Significance
Genomic instability fuels tumor progression and treatment resistance in cancer. Causative factors and means for clinical detection of genomic instability in the bulk of gliomas have been inadequately explored. We identify a conserved gene coexpression module centered on CDC20 (CDC20-M) in glioma transcriptomes that encompasses broad activities of cell proliferation and genome maintenance. We show that high CDC20-M expression robustly predicts poor prognosis and severe chromosomal instability and mutation burden in glioma. Core members of CDC20-M, including AURKA, are dysregulated and may provide new therapeutic targets in glioma. These data suggest that the CDC20-M signature could be applied for risk stratification and for the development of treatment protocols for glioma and other cancers.
Keywords: glioma, genomic instability, CDC20-M, gene coexpression module, MLN8237
Abstract
Genomic instability (GI) drives tumor heterogeneity and promotes tumor progression and therapy resistance. However, causative factors underlying GI and means for clinical detection of GI in glioma are inadequately identified. We describe here that elevated expression of a gene module coexpressed with CDC20 (CDC20-M), the activator of the anaphase-promoting complex in the cell cycle, marks GI in glioma. The CDC20-M, containing 139 members involved in cell proliferation, DNA damage response, and chromosome segregation, was found to be consistently coexpressed in glioma transcriptomes. The coexpression of these genes was conserved across multiple species and organ systems, particularly in human neural stem and progenitor cells. CDC20-M expression was not correlated with the morphological subtypes, nor with the recently defined molecular subtypes of glioma. CDC20-M signature was an independent and robust predictor for poorer prognosis in over 1,000 patients from four large databases. Elevated CDC20-M signature enabled the identification of individual glioma samples with severe chromosome instability and mutation burden and of primary glioma cell lines with extensive mitotic errors leading to chromosome mis-segregation. AURKA, a core member of CDC20-M, was amplified in one-third of CDC20-M–high gliomas with gene-dosage–dependent expression. MLN8237, a Food and Drug Administration-approved AURKA inhibitor, selectively killed temozolomide-resistant primary glioma cells in vitro and prolonged the survival of a patient-derived xenograft mouse model with a high–CDC20-M signature. Our findings suggest that application of the CDC20-M signature may permit more selective use of adjuvant therapies for glioma patients and that dysregulated CDC20-M members may provide a therapeutic vulnerability in glioma.
Glioblastomas (GBM) constitute the major fraction of primary tumors in the adult central system. Despite intensive research, the prognosis of GBM is poor, and the median survival of GBM patients remains less than 2 y (1). GBMs constantly evolve and show extensive cellular and genetic heterogeneity (2). Enhanced replication stress and DNA damage response (DDR) are constitutively active in GBM (3). Genomes of GBM harbor numerous single-nucleotide exchanges. They are also subject to chromosomal instability (CIN), frequently generating an abnormal number of chromosomes (aneuploidy) and chromosomal structural abnormalities (4, 5). This genomic instability (GI) leads to new clonal variants among individual GBM cells (6, 7) and may thereby drive glioma progression and cause resistance to therapy (8–14).
GBM and lower-grade glioma may originate from or acquire features of neural stem or progenitor cells (termed NPC) (15). Mechanisms regulating chromosome segregation critically regulate self-renewal and differentiation of NPCs. Defects in several centrosome- and microtubule-regulating genes, including ASPM, NDE1, CEP120, CENPF, or TACC, cause severe defects in cell-fate decision and differentiation in the NPC pool during neocortical development (16–22). On the other hand, a number of comprehensive genomic studies have made important steps forward in understanding the causes and implications of GI in glioma. Singh et al. (5) found that a fusion between the fibroblast growth factor receptor (FGFR) gene and the transforming acidic coiled-coil (TACC) gene causes CIN in <5% of the GBMs. The Cancer Genome Atlas (TCGA) recently reported that 16% of low-grade gliomas and GBMs harbor mutations and/or somatic copy-number alterations (SCNA) in multiple genes involved in the cohesin complex, including NIPBL and STAG2 (23, 24). Carter et al. (25) developed an aneuploidy-correlated CIN70 signature, which was predictive of poor outcome in glioma and several other cancer types. Based on the SNP6.0 data, TCGA recently reported an aneuploidy score, which correlates with TP53 mutations and the overall mutation rate (26). While these reports highlight the implications of GI in glioma etiology, causative mechanisms and markers of GI remain poorly defined.
We hypothesized that aberrant activities in a gene network, which is involved in maintaining genomic stability in NPCs, may play a causative role in glioma GI. Accordingly, we aimed to develop a GI signature in glioma to fulfill the following features: (i) to include genome maintenance functions the activities or dysregulation of which may cause or be associated with GI; (ii) to make a prognosis; (iii) to contain potential therapeutic targets; and (iv) to be applicable to the majority of gliomas (and nonglioma cancers).
To test this hypothesis, we screened glioma transcriptome for gene coexpression modules centered on the regulators of NPC development and on the known activities associated with CIN. We here report the identification of a gene coexpression module around CDC20 (CDC20-M), the activator of anaphase-promoting complex (APC) of the cell cycle. Members of CDC20-M contain key regulators of cell proliferation, DNA replication, DDR, and chromosome segregation. Our findings demonstrate that the CDC20-M signature permits the detection of gliomas with severe GI and poor prognosis, and core members of CDC20-M may provide potential therapeutic targets for treatment of glioma.
Results
Identification of a Conserved, Stem/Progenitor Cell-Enriched CDC20-M in the Glioma Transcriptome.
