Significance
Photosynthesis has been gained many times in eukaryotic evolution via endosymbiosis. It has also been lost many times, including multiple occasions in the chrysophyte algae, a lineage of unicellular algae related to diatoms. This study reveals the functions of nonphotosynthetic chrysophyte plastids in six lineages that have lost photosynthesis independently. We see a remarkable degree of convergence in retained functions among these chrysophyte lineages. Moreover, the retained functions are highly similar to those of apicomplexans such as the malaria parasite Plasmodium. The shared losses of function provide insight into the principles of and constraints on plastid reductive evolution, not only within chrysophytes, but across photosynthetic and secondarily nonphotosynthetic eukaryotes.
Keywords: heterotrophy, ochrophyte, phylogenomics, dual targeting, protist
Abstract
The division of life into producers and consumers is blurred by evolution. For example, eukaryotic phototrophs can lose the capacity to photosynthesize, although they may retain vestigial plastids that perform other essential cellular functions. Chrysophyte algae have undergone a particularly large number of photosynthesis losses. Here, we present a plastid genome sequence from a nonphotosynthetic chrysophyte, “Spumella” sp. NIES-1846, and show that it has retained a nearly identical set of plastid-encoded functions as apicomplexan parasites. Our transcriptomic analysis of 12 different photosynthetic and nonphotosynthetic chrysophyte lineages reveals remarkable convergence in the functions of these nonphotosynthetic plastids, along with informative lineage-specific retentions and losses. At one extreme, Cornospumella fuschlensis retains many photosynthesis-associated proteins, although it appears to have lost the reductive pentose phosphate pathway and most plastid amino acid metabolism pathways. At the other extreme, Paraphysomonas lacks plastid-targeted proteins associated with gene expression and all metabolic pathways that require plastid-encoded partners, indicating a complete loss of plastid DNA in this genus. Intriguingly, some of the nucleus-encoded proteins that once functioned in the expression of the Paraphysomonas plastid genome have been retained. These proteins were likely to have been dual targeted to the plastid and mitochondria of the chrysophyte ancestor, and are uniquely targeted to the mitochondria in Paraphysomonas. Our comparative analyses provide insights into the process of functional reduction in nonphotosynthetic plastids.
Although photosynthesis should be beneficial to any organism by converting solar power to available ATP and NADPH, some algal and plant lineages have lost their ability to photosynthesize. Some secondarily nonphotosynthetic taxa are parasites, including the apicomplexans (e.g., the malaria pathogen Plasmodium falciparum) and several parasitic plant lineages (1, 2). Others are phagotrophs or osmotrophs, including many lineages descended from microalgae including green algae, euglenophytes, dinoflagellates, cryptophytes, and the “ochrophyte” group that includes diatoms and chrysophytes (sometimes called “golden algae”) (3–8).
Regardless of their current ecological modes, secondarily nonphotosynthetic organisms frequently retain nonphotosynthetic organelles descended from their original chloroplasts. These organelles typically no longer perform the metabolic functions associated with photosynthesis, or dependent on photosynthetic ATP and NAD(P)H production (2, 9). However, these plastids may still perform essential nonphotosynthetic functions, such as cofactor biosynthesis (e.g., Fe–S cluster and haem), fatty acid and isoprenoid biosynthesis, and aspects of respiratory carbon metabolism (9). These processes may depend both on plastid-targeted proteins, encoded in the nucleus, and on proteins encoded within the plastid itself (e.g., the cysteine desulfurase gene sufB for Fe–S complex assembly) (1). Thus, most nonphotosynthetic plastids retain their own genomes as well as the genes essential for the maintenance and expression of those genomes (2). Some exceptions to these rules have been documented. For example, some nonphotosynthetic plants and algae retain genes in their nuclear and plastid genomes previously associated with photosynthesis (2, 3, 10). Conversely, some other secondarily nonphotosynthetic organisms have either completely lost their plastid [e.g., the dinoflagellate Hematodinium; the apicomplexan Cryptosporidium (11)] or retain a plastid that lacks an associated genome (e.g., the green alga Polytomella; and the dinoflagellate relative Perkinsus) (1, 6).
Although multiple secondarily nonphotosynthetic plastids have been studied, it is not clear to what extent these plastids converge on the same functions. Why are plastid genomes retained versus lost after loss of photosynthesis? What becomes of the plastid-targeted proteins in different secondarily nonphotosynthetic lineages? To close these knowledge gaps, we have investigated plastid-related pathways in chrysophyte algae. The chrysophytes (including synurophytes) are part of the ochrophyte group, containing plastids derived from the secondary endosymbiosis of a red alga, and are functionally and trophically diverse, including swimming, sessile, single-celled, and colonial forms; unarmored and silicified cells; and obligate phototrophs, photo-mixotrophs, and obligate heterotroph lineages (5, 12–14). The obligate heterotrophic species are globally distributed and abundant components of freshwater, pelagic, and benthic microbial communities (14, 15). These species are typically phagotrophic predators, consuming bacteria and in certain cases microbial eukaryotes (16, 17), using their long flagellum to selectively sweep prey into endocytotic “feeding cups” that form on the cell surface (18, 19). An alternative phagocytotic feeding strategy, using rhizopodia, has been inferred in the genus Chrysamoeba, and some chrysophytes can supplement their phagocytotic activities via the osmotrophic uptake of dissolved organic vitamin B12 and biotin (18); however, to date, no parasitic or solely osmotrophic nonphototrophic chrysophytes have been described (14). Plastid-derived organelles have been identified by microscopy in multiple nonphotosynthetic chrysophytes [e.g., Paraphysomonas spp. (20) and “Spumella” sp. NIES-1846 (16)], confirming their utility for exploring nonphotosynthetic plastid evolution.
In this study, we demonstrate that photosynthesis has been lost multiple times in chrysophytes, as revealed by multigene and taxon-dense 18S rRNA gene phylogenies. We present a sequenced plastid genome of a nonphotosynthetic chrysophyte, “Spumella” sp. NIES-1846, alongside transcriptome-based reconstructions of the plastid proteomes of a diverse range of nonphotosynthetic chrysophytes. Our data reveal that nonphotosynthetic chrysophyte species show a high degree of convergence with respect to which plastid functions have been retained, both to each other, and to apicomplexan parasites. However, we also identify exceptions, including limited reductive evolution in the nonphotosynthetic plastids of Cornospumella, and present evidence for a dramatically reduced plastid that lacks DNA in Paraphysomonas. Finally, we show that proteins that historically functioned in the expression of the Paraphysomonas plastid genome now solely support the biology of the mitochondria. Our data provide insights into the evolutionary mechanisms that surround the loss of photosynthesis, and the long-term interactions between different cellular compartments in plastid-bearing organisms.
Results
Multigene Phylogeny of Chrysophytes.
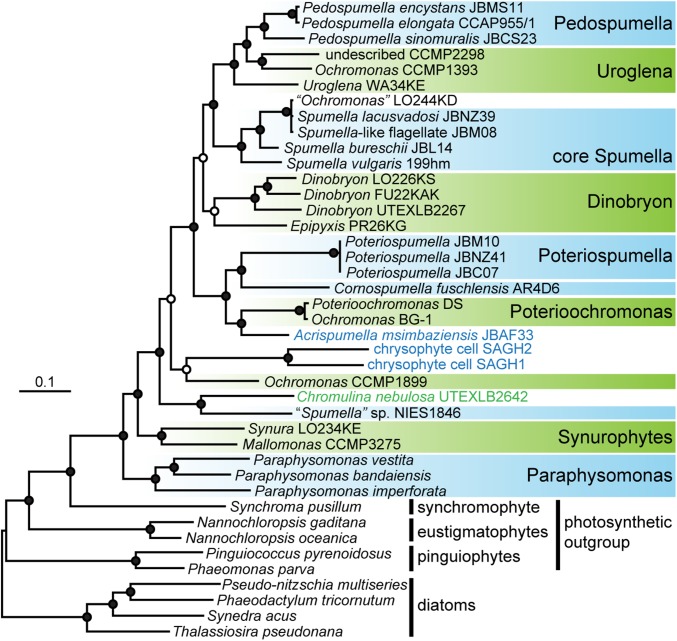
To trace how frequently photosynthesis has been lost in chrysophytes, we constructed phylogenies from a 41-taxa × 17,439-amino acid dataset, including our newly assembled transcriptome data of “Spumella” sp. NIES-1846 and previously published chrysophyte sequence libraries (12, 15, 21). Both maximum likelihood and Bayesian analyses provided well-resolved trees containing strongly supported clades (Fig. 1).
Fig. 1.
Multigene Bayesian consensus phylogeny of chrysophytes inferred from 17,439 amino acid sites under three substitution models. Photosynthetic and secondarily nonphotosynthetic chrysophyte lineages are indicated in green and blue, respectively; the trophic status of Ochromonas LO244KD remains debated (15, 21). Taxa used for subsequent plastid metabolism comparisons are contained in boxes. Filled circles at nodes indicate Bayesian posterior probabilities of 1.0 and maximum likelihood bootstrap support greater than 80% for all analyses. Open circles at nodes indicate Bayesian posterior probabilities greater than 0.8 for all analyses. The topology is displayed following (5), with the root between the diatoms and the PESC clade (pinguiophytes, eustigmatophytes, synchromophytes, and chrysophytes). A corresponding 18S rDNA tree is shown in SI Appendix, Fig. S1.
Our tree contains seven distinct clades of nonphotosynthetic chrysophytes, separated from one another by photosynthetic lineages (Fig. 1, blue boxes plus Acrispumella and SAGH cells). We additionally analyzed a more taxon-rich 18S rRNA gene phylogeny, identifying multiple distinct nonphotosynthetic clades in total, of which 13 could be resolved with strong support (Bayesian PP >0.9; RAxML bootstrap >60% for three different alignments, with 50%, 80%, and 90% occupancy at each site, respectively; SI Appendix, Fig. S1). We found that Cornospumella and Poteriospumella are separated by photosynthetic lineages in the 18S rRNA tree with high statistical support, indicative of eight independent lineages of nonphotosynthetic chrysophytes for which transcriptome data are available (Fig. 1 and SI Appendix, Fig. S1).
