Significance
Malaria and cryptosporidiosis are major burdens to both global health and economic development in many countries. Malaria caused >400,000 deaths in 2017, and cryptosporidiosis is estimated to cause >200,000 deaths a year. The spread of drug resistance is a growing concern for malaria treatment, and there is no effective treatment for malnourished or immunocompromised children infected with Cryptosporidium. New treatments with novel mechanisms of action are needed for both diseases. We present a selective inhibitor of both Plasmodium and Cryptosporidium lysyl-tRNA synthetase capable of clearing parasites from mouse models of malaria and cryptosporidiosis infection. This provides very strong validation of lysyl-tRNA synthetase as a drug target in these organisms and a lead for further drug discovery.
Keywords: malaria, cryptosporidiosis, tRNA synthetase
Abstract
Malaria and cryptosporidiosis, caused by apicomplexan parasites, remain major drivers of global child mortality. New drugs for the treatment of malaria and cryptosporidiosis, in particular, are of high priority; however, there are few chemically validated targets. The natural product cladosporin is active against blood- and liver-stage Plasmodium falciparum and Cryptosporidium parvum in cell-culture studies. Target deconvolution in P. falciparum has shown that cladosporin inhibits lysyl-tRNA synthetase (PfKRS1). Here, we report the identification of a series of selective inhibitors of apicomplexan KRSs. Following a biochemical screen, a small-molecule hit was identified and then optimized by using a structure-based approach, supported by structures of both PfKRS1 and C. parvum KRS (CpKRS). In vivo proof of concept was established in an SCID mouse model of malaria, after oral administration (ED90 = 1.5 mg/kg, once a day for 4 d). Furthermore, we successfully identified an opportunity for pathogen hopping based on the structural homology between PfKRS1 and CpKRS. This series of compounds inhibit CpKRS and C. parvum and Cryptosporidium hominis in culture, and our lead compound shows oral efficacy in two cryptosporidiosis mouse models. X-ray crystallography and molecular dynamics simulations have provided a model to rationalize the selectivity of our compounds for PfKRS1 and CpKRS vs. (human) HsKRS. Our work validates apicomplexan KRSs as promising targets for the development of drugs for malaria and cryptosporidiosis.
Malaria is caused by Plasmodium spp.; the most significant species from a disease perspective are Plasmodium falciparum and Plasmodium vivax. In 2017, there were estimated to be 219 million clinical cases of malaria and 435,000 deaths from the disease (1, 2). There is an urgent need for new drugs for malaria to deal with the constant threat of drug resistance and to provide new drugs for chemoprotection, prevention of transmission, and treatment of relapsing (vivax) malaria (2). In humans, cryptosporidiosis is predominantly caused by Cryptosporidium hominis and Cryptosporidium parvum. The recent Global Enteric Multicenter Study has highlighted cryptosporidiosis as a leading cause of moderate-to-severe diarrheal diseases in infants. The association of cryptosporidiosis with death was the highest for any pathogen in 6- to 18-mo-old children with moderate-to-severe diarrhea (3–5). Cryptosporidiosis is estimated to lead to >200,000 deaths a year and is also associated with malnutrition, stunted growth, and cognitive-development problems in children (6). The currently approved drug nitazoxanide has poor efficacy, particularly in the case of immune-compromised patients and malnourished children, where there is no effective treatment (7, 8).
In the last decade, Plasmodium aminoacyl-tRNA synthetases have received increased attention as new targets for antimalarial drug discovery (9). Aminoacyl-tRNA synthetases catalyze aminoacylation of tRNAs with their cognate amino acids in two stages (10). First, the amino acid is activated by ATP to yield the AMP-activated amino acid, with loss of pyrophosphate, followed by transfer of the amino acid onto the tRNA. By way of example, a series of novel antimalarial bicyclic azetidines, identified by phenotypic screening, were found to inhibit cytosolic Plasmodium phenylalanyl-tRNA synthetase (11). These compounds showed activity across multiple life stages of the parasite and in vivo efficacy in a malaria mouse model. Malaria parasite genomes encode two different lysyl-tRNA synthetases (KRSs) that play a role in translation in either the cytoplasm (PfKRS1) or in the apicoplast (PfKRS2) (9, 12, 13), while Cryptosporidium parasites and humans encode one copy. Human KRS (HsKRS) is found in both the cytosol and mitochondrion and has additional roles within human cells (14).
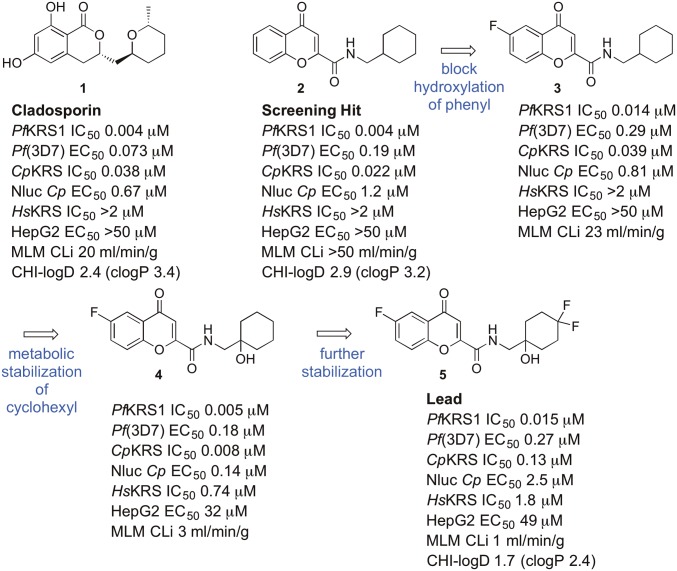
Hoepfner et al. (15) discovered that the fungal secondary metabolite cladosporin 1 (Fig. 1) was a nanomolar inhibitor of parasite growth in both blood and liver stages of Plasmodium. They demonstrated that cladosporin inhibited cytosolic PfKRS1 with >100-fold selectivity compared with HsKRS. Unfortunately, cladosporin is not amenable to development as a drug lead itself because of high metabolic instability (see data we generated in Fig. 1), which would mean that it would not be significantly orally bioavailable. In this work, we report the discovery and optimization of drug-like inhibitors against PfKRS1 and CpKRS, which showed oral activity in mouse models of malaria and cryptosporidiosis.