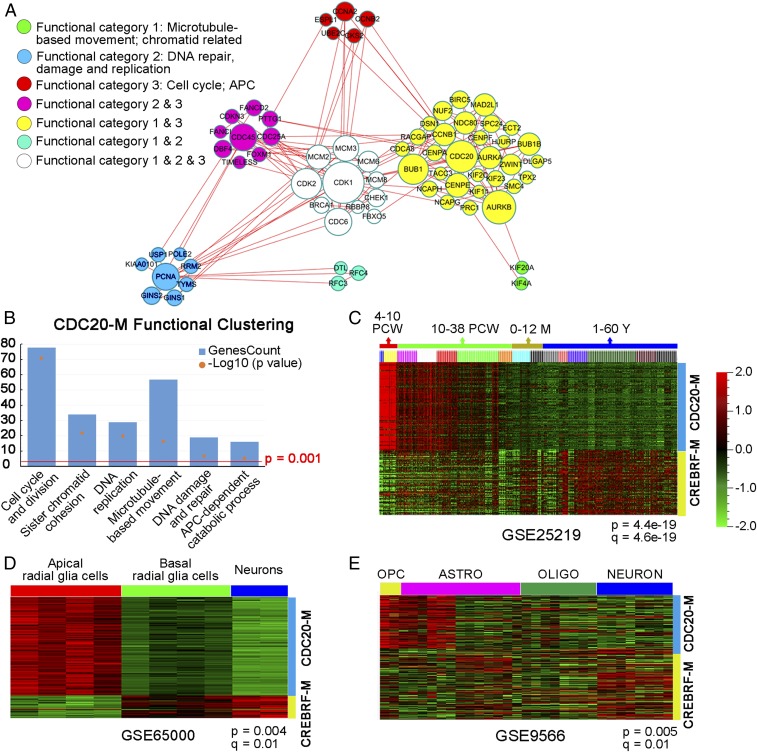
Using Pearson correlation coefficient analysis in glioma transcriptome databases (SI Appendix, Table S1), we set out to screen for coexpressed gene modules centered around known regulators of NPC self-renewal or CIN as a means to cluster gliomas into subgroups with distinct severities of GI. To this end, CDC20-M (SI Appendix, Table S2) was selected because of the following features: (i) the 139 genes in CDC20-M form a large protein–protein association network (Fig. 1A); (ii) in addition to genes involved in the cell cycle and division (e.g., CCNB1, CDK1, CDK2, CDC45, CKS2, MCM2, and PCNA), DNA replication (e.g., CDC25A, TIMELESS, and RFC2), replication stress and DDR (e.g., BRCA1, CHEK1, FANCI, and TOP2A), sister chromatid cohesion (e.g., PTTG1/SECURIN), spindle organization and biogenesis/microtubule-based movement (e.g., ASPM, KIF11, and KIF14), and APC-dependent catabolic process (e.g., CCNA2, UBE2C, UBE2S, and ESPL1) (Fig. 1B), gene ontology analysis shows that CDC20-M members are also involved in centrosome duplication (e.g., AURKA, NEK2, STIL, and TACC3), kinetochore microtubule dynamics and attachment stability (members of the CENP family) and the spindle assembly checkpoint (BUB1, BUBR1, CENP-E, CDC20, MAD2L1, NDC80, and RAE1) (27–34); (iii) loss of function in CDC20-M members ASPM, CENPF, and TACC3 causes defects of chromosome segregation in NPCs (17, 21, 22); and (iv) aberrant activities of CDC20-M members, such as AURKA, BUB1, BUBR1, CENPE, CDC20, NDC80, and RAE1, induce aneuploidy and tumorigenesis (35–39). A small subset of the CDC20-M members (37 of the 139 genes) overlapped with the previously reported CIN70 signature (25); however, hierarchy clustering analysis in glioma transcriptomes suggests that nearly 18% of CIN70 genes were not coexpressed with the remaining members (SI Appendix, Fig. S1).
Fig. 1.
Identification of CDC20-M in the glioma transcriptome and its expression profile in brain development. (A) Depiction of the protein–protein association network among CDC20-M members. Sixty-eight CDC20-M members with a combined association score >0.99 in the STRING database were used to construct this network using Cytoscape. The nodes are color-coded according to the functional categories in DAVID bioinformatics resources, and their sizes correspond to the extent of association. (B) Enrichment of gene ontology terms in CDC20-M as analyzed in DAVID bioinformatics resources. The −log10 P value and gene number of the top six functional clusters of the CDC20-M are shown. (C) Enriched CDC20-M expression during early development of the human brain. CDC20-M was particularly highly expressed during the first 4–10 PCW of human brain development; CREBRF-M showed enriched expression in brain tissues from 12 to 60 postnatal years. M: postnatal months; Y: postnatal years. Hierarchical clustering for the expression profiles of CDC20-M and CREBRF-M was performed in GSE25219 under the supervision of brain development stages. Heatmap at P = 4.4e-19 and q = 4.6e-19 is shown. (D) Enriched CDC20-M signature in human apical radial glia cells. Compared with basal radial glia cells and neurons, apical radial glia cells showed the highest expression of CDC20-M. Heatmap of supervised hierarchical clustering in GSE65000 for the expression profiles of CDC20-M and CREBRF-M at P = 0.004 and q = 0.01 is shown. (E) Enriched CDC20-M signature in murine OPCs. Heatmap of supervised hierarchical clustering of CDC20-M and CREBRF-M expression in the transcriptome of purified cell types of mouse brain (GSE9566; P = 0.005 and q = 0.01) is shown.