Convergent Evolution of Nonphotosynthetic Chrysophyte and Apicomplexan Plastid Genomes.
As a first investigation into plastid function in nonphotosynthetic chrysophytes, we sequenced the plastid genome of “Spumella” sp. NIES-1846. This strain forms the second-deepest branch of nonphotosynthetic chrysophytes for which extensive sequence data are available (Fig. 1), and was selected for study because of its rapid growth under laboratory conditions, because its nonphotosynthetic plastid has been extensively morphologically well characterized (16), and because our transcriptomic survey indicates that it has a typically reduced plastid-associated metabolism for a nonphotosynthetic chrysophyte (discussed below). The “Spumella” sp. NIES-1846 plastid genome is a circularly mapping molecule, containing two inverted repeats and small and large single copy regions (SI Appendix, Fig. S2). It is 53,209 bp in length and contains 45 protein-coding genes. This is 2.5 times smaller than the plastid genome of the photosynthetic chrysophyte Ochromonas sp. CCMP1393 (22) in both size (126,750 bp) and the number of encoded proteins (124; Fig. 2A), mainly due to the loss of photosynthesis-related functions. We found only one photosystem subunit (ferredoxin, petF), and none of the genes for carbon fixation, chlorophyll biosynthesis, cytochrome biogenesis (ccs), or acetolactate synthesis (ilv) in the “Spumella” sp. NIES-1846 plastid genome, that are typically retained on the plastid genomes of photosynthetic chrysophytes [e.g., Ochromonas sp. CCMP1393, Mallomonas splendens (22)]. We also could not find genes encoding sec and tat complexes for protein transport into the thylakoid lumen, consistent with previous reports noting the lack of thylakoid-like structures inside the “Spumella” sp. NIES-1846 plastid (16).
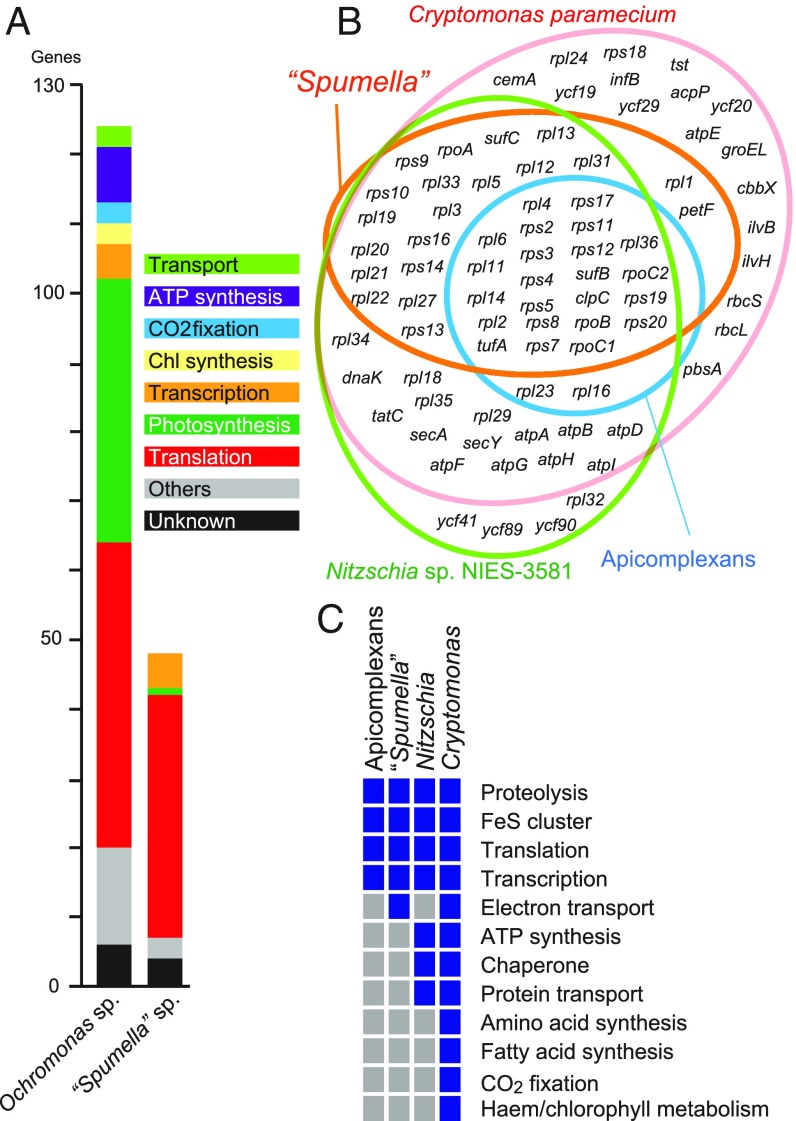
Fig. 2.
Plastid genome of “Spumella” sp. NIES-1846. (A) Comparison of protein-coding genes between a photosynthetic chrysophyte and “Spumella” sp. NIES-1846 (22). Each color bar shows a functional category. (B) Venn diagram of protein-coding genome contents in various nonphotosynthetic, red-alga–derived plastid lineages. Nitzschia sp. NIES-3581 (3) and Cryptomonas paramecium (4) are used as representatives for nonphotosynthetic diatoms and cryptomonads. (C) Plastid-encoded functions in four nonphotosynthetic, red-alga–derived plastid lineages. Blue and gray boxes indicate presence and absence, respectively. A complete plastid genome map is provided in SI Appendix, Fig. S2; comparisons of plastid coding content in a wider range of nonphotosynthetic species are shown in SI Appendix, Fig. S3; and schematic reconstructions and exemplar localizations of plastid metabolism pathways are in SI Appendix, Figs. S4–S6.
A set of genes with photosynthesis-independent functions is retained in the “Spumella” sp. NIES-1846 plastid genome, including two rRNA and 25 tRNA genes, 36 genes encoding proteins for translation (rps, rpl, tufA), five for transcription (rpo), two for Fe–S cluster assembly (suf), and one for proteolysis (clpC). Four ORFs had no obvious homologs to other organisms, as assessed by BLAST search with threshold e value of ≤10−5. We compared the coding content of the “Spumella” sp. NIES-1846 plastid to that of other red alga-derived, secondarily nonphotosynthetic plastids within the diatoms (Nitzschia) (3), cryptomonads (Cryptomonas) (4), and apicomplexans (1) (Fig. 2 B and C), and to a broader set of nonphotosynthetic plastids including those of plants and green algae (SI Appendix, Fig. S3). The “Spumella” sp. NIES-1846 plastid genome retains fewer protein-coding genes and encodes a narrower range of functions than the nonphotosynthetic diatom and cryptophyte plastid genomes. However, the “Spumella” sp. NIES-1846 plastid genome encodes a remarkably similar set of functions to the plastid genomes found in apicomplexan plastid lineages, that is, gene expression, protein import, and Fe–S cluster biosynthesis, with the only difference being the retention of petF in “Spumella” sp. NIES-1846 (Fig. 2C and SI Appendix, Fig. S3). This is indicative of convergence in function between nonphotosynthetic chrysophyte and apicomplexan plastids.
Reductive Evolution of “Spumella” sp. NIES-1846 Plastid Metabolism.
Next, we considered which plastid-targeted proteins support the biology of the “Spumella” sp. NIES-1846 plastid. We searched our “Spumella” sp. NIES-1846 transcriptome for enzymes frequently associated with nonphotosynthetic plastids, including haem, carbon, isopentenyl pyrophosphate (IPP), and lipid metabolism (1). For example, we found 8 of the 10 plastid-targeted proteins required for haem biosynthesis, of which 5 possessed detectable plastid-targeting signals (SI Appendix, Fig. S4). We created a recombinant plasmid encoding the N-terminal region of “Spumella” sp. NIES-1846 ferrochelatase [which catalyzes the last haem biosynthesis step (5)] with a C-terminal GFP fusion and expressed it in the diatom Phaeodactylum tricornutum. We observed GFP colocalizing with chlorophyll autofluorescence, indicating that haem biosynthesis occurs in the “Spumella” sp. NIES-1846 plastid (SI Appendix, Fig. S5A).
We detected evidence for a modified plastid carbon metabolism in “Spumella” sp. NIES-1846. This consists of plastid-targeted proteins that would enable the glycolytic conversion of triose phosphate into phospho-enol-pyruvate (i.e., glyceraldehyde 3-phosphate dehydrogenase, triose phosphate isomerase, phosphoglycerate kinase, phosphoglycerate mutase, and enolase) (SI Appendix, Fig. S4) (5). We could not detect plastid-targeted enzymes involved in the reductive pentose phosphate pathway (i.e., the “Calvin cycle”; SI Appendix, Fig. S4), enzymes that catalyze the interconversion of phosphoenolpyruvate and pyruvate (pyruvate kinase and pyruvate phosphate dikinase), or a plastid-targeted pyruvate dehydrogenase complex (SI Appendix, Fig. S4). This suggests that the “Spumella” sp. NIES-1846 plastid engages in fermentative carbon metabolism but does not synthesize pyruvate.