Fig. 1.
The structure of cladosporin 1, evolution of the lead 5 from the screening hit 2. Enzyme data were obtained with the Kinase-Glo assay. MLM, mouse liver microsomes.
Results and Discussion
Characterization of KRS Enzymes.
We produced recombinant PfKRS1 (77–583 and 80–583), CpKRS (46-end), and HsKRS (full-length) proteins and developed biochemical assays based on the luciferase ATP consumption assay (Kinase-Glo; Promega) (16), which was suitable for the high-throughput screening, and the pyrophosphate generation assay (EnzChek) (17) format for kinetic characterization of the enzymes. The activities of recombinant enzymes were analyzed by monitoring only the first stage of the aminoacylation reaction. This reaction is suitable for high-throughput screening campaigns and makes the reaction more amenable for steady-state kinetic studies. By using the EnzChek assay, which monitors the production of pyrophosphate (when coupled with pyrophosphatase), Km values were obtained for PfKRS1, CpKRS, and HsKRS for ATP and l-lysine (Table 1 and SI Appendix, Fig. S1 and Table S1). The Km values for the human enzyme are significantly smaller than for the parasite enzymes. This may be due to structural differences (see below) between the active sites of the parasite and human enzymes. Even so, the Km values for ATP and l-lysine obtained are comparable with KRSs from other species (https://www.brenda-enzymes.org/) (18).
Table 1.
Kinetic parameters for KRS determined by using EnzCheck
| Enzyme | Km ATP, µM* | HillATP | Km Lys, µM† | Hilll-Lys |
| PfKRS1 | 68 ± 3 | — | 413 ± 37 | 0.89 ± 0.04 |
| CpKRS | 346 ± 128 | 0.71 ± 0.09 | 1,045 ± 640 | 0.49 ± 0.06 |
| HsKRS | 2.22 ± 0.44 | — | 1.92 ± 0.37 | — |
Determined in the presence of saturating concentration of the cosubstrate l-lysine, 5 mM l-lysine for Pf and Cp and 0.075 mM for HsKRS.
Determined in the presence of saturating concentration of the cosubstrate ATP, 0.5 mM ATP for Pf, 2 mM for Cp, and 0.1 mM for HsKRS.
Hit and Lead Discovery.
By using the luciferase ATP consumption (Kinase-Glo) assay platform with sub-Km substrate concentrations (thus biasing the assay toward identifying ATP-competitive inhibitors), the GlaxoSmithKline malaria actives set of ∼13,000 compounds (the Tres Cantos Antimalarial Set) (19) was screened against recombinant PfKRS1, leading to the discovery of a PfKRS1 inhibitor, compound 2 (Fig. 1). Compound 2 displayed similar levels of inhibition of PfKRS1 and growth of P. falciparum to cladosporin 1. Compound 2 suffered from high metabolic instability (CLi > 50 mL⋅min−1⋅g−1 in mouse liver microsomes); however, in contrast to cladosporin 1, it was chemically tractable.
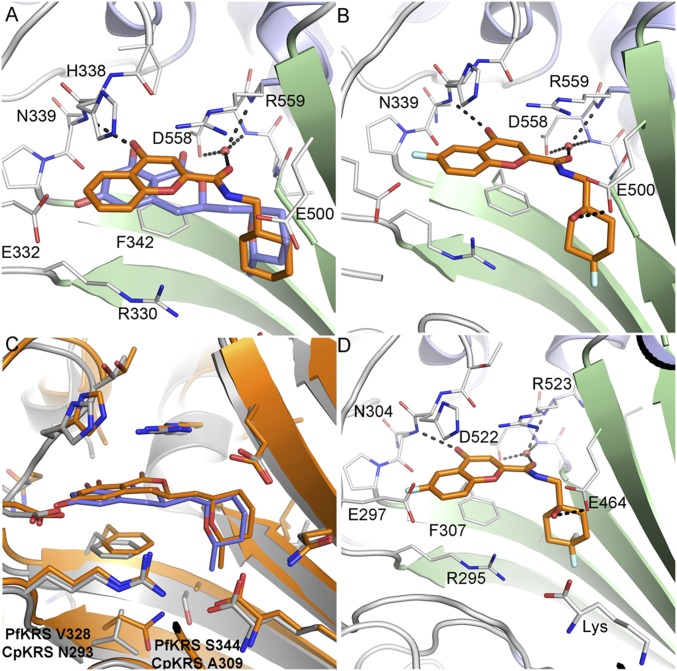
In the published structure of cladosporin bound to PfKRS1, cladosporin binds within the ATP binding pocket (20). The isocoumarin moiety occupies the same space as the adenine ring of ATP, and the pyran system occupies the same position as the ribose ring of ATP. The two phenolic hydroxy groups of the isocoumarin ring form hydrogen bonds with the side chain of E332 and the backbone NH of N339, while the carbonyl interacts with a highly coordinated conserved water molecule (20). Screening hit 2 was cocrystallized with PfKRS1 and also binds in the ATP binding pocket (Fig. 2A), in a similar fashion to cladosporin, although the bicyclic core is rotated 30° with respect to cladosporin. The chromone core stacks between the side chain of F342 on one face and the side chains of H338 and R559 on the other. The ring carbonyl forms an H-bond to the backbone NH of N339, mimicking the N1 of adenine and the O1 OH of cladosporin. The amide carbonyl H-bonds to a highly conserved water molecule coordinated by the side chain of D558 and the backbone NHs of D558 and R559. The cyclohexyl moiety projects into a pocket formed by the side chains of R330, F342, and S344 and the backbone of L555 and G556. This pocket is completed by the substrate lysine and is similar to that occupied by the pyran ring of cladosporin, except that the cyclohexyl ring probes deeper into the pocket (Fig. 2A).