We first analyzed the pattern of CDC20-M expression in brain development and homeostasis. For defining stable expression clusters, a nontumor brain-tissue–enriched, CDC20-negatively correlated CREBRF coexpression module (CREBRF-M, containing 120 genes, SI Appendix, Fig. S2 and Table S3) was identified in glioma transcriptome. CREBRF is ubiquitously expressed in various mature tissues as a negative regulator of the unfolded protein response (40). In the transcriptome dataset of human brain development and maturation [National Center for Biotechnology Information Gene Expression Omnibus no. GSE25219 (41)], CDC20-M was highly expressed in brain tissues at the embryonic and early fetal development stages, which are marked by extensive proliferation of neural stem cells and by the initiation of neurogenesis [4–10 postconceptional weeks (PCW)] (Fig. 1C). Among the purified cell populations from the developing human neocortex [GSE65000 (42)], CDC20-M was enriched in the apical radial glial cells, the well-recognized NPCs in human brain (Fig. 1D). Compared with the mature neuron and oligodendrocytes [GSE9566 (43)], murine oligodendrocyte progenitor cells (OPC) showed enriched CDC20-M expression (Fig. 1E). Enriched CDC20-M expression was also found in hematopoietic progenitor cells. In steady-state human hematopoiesis [GSE24759 (44)], progenitor cells in the megakaryocyte/erythroid and granulocyte lineages and pro-B progenitors showed the highest CDC20-M expression (SI Appendix, Fig. S3A). In murine hematopoiesis [GSE77098 (45)], erythroid progenitors and pro-B progenitors remained as the cell types with the highest CDC20-M expression, whereas the mature cell types and long-term hematopoietic stem cells showed relatively low CDC20-M signature (SI Appendix, Fig. S3B). CDC20-M was also coexpressed in the transcriptomes of Caenorhabditis elegans (46) (SI Appendix, Fig. S3C) and Drosophila (47) (SI Appendix, Fig. S3D). These findings together suggest that, although CDC20-M was identified in glioma transcriptomes, it constitutes a conserved activity required for or associated with cell proliferation, particularly in the stem/progenitor cell pool.
The CDC20-M Signature Is a Robust Marker of Poor Prognosis for Adult Diffuse Glioma.
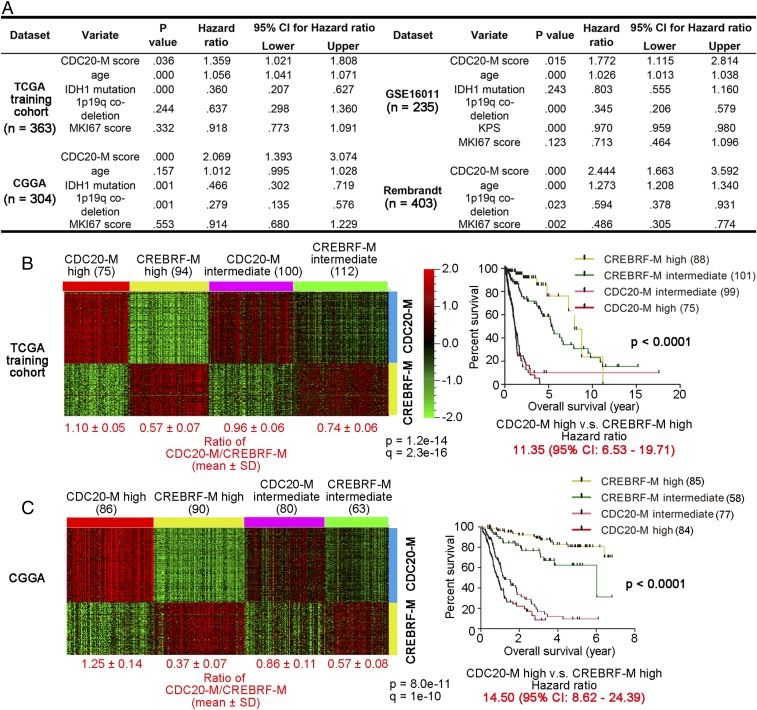
To assess the relevance of the CDC20-M signature in glioma biology, we first assessed whether the CDC20-M signature enables the risk stratification of adult diffuse gliomas of all morphological subtypes and grades. In 1,305 patients from four datasets with known survival information, generated in three different continents, multivariate Cox regression analysis showed that the CDC20-M score (the average expression of CDC20-M) is the only marker consistently associated with poor prognosis (Fig. 2A), which is complementary to IDH mutation and 1p19q codeletion as indicators of good prognosis (48, 49). CIN70 signature was prognostic only in the Chinese Glioma Genome Atlas (CGGA) and REMBRANDT datasets (SI Appendix, Table S4).
Fig. 2.