We did not find genes for plastid fatty acid metabolism in “Spumella” sp. NIES-1846. This is consistent with the carbon metabolism pathways observed, as plastid fatty acid biosynthesis begins with pyruvate (SI Appendix, Fig. S4) (5). Even though several homologs for fatty acid biosynthesis were detected in the transcriptome data, they were all related to nonplastid enzymes of other ochrophyte lineages, and none was found to possess explicit plastid targeting signals, instead being predicted to localize variously to the mitochondria, endomembrane system, or cytoplasm (SI Appendix, Fig. S6). We confirmed the completeness of three fatty acid synthesis genes (FabD, FabF, and FabG) by 5′-rapid amplification of cDNA ends (5′-RACE) analyses (SI Appendix, Fig. S5B). We also tested the localization of FabG by expression of the N-terminal region fused with GFP in Phaeodactylum and observed colocalization with MitoTracker Orange, indicating a mitochondrial localization (SI Appendix, Fig. S5C). Thus, our data support a complete loss of fatty acid synthesis from the “Spumella” sp. NIES-1846 plastid. We did identify putative plastid-associated copies of enzymes (i.e., genes encoding plastid-targeting sequences, which resolved phylogenetically with other ochrophyte plastid-targeted enzymes) involved in lipid head group metabolism (diacylglycerol acetyltransferase, glycerol-3-phosphate acetyltransferase, and phosphatidate cytidyltransferase), indicating that the “Spumella” sp. NIES-1846 plastid may still utilize this compartment for the synthesis of triglycerides from free fatty acids and glycerol, as in other ochrophytes (SI Appendix, Fig. S6). We additionally note that at least one of the enzymes associated with fatty acid catabolism in “Spumella” sp. NIES-1846, lysophospholipase, has an inferred mitochondrial localization, suggesting that in this species certain steps of fatty acid synthesis and catabolism may occur in the same subcellular compartment (SI Appendix, Fig. S6).
Convergent Evolution of Plastid Metabolism in Nonphotosynthetic Chrysophytes.
Photosynthesis has been independently lost in many distinct chrysophyte lineages (Fig. 1 and SI Appendix, Fig. S1). We wanted to determine whether different secondarily nonphotosynthetic chrysophytes retain similar plastid functions to “Spumella” sp. NIES-1846 or follow distinct trajectories of plastid reduction. We used a published protocol, based on BLAST searches with floating e-value thresholds (5), to identify orthologs of 9,531 ochrophyte plastid-targeted proteins in photosynthetic and nonphotosynthetic members of the “PESC” clade (pinguiophytes, eustigmatophytes, synchromophytes, and chrysophytes) (5, 12, 15, 21) (Dataset S1). We used custom in silico prediction thresholds, guided by experimental data, to infer localizations of each protein (Materials and Methods).
We noted a drastic reduction in the sizes of the plastid proteomes of nonphotosynthetic chrysophytes, both in terms of the number of query proteins for which homologs could be found and the proportion of these homologs inferred to possess plastid-targeting sequences (SI Appendix, Fig. S7). To understand what underpins these reductions in plastid proteome content, we constructed phylogenies to infer the presence of 303 core plastid metabolism and biogenesis proteins (5) across the tree of chrysophytes. We used our phylogenetic analyses to define monophyletic groups of photosynthetic species and groups of nonphotosynthetic species that are derived from a single loss of photosynthesis (Fig. 1, green and blue taxa, respectively). We pooled transcriptomes within each group to reduce the likelihood of falsely inferring protein losses from incomplete transcriptomes. A total of six of the pooled libraries of photosynthetic lineages and six of the pooled libraries of nonphotosynthetic lineages were inferred to cover more than 65% of the query proteins in the BUSCO, version 2, library and were retained for further analysis (Fig. 1 and SI Appendix, Fig. S7B).
Several plastid-targeted proteins were commonly missing across the six investigated nonphotosynthetic lineages (Fig. 3, SI Appendix, Fig. S8, and Dataset S1). These include photosystem subunits and light-harvesting complex proteins; the reductive pentose phosphate pathway; chlorophyll synthesis; the nonmevalonate pathway for IPP synthesis and carotenoid metabolism; core plastid amino acid synthesis (glutamine/glutamate, lysine, branched-chain and aromatic amino acids); and thylakoid protein import and biogenesis proteins (Fig. 3). Several of these losses have been documented in a smaller-scale study of Poteriospumella lacustris, Pedospumella encystans, and Spumella vulgaris (12). These pathways are almost universally detected in photosynthetic PESC clade members, suggesting that they have been lost independently alongside the loss of photosynthesis (Fig. 3 and SI Appendix, Fig. S8).
Fig. 3.
Plastid-targeted proteome content across chrysophyte lineages. This heatmap shows the functional distributions of proteins inferred to be orthologous to other ochrophyte plastid proteins, via a phylogenetic approach, in published PESC clade sequence libraries (5). Cells are shaded purple if at least one member of the pathway was identified to possess a plastid-targeting sequence; orange if at least one member was identified to possess a mitochondrial-targeting sequence; and gray if homologs were found; but none of these possessed organelle-targeting sequences, in other words, correspond to cytoplasmic, or N-incomplete homologs. Coverage statistics for each library are considered in SI Appendix, Fig. S7; detailed outputs for individual proteins in each pathway are shown in SI Appendix, Fig. S8; an analogous map of orthologs of plastid-encoded genes in chrysophyte transcriptomes is shown in SI Appendix, Fig. S9; and exemplar tree topologies are shown in SI Appendix, Figs. S10–S12.
Other plastid-targeted proteins are broadly conserved across nonphotosynthetic chrysophytes. These include proteins associated with oxidative carbon metabolism and plastid glycolysis: haem synthesis, fatty acid and steroid metabolism, cysteine and Fe–S cluster synthesis, and plastid protein import, division, and genome expression proteins (Fig. 3 and SI Appendix, Fig. S8). Because each nonphotosynthetic chrysophyte group has independently lost photosynthesis, these observations represent convergent evolution in plastid function (Figs. 1 and 3 and SI Appendix, Fig. S8).
Alongside this, we searched each PESC clade transcriptome library for orthologs of genes known to be encoded in other chrysophyte plastids (SI Appendix, Fig. S9 and Dataset S1) (22, 23). These sequences might represent fragments of plastid transcript sequence that survived poly(A) RNA enrichment, or alternatively transcripts of genes relocated from the plastid to the nucleus of individual chrysophytes, which may be inferred by the presence of N-terminal targeting sequences (5). We uncovered sporadic evidence for plastid DNA-derived sequences in both photosynthetic and nonphotosynthetic chrysophyte species, including three proteins likely to be the plastid-targeted expression products of genes that had been relocated to the nuclei of specific PESC clade species: tsf (encoding elongation factor Ts) in Poterioochromonas and Epipyxis sp., and petJ (cytochrome c6) in Pinguiococcus pyrenoidosus (SI Appendix, Fig. S9).
Cornospumella and Paraphysomonas Represent Extremes in Chrysophyte Plastid Evolution.
Alongside convergent changes, we noted lineage-specific plastid functions in nonphotosynthetic chrysophytes (Fig. 3 and SI Appendix, Fig. S8). These include the absence of plastid fatty acid synthesis from “Spumella” sp. NIES-1846, which we detected in all other nonphotosynthetic chrysophytes surveyed (Fig. 3 and SI Appendix, Figs. S6 and S8E). The least reduced plastid belongs to Cornospumella fuschlensis (Fig. 3). C. fuschlensis retains multiple plastid-targeted proteins associated with photosystems (e.g., PsbP) and light-harvesting complexes (e.g., LI818/Lhcx) of clear chrysophyte origin (SI Appendix, Figs. S10 and S11). The only plastid functions not detected at all in Cornospumella were plastid-targeted proteins for reductive carbon metabolism, and all plastid amino acid biosynthesis pathways, except for cysteine synthesis (Fig. 3 and SI Appendix, Fig.S8 B and D). Thus, Cornospumella might have lost its plastid carbon fixation and nitrogen metabolism pathways before losing proteins directly involved in photosynthesis.
In contrast to Cornospumella, we found extensive reduction in the plastid proteome of Paraphysomonas. Its plastid is left with only haem, lipid and steroid synthesis, glycolysis, and plastid protein import and division (Fig. 3). We verified the evolutionary affinities of key enzymes within these pathways phylogenetically (e.g., ferrochelatase, SI Appendix, Fig. S12), and localized components of plastid metabolism (SI Appendix, Fig. S13A) and plastid protein import complexes (SI Appendix, Fig. S13B) to the plastid and periplastid compartment, through the heterologous expression of GFP-linked N-terminal constructs in Phaeodactylum. Considered alongside previous microscopy studies (20), these data show that Paraphysomonas retains a functional plastid, albeit one with a minimal proteome.
A Probable Plastid Genome Loss in Paraphysomonas.
In contrast to all other nonphotosynthetic chrysophytes, we could not detect transcripts in Paraphysomonas for plastid-targeted proteins that interact with proteins typically encoded on nonphotosynthetic plastid genomes (Fig. 3). These include an absence of plastid-targeted proteins associated with Fe–S cluster and cysteine biosynthesis pathways, which would typically interact with the plastid-encoded cysteine desulfurase sufB (SI Appendix, Fig. S8 D and E); Clp chaperones that interact with plastid-encoded clpC subunits (SI Appendix, Fig. S8G); and glutamyl-tRNA synthetase, which is used alongside plastidic tRNA-Glu for the C5 haem synthesis pathway (otherwise well conserved in Paraphysomonas; SI Appendix, Figs. S8C and S12). More broadly, we could not identify any plastid-targeted proteins associated with plastid DNA replication or expression in Paraphysomonas, assessed both by phylogeny and eukaryotic orthologous group (KOG) annotation of Paraphysomonas transcriptomes (Fig. 3 and SI Appendix, Fig. S14A).
Previous studies have noted losses of plastid clp, Fe–S, and other plastid DNA-associated proteins in taxa inferred to have either completely lost a plastid (Cryptosporidium, Hematodinium) (1, 11) or to have lost the plastid genome (Perkinsus, Polytomella) (1, 6). To explore the possibility of plastid genome loss in Paraphysomonas, we first performed a next-generation sequencing survey of genomic DNA from Paraphysomonas bandaiensis RCC383. This yielded 4.6 Mbp of assembled sequence data of clear chrysophyte origin, including 6895 bp of mitochondrial DNA, but no contigs corresponding to plastid DNA. We additionally did not detect plastid genome-related sequences in any Paraphysomonas transcriptome library (SI Appendix, Fig. S9).