Fig. 2.
Binding modes of ligands bound to PfKRS1 and CpKRS. (A) PfKRS1:Lys:2 showing the binding mode of 2 (C atoms, gold) bound to the ATP site of PfKRS1 (PDB ID code 6AGT) superimposed upon PfKRS1:Lys:cladosporin (cladosporin C atoms, slate; PDB ID code 4PG3). (B) PfKRS1:5 showing binding mode of 5 bound to PfKRS1 (PDB ID code 6HCU). (C) Overlay of CpKRS:Lys:cladosporin (C atoms, gold; PDB ID code 5ELO) compared with PfKRS1:Lys:cladosporin (C atoms, gray; PDB ID code 4PG3). Nonconserved residues within the ligand binding site are labeled. (D) CpKRS:Lys:5 showing binding mode of 5 (C atoms, gold) in complex with CpKRS:Lys (C atoms, gray; PDB ID code 6HCW). H-bonds are shown as dashed lines, and key residues are labeled for clarity.
Metabolite-identification studies suggested that hydroxylation occurred in both the phenyl ring of the chromone and the cyclohexyl ring. By preparing several potential metabolites, we identified the major site of hydroxylation as carbon-6 at the phenyl ring. Addition of a fluorine in the phenyl ring at C-6 blocked hydroxylation at the phenyl ring of the chromone (compound 3; Fig. 1), and introduction of a hydroxyl at the bridgehead of the cyclohexyl substituent was tolerated without loss of potency, while reducing lipophilicity and intrinsic clearance (compound 4). The cocomplex of 4 bound to PfKRS1 showed that the ligand retained the binding mode of 2, conserving key H-bonds from the core scaffold to the protein. The addition of the 6F atom did not afford new interactions with the protein or ordered solvent. The bridgehead hydroxyl was close to the side chain of E500, forming a weak interaction (3.4 Å) and interacting with the ordered water network (SI Appendix, Fig. S7). Addition of fluorines on the 4-position of the cyclohexyl ring in 5 was tolerated and led to excellent metabolic stability, both in mouse and human liver microsomes. (Fig. 1B and Table 2). The complex of 5 bound to PfKRS1 showed that the addition of the difluoro moiety on the cyclohexyl ring had minimal effect upon the position of the ligand within the binding site with respect to 4, and there was no evidence of protein rearrangement. In this complex, all polar interactions were retained, although the H-bond between the bridge-head OH and the side chain of Glu-500 had shortened to 3.0 Å (Fig. 2B).
Table 2.
In vitro activity and DMPK profile of lead compound 5
| Assay | Data |
| P. falciparum 3D7 EC50 | 0.27 μM |
| P. vivax liver schizonts/hypnozoites EC50 (prophylactic mode) | 0.95 μM/>10 μM |
| P. berghei liver schizonts EC50 | 0.9 μM |
| P. falciparum stage V gametocytes EC50 | 9.9 μM |
| P. falciparum male/female gamete formation EC50 | >1 μM |
| FaSSIF solubility | 255 μM |
| Microsomal stability CLint | 1 (mouse), <0.5 (human) mL⋅min−1⋅g−1 |
| Hepatocyte stability CLint | 0.5 (mouse), <0.5 (human) mL⋅min−1⋅g−1 |
| CYP inhibition (CYP1A2, 2D6, 2C9, 2C19, 3A4) IC50 | >10 μM |
| Mouse PK intravenously (dose, Clb, AUC, T1/2, Vdss) | 3 mg/kg, 3.4 mL⋅min−1⋅kg−1, 890 μg⋅min/mL, 2.5 h, 1 L/kg |
| Mouse PK orally (dose, Tmax, Cmax, AUC0–1440, F%) | 10 mg/kg, 2 h, 5.4 μg/mL, 1,300–3,000 μg⋅min/mL, 100% |
Enzymatic studies of the inhibition of PfKRS1 by compound 5 were performed by using the pyrophosphate generation (EnzChek) platform. In the presence of saturating concentrations of both substrates, an IC50 of 210 nM was obtained (SI Appendix, Fig. S2A). To study the mechanism of inhibition by compound 5, single-inhibition measurements were performed at a fixed saturating concentration of one substrate and fixed variable concentrations of the second substrate. Under our experimental conditions, results showed a linear competitive inhibition vs. ATP with a Ki of 32 nM and a linear uncompetitive inhibition vs. l-lysine with a Ki of 212 nM (SI Appendix, Fig. S2 B and C and Table S4). These results indicate that compound 5 competes with ATP for the same binding site and only binds in the presence of l-lysine, also suggesting a sequential ordered kinetic mechanism where l-lysine is the first substrate to bind. The results also show that, in the presence of high concentrations of ATP, the binding affinity of compound 5 is reduced, whereas in the presence of high concentrations of l-lysine, it is increased. Because the mode of inhibition studies are performed at saturating concentration of the cosubstrate, this leads to a lower, more potent Ki against ATP (l-lysine is saturating) and a higher, less potent Ki against l-lysine (ATP is saturating). It is noteworthy that the selectivity ratio for HsKRS/PfKRS1 (120-fold in Kinase-Glo) is similar to the 180-fold cellular selectivity observed between P. falciparum parasites and human HepG2 cells.