Elevated CDC20-M expression is a superior and robust signature of poor prognosis. (A) Results of multivariate Cox regression analysis for CDC20-M score, age at diagnosis, IDH1 mutation, 1p19q codeletion, and MKI67 score (the log2 RNA expression of Ki-67) in four large glioma datasets. Only the CDC20-M score showed consistent correlation with patient survival. (B, Left) Heatmap of unsupervised consensus clustering of the CDC20-M and CREBRF-M signature in the TCGA training cohort. The ratios of average expression of CDC20-M and CREBRF-M are shown under each subgroup. (Right) Kaplan–Meier plot of the overall survival for patients from each molecular subtype is shown. The overall survival data were analyzed using log-rank tests. The hazard ratio between patients with high CDC20-M glioma and patients with high CREBRF-M gliomas is shown under the survival curves. (C, Left) Heatmap of the CDC20-M and CREBRF-M signature in the CGGA glioma dataset supervised by SSP prediction of the CDC20-M/CREBRF-M subtypes. Using Spearman correlation, CGGA glioma samples were assigned to the nearest centroid of the four CDC20-M/CREBRF-M subtypes defined in the TCGA training cohort. The ratios of average expression of CDC20-M and CREBRF-M are shown under each subgroup. (C, Right) Kaplan–Meier plot of the overall survival for patients from each molecular subtype is shown. The overall survival data were analyzed using log-rank tests. The hazard ratio between patients with high CDC20-M glioma and patients with high CREBRF-M gliomas is shown under the survival curves.
In the context of morphological diagnosis, the majority of GBMs showed enriched CDC20-M expression; small subsets of grade II/III astrocytoma and oligodendroglioma also showed relatively enriched CDC20-M expression (SI Appendix, Fig. S4A). Among the grade II/III astrocytomas, gliomas with low CDC20-M but high CREBRF-M expression were associated with good prognosis compared with those with relatively enriched CDC20-M expression (SI Appendix, Fig. S4B). Based on the pathways conserved between brain development and glioma pathogenesis, we have recently identified gene coexpression modules around epidermal growth factor receptor (EGFR) (EGFR module, EM) or platelet-derived growth factor receptor A (PDGFRA) (PDGFRA module, PM) in the glioma transcriptome and defined an EM/PM classification scheme that assigns all adult diffuse gliomas to either the EM or the PM subtype; the EM and PM gliomas showed transcriptomic similarities to neural stem cells and OPCs, respectively (50). The majority of EM gliomas and a subset of PM gliomas were enriched in CDC20-M expression (SI Appendix, Fig. S4C). Compared with the PM gliomas with low CDC20-M expression, PM gliomas expressing high CDC20-M showed a poorer prognosis (SI Appendix, Fig. S4D).
We performed unsupervised consensus clustering in the TCGA training cohort (containing both low- and high-grade gliomas) to define stable molecular clusters with a unique pattern of CDC20-M and CREBRF-M expression (SI Appendix, Fig. S5A). Independent of the morphological diagnosis, gliomas were assigned to stable subgroups with high expression of CDC20-M or CREBRF-M or with intermediate expression of CDC20-M or CREBRF-M (Fig. 2B and SI Appendix, Fig. S5B). Compared with gliomas with high or intermediate expression of CREBRF-M, gliomas with high or intermediate CDC20-M expression showed poorer prognosis (Fig. 2B). A hazard ratio of 11.35 [95% confidence interval (CI): 6.53–19.71] between patients with CDC20-M–high gliomas and patients with CREBRF-M–high gliomas was observed. Patients with CDC20-M–high gliomas were older at diagnosis compared with the patients with high CREBRF-M expression. Furthermore, mutations in ATRX and IDH1 and codeletion of chromosome 1p/19q occurred predominantly in gliomas with high CREBRF-M expression (SI Appendix, Fig. S5B).
Using unsupervised clustering analysis between glioma samples and the signature of the 2,275 most variably expressed genes, TCGA recently defined the LGr1-4 whole-transcriptome clusters with LGr1-3 enriched in IDH1 or IDH2 mutated grade II–III gliomas and LGr4 in IDH1/2 wild-type GBMs (23). Within the LGr1 and LGr3 clusters, the CDC20-M/CREBRF-M signature-based clustering clearly distinguished the lower-grade gliomas with a poorer prognosis from those with a better prognosis (SI Appendix, Fig. S6).
Next, we applied the single sample predictor (SSP) approach (51) to assign individual glioma samples to the CDC20-M/CREBRF-M–defined subtypes. Based on the data of the TCGA training cohort, a mean expression profile, the centroid, was created and fixed for each CDC20-M/CREBRF-M subtype, 301 grade II–III gliomas from the TCGA validation cohort, and 319 grade II–IV gliomas from the CGGA dataset were then compared with each centroid and assigned to the nearest centroid using Spearman correlation (Fig. 2C and SI Appendix, Fig. S5 C and D). In both the TCGA validation cohort and the CGGA dataset, the CDC20-M/CREBRF-M subtypes identified by the SSP showed similar differences in overall survival, age at diagnosis, and frequencies of IDH1/IDH2 mutation and 1p/19q codeletion, as in the TCGA training cohort (Fig. 2C and SI Appendix, Fig. S5 C and D).
Notably, in the TCGA validation cohort, the CDC20-M signature showed a particularly strong prognostic effect in the lower-grade gliomas, and a hazard ratio of 700 (95% CI: 89.8–5470) was observed between the patients with gliomas of high CDC20-M signature compared with patients with gliomas of high CREBRF-M signature (SI Appendix, Fig. S5C). Patients with grade II gliomas are generally associated with a prognosis of >5 y; however, some patients progress rapidly to recurrence or death due to yet poorly characterized mechanisms (52). In the CGGA, GSE16011, and REMBRANDT datasets, we consistently found that grade II astrocytomas with high CDC20-M expression progressed more rapidly compared with those grade II astrocytomas with high CREBRF-M expression (SI Appendix, Fig. S7). The four grade II astrocytomas from CGGA and one grade II astrocytoma from GSE16011 with poor prognosis harbored the IDH1 mutation. However, subtypes defined by CIN70 signature-based SSP were less distinct in terms of overall survival compared with the CDC20-M/CREBRF-M subtypes (SI Appendix, Fig. S8).