We supplemented this initial sequencing data with a more directed test for the presence of plastid DNA, performing PCRs using P. bandaiensis gDNA, and primers designed for representative chrysophyte plastid (16S, 23S), mitochondrial (16S, 23S, coxI), and nuclear (18S, ITS1) genes. The plastid primer combinations were designed over regions of sequence that are conserved in all known chrysophyte, eustigmatophyte, and pinguiophyte sequences, and therefore should be present in the Paraphysomonas plastid genome if present; but are not found in the corresponding sequences from either the food substrate (rice) or bacterial commensals in the culture medium (17). These primers indeed amplified the expected DNA regions for gDNA harvested from the nonphotosynthetic chrysophyte Spumella elongata CCAP 955/1 (SI Appendix, Fig. S14B). In contrast, only the nuclear and mitochondrial primers amplified Paraphysomonas genes, while the plastid consensus primers amplified bacterial contaminants (SI Appendix, Fig. S14B). The specific amplification of bacteria in lieu of plastid DNA indicates that no sequences matching the chrysophyte plastid consensus sequence were present, strongly suggesting that plastid DNA has been lost from this lineage.
Paraphysomonas Retains Mitochondria-Targeted Copies of Plastid Genome-Associated Proteins.
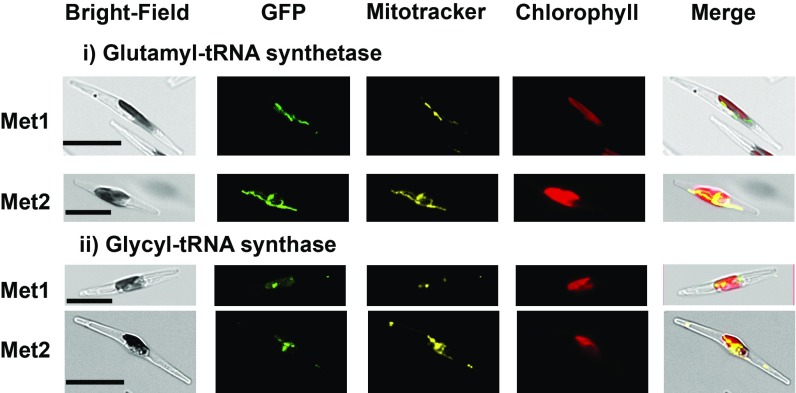
We noted the presence of some proteins orthologous to plastid genome-associated factors in Paraphysomonas (Fig. 3). None of these proteins possesses detectable plastid-targeting sequences, but instead possesses mitochondrial targeting peptides or has missing or ambiguous targeting signals (Fig. 3 and SI Appendix, Fig. S8 E and F). We selected five Paraphysomonas aminoacyl-tRNA synthetase genes (GluRS, GlyRS, AspRS, IleRS, and MetRS) for experimental characterization. We sequenced the 5′-ends of each transcript using thermal asymmetric interlaced PCR (TAiL-PCR) (24) (SI Appendix, Fig. S15) and localized the encoded proteins using GFP-linked N-terminal constructs expressed in Phaeodactylum (Fig. 4 and SI Appendix, Fig. S16). Each construct localized to the mitochondria, confirmed using MitoTracker Orange.
Fig. 4.
Mitochondrial localization of Paraphysomonas proteins evolved from plastid-targeted ancestors. This figure shows constructs for N-terminal regions of Paraphysomonas orthologs of plastid genome-associated proteins (i, glutamyl-tRNA synthetase; ii, glycyl tRNA-synthetase) that localize to the mitochondria, stained with MitoTracker Orange. Both genes have two possible methionine initiator codons (Met1 and Met2) that produce identical localizations. (Scale bars, 10 μm.) Additional GFP images for Paraphysomonas plastid proteins and proteins historically associated with the plastid genome are shown respectively in SI Appendix, Figs. S13 and S16; experimental evidence for the loss of the plastid genome is shown in SI Appendix, Fig. S14; TAiL-PCR alignments of genes that once functioned to supported the plastid genome are shown in SI Appendix, Fig. S15; consensus Bayesian topologies for these genes are shown in SI Appendix, Figs. S17 and S18; and an overview of the evolutionary fates of plastid-genome associated proteins in Paraphysomonas is shown in SI Appendix, Fig. S19.
We next investigated the evolutionary history of proteins that were once associated with the Paraphysomonas plastid genome. We searched Paraphysomonas transcriptomes for homologs of 34 genes related to plastid genome expression, including all aminoacyl-tRNA synthetases, select ribosomal proteins, and translation factors (5). Copies of these genes are required for gene expression pathways in the nucleus and mitochondria as well as the plastid, and accordingly ochrophytes may possess multiple copies. These include at least one gene encoding a cytoplasmic protein plus either two separate genes for mitochondria- and plastid-targeted proteins, or a single gene that encodes a dual-targeted protein that functions in both organelles (5, 25).
All of the proteins once associated with the Paraphysomonas plastid genome that have been retained have probable dual-targeted ancestries, including proteins of clear red algal (i.e., plastid) origin such as glutamyl-tRNA synthetase (SI Appendix, Figs. S17–S19). Thus, Paraphysomonas has converted dual-targeted proteins, which historically supported both its plastid and mitochondrial genomes, into proteins with solely mitochondrial localizations. In contrast, plastid genome-associated proteins for which a separate, mitochondrial, copy exists have been lost from Paraphysomonas (SI Appendix, Fig. S19).
Discussion
Rampant Losses of Photosynthesis Within Chrysophyte Algae.
In this study, we use high-throughput sequence analysis, phylogenetics, and fluorescence microscopy to illuminate the diversity of nonphotosynthetic plastids in chrysophyte algae. We show that photosynthesis has been lost on multiple occasions within chrysophyte evolution (Fig. 1 and SI Appendix, Fig. S1). Photosynthetic chrysophytes are generally mixotrophic (with the exception of synurophytes), feeding both through phototrophy and phagotrophy (12, 13). Mixotrophic lifestyles allow chrysophytes to acquire fixed carbon and other essential organic nutrients in the absence of photosynthesis, and previous studies have suggested that some mixotrophic chrysophytes have lost components of their plastid metabolism (12, 13).
Notably, all of the documented obligate heterotrophs within the chrysophytes are phagotrophic predators, with none known to subsist solely on osmotrophy (14, 17, 18). It has been suggested that the efficient feeding of these phagotrophic chrysophytes on very tiny bacteria can select for plastid reduction in nutrient-poor environments (26). All of the nonphotosynthetic chrysophytes considered in this study lack plastid-targeted amino acid synthesis pathways other than for cysteine (along with the retention of isolated enzymes involved in lysine and aromatic amino acids in “Spumella” sp. NIES-1846, and members of the core Spumella group; Fig. 3 and SI Appendix, Fig. S8). This may reflect the bacteriovorous lifestyle of these chrysophytes. Amino acids may be acquired through phagotrophy, thus releasing these organisms from their dependence on nitrogen assimilation, and major plastid biosynthesis of amino acids.
Previously, we proposed that, in contrast to the situation for lineages such as chrysophytes that engage in phagotrophy, extracellular metabolites that are synthesized in plastids (e.g., fatty acids, amino acids, and haem) are difficult to acquire through osmotrophic lifestyles (3, 27). The osmotrophic diatom Nitzschia sp. NIES-3581 still retains most of its plastid biosynthetic pathways after loss of photosynthesis (27). Consistent with this, dramatic cases of plastid reduction, such as the complete loss of an entire plastid, has only previously been documented in parasites (Cryptosporidium; Haematodinium; and red algal adelphoparasites) (1, 7, 11).
Convergent Evolution of Nonphotosynthetic Secondary Red Plastids.
We present the 53-kb-long plastid genome of “Spumella” sp. NIES-1846, which retains only genes for expression, ferredoxin-mediated electron transport, Clp protease, and Fe–S cluster assembly (Fig. 2). Notably, this genome encodes an extremely similar set of functions as the plastid genomes of apicomplexan parasites (Fig. 2 and SI Appendix, Fig. S3). This supports previous suggestions that this functional gene set represents the minimum required for the retention of red-algal–derived plastid genomes (1). Different trends may apply in other plastid lineages: for example, the plastid genomes of the parasitic plants Epifagus and Balanophora do not encode Fe–S cluster synthesis proteins, which are nucleus encoded in plants (2, 28, 29), but do encode the fatty acid synthesis subunit accD, which is nucleus encoded in ochrophytes (5) (and is absent from “Spumella” sp. NIES-1846, per the lack of plastid fatty acid synthesis in this species; SI Appendix, Figs. S5 and S6). Similarly, the nonphotosynthetic euglenophyte Euglena longa retains a highly divergent plastid rubisco large subunit gene (rbcL) of undetermined function (7, 30) (SI Appendix, Fig. S3).
We additionally find evidence for convergent evolution in the plastid proteomes of independently evolved nonphotosynthetic chrysophyte lineages (Figs. 1 and 3). This includes a shared set of plastid-targeted proteins, that is, glycolytic carbon metabolism; cofactor biosynthesis; and essential proteins for plastid biogenesis and gene expression (Fig. 3). We note some exceptions to this rule, for example, the retention of a number of plastid-derived photosynthesis-related proteins in Cornospumella (Fig. 3 and SI Appendix, Figs. S8, S10, and S11). This phenomenon is similar to the retention of plastid chlorophyll biosynthesis-related enzymes in multiple groups of parasitic plants (10, 31) and might plausibly reflect a very recent loss of photosynthesis in this lineage, given its close phylogenetic relationship to the photosynthetic Ochromonas sp. TCS-2004 (SI Appendix, Fig. S1). Overall, our data support the idea that independently evolved nonphotosynthetic plastids converge on similar genome architectures and metabolic functions, albeit with different intermediate levels of reduction.