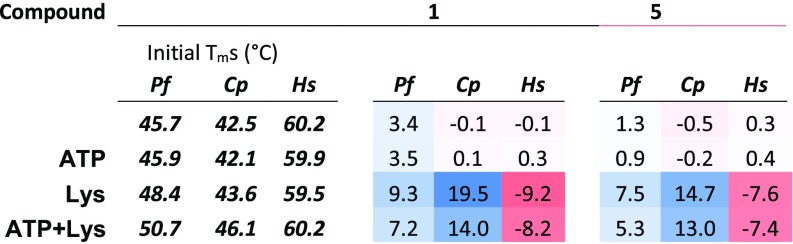
It was reported that cladosporin binds to PfKRS1 in a cooperative manner with l-lysine, leading to significant thermal stabilization (increased melting temperature, Tm) (21). Notably, this stabilization effect was not observed in the human counterpart. To determine whether the chromone series retained a similar stabilization effect, KRS enzymes were incubated with inhibitor and substrates (ATP and l-lysine) in various combinations and gradually heated for observable shifts in Tm. For both PfKRS and CpKRS enzymes, a marked shift (>2 °C) was observed when l-lysine was present, suggesting an analogous codependent binding mode (Fig. 3). This agrees with the results of the study of the mechanism of inhibition of PfKRS1 by compound 5, in which there is a higher Ki determined for this compound in the presence of l-lysine. In contrast, the HsKRS exhibited a reduction in Tm in the presence of inhibitor and l-lysine.
Fig. 3.
Heatmap showing effects of compounds 1 and 5 on the melting temperature (ΔTm) of KRS enzymes.
Lead compound 5 showed good systemic exposure after oral dosing with excellent oral bioavailability (F = 100%) and moderate half-life (T1/2 = 2.5 h) (Table 2). A preliminary selectivity study in a 44 receptor/enzyme panel showed no activity at a concentration of 10 μM. The compound did not show inhibition of a range of cytochrome P450 enzymes and did not inhibit hERG (EC50 > 100 μM). While the compound has a good profile in in vitro assays, compound 5 showed toxicity in mice at higher doses (50 mg/kg orally) and was itself not suitable for further progression. It is likely that the toxicity at higher doses is due to inhibition of mammalian KRS. Indeed, at a dose of 50 mg/kg, the blood concentration of compound 5 in mice reached the EC50 for HepG2 cells. Nonetheless, this compound is a drug-like tool compound to explore KRSs as drug targets.
Profile in Malaria.
Compound 5 was active against both PfKRS1 (IC50 = 0.015 μM) and whole-cell bloodstream P. falciparum 3D7 (EC50 = 0.27 μM) and was selective compared with both the HsKRS (IC50 = 1.8 μM) and HepG2 cells (EC50 = 49 μM). The drop-off from enzyme to cell is probably due to multiple factors—but the two most likely are that high levels of enzyme inhibition may be required for a phenotypic response and the 1,000-fold increase in the concentration of ATP between the enzyme assay and within the parasite cell (22), given that ATP competes for binding with our inhibitors. The activity of 5 against parasites resistant to chloroquine Pf(K1) (EC50 = 0.51 μM) or atovaquone Pf(TM90C2B) (EC50 = 0.52 μM) is similar to the drug-sensitive strain Pf(NF54) (EC50 = 0.39 μM). We investigated the activity against different life-cycle stages of malaria (Table 2). The lead compound 5 showed comparable activity in liver schizonts (P. vivax liver schizont EC50 = 0.95 μM) to asexual blood stages. The in vitro parasite reduction ratio (PRR) assay (23) identified 5 as a compound with a slow rate of killing, displaying an overall biological profile similar to other Plasmodium protein-synthesis inhibitors acting on cytosolic targets and to atovaquone (24) (SI Appendix, Fig. S3).
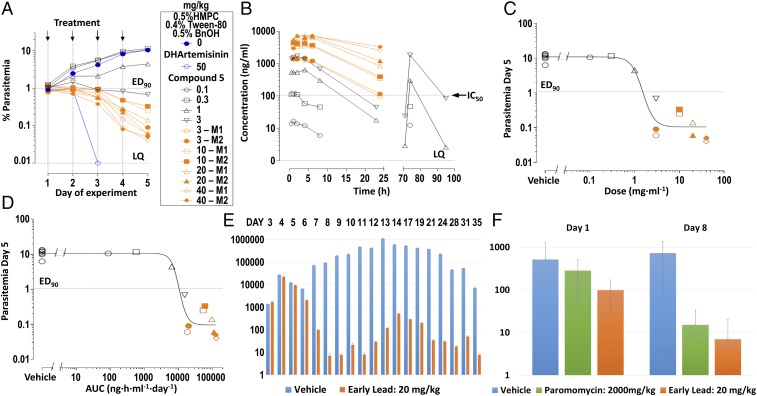
The biological and pharmacokinetic profile was sufficient to justify a rodent efficacy study. Compound 5 was evaluated in vivo against P. falciparum parasites grown in the peripheral blood of NODscidIL2Rγnull mice (SCID), engrafted with human erythrocytes (25). Three days after infection, mice were dosed orally once a day for 4 d with 5, at concentrations up to 40 mg/kg (Fig. 4A). From dose–response studies, a daily oral dose of ED90 = 1.5 mg⋅kg−1 (1.0–2.3 mg⋅kg−1) (Fig. 4C), or its equivalent estimated daily exposure in blood AUCED90 = 11,000 ng⋅h⋅ml−1⋅d−1 (6,900–14,000 ng⋅h⋅ml−1⋅d−1) (Fig. 4D), reduced parasitemia by 90% at day 5 of the study. The rate of parasite clearance in vivo is consistent with the PRR data in vitro.