The ability of the CDC20-M signature to predict patient prognosis outperformed that of Ki-67 staining. The Ki-67 staining data of 239 gliomas of the CGGA dataset were available for analysis. Whereas gliomas with very low Ki-67 staining (Ki-67 intensity grade = 0) showed good prognosis compared with the gliomas with extensive Ki-67 staining (Ki-67 intensity grades of 1, 2, or 3), prognostic difference was not found in gliomas with Ki-67 intensity grades of 1, 2, or 3 (SI Appendix, Fig. S9A). In subgroups with Ki-67 staining intensity grades of 0–2, gliomas with high CDC20-M expression consistently showed poorer prognosis compared with those with high CREBRF-M expression (SI Appendix, Fig. S9B).
In summary, these findings show that, whereas IDH mutation and 1p19q codeletion predict good prognosis, the CDC20-M signature is a distinct molecular signature that robustly predicts poor prognosis and rapid progression of adult diffuse glioma.
Elevated CDC20-M Expression Marks Severe GI in Glioma.
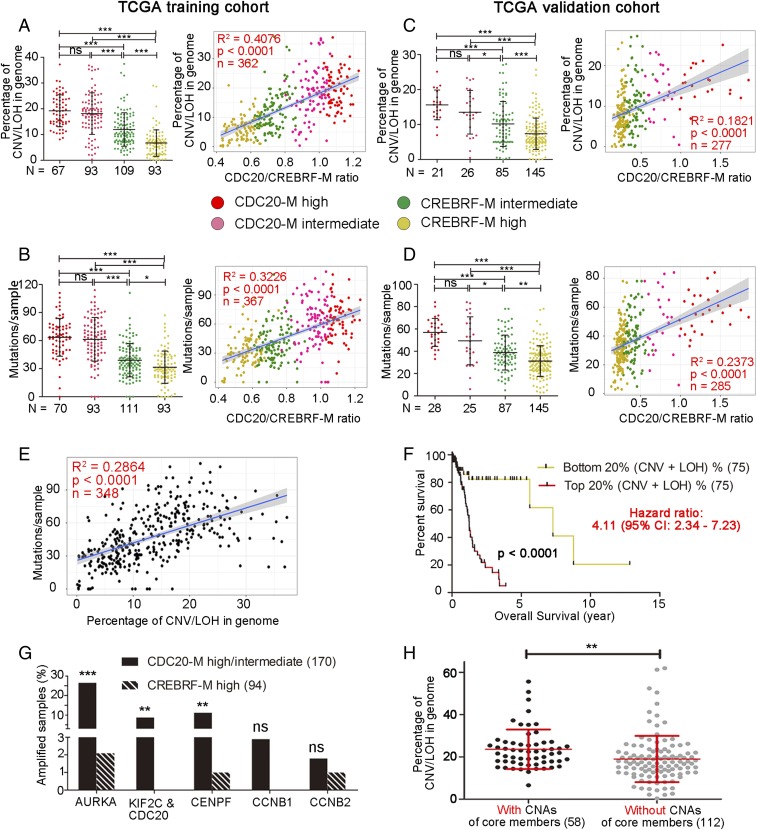
Because CDC20-M members are critically involved in DNA replication, DDR, and chromosome segregation, their aberrant activity may indicate GI. Leveraging the SNP6.0 and whole-exome sequencing data of the 682 glioma samples from TCGA [among them, one sample (TCGA-27-1835), harbored FGFR-TACC fusion], we first compared the extent of chromosome copy number variation (CNV), loss of heterozygosity (LOH), and mutation burden according to CDC20-M/CREBRF-M clustering in the TCGA training cohort. Gliomas with high or intermediate CDC20-M expression harbored more CNV and LOH events and mutation burdens compared with gliomas with high or intermediate CREBRF-M expression (Fig. 3 A and B, Left). The ratio between CDC20-M and CREBRF-M expression correlated to the extent of chromosomal abnormalities (Fig. 3A, Right; more closely to arm-level than to focal CNV, SI Appendix, Fig. S10 A and B), to total mutations, and to nonsynonymous mutations (Fig. 3B, Right, and SI Appendix, Fig. S10C). We also compared the aneuploidy score [reflecting the total arm-level copy-number alterations in a sample (53)] between the CDC20-M/CREBRF-M signature-defined subtypes, and the higher ratio between CDC20-M and CREBRF-M expression correlated to a higher aneuploidy score (SI Appendix, Fig. S10D). Similar results were found in the TCGA validation cohort (Fig. 3 C and D). Furthermore, concordant severities in chromosomal abnormality and mutation burden were observed in the vast majority of the samples (Fig. 3E). Compared with the subtypes defined by the CDC20-M/CREBRF-M signature, glioma subtypes defined by the CIN70 signature were less distinct in the extent of their chromosomal abnormalities and mutation burden (SI Appendix, Fig. S11). Gliomas with the top 20% most severe chromosomal abnormalities were associated with a significantly poorer prognosis compared with gliomas with the 20% least severe chromosomal abnormalities, reaffirming the association between genomic instability and prognosis in glioma (Fig. 3F).
Fig. 3.