Evolution of a Minimal Plastid Proteome in Paraphysomonas.
In stark contrast to all of other nonphotosynthetic chrysophytes, Paraphysomonas spp. do not retain plastid genomes, as evidenced by the lack of plastid-targeted proteins with genome-related functions (Fig. 3 and SI Appendix, Fig. S14A) and the absence of identifiable plastid DNA from next-generation sequencing surveys, or targeted PCR, of this lineage (SI Appendix, Fig. S14B). This is likely to be an exceptional feature within nonphotosynthetic chrysophyte plastids, as evidenced by the detection of plastid DNA through next-generation sequencing in “Spumella” sp. NIES-1846 (Fig. 2), PCR in Spumella elongata (SI Appendix, Fig. S14B), and the identification of plastid genome-derived sequences in five other nonphotosynthetic chrysophyte transcriptomes (SI Appendix, Fig. S9). Paraphysomonas thus joins a growing list of nonphotosynthetic eukaryotes, including green algae (Polytomella) and dinoflagellates (Perkinsus), inferred to have dispensed with a plastid genome (1, 6).
It is intriguing to consider what factors have allowed complete plastid genome loss in Paraphysomonas. Paraphysomonas does retain a plastid, detectable through microscopy (20), along with plastid-targeted proteins for core plastid metabolism and biogenesis pathways (Fig. 3 and SI Appendix, Figs. S8 and S13). Nucleus-encoded and plastid-targeted proteins that typically interact with plastid genes (Clp protease, which interacts with plastid-encoded clpC; Fe–S cluster and cysteine biosynthesis, which interacts with sufB; and glutamyl-tRNA synthetase, which interacts with glu-tRNA) are all absent from Paraphysomonas transcriptomes. Thus, these plastid metabolic functions have been completely lost from Paraphysomonas.
In the case of Fe–S synthesis, we presume that Paraphysomonas no longer retains ferredoxin (as it is not detected as a nucleus-encoded and plastid-targeted protein, and is retained in “Spumella” sp. NIES-1846 on the plastid genome; Fig. 2 and SI Appendix, Fig. S8), so no longer requires Fe–S cofactors for ferredoxin-dependent plastid metabolism (32). We additionally note that chrysophytes are thiamine auxotrophs, neither retaining plastid-encoded thiG and thiS genes (“Spumella” sp. NIES-1846 and Ochromonas sp. CCMP1393; Fig. 2; ref. 22), nor possessing plastid-targeted copies (SI Appendix, Fig. S8), and therefore Paraphysomonas should not need plastid cysteine desulfurase activity to liberate activated sulfur for thiamine biosynthesis. The function of Clp protease remains unclear (33), although in plants it increases the efficiency of protein import into the plastid stroma and regulates the expression of the plastid genome. The latter function is not relevant to Paraphysomonas; the precise plastid protein import strategies used by Paraphysomonas remain to be determined.
Most intriguing is the absence of plastidial glutamyl-tRNA and glutamyl-tRNA synthetase from Paraphysomonas. We detected the remainder of the plastidial C5 haem biosynthesis pathway, including a plastid-targeted glutamyl tRNA reductase (SI Appendix, Fig. S8C), and a ferrochelatase that localizes to the periplastid compartment and resolves phylogenetically with plastidial enzymes from other eukaryotes, as opposed to with isoforms used in the mitochondrial/cytoplasmic haem biosynthesis pathway (SI Appendix, Figs. S12 and S13). Thus, Paraphysomonas performs haem synthesis in its plastid but presumably imports glutamyl-tRNA of an external origin, as has previously been proposed to occur in Polytomella (6) and as has been shown to occur in some parasitic plants (7). It remains to be determined whether Paraphysomonas imports glutamyl-tRNA from the cytoplasm or mitochondria into the plastid (6, 34).
A Plastidial Imprint on the Paraphysomonas Mitochondrion.
Finally, we show that aminoacyl tRNA synthetases that were previously dual targeted to the plastid and mitochondria in the chrysophyte ancestor are retained in Paraphysomonas, following the loss of the plastid genome. These proteins now solely support the biology of the mitochondria and are no longer imported into the plastid (Fig. 4 and SI Appendix, Figs. S16–S19). Typically, dual-targeted proteins in ochrophytes either possess a plastid-targeting sequence with an internal initiation codon allowing the translation of an alternative mitochondria-targeting sequence, or they possess an ambiguous targeting sequence that may be recognized by both the endomembrane and mitochondrial protein import machineries (5, 25). These targeting sequences could be easily converted into a solely mitochondria-targeting sequence, respectively by preventing expression of the upstream signal peptide (and allowing translation initiation only from the internal mitochondria-targeting sequence) or by mutation of the key residues [e.g., ASAFAP motif (5)] allowing recognition of the plastid-targeting sequence. This seems a much less complicated scenario than the complete loss of a dual-targeted protein, which would require the compensatory evolution of a mitochondria-targeted enzyme to replace the dual-targeted isoform. It remains to be determined to what extent proteins are exchanged over evolutionary timescales between ochrophyte plastids and mitochondria, and why aminoacyl tRNA synthetases are particularly prone to dual targeting in these and in other taxa [e.g., plants (5, 25, 34)].
These data provide a complex portrait of the processes underpinning organelle evolution. Previously, we and others have shown that plastids are complex mosaics, utilizing nucleus-encoded proteins of diverse evolutionary origin including the plastid endosymbiont, the host, and the expression products of genes obtained through horizontal gene transfer, or retained from previous endosymbioses (5, 24). This mosaic evolution may be particularly important to the evolution of nonphotosynthetic plastids following the loss of photosynthetic capacity, as plastid-targeted proteins of bacterial origin (e.g., rpl26 family proteins in the euglenid Euglena longa) (8), or plastid-targeted proteins recruited from the host cytoplasm (e.g., transaldolase in the diatom Nitzschia sp. NIES-3581) (27), may supplement or compensate for reductive evolution in these plastid lineages. Here, we show that a defunct plastid genome has left an imprint on the biology of the mitochondria via the evolution of dual-targeted proteins, which have displaced the endogenous mitochondria-targeted enzymes. Further sampling of secondarily heterotrophic lineages across the tree of life will further illuminate the ecological and evolutionary processes that underpin the loss of photosynthesis, and the interactions between chloroplasts and other organelles over their evolutionary history.
Materials and Methods
Cultures.
“Spumella” sp. NIES-1846 was purchased from and cultured according to the instruction by National Institute for Environmental Studies, Japan (NIES). Paraphysomonas bandaiensis RCC383 and Spumella elongata CCAP955/1 were grown in filtered sea water (Roscoff) and mineral water (Volvic), respectively, equilibrated to pH 8, to which 40 grains of rice per L (Biocoop) were added. Cultures were maintained at a constant temperature of 19 °C, without shaking, until ready to harvest.
Phaeodactylum tricornutum CCAP1055/2 and UTEX 642 were maintained in enhanced seawater medium as previously described in refs. 5 and 35, respectively.
Molecular Biology.
Cultures were harvested in late-log phase (∼3 mo postinoculation) for nucleic isolation. Total cultures were initially filtered using 22-μm-diameter Miracloth (Millipore) and washed with sterile growth medium three times to remove excess rice particles. Genomic DNA and total cellular RNA were extracted from P. bandaiensis and S. elongata using previously described techniques (24). Precipitation steps were performed with ethanol or isopropanol (as appropriate), equilibrated with 10% 3 M NaCl to prevent the precipitation of residual starch particles in the filtrate. “Spumella” sp. NIES-1846 DNA was extracted using a Plant DNA extraction kit (Jena BioSciences), and total cellular RNA was extracted by TRIzol (Sigma) following the manufacturer’s instructions.
RT-PCR and PCR reactions were performed as previously described (24). For degenerate PCRs, chloroplast and mitochondria primers were designed over regions with maximal conservation across published chrysophyte, eustigmatophyte, and pinguiophyte sequences for select genes, and that were neither found in rice nor bacterial sequences, and therefore should specifically amplify chrysophyte template from the DNA template (22). Consensus nuclear gene primers were obtained from a previous study (24). Each PCR was performed multiple times, including in highly permissive reaction conditions (annealing temperatures between 5° and 15° below primer melt temperatures; or PCR reamplifications, in which the primary reaction product was used as a template for a second series of PCR cycles) to maximize the probability of successful amplification. N-terminal regions of Paraphysomonas genes of interest were verified by TAiL-PCR as previously described (24). 5′-RACE analyses of “Spumella” sp. NIES-1846 Fab mRNAs were conducted using a 5′-RACE kit (Invitrogen), following the manufacturer’s instructions.
The 5′-terminal sequences, including the N-terminal organellar targeting regions for transcripts encoding Paraphysomonas and “Spumella” sp. NIES-1846 plastid-targeted proteins, were expressed in Phaeodactylum using previously described techniques (5, 27). Briefly, constructs for which an N-terminal methionine was confirmed (“Spumella” sp. NIES-1846: mRNA 5′-end sequence confirmed by 5′-RACE; Paraphysomonas: the inferred 5′ methionine identified by TAiL-PCR to be immediately downstream of an in-frame 5′-UTR stop codon; SI Appendix, Figs. S5 and S15) were cloned into pPhat-eGFP vectors under either the NR (“Spumella” sp. NIES-1846) or FcpA promoters (Paraphysomonas). Where a sequence possessed multiple possible initiator methionines (defined as an in-frame methionine upstream of the CDD, and downstream of the first in-frame stop codon in the UTR), each methionine was used to design a separate GFP construct and independently localized. All construct sequences and primer combinations used are described in Dataset S2, sheet 1.
pPHA-FcpA constructs were introduced into P. tricornutum CCAP1055/2 by biolistic transformation using a Bio-Rad gene gun, following the manufacturer’s instructions. Similarly, pPHA-NR constructs were introduced into P. tricornutum UTEX642 using a NEPA21 gene gun (NEPAGENE) (35).
