. COS7 cells were transfected using calcium phosphate precipitation method. IP accumulation assays were conducted as described by Hauge et al. (7). Competition binding assays were performed by adding increasing doses of TAK-875 and Compound 1 to the binding buffer (Hepes wash buffer + 100 µg/mL bacitracin), and immediately after, 50 µL tracer solution containing 3H-TAK-875 (5,000 cpm/well) was added. Plates were incubated for 3 h and washed twice, and γ-radiation was counted on a Packard Top Count NXT counter. Additional details are included in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Susanne Hummelgaard for performing the experimental compound screening. We would further like to acknowledge an anonymous referee for proposing additional MD simulations to further substantiate our conclusions about the allosteric mechanism between the different FFAR1 binding sites. The Novo Nordisk Foundation Center for Basic Metabolic Research (NNF10CC1016515) and the Novo Nordisk Foundation Center for Protein Research (NNF14CC0001) are supported by an unconditional grant from the Novo Nordisk Foundation to the University of Copenhagen. M.L. received a faculty scholarship from the Faculty of Health and Medical Sciences, University of Copenhagen. The work is also supported by Challenge Grant NNF14OC0013655 from the Novo Nordisk Foundation (T.W.S). K.L.-L. and J.M. were supported by a Sapere Aude Starting grant from the Independent Research Fund Denmark (DFF).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811066116/-/DCSupplemental.
References
- 1.Christiansen E, et al. Activity of dietary fatty acids on FFA1 and FFA4 and characterisation of pinolenic acid as a dual FFA1/FFA4 agonist with potential effect against metabolic diseases. Br J Nutr. 2015;113:1677–1688. doi: 10.1017/S000711451500118X. [DOI] [PubMed] [Google Scholar]
- 2.Itoh Y, et al. Free fatty acids regulate insulin secretion from pancreatic beta cells through GPR40. Nature. 2003;422:173–176. doi: 10.1038/nature01478. [DOI] [PubMed] [Google Scholar]
- 3.Kotarsky K, Nilsson NE, Flodgren E, Owman C, Olde B. A human cell surface receptor activated by free fatty acids and thiazolidinedione drugs. Biochem Biophys Res Commun. 2003;301:406–410. doi: 10.1016/s0006-291x(02)03064-4. [DOI] [PubMed] [Google Scholar]
- 4.Flodgren E, et al. GPR40 is expressed in glucagon producing cells and affects glucagon secretion. Biochem Biophys Res Commun. 2007;354:240–245. doi: 10.1016/j.bbrc.2006.12.193. [DOI] [PubMed] [Google Scholar]
- 5.Shapiro H, Shachar S, Sekler I, Hershfinkel M, Walker MD. Role of GPR40 in fatty acid action on the beta cell line INS-1E. Biochem Biophys Res Commun. 2005;335:97–104. doi: 10.1016/j.bbrc.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, et al. Free fatty acid receptor agonists for the treatment of type 2 diabetes: Drugs in preclinical to phase II clinical development. Expert Opin Investig Drugs. 2016;25:871–890. doi: 10.1080/13543784.2016.1189530. [DOI] [PubMed] [Google Scholar]
- 7.Hauge M, et al. GPR40 (FFAR1)–Combined Gs and Gq signaling in vitro is associated with robust incretin secretagogue action ex vivo and in vivo. Mol Metab. 2014;4:3–14. doi: 10.1016/j.molmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin DC-H, et al. Identification and pharmacological characterization of multiple allosteric binding sites on the free fatty acid 1 receptor. Mol Pharmacol. 2012;82:843–859. doi: 10.1124/mol.112.079640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava A, et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature. 2014;513:124–127. doi: 10.1038/nature13494. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, et al. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat Struct Mol Biol. 2017;24:570–577. doi: 10.1038/nsmb.3417. [DOI] [PubMed] [Google Scholar]