Elevated CDC20-M signature marks extensive chromosome abnormalities and mutation burden in glioma. (A and B, Left) The extent of chromosomal abnormalities (A) or mutation burden (B) in the CDC20-M–defined glioma subgroups in the TCGA training cohort. ***P < 0.001; ns: P > 0.05 as analyzed in one-way ANOVA test. (Right) Linear regression analysis between the ratio of CDC20-M/CREBRF-M and the extent of chromosomal abnormalities or mutation burden in glioma samples. The same color codes were used in all panels. Regression coefficient (R2), P value, and number of samples (n) are indicated. (C and D, Left) The extent of chromosomal abnormalities (C) or mutation burden (D) in the SSP-defined CDC20-M glioma subgroups in the TCGA validation cohort. ***P < 0.001; **P < 0.01; *P < 0.05; ns: P > 0.05 as analyzed in one-way ANOVA test. (Right) Linear regression analysis between the ratio of CDC20-M/CREBRF-M and the extent of chromosomal abnormalities or mutation burden in glioma samples. (E) Linear regression analysis between the extent of chromosomal abnormalities and mutation burdens per glioma sample in the TCGA training cohort. Regression coefficient (R2), P value, and number of samples (n) are indicated. (F) Survival comparison between the patients with gliomas with the top 20% most severe chromosomal abnormalities and the patients with gliomas with the lowest 20% chromosomal abnormalities in the TCGA training cohort. (G) Percentage of glioma samples with amplifications in the six core members of CDC20-M in the TCGA training cohort. Copy numbers were analyzed using GISTIC 2.0. ***P < 0.001, **P < 0.01, ns: P > 0.05. Fisher’s exact test was applied to compare CDC20-M–high/intermediate with CREBRF-M–high samples. (H) Comparison of the extent of chromosomal abnormalities between samples harboring amplifications of any of the core members (AURKA, CDC20, KIF2C, CENPF) and samples without amplification of these core members in the glioma with high or intermediate CDC20-M expression. **P < 0.01, unpaired two-sided Student’s t test.
Next, we assessed the pattern of CNV and LOH between the CDC20-M/CREBRF-M signature-defined subtypes. Among the CDC20-M–high gliomas, 81% harbored chromosome 7 gain and chromosome 10 loss, 19% harbored complex CNVs with alterations in most chromosomes such that no particular patterns could be summarized, and 9% harbored chromothripsis events that were not observed in the CREBRF-M group (SI Appendix, Fig. S12A). Among the CREBRF-M–high gliomas, 37% harbored 1p and 19q codeletions, 31% harbored 17p LOH or 17p loss (where TP53 is located), 11% harbored relatively complex chromosome abnormalities, 14% had less than three chromosomes CNV, and 7% harbored no detectable CNV (SI Appendix, Fig. S12B). Thus, CDC20-M–high glioma samples harbored more complex CNV and LOH events compared with CREBRF-M–high glioma samples.
Notably, somatic alterations (CNV, nonsynonymous and indels/frameshifts mutations) in the core components of the TP53 pathway were differentially involved in CDC20-M/CREBRF-M classified gliomas (P < 0.0001, x2 test), with a lower frequency of somatic alterations in gliomas with high or intermediate CREBRF-M expression (50.0 and 70.9%, respectively) and a higher frequency in gliomas with high or intermediate CDC20-M expression (98.6 and 88.4%, respectively) (SI Appendix, Tables S5–S8), suggesting that inactivation of the TP53 pathway is a core requirement for a high proliferation rate of CDC20-M–high gliomas.
Gene-set enrichment analysis (54) identified significant enrichment of DDR signatures in CDC20-M–high glioma subtypes (SI Appendix, Fig. S13). In the CDC20-M–high glioma samples, immunohistochemical staining showed high expression of CDC20 and p-AURKA. Notably, the DDR marker phospho-histone H2AX (p-H2A.X) was more intense in CDC20-M–high samples compared with CREBRF-M–high samples (SI Appendix, Fig. S14), indicating elevated replication stress and ongoing DDR in CDC20-M–high gliomas.
To classify glioma cell lines according to the extent of CDC20-M expression, we first identified AURKA (20q13), KIF2C/CDC20 (both at 1p34), CENPF (1q41), CCNB1 (5q12), and CCNB2 (15q22.2) as the core members of CDC20-M, since they were consistently among the top 20 transcripts most correlated to the CDC20 expression in three large glioma transcriptome datasets (SI Appendix, Table S2). GISTIC analyses of the SNP6.0 data showed that, among these genes, AURKA, KIF2C/CDC20, and CENPF were frequently amplified in gliomas with high or intermediate CDC20-M expression (Fig. 3G). Among the gliomas with high or intermediate CDC20-M expression, higher percentages of genome changes were found in those samples harboring amplifications in any of these four genes compared with the remaining samples without amplifications in these genes (Fig. 3H). Gene dosage-dependent expression in gliomas with high or intermediate CDC20-M expression was found for the amplifications of AURKA, CDC20, KIF2C, and CENPF (SI Appendix, Fig. S15).