Actively growing P. tricornutum transformants in a zeocin-based selection medium were observed with an Olympus BX51 fluorescent microscope (Olympus) equipped with an Olympus DP72 CCD color camera (Olympus) to observe localization of the GFP recombinant proteins in P. tricornutum cells. Mitochondrial and chloroplast endoplasmic reticulum localizations were confirmed using MitoTracker Orange and DAPI staining, respectively, as previously described (5, 36). GFP fluorescence was detected with a 510- to 550-nm filter by 485-nm excitation, MitoTracker fluorescence was detected with a 575- to 590-nm filter by 548-nm excitation, DAPI fluorescence was detected with a 400- to 460-nm filter by 368-nm excitation, and chlorophyll autofluorescence was detected with a 610- to 680-nm filter. All microscopy experiments were performed using relevant control lines to identify optimal detection exposure times: wild-type cells for GFP fluorescence, and unstained lines for MitoTracker and DAPI visualization (SI Appendix, Figs. S5 and S13).
Next-Generation Sequencing.
“Spumella” sp. NIES-1846 genomic DNA was sent to Hokkaido System Science Company to be subjected to 350-bp insert library preparation by TruSeq Nano DNA Library Prep Kit followed by sequencing with HiSeq 2500 (Illumina), resulting in 47.1 million of 100-bp paired-end reads. Adapter sequences were trimmed using cutadapt 1.1 (37) followed by Trimmomatic 0.32 (38) (trimming reads with length of <50 bp, mean QVs of <20 in a window size of 20 bp), resulting in 45.8 million paired-end reads. Reads were assembled using Velvet 1.2.08 (39) with hash length of 65. Plastid DNA-derived contigs were detected by homology-based surveys using the plastid genome gene sequence of Ochromonas sp. CCMP1393 (22). Six contigs, one of which carries an rRNA operon, were identified. All of the contigs had between 18.7- and 20.3-fold average coverage, except for the rRNA-carrying contig, which had 44.2-fold average coverage (approximately twice the coverage of other regions), indicating that it likely exists as a repeat region. Gap filling was performed by PCR and Sanger sequencing, confirming a tetrapartite structure with inverted repeat regions containing an rRNA operon. The genome was annotated using Mfannot (40), tRNA-scan (41), and blastX searches against the nonredundant NCBI (nr) protein database.
“Spumella” sp. NIES-1846 total cellular RNA was sent to Hokkaido System Science Company for library preparation with TruSeq RNA sample Prep Kit, version 2, followed by Illumina HiSeq 2500 sequencing as before, resulting in 44.8 million of 100-bp paired-end reads. Adapter trimming and quality checking were performed as described above, generating 44.5 million paired-end reads. Contigs were assembled using Trinity 2.0.6 (42) with the default settings, resulting in 22,344 contigs.
P. bandaiensis RCC383 genomic DNA was sent to the Office of Knowledge Enterprise Development (OKED) sequencing core at Arizona State University for library preparation with LTP library prep kit (KAPA Biosystems) followed by sequencing on the Illumina MiSeq platform, yielding 9.3 million 250-bp read pairs. The raw reads were quality checked using FastQC, version 0.10.1, followed by adapter trimming and quality filtering by Trimmomatic 0.35 (38) (trimming reads with length of <150 bp or mean QVs of <18 in a window size of 4 bp), resulting in 5.6 million paired-end reads. Clean read pairs were assembled using Spades 3.11.1 with mismatch corrector mode applied, using k-mer size 127 as optimized using Quast 4.5 (43, 44). Sequences of clear chrysophyte origin were identified within this library via reciprocal BLAST, with threshold e value of 1 × 10−05, against a composite library of the Paraphysomonas bandaiensis MMETSP transcriptome (MMETSP1103); the Oryza sativa ssp. indica genome; and genome sequences from two bacterial commensals (Labrenzia sp., Marinobacter sp.) identified in the culture by 16S rDNA sequencing (SI Appendix, Fig. S14) (45–48). Mitochondrial DNA contigs from this assembly are provided in Dataset S2, sheet 2.
Assembly of a Multispecies Chrysophyte Transcriptome Dataset.
PESC clade (pinguiophytes, eustigmatophytes, synchromophytes, and chrysophytes) (5, 9) nucleotide and peptide sequence libraries were downloaded from the Marine Microbial Eukaryotes Transcriptome Sequencing Project (MMETSP) (45), along with libraries for independent transcriptomes (12, 21), genomes (49, 50), advance access genome drafts (https://genome.jgi.doe.gov/Ochro2298_1/Ochro2298_1), and single-celled genomes libraries (15) housed on the jgi genomes and Uniref portals. ORF conceptual translations were generated for each nucleotide sequence library using a conventional translation table and EMBOSS (51). The longest N-terminal complete ORF (i.e., the longest ORF starting in a methionine codon) was used for subsequent analyses, alongside conceptual protein translations previously generated for MMETSP and genomes libraries. The evolutionary origin of each transcriptome was verified by 18S sequence analysis; the previously identified Apoikiospumella mondseensis JBM08 transcriptome (12, 21) was found through this approach to instead correspond the nonphotosynthetic chrysophyte Spumella lacusvadoi JBNZ39 (Dataset S2, sheet 3).
Technical contamination, resulting from the mixing of RNA samples from different species in the same sequencing microchip, was removed from the MMETSP and uniref transcriptomes using a previously defined pipeline (5, 52). Briefly, each library generated within a particular study [MMETSP (45) or an independent transcriptomic survey of chrysophyte diversity (21)] was searched against the others using BLASTp. The percentage identities between each BLAST top hit obtained were used to build a frequency distribution, for which an inflection point n (defined as the highest percentage sequence identity for which fewer top hits could be found than the number of top hits with n-1 and n-2 percent sequence identity) were defined. All sequences yielding BLAST top hits above this threshold percentage identity were removed, except in the case of BLAST searches performed between two species within the same genus. Exemplar frequency distributions, and a heatmap showing the proportion of sequences removed from different uniref chrysophyte transcriptomes (21), are shown in SI Appendix, Fig. S20 A and B, respectively.
Identification of Mitochondria- and Plastid-Targeting Peptides.
Possible targeting sequences were identified for each N-complete peptide sequence using ASAFind, version 2.0, used in conjunction of SignalP, version 3.0 (53, 54); HECTAR, integrated into the Galaxy cluster (55, 56); TargetP, version 1.1 (57); PredSL (58); and MitoFates (59). Proteins were annotated as being plastid-targeted if they were identified to possess plastid targeting sequences using either ASAFind or HECTAR; and mitochondria-targeted if they were identified to possess mitochondria targeting sequences by two of TargetP, MitoFates, and HECTAR.
To avoid mispredictions of the localization of proteins within each sequence library, due to divergent evolution in the targeting sequences recognized by protein import machinery PESC clade members (5, 9), custom targeting thresholds were designed for each predictor (Dataset S3). Briefly, this involved assembling a dataset of proteins experimentally verified to localize to different subcellular compartments in chrysophytes (Paraphysomonas bandaiensis, “Spumella” sp. NIES-1846) and eustigmatophytes (Nannochloropsis gaditana, N. salina, and N. oceanica) through GFP localization, derived from this (Fig. 4 and SI Appendix, Figs. S5, S13, and S16) and previous studies (5, 60–64). The threshold values were manipulated for each predictor to define optimal ones for the detection of experimental localizations, with the best trade-offs between sensitivity and specificity identified for the following: the default conditions for SignalP and ASAFind; modified mitochondria targeting thresholds of 0.7 and 0.35 for TargetP and MitoFates, respectively, as previously published (5, 9); and a substantially lower chloroplast targeting threshold (0.0855, defined as the midpoint between the 90% sensitivity and 90% stringency; with the additional stipulation that the chloroplast targeting value be greater than the signal anchor targeting value) for HECTAR (SI Appendix, Fig. S21A). The increased efficacy of the modified prediction values was finally validated using an independently sourced dataset of 48 proteins identified for “Spumella” sp. NIES-1846 that had previously been confirmed phylogenetically to resolve with other ochrophyte plastid proteins (SI Appendix, Fig. S21B).
BLAST-Based Identification of Homologs to Known Plastid Proteins.
A query library of 9,531 evolutionarily redundant plastid-targeted proteins were assembled from publicly accessible ochrophyte, cryptomonad, and haptophyte genomes, using a previously defined technique (5). Briefly, this involved identifying plastid-targeted proteins from each genome using in silico prediction, searching these proteins against each other, and a modified version of uniref (downloaded June 2015) from which all species with a suspected history of secondary endosymbiosis (cryptomonads, haptophytes, ochrophytes, dinoflagellates, apicomplexans, chlorarachniophytes, and euglenids) had been removed; and then reducing each group of homologous query sequences, defined as those that yielded BLAST hits against another with a smaller e value than the corresponding uniref top hit, to one type query sequence.