Fig. 4.
In vivo efficacy of compound 5 in mouse models of malaria and cryptosporidiosis infections. (A) The in vivo efficacy data for compound 5 in P. falciparum-infected SCID mice. (B) The levels of compound 5 in blood of the mice during the malaria efficacy experiment, 24 h after the first oral dose and 24 h after the administration of the last dose on day 4. The symbols represent the same individuals depicted in plot A. (C) Determination of daily dose to reduce P. falciparum parasitemia by 90% at day 5 of the experiment. (D) Determination of daily exposure in blood to reduce P. falciparum parasitemia by 90% at day 5 of the experiment. (E) Efficacy of compound 5 in the Nluc-cryptosporidiosis INF-γ–knockout mouse model when dosed orally at a concentration of 20 mg/kg once a day for 7 d. Orange bars show fecal parasite levels for mice treated with compound 5 and blue bars for the vehicle-treated control mice. At day 8, there is a 10,000-fold reduction in parasite levels compared with control. n = 4 mice per group. (F) Efficacy of compound 5 in the cryptosporidiosis NOD SCID gamma mouse model when dosed orally at a concentration of 20 mg/kg once a day for 7 d. Orange bars represent fecal parasite levels for mice treated with compound 5, green bars mice treated with paromomycin (2,000 mg/kg), and blue bars for vehicle-treated control mice. At day 8, there is a 100-fold reduction in parasite levels compared with control.
Pathogen Hopping: Cryptosporidiosis.
There is a high level of sequence identity within the active-site region of PfKRS1 and CpKRS (96% identity) and an overall sequence identity of 47.7% and similarity of 64.6% across the entire protein. Furthermore, structurally, the active sites are very similar. Therefore, we tested cladosporin, the screening hit 2, and the lead compound 5 in a cellular assay against C. parvum. The three compounds showed inhibition of parasite growth with EC50 of 0.7, 1.2, and 2.5 µM, respectively. Our lead 5 is similarly active against a small panel of isolates: C. hominis (TU502) (EC50 = 6.0 µM) and the C. parvum Iowa strain (EC50 = 1.3 µM). In time-kill curve studies conducted by using C. parvum in the HCT-8 cell-culture system (26), both cladosporin and compound 5 eliminated parasites at an exponential rate, consistent with other protein synthesis inhibitors studied to date (SI Appendix, Fig. S4). Subsequently, a crystal structure was determined of CpKRS in complex with cladosporin and l-lysine [Protein Data Bank (PDB) ID code 5ELO] (Fig. 2C), showing retention of the ligand binding mode compared with the PfKRS1 structure and the high level of sequence conservation within the active site, with only two sequence differences at the base of the ligand binding site (PfKRS1 V328 S344 and CpKRS N293 A309). Further CpKRS:ligand structures were obtained for several compounds within the chromone series, including CpKRS:5 (Fig. 2D), showing 5 to bind in an identical manner to CpKRS as to PfKRS1. In cryptosporidiosis, the parasite is found predominantly in the epithelial cells (enterocytes) in the gastrointestinal tract (8), although it is thought that there may also be some parasites present in the biliary tract. Therefore, it is likely that a compound used for treating cryptosporidiosis would need to have a good exposure in the gastrointestinal tract and possibly also some systemic exposure (27). After oral dosing, compound 5 was completely bioavailable. However, some compound was present in mouse stools (17% of oral dose), suggesting that some biliary excretion had occurred. This raises the possibility of deliberately utilizing enterohepatic recirculation to maintain both gastrointestinal and systemic exposure. Compound 5 showed in vivo efficacy in two different Cryptosporidium mouse models, the NOD SCID gamma and INF-γ–knockout mouse models.
INF-γ–knockout mice (28, 29) were infected orally with Nluc-expressing transgenic C. parvum oocysts. Treatment started upon patency 4 d postinfection (p.i.), and mice were treated orally once a day for 7 d. Infection was monitored daily by luciferase measurements in pooled feces of the entire cage. Mice were followed for 3 wk after completion of drug treatment. Compound 5, when dosed orally at 20 mg/kg once a day for 7 d, reduced parasite shedding below detection level, and this reduction was sustained for 3 wk after treatment had stopped (Fig. 4E). NOD SCID gamma mice were infected with C. parvum oocysts (26). Treatment started 7 d p.i., and mice were treated orally once a day for 7 d. The study was run with four mice per cage; infection was monitored by quantitative PCR on day eight for individual mice, and data are shown as oocysts per milligram of feces. Compound 5 dosed orally at a concentration of 20 mg/kg once a day for 7 d showed 96% reduction of parasite shedding comparable to paromomycin (Fig. 4F).
Molecular Basis of PfKRS1 Inhibitor Selectivity.
Molecular dynamics (MD) simulations were successful in reproducing the binding pose and interactions of compound 5 observed in the co-crystal structure of PfKRS1 (SI Appendix, Fig. S5A). In addition to those interactions, the inhibitor was found to be stabilized by hydrophobic contacts established between the cyclohexyl moiety and the bound substrate l-lysine. Simulations performed in the absence of l-lysine showed a notable destabilization of compound 5, suggesting a key role of l-lysine in the binding of PfKRS1 inhibitors (SI Appendix, Fig. S5B). This was confirmed by both the structural information and the thermal shift assays (Figs. 2 and 3).