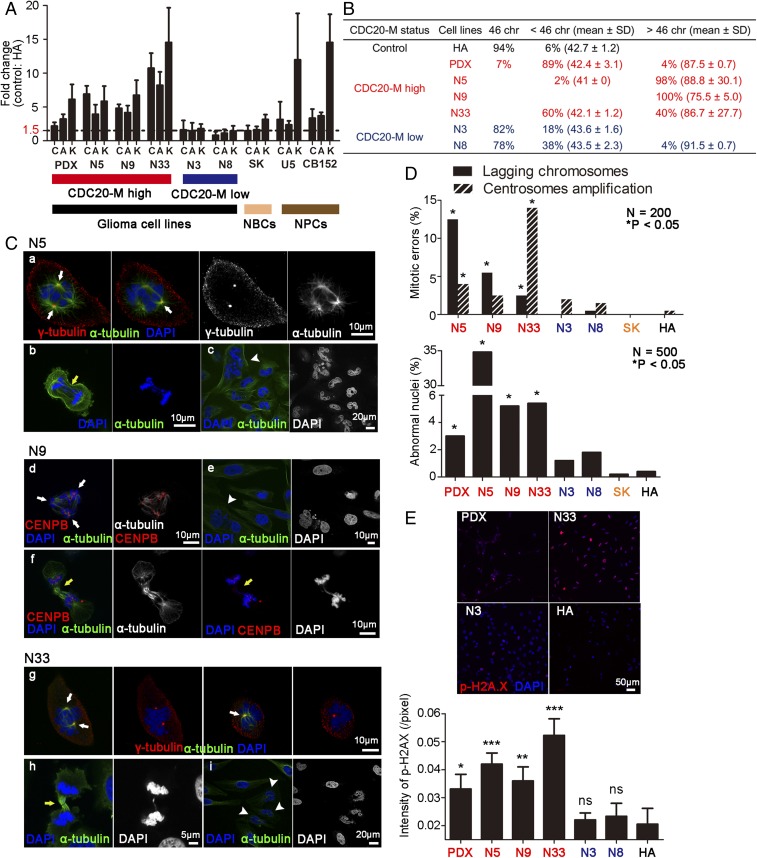
We then applied real-time RT-PCR to assess the expression of CDC20, AURKA, and KIF2C in five primary glioma cell lines (N5, N9, N33, N3, and N8), one patient-derived xenograft cell line (PDX) (SI Appendix, Table S9, all IDH1/IDH2 wild type), one neuroblastoma cell line (SK), and two human NPC lines (U5 and CB152). FGFR3-TACC3 fusion was not detected in all glioma cell lines tested (SI Appendix, Fig. S16). The PDX, N5, N9, and N33 lines showed higher expression of CDC20-M core members compared with N3, N8, and SK lines (Fig. 4A). Notably, the two human NPC lines also showed relatively high expression of AURKA, CDC20, and KIF2C. Karyotypic diversity can arise from CIN caused by enhanced replication stress or directly from chromosome mis-segregation (6, 55). For all karyotypes analyzed, human astrocytes (HA) had a modal chromosome number (46 chromosomes) and a low percentage of mode deviation (6%), as expected for untransformed cells. In contrast, all of the CDC20-M high cell lines contained a high fraction of cells in the S-phase, were aneuploid in most cells, and exhibited a high degree of near tetraploid unstable aneuploidy with a broad distribution of chromosome counts (Figs. 4B and 5C and SI Appendix, Fig. S18C). These findings suggest that glioma cell lines with high CDC20-M expression have more diverse karyotypes compared with glioma cell lines with low CDC20-M expression.
Fig. 4.
On-going CIN in glioma cells with high CDC20-M expression. (A) Real-time RT-PCR assessment of CDC20 (C), AURKA (A), and KIF2C (K) expression in six glioma cell lines (PDX, N5, N9, N33, N3, and N8), one neuroblastoma cell line (SK), and two human NPCs (U5 and CB152). The expression levels were normalized against invariably expressed ubiquitin C and subsequently compared with untransformed HA. Mean fold changes and their standard deviations from three independent experiments are shown. (B) Unstable aneuploidy in glioma cell lines with high expression of CDC20-M members. Chromosome counts of 50 karyotypes for each cell line were analyzed. (C) Representative images of CDC20-M–high glioma cells with lagging chromosome, centrosome amplification, or abnormal nuclei. Representative images of staining for DNA (blue), centrosome or centromere (red), and microtubule (green) are shown. Centrosome amplification, lagging chromosomes, and abnormal nuclei are depicted by white arrows, yellow arrows, and white arrowheads, respectively. (D) Percentage of mitotic errors and abnormal nuclei in cell lines analyzed. For the mitotic error count, ≥3 coverslips were analyzed for 200 dividing cells; for the abnormal nuclei count, ≥3 coverslips were analyzed for 500 cells. *P < 0.05, Fisher’s exact test with HA cells as the control. (E, Upper) Representative images of fluorescence labeling of p-H2AX in PDX, N33, N3, and HA cells. Images of staining for DNA (blue) and p-H2A.X (red) are shown. (E, Bottom) The intensities of p-H2A.X staining in glioma cell lines and the control HA cells are shown (n = 3, mean ± SD). ***P < 0.001; **P < 0.01, and *P < 0.05, Fisher’s exact test with HA cells as the control.
Fig. 5.