Possible homologs to each query sequence were identified in each PESC clade library using two BLAST-based techniques. First, each query sequence was searched against each PESC clade library using reciprocal BLAST best hit with threshold e value 1 × 10−05 (65). Next, each query sequence was searched against each PESC clade library using a floating e-value threshold, defined by the e value obtained for the uniref BLAST best hit in the search above, following previous methodology (5). To account in this latter search for C-terminally incomplete sequences, which may lack the number of required identities to yield an e value below the threshold, a modified threshold was defined for each protein as follows:
This was defined by considering the e-value relationship best able to recover a phylogenetically verified set of 165 proteins with ochrophyte plastid orthology from “Spumella” sp. NIES-1846, without admitting a dataset of 95 negative controls, consisting of mitochondria-targeted homologs of protein complexes common to both photosynthetic and respiratory electron transport chains, and cytoplasmic homologs of proteins inferred to be retained in both the “Spumella” sp. NIES-1846 and Ochromonas sp. CCMP1393 plastid genomes (22). Detailed outputs of these tests are provided in Dataset S4, and a scatterplot of the uniref threshold and control protein BLAST top hit e values are provided in SI Appendix, Fig. S22.
All quantitative analysis of the plastid proteome content of each PESC clade species (i.e., total numbers of homologs; total proportion of homologs with plastid targeting sequences; KOG annotations; SI Appendix, Figs. S7 and S14) were performed on the subset of proteins identified to be homologs through both the reciprocal and floating BLAST searches.
Homologs of proteins encoded in the Nannochloropsis gaditana, Ochromonas sp. CCMP1393, and Mallomonas splendens plastid genomes were identified in each library by BLASTp searches with threshold values 1 × 10−05 (22, 23, 50). Matching sequences were manually verified by reciprocal tBLASTn searches against the nr database, with only proteins that gave top hits against other chrysophyte plastid sequences retained for further analysis. Finally, each protein sequence was aligned against a composite library of protein sequences from a further 38 completed ochrophyte plastid genomes (9) (Dataset S1) and used for the generation of single-gene trees, using methodology described below. Sequences that failed to form a monophyletic clade with other PESC clade plastid-encoded sequences were rejected, yielding 71 remaining sequences of probable plastid origin. The alignments obtained were also used for the inspection of predicted plastid-targeted homologs identified through this approach, with sequences only inferred to be plastid-targeted if (i) the encoded protein contained a predicted plastid-targeting peptide, as inferred with ASAFind or HECTAR, and (ii) the encoded protein sequence contained an N-terminal extension of at least 30 aa, upstream of the region conserved with plastid-encoded equivalents.
Phylogenetic Techniques.
The distribution and localization of homolog of 303 central components of ochrophyte plastid proteomes (SI Appendix, Fig. S8) were resolved using a modified version of a previously published phylogenetic pipeline (5). For this, each protein sequence within the dataset of 9,531 query plastid proteins with a given functional annotation was searched against 144 different libraries, generated from uniref, jgi, MMETSP and other genome and transcriptome data across the tree of life (taxonomic divisions and their constituent libraries are listed in Dataset S5). The top hits to each protein were extracted and combined with all possible PESC clade homologs identified using either the floating or reciprocal BLAST hit searches. Finally, each cluster was enriched with a previously defined set of plastid-targeted protein homologs (“HPPG”) from a wide range of ochrophyte lineages (5).
Each assembled cluster of proteins were aligned sequentially using MAFFT, version 7.409, MUSCLE, version 8.0, and the in-built alignment program in GeneIOUS, version 4.76 (66–68). At each stage, poorly aligned sequences and sequences that were clearly identified as N-terminally truncated (based on incomplete coverage of a conserved region shared by most other sequences in the alignment) were manually identified and removed. Each curated alignment was finally trimmed (at the N and C termini to the first and last residues, respectively, with >50% identity; and then internally using trimAl with the –gt 0.5 option) (69); and single-gene trees were built with RAxML, version 8.1, the GTR + Γ substitution model, and automatic bootstopping (70).
PESC clade proteins with orthology to other ochrophyte plastid proteins were identified manually from each RAxML best-scoring tree. Proteins were annotated as being of ochrophyte plastid origin if they resolved within a cluster of proteins containing two or more ochrophyte plastid-targeted sequences (either from PESC clade taxa, or the HPPG proteins enriched in each cluster), with no inclusion of proteins from prokaryotes or from eukaryotic lineages without a suspected history of secondary endosymbiosis (i.e., red algae, green algae, glaucophytes, opisthokonts, and plastid-lacking members of the excavate and SAR groups) (5).
All exemplar trees (i.e., multigene and 18S tree topologies, and the exemplar plastid-targeted proteins; Fig. 1 and SI Appendix, Figs. S1, S10–S12, S17, and S18) were inferred using the MrBayes and RAxML programs incorporated into the CIPRES server, using previously defined parameters (5, 71). Tree outputs in each case are provided via the University of Cambridge dSpace server (https://www.repository.cam.ac.uk/handle/1810/284182) (72).
Data Deposition.
The “Spumella” sp. NIES-1846 plastid genome and transcriptome data are provided through DNA Data Bank of Japan (accession no. AP019363 and BioProject no. PRJDB7829, respectively). The P. bandaiensis genome sequence survey was deposited to the National Center for Biotechnology Information Short Read Archive under accession no. 1067 SAMN10793136 and BioProject under accession no. PRJNA453414. Additional data for this project, consisting of decontaminated chrysophyte transcriptome datasets, clusters of evolutionarily distinct plastid-targeted proteins, and alignments and RAxML trees of select clusters, are available through the University of Cambridge dSpace server: https://www.repository.cam.ac.uk/handle/1810/284182 (72).
Supplementary Material
Acknowledgments
We thank the OKED Genomics Core at Arizona State University for assistance with next-generation sequencing, Catherine Cantrel (CNRS and École Normale Supérieure) for assistance with diatom transformations, and Louis Graf (Sungkyunkwan University) for assistance with phylogenetic analyses. We thank Virginia Armbrust (University of Washington), Jackie Collier (Stony Brook University), and Connie Lovejoy (Université Laval), respectively, for the advance provision of genome sequence data from Pseudo-nitzschia multiseries, four species of Arctic microalgae (CCMP2298, CCMP2097, CCMP2436, and CCMP2293), and three species of marine labyrinthulomycetes (Aplanochytrium kerguelensis, Aurantiochytrium limnaticum, and Schizochytrium aggegatum), used to enrich the phylogenetic reference datasets used in this study. These sequence data were produced by the US Department of Energy Joint Genome Institute (https://www.jgi.doe.gov/), a Department of Energy Office of Science User Facility, supported by the Office of Science of the US Department of Energy under Contract DE-AC02-05CH11231, in collaboration with the user community. This work was supported by a Grant-in-Aid for Young Scientists (A) from the Japan Society for the Promotion of Sciences (JSPS) (15H05606; awarded to R.K.); a JSPS Research Fellowship for Young Scientist (18J00886; awarded to M.N.); an EMBO Early Career Fellowship (Fellowship 1124/2014) and CNRS Momentum Fellowship (2018 competition) (both awarded to R.G.D.); and funding from the French Government “Investissements d’Avenir” Programmes MEMO LIFE Grant ANR-10-LABX-54, Université de Recherche Paris Sciences et Lettres (PSL) Grant ANR-1253 11-IDEX-0001-02, and OCEANOMICS Grant ANR-11-BTBR-0008 (awarded to C.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.M.A. is a guest editor invited by the Editorial Board.
Data deposition: The “Spumella” sp. NIES-1846 plastid genome and transcriptome data have been deposited in the DNA Data Bank of Japan (accession no. AP019363) and BioProject database (accession no. PRJDB7829). The Paraphysomonas bandaiensis genome sequence survey has been deposited in the National Center for Biotechnology Information Short Read Archive database (accession no. SAMN10793136) and BioProject (accession no. PRJNA453414). Additional data for this project, consisting of decontaminated chrysophyte transcriptome datasets, clusters of evolutionarily distinct plastid-targeted proteins, and alignments and RAxML trees of select clusters, are available through the University of Cambridge dSpace server: https://www.repository.cam.ac.uk/handle/1810/284182.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819976116/-/DCSupplemental.
References
- 1.Janouškovec J, et al. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc Natl Acad Sci USA. 2015;112:10200–10207. doi: 10.1073/pnas.1423790112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krause K. Piecing together the puzzle of parasitic plant plastome evolution. Planta. 2011;234:647–656. doi: 10.1007/s00425-011-1494-9. [DOI] [PubMed] [Google Scholar]
- 3.Kamikawa R, et al. Proposal of a twin arginine translocator system-mediated constraint against loss of ATP synthase genes from nonphotosynthetic plastid genomes. Mol Biol Evol. 2015;32:2598–2604. doi: 10.1093/molbev/msv134. [DOI] [PubMed] [Google Scholar]
- 4.Donaher N, et al. The complete plastid genome sequence of the secondarily nonphotosynthetic alga Cryptomonas paramecium: Reduction, compaction, and accelerated evolutionary rate. Genome Biol Evol. 2009;1:439–448. doi: 10.1093/gbe/evp047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorrell RG, et al. Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. eLife. 2017;6:e23717. doi: 10.7554/eLife.23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith DR, Lee RW. A plastid without a genome: Evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 2014;164:1812–1819. doi: 10.1104/pp.113.233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadariová L, Vesteg M, Hampl V, Krajčovič J. Reductive evolution of chloroplasts in non-photosynthetic plants, algae and protists. Curr Genet. 2018;64:365–387. doi: 10.1007/s00294-017-0761-0. [DOI] [PubMed] [Google Scholar]
- 8.Záhonová K, et al. Peculiar features of the plastids of the colourless alga Euglena longa and photosynthetic euglenophytes unveiled by transcriptome analyses. Sci Rep. 2018;8:17012. doi: 10.1038/s41598-018-35389-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorrell RG, Bowler C. Secondary plastids of stramenopiles. In: Hirakawa Y, editor. Advances in Botanical Research: Secondary Endosymbiosis. Vol 84. Academic; San Diego: 2017. pp. 59–103. [Google Scholar]
- 10.Wickett NJ, et al. Transcriptomes of the parasitic plant family Orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr Biol. 2011;21:2098–2104. doi: 10.1016/j.cub.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Gornik SG, et al. Endosymbiosis undone by stepwise elimination of the plastid in a parasitic dinoflagellate. Proc Natl Acad Sci USA. 2015;112:5767–5772. doi: 10.1073/pnas.1423400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graupner N, et al. Evolution of heterotrophy in chrysophytes as reflected by comparative transcriptomics. FEMS Microbiol Ecol. 2018;94:fiy039. doi: 10.1093/femsec/fiy039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lie AAY, et al. A tale of two mixotrophic chrysophytes: Insights into the metabolisms of two Ochromonas species (Chrysophyceae) through a comparison of gene expression. PLoS One. 2018;13:e0192439. doi: 10.1371/journal.pone.0192439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walker G, Dorrell RG, Schlacht A, Dacks JB. Eukaryotic systematics: A user’s guide for cell biologists and parasitologists. Parasitol. 2011;138:1638–1663. doi: 10.1017/S0031182010001708. [DOI] [PubMed] [Google Scholar]
- 15.Seeleuthner Y, et al. Tara Oceans Coordinators Single-cell genomics of multiple uncultured stramenopiles reveals underestimated functional diversity across oceans. Nat Commun. 2018;9:310. doi: 10.1038/s41467-017-02235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yubuki N, Nakayama T, Inouye I. A unique life cycle and perennation in a colourless chrysophyte Spumella sp. J Phycol. 2008;44:164–172. doi: 10.1111/j.1529-8817.2007.00441.x. [DOI] [PubMed] [Google Scholar]
- 17.Raven J. Phagotrophy in phototrophs. Limnol Oceanogr. 1997;42:198–205. [Google Scholar]
- 18.Moestrup Ø, Andersen R. 1991. Organization of heterotrophic heterokonts The Biology of Free-Living Heterotrophic Flagellates, ed Patterson DJ, Larsen J (Clarendon, Oxford), Vol 43, pp 333–360.