- 11.Zhang K, et al. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature. 2014;509:115–118. doi: 10.1038/nature13083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu H, et al. Structure of the human κ-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller RL, et al. The importance of ligand-receptor conformational pairs in stabilization: Spotlight on the N/OFQ G protein-coupled receptor. Structure. 2015;23:2291–2299. doi: 10.1016/j.str.2015.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dror RO, et al. Activation mechanism of the β2-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nygaard R, et al. The dynamic process of β(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen SGF, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laio A, Parrinello M. Escaping free-energy minima. Proc Natl Acad Sci USA. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tribello GA, Bonomi M, Branduardi D, Camilloni C, Bussi G. PLUMED 2: New feathers for an old bird. Comput Phys Commun. 2014;185:604–613. [Google Scholar]
- 19.Frimurer TM, et al. Model-based discovery of synthetic agonists for the Zn2+-sensing G-protein-coupled receptor 39 (GPR39) reveals novel biological functions. J Med Chem. 2017;60:886–898. doi: 10.1021/acs.jmedchem.6b00648. [DOI] [PubMed] [Google Scholar]
- 20.Auld DS, Inglese J, Dahlin JL. 2004 Assay interference by aggregation. Assay Guidance Manual, eds Sittampalam GS et al. (Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda, MD). Available at https://www.ncbi.nlm.nih.gov/books/NBK442297/. Accessed December 18, 2018.
- 21.Burger WAC, Sexton PM, Christopoulos A, Thal DM. Toward an understanding of the structural basis of allostery in muscarinic acetylcholine receptors. J Gen Physiol. 2018;150:1360–1372. doi: 10.1085/jgp.201711979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thal DM, et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016;531:335–340. doi: 10.1038/nature17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz TW. Locating ligand-binding sites in 7TM receptors by protein engineering. Curr Opin Biotechnol. 1994;5:434–444. doi: 10.1016/0958-1669(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 24.Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson N, et al. Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature. 2018;553:111–114. doi: 10.1038/nature25025. [DOI] [PubMed] [Google Scholar]
- 26.Oswald C, et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature. 2016;540:462–465. doi: 10.1038/nature20606. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz TW, Rosenkilde MM. Is there a ‘lock’ for all agonist ‘keys’ in 7TM receptors? Trends Pharmacol Sci. 1996;17:213–216. doi: 10.1016/0165-6147(96)10017-1. [DOI] [PubMed] [Google Scholar]
- 28.Tunaru S, et al. 20-HETE promotes glucose-stimulated insulin secretion in an autocrine manner through FFAR1. Nat Commun. 2018;9:177. doi: 10.1038/s41467-017-02539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trauelsen M, Lückmann M, Frimurer TM, Schwartz TW. The HETE is on FFAR1 and pancreatic islet cells. Cell Metab. 2018;27:273–275. doi: 10.1016/j.cmet.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 30.An J, Totrov M, Abagyan R. Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol Cell Proteomics. 2005;4:752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang D, et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature. 2015;520:317–321. doi: 10.1038/nature14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller-Gallacher JL, et al. The 2.1 Å resolution structure of cyanopindolol-bound β1-adrenoceptor identifies an intramembrane Na+ ion that stabilises the ligand-free receptor. PLoS One. 2014;9:e92727. doi: 10.1371/journal.pone.0092727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghislain J, Poitout V. The Role and Future of FFA1 as a Therapeutic Target. Springer International Publishing; New York: 2016. pp. 159–180. [DOI] [PubMed] [Google Scholar]
- 34.Keov P, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors: A pharmacological perspective. Neuropharmacology. 2011;60:24–35. doi: 10.1016/j.neuropharm.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Abraham MJ, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 36.Huang J, MacKerell AD., Jr CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J Comput Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf MG, Hoefling M, Aponte-Santamaría C, Grubmüller H, Groenhof G. g_membed: Efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J Comput Chem. 2010;31:2169–2174. doi: 10.1002/jcc.21507. [DOI] [PubMed] [Google Scholar]
- 38.Sterling T, Irwin JJ. ZINC 15–Ligand discovery for everyone. J Chem Inf Model. 2015;55:2324–2337. doi: 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain AN. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.