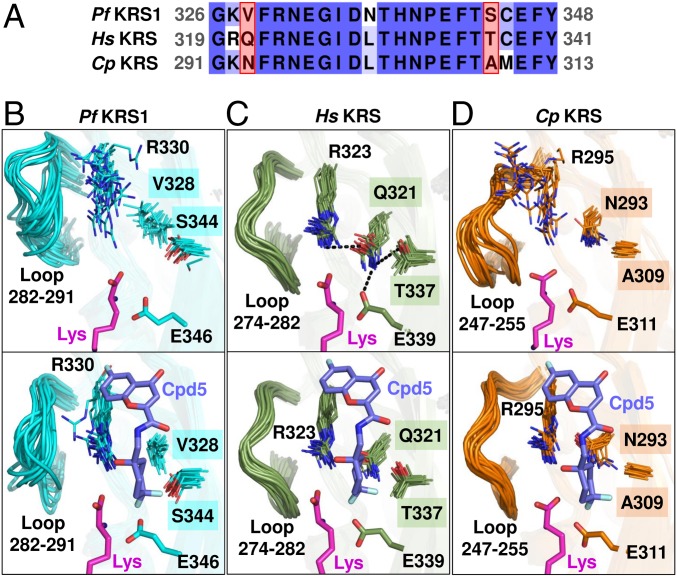
MD simulations of HsKRS predicted a similar binding mode of compound 5 to that in PfKRS1, and the per-residue contributions to the ligand binding energy also showed very similar results (SI Appendix, Fig. S5 C and E). However, despite the overall high sequence and structural similarity between PfKRS1 and HsKRS, two nonconserved residues were present within the active site: V328 and S344 in PfKRS1 correspond to bulkier residues Q321 and T337 in HsKRS (Fig. 5 A–C). To investigate whether this subtle difference might have an impact on the binding process of compound 5, both enzymes were also simulated in the absence of inhibitor (apo systems), with the main focus placed on the conformational features of the binding pocket. In apo-PfKRS1, due to the smaller size of V328 and S344 side chains, the ligand binding pocket remained accessible to compound 5 (Fig. 5B). Conversely, in apo-HsKRS, the binding site remained partially inaccessible due the extended side chain of Q321, which formed a hydrogen-bond network with R323, T337, and E339 (Fig. 5C and SI Appendix, Fig. S6A). Hence, a disruption of the hydrogen-bond interactions within the active site of HsKRS is required to enable the binding of the inhibitor.
Fig. 5.
Differential binding mode of compound 5 to PfKRS1, HsKRS, and CpKRS. (A) Sequence alignment of the active sites of PfKRS1, HsKRS, and CpKRS. Two nonconserved residues suggested to be responsible for the selectivity of compound 5 are highlighted in red boxes. (B–D) Multiframe representations of the active site from the MD simulations of the active sites of PfKRS1 (B) (cyan), HsKRS (C) (green), and CpKRS (D) (orange) in the absence (Upper) and presence (Lower) of compound 5. Lysine is shown in magenta, compound 5 is shown in purple, and nonconserved residue labels are highlighted in boxes.
The comparison of apo and holo PfKRS1 and HsKRS systems also showed significant differences in the dynamics of residues neighboring the active site. In apo-PfKRS1, R330 was highly flexible and was stabilized only after binding of the inhibitor. Similar behavior was observed for the loop 282–291, which was highly disordered in the absence of the ligand, but became ordered upon ligand binding (Fig. 5B and SI Appendix, Fig. S6C). This was corroborated by the marked positive shift in PfKRS1’s Tm in the presence of inhibitor and l-lysine compared with the apo state (Fig. 3). On the other hand, in HsKRS, the equivalent R323 and loop 274–282 remained highly stable, regardless of the presence of the ligand (Fig. 5C and SI Appendix, Fig. S6 B and C). Such ligand-induced stabilization observed for the mobile loop and residues near the PfKRS1 active site could potentially account for a more favorable binding of compound 5 to PfKRS1 with respect to HsKRS. The CpKRS system exhibited behavior similar to PfKRS1: an accessible binding site and a high degree of flexibility of the loop and R295 in the apo-state and dramatic stabilization upon ligand binding, which, again, was supported by the large observed shift in Tm from the apo state to the l-lysine plus inhibitor state (Figs. 3 and 5D and SI Appendix, Fig. S6 B and C). This provided a rationale for the compound 5 affinity toward CpKRS.
The results of MD simulations suggested that the parasite KRS selectivity vs. HsKRS observed for compound 5 was due to a combination of two factors: (i) a more favorable (i.e., more accessible) configuration of the binding site in the parasite enzyme, and (ii) a higher degree of stabilization for the PfKRS1 and CpKRS residues upon ligand binding. Our findings are in good agreement with previous experimental and structural studies that reported an increased flexibility of PfKRS1 over HsKRS and suggested that the active-site loops in aminoacyl-RNA transferases are likely to have a critical role in specific ligand recognition (21, 30, 31). It is likely that full understanding of the observed selectivity can only be obtained by reproducing the entire process of ligand binding to KRS.
In conclusion, identification of the molecular targets of phenotypic hits and subsequent target-based approaches to these targets is a promising way to develop new antiinfective agents. PfKRS1 was shown to be the target of the natural product cladosporin, which was found to be active against P. falciparum in cell culture. Cladosporin itself is not suitable for progression to animal studies, as it is not metabolically stable or orally bioavailable. Given that cladosporin has a complex synthesis with low overall yield (eight steps with an overall yield of 8%) (32), chemical modification to improve the metabolic stability looked challenging, and a long synthesis means that the cost of goods would likely fall outside the Target Product Profiles for malaria and cryptosporidiosis. Therefore, we carried out a small-molecule screen against PfKRS1 to find an alternate chemotype for optimization. Following optimization of a hit molecule (2), we identified a metabolically stable and orally bioavailable compound (5) which inhibited PfKRS1 selectively. A low oral dose (1.5 mg/kg once a day for 4 d) of our KRS inhibitor, compound 5, reduced parasitemia by 90% in the malaria SCID mouse model. These results represent an in vivo validation of PfKRS1 as a promising antimalarial target for drug development. We have also successfully undertaken pathogen-hopping (33), demonstrating that this compound series inhibits CpKRS and the growth of C. parvum in vitro. Moreover, compound 5 showed a reduction of parasite burden by two orders of magnitude when dosed orally for 7 d in two different mouse models of cryptosporidiosis. There are very few validated targets for cryptosporidiosis, and here we chemically validated in vitro and in vivo CpKRS as a target for drug development.