- 19.Wetherbee R, Andersen R. Flagella of a chrysophycean alga play an active role in prey capture and selection: Direct observations on Epipyxis pulchra using image enhanced video microscopy. Protoplasma. 1992;166:1–7. [Google Scholar]
- 20.Preisig HR, Hibberd DJ. Virus-like particles and endophytic bacteria in Paraphysomonas and Chromophysomonas (Chrysophyceae) Nord J Bot. 1984;4:279–285. [Google Scholar]
- 21.Beisser D, et al. Comprehensive transcriptome analysis provides new insights into nutritional strategies and phylogenetic relationships of chrysophytes. PeerJ. 2017;5:e2832. doi: 10.7717/peerj.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ševčíková T, et al. Updating algal evolutionary relationships through plastid genome sequencing: Did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci Rep. 2015;5:10134. doi: 10.1038/srep10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim JI, et al. Comparative plastid genomics of Synurophyceae: Inverted repeat dynamics and gene content variation. BMC Evol Biol. 2019;19:20. doi: 10.1186/s12862-018-1316-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorrell RG, Howe CJ. Functional remodeling of RNA processing in replacement chloroplasts by pathways retained from their predecessors. Proc Natl Acad Sci USA. 2012;109:18879–18884. doi: 10.1073/pnas.1212270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gile GH, Moog D, Slamovits CH, Maier UG, Archibald JM. Dual organellar targeting of aminoacyl-tRNA synthetases in diatoms and cryptophytes. Genome Biol Evol. 2015;7:1728–1742. doi: 10.1093/gbe/evv095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Castro F, Gaedke U, Boenigk J. Reverse evolution: Driving forces behind the loss of acquired photosynthetic traits. PLoS One. 2009;4:e8465. doi: 10.1371/journal.pone.0008465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamikawa R, et al. A Non-photosynthetic diatom reveals early steps of reductive evolution in plastids. Mol Biol Evol. 2017;34:2355–2366. doi: 10.1093/molbev/msx172. [DOI] [PubMed] [Google Scholar]
- 28.Su HJ, et al. Novel genetic code and record-setting AT-richness in the highly reduced plastid genome of the holoparasitic plant Balanophora. Proc Natl Acad Sci USA. 2019;116:934–943. doi: 10.1073/pnas.1816822116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu X, et al. The iron-sulfur cluster biosynthesis protein SUFB is required for chlorophyll synthesis, but not phytochrome signaling. Plant J. 2017;89:1184–1194. doi: 10.1111/tpj.13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Záhonová K, Füssy Z, Oborník M, Eliáš M, Yurchenko V. RuBisCO in non-photosynthetic alga Euglena longa: Divergent features, transcriptomic analysis and regulation of complex formation. PLoS One. 2016;11:e0158790. doi: 10.1371/journal.pone.0158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schelkunov MI, Penin AA, Logacheva MD. RNA-seq highlights parallel and contrasting patterns in the evolution of the nuclear genome of fully mycoheterotrophic plants. BMC Genomics. 2018;19:602. doi: 10.1186/s12864-018-4968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ralph SA, et al. Tropical infectious diseases: Metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 33.Olinares PD, Kim J, van Wijk KJ. The Clp protease system; a central component of the chloroplast protease network. Biochim Biophys Acta. 2011;1807:999–1011. doi: 10.1016/j.bbabio.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Duchêne AM, Pujol C, Maréchal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- 35.Miyahara M, Aoi M, Inoue-Kashino N, Kashino Y, Ifuku K. Highly efficient transformation of the diatom Phaeodactylum tricornutum by multi-pulse electroporation. Biosci Biotechnol Biochem. 2013;77:874–876. doi: 10.1271/bbb.120936. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka A, et al. Ultrastructure and membrane traffic during cell division in the marine pennate diatom Phaeodactylum tricornutum. Protist. 2015;166:506–521. doi: 10.1016/j.protis.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Didion JP, Martin M, Collins FS. Atropos: Specific, sensitive, and speedy trimming of sequencing reads. PeerJ. 2017;5:e3720. doi: 10.7717/peerj.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valach M, Burger G, Gray MW, Lang BF. Widespread occurrence of organelle genome-encoded 5S rRNAs including permuted molecules. Nucleic Acids Res. 2014;42:13764–13777. doi: 10.1093/nar/gku1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics. 2018;34:i142–i150. doi: 10.1093/bioinformatics/bty266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bankevich A, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keeling PJ, et al. The marine microbial eukaryote transcriptome sequencing project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song L, et al. Complete genome sequence of Marinobacter sp. BSs20148. Genome Announc. 2013;1:e00236-13. doi: 10.1128/genomeA.00236-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiebig A, et al. Genome of the R-body producing marine alphaproteobacterium Labrenzia alexandrii type strain (DFL-11(T)) Stand Genomic Sci. 2013;7:413–426. doi: 10.4056/sigs.3456959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 49.Wang D, et al. Nannochloropsis genomes reveal evolution of microalgal oleaginous traits. PLoS Genet. 2014;10:e1004094. doi: 10.1371/journal.pgen.1004094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Radakovits R, et al. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nat Commun. 2013;4:686. doi: 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mullan LJ, Bleasby AJ. Short EMBOSS user guide. European Molecular Biology open software suite. Brief Bioinform. 2002;3:92–94. doi: 10.1093/bib/3.1.92. [DOI] [PubMed] [Google Scholar]
- 52.Marron AO, et al. The evolution of silicon transport in eukaryotes. Mol Biol Evol. 2016;33:3226–3248. doi: 10.1093/molbev/msw209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gruber A, Rocap G, Kroth PG, Armbrust EV, Mock T. Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 2015;81:519–528. doi: 10.1111/tpj.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 55.Gschloessl B, Guermeur Y, Cock JM. HECTAR: A method to predict subcellular targeting in heterokonts. BMC Bioinformatics. 2008;9:393. doi: 10.1186/1471-2105-9-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Afgan E, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 58.Petsalaki EI, Bagos PG, Litou ZI, Hamodrakas SJ. PredSL: A tool for the N-terminal sequence-based prediction of protein subcellular localization. Genomics Proteomics Bioinformatics. 2006;4:48–55. doi: 10.1016/S1672-0229(06)60016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fukasawa Y, et al. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 2015;14:1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gee CW, Niyogi KK. The carbonic anhydrase CAH1 is an essential component of the carbon-concentrating mechanism in Nannochloropsis oceanica. Proc Natl Acad Sci USA. 2017;114:4537–4542. doi: 10.1073/pnas.1700139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma X, et al. RNAi-mediated silencing of a pyruvate dehydrogenase kinase enhances triacylglycerol biosynthesis in the oleaginous marine alga Nannochloropsis salina. Sci Rep. 2017;7:11485. doi: 10.1038/s41598-017-11932-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nobusawa T, Hori K, Mori H, Kurokawa K, Ohta H. Differently localized lysophosphatidic acid acyltransferases crucial for triacylglycerol biosynthesis in the oleaginous alga Nannochloropsis. Plant J. 2017;90:547–559. doi: 10.1111/tpj.13512. [DOI] [PubMed] [Google Scholar]
- 63.Wei H, et al. A type-I diacylglycerol acyltransferase modulates triacylglycerol biosynthesis and fatty acid composition in the oleaginous microalga, Nannochloropsis oceanica. Biotechnol Biofuels. 2017;10:174. doi: 10.1186/s13068-017-0858-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moog D, Stork S, Reislöhner S, Grosche C, Maier UG. In vivo localization studies in the stramenopile alga Nannochloropsis oceanica. Protist. 2015;166:161–171. doi: 10.1016/j.protis.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- 66.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. September 6, 2017 doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kearse M, et al. Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller MA, et al. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol Bioinform Online. 2015;11:43–48. doi: 10.4137/EBO.S21501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dorrell R, et al. 2018 Chrysophyte plastid evolution dataset. University of Cambridge Apollo Data Repository. Available at https://www.repository.cam.ac.uk/handle/1810/284182. Deposited October 21, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.