We have carried out MD simulations to understand the observed selectivity of compound 5 and other analogs for PfKRS1 and CpKRS vs. HsKRS. MD simulations suggest that the selectivity observed for compound 5 is due to a combination of a more favorable configuration of the binding site and a higher degree of stabilization upon ligand binding in the parasite enzymes. This work represents a strong validation of lysyl tRNA synthetase as a drug target in both malaria and cryptosporidiosis in animal models. We have identified a valuable tool compound, although further optimization is required in terms of both potency and selectivity to obtain a preclinical candidate.
Materials and Methods
Full details are in SI Appendix. This includes the following information: (i) the chemical synthesis of compounds described in the paper; (ii) the methods for protein expression and purification, kinetic characterization of enzymes, screening of library, and mode of inhibition studies; (iii) the methods for parasite assays using the different life-cycle stages of P. falciparum and different species of Cryptosporidium; (iv) methods for in vitro drug metabolism and pharmacokinetic assay; (v) methods for in vivo pharmacokinetics and efficacy studies; (vi) details for molecular modeling and dynamics simulations; (vii) details of X-ray crystallography; (viii) ethical use of animals; and (ix) detailed author contributions. Ethical approval for rodent experiments was given by the University of Vermont Institutional Animal Care and Use Committee, The Art of Discovery Institutional Animal Care and Use Committee (TAD-IACUC), Veterinäramt Basel Stadt, the University of Dundee “Welfare and Ethical Use of Animals Committee,” and the University of Georgia Animal Care and Use Committee. Human biological samples were sourced ethically and used under informed consent.
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility for beamtime, highlighting the staff of beamlines BM14 and ID29; Diamond Light Source for beamtime (proposal mx10071); the staff of beamline I24 for assistance with crystal testing and data collection; the entire Seattle Structural Genomics Center for Infectious Disease team; the Division of Biological Chemistry and Drug Discovery Protein Production Team; GlaxoSmithKline for the Tres Campos Antimalarial screening set; the Scottish Blood Transfusion Centre (Ninewells Hospital, Dundee) for providing human erythrocytes; Christoph Fischli and Sibylle Sax at the SwissTPH for technical assistance with the SCID mouse model; and Anja Schäfer for technical assistance with the in vitro antimalarial activity testing. The Art of Discovery thanks Dr. Cristina Eguizabal and the Basque Center of Transfusion and Human Tissues (Galdakao, Spain) and the Bank of Blood and Tissues (Barcelona, Spain) for providing human blood. The University of California, San Diego thanks Jenya Antonova-Koch for help. This work was supported by the Bill and Melinda Gates Foundation through Grant OPP1032548 to the Structure-Guided Drug Discovery Coalition and OPP1134302 (to B.S.). This work was also supported in part from federal funds, from the NIH/National Institute of Allergy and Infectious Diseases Grant R21AI123690 (to K.K.O.) and Contracts HHSN272201200025C and HHSN272201700059C (to P.J.M.); Medicines for Malaria Venture (through access to assays to I.H.G. and through RD/08/2800 to J.B.); Wellcome Trust for support of the X-ray Crystallography Facility 094090, IT support Grant 105021 (to I.H.G.), and Institutional Strategic Support Fund 204816 (to A.V.P.), all at the University of Dundee and for Investigator Award 100993 (to J.B.).
Footnotes
Conflict of interest statement: A patent relating to this work has been filed (PCT/GB2017/051809). F.-J.G. and L.M.S. are employees of GlaxoSmithKline and own shares of the company. M.B.J.-D. and I.A.-B. have shares in The Art of Discovery. Editor D.E.G. is a recent coauthor with two authors of this paper. He published a research article with M.A. in 2015. With E.A.W. he published two research articles in 2016, one research article in 2018, and coauthored a research article forthcoming in 2019. D.E.G. is a coinvestigator with E.A.W. on a 2012–2019 grant.
This article is a PNAS Direct Submission. D.E.G. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6AGT, 6HCU, 6HCV, 5ELN, 5ELO, and 6HCW).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814685116/-/DCSupplemental.
References
- 1.De Rycker M, Baragaña B, Duce SL, Gilbert IH. Challenges and recent progress in drug discovery for tropical diseases. Nature. 2018;559:498–506. doi: 10.1038/s41586-018-0327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . World Malaria Report 2018. WHO; Geneva: 2018. [Google Scholar]
- 3.Kotloff KL, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: A reanalysis of the GEMS case-control study. Lancet. 2016;388:1291–1301. doi: 10.1016/S0140-6736(16)31529-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sow SO, et al. The burden of cryptosporidium diarrheal disease among children < 24 months of age in moderate/high mortality regions of sub-Saharan Africa and south Asia, utilizing data from the Global Enteric Multicenter Study (GEMS) PLoS Negl Trop Dis. 2016;10:e0004729. doi: 10.1371/journal.pntd.0004729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MAL-ED Network Investigators Childhood stunting in relation to the pre- and postnatal environment during the first 2 years of life: The MAL-ED longitudinal birth cohort study. PLoS Med. 2017;14:e1002408. doi: 10.1371/journal.pmed.1002408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amadi B, et al. High dose prolonged treatment with nitazoxanide is not effective for cryptosporidiosis in HIV positive Zambian children: A randomised controlled trial. BMC Infect Dis. 2009;9:195. doi: 10.1186/1471-2334-9-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manjunatha UH, Chao AT, Leong FJ, Diagana TT. Cryptosporidiosis drug discovery: Opportunities and challenges. ACS Infect Dis. 2016;2:530–537. doi: 10.1021/acsinfecdis.6b00094. [DOI] [PubMed] [Google Scholar]
- 9.Manickam Y, et al. Drug targeting of one or more aminoacyl-tRNA synthetase in the malaria parasite Plasmodium falciparum. Drug Discov Today. 2018;23:1233–1240. doi: 10.1016/j.drudis.2018.01.050. [DOI] [PubMed] [Google Scholar]
- 10.Fang P, Guo M. Evolutionary limitation and opportunities for developing tRNA synthetase inhibitors with 5-binding-mode classification. Life (Basel) 2015;5:1703–1725. doi: 10.3390/life5041703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato N, et al. Diversity-oriented synthesis yields novel multistage antimalarial inhibitors. Nature. 2016;538:344–349. doi: 10.1038/nature19804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhatt TK, et al. A genomic glimpse of aminoacyl-tRNA synthetases in malaria parasite Plasmodium falciparum. BMC Genomics. 2009;10:644. doi: 10.1186/1471-2164-10-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoen R, et al. Selective inhibition of an apicoplastic aminoacyl-tRNA synthetase from Plasmodium falciparum. ChemBioChem. 2013;14:499–509. doi: 10.1002/cbic.201200620. [DOI] [PubMed] [Google Scholar]
- 14.Fang P, Guo M. Structural characterization of human aminoacyl-tRNA synthetases for translational and nontranslational functions. Methods. 2017;113:83–90. doi: 10.1016/j.ymeth.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Hoepfner D, et al. Selective and specific inhibition of the plasmodium falciparum lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin. Cell Host Microbe. 2012;11:654–663. doi: 10.1016/j.chom.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cali JJ, et al. Bioluminescent assays for ADMET. Expert Opin Drug Metab Toxicol. 2008;4:103–120. doi: 10.1517/17425255.4.1.103. [DOI] [PubMed] [Google Scholar]
- 17.Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci USA. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Placzek S, et al. BRENDA in 2017: New perspectives and new tools in BRENDA. Nucleic Acids Res. 2017;45:D380–D388. doi: 10.1093/nar/gkw952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gamo FJ, et al. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- 20.Khan S, Sharma A, Belrhali H, Yogavel M, Sharma A. Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin. J Struct Funct Genomics. 2014;15:63–71. doi: 10.1007/s10969-014-9182-1. [DOI] [PubMed] [Google Scholar]
- 21.Fang P, et al. Structural basis for specific inhibition of tRNA synthetase by an ATP competitive inhibitor. Chem Biol. 2015;22:734–744. doi: 10.1016/j.chembiol.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saliba KJ, Kirk K. pH regulation in the intracellular malaria parasite, Plasmodium falciparum. H(+) extrusion via a V-type H(+)-ATPase. J Biol Chem. 1999;274:33213–33219. doi: 10.1074/jbc.274.47.33213. [DOI] [PubMed] [Google Scholar]
- 23.Sanz LM, et al. P. falciparum in vitro killing rates allow to discriminate between different antimalarial mode-of-action. PLoS One. 2012;7:e30949. doi: 10.1371/journal.pone.0030949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baragaña B, et al. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature. 2015;522:315–320. doi: 10.1038/nature14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiménez-Díaz MB, et al. Improved murine model of malaria using Plasmodium falciparum competent strains and non-myelodepleted NOD-scid IL2Rgammanull mice engrafted with human erythrocytes. Antimicrob Agents Chemother. 2009;53:4533–4536. doi: 10.1128/AAC.00519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jumani RS, et al. A novel piperazine-based drug lead for cryptosporidiosis from the medicines for malaria venture open-access malaria box. Antimicrob Agents Chemother. 2018;62:e01505-17. doi: 10.1128/AAC.01505-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold SLM, et al. Necessity of bumped kinase inhibitor gastrointestinal exposure in treating Cryptosporidium infection. J Infect Dis. 2017;216:55–63. doi: 10.1093/infdis/jix247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vinayak S, et al. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature. 2015;523:477–480. doi: 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manjunatha UH, et al. A Cryptosporidium PI(4)K inhibitor is a drug candidate for cryptosporidiosis. Nature. 2017;546:376–380. doi: 10.1038/nature22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bilokapic S, et al. Structure of the unusual seryl-tRNA synthetase reveals a distinct zinc-dependent mode of substrate recognition. EMBO J. 2006;25:2498–2509. doi: 10.1038/sj.emboj.7601129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yaremchuk A, Tukalo M, Grøtli M, Cusack S. A succession of substrate induced conformational changes ensures the amino acid specificity of Thermus thermophilus prolyl-tRNA synthetase: Comparison with histidyl-tRNA synthetase. J Mol Biol. 2001;309:989–1002. doi: 10.1006/jmbi.2001.4712. [DOI] [PubMed] [Google Scholar]
- 32.Zheng H, et al. Asymmetric total synthesis of cladosporin and isocladosporin. J Org Chem. 2012;77:5656–5663. doi: 10.1021/jo300805n. [DOI] [PubMed] [Google Scholar]
- 33.Jain V, Sharma A, Singh G, Yogavel M, Sharma A. Structure-based targeting of orthologous pathogen proteins accelerates antiparasitic drug discovery. ACS Infect Dis. 2017;3:281–292. doi: 10.1021/acsinfecdis.6b00181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.