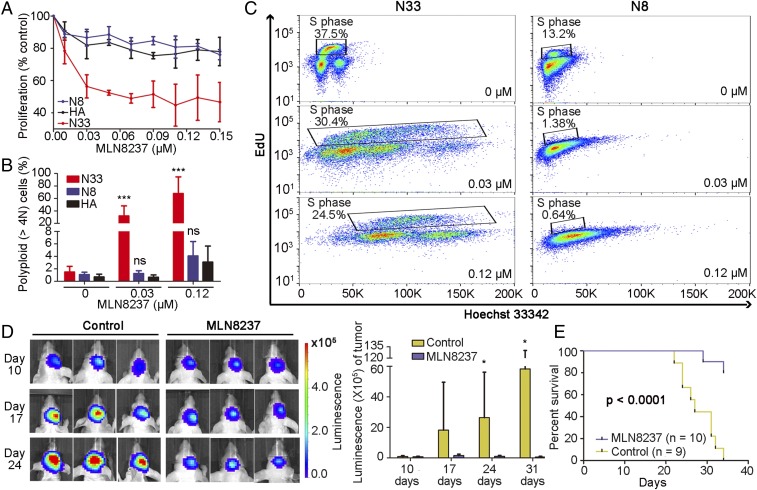
Inhibition of CDC20-M core member AURKA suppresses glioma cell growth in vitro and in vivo. (A) MLN8237 treatment strongly inhibited cell growth in CDC20-M–high N33 cells, but not in CDC20-M low N8 cells and HA cells. (B) MLN8237 induced a dose-dependent polyploidization in CDC20-M–high N33 cells. Quantification of three independent experiments is shown. Mean ± SD of three independent experiments is shown. ***P < 0.001 and ns: P > 0.05, compared with HA in each concentration using two-way ANOVA test. (C) Cell proliferative index (% of EdU-positive S-phase cells) and polyploidization in CDC20-M–high N33 and CDC20-M low N8 cells with or without MLN8237 treatment are shown. Results are representative of technical triplicates. (D, Left) Tumor volume of vehicle- or MLN8237-treated mice at 10, 17, and 24 d after tumor implantation was determined. (Right) Histogram shows the intensity of bioluminescence in mice. *P < 0.05, unpaired two-sided Student’s t test. (E) Survival comparison of vehicle- or MLN8237-treated mice analyzed in log-rank test. Results are representative of two independent experiments.
We further analyzed on-going CIN events in these cell lines. First, we measured the frequency of chromosome segregation defects including lagging chromosomes and centrosome amplification in these cells (Fig. 4 C and D). These defects were found in up to 3–16.5% of the mitotic cells in the CDC20-M high glioma cell lines, whereas HA, SK, and the CDC20-M low glioma cell lines had fewer than 2% of mitotic cells with chromosome segregation defects. The number of lagging chromatids per anaphase in CDC20-M–high glioma cells was between 2.5- and 12.5-fold higher than chromosomally stable HA cells, and centrosome amplification events were up to 14-fold more frequent in CDC20-M–high glioma cells. Chromosome segregation errors can lead to the formation of abnormal nuclei (56). PDX, N5, N9, and N33 cultures contained more cells with abnormal nuclei compared with N3, N8, SK, and HA. In particular, 35% of N5 cells contained abnormal nuclei. N5 cells also harbored the largest number of lagging chromosomes events (12.5%) among all cell lines. Elevated staining intensities of p-H2A.X were found in the CDC20-M–high PDX, N5, N9, and N33 cells compared with the CDC20-M low glioma cell lines (Fig. 4E).
Together, our findings show that high CDC20-M expression marks severe GI in gliomas, reflected in a high degree of CIN and an elevated mutation burden.
Core Members of CDC20-M Provide Potential Therapeutic Vulnerability in Glioma.
The activities of CDC20-M members across the cell-cycle phases are precisely regulated (57). However, 93.5% of the gliomas with high or intermediate CDC20-M expression harbored copy-number amplifications in 18 ± 13 CDC20-M members, and 24.7% of these gliomas harbored mutations in at least one CDC20-M member (SI Appendix, Tables S10 and S11). Thus, cell-cycle progression in these gliomas is likely vulnerable to further damage targeting CDC20-M members. We tested whether inhibition of CDC20-M core members provides a therapeutic vulnerability in glioma. All glioma cell lines tested showed considerable resistance toward temozolomide (the current standard chemotherapeutic for GBM), and their IC50 concentrations were >450 µM (SI Appendix, Fig. S17A). To suppress the activity of CDC20, we used proTAME, a prodrug of TAME, which binds to the APC and displaces the C-terminal isoleucine-arginine tail of CDC20, thereby preventing APC activation and thus inducing mitotic arrest (58). In the CDC20-M–high N5, N9, and N33 cell lines, proTAME treatment resulted in mitotic arrest-mediated cell death in 24 h with an IC50 concentration <20 µM. Consistent with this, proTAME-induced mitotic arrest requires sustained spindle assembly checkpoint activity (58), and N3 and N8 cell lines showed relatively higher IC50 concentrations compared with those of CDC20-M–high cell lines. HA cells were nearly unaffected under the same conditions, with an IC50 concentration >50 µM (SI Appendix, Fig. S17B).
We also used a selective AURKA inhibitor, the Food and Drug Administration-approved MLN8237 (Alisertib), with an indicative inhibitory effect in a variety of malignancies including GBM (59–63). The IC50 concentrations of MLN8237 in the CDC20-M–high N5, N9, and N33 glioma cell lines were about 100-fold lower than that of proTAME. MLN8237 inhibited the growth of the CDC20-M–high glioma cell lines, and only marginal effects were observed in CDC20-M–low glioma cell lines and in HA at the same concentrations (Fig. 5A and SI Appendix, Fig. S18A). Moreover, DNA staining showed that MLN8237 mediated strong cell polyploidization in the CDC20-M–high glioma cell lines, but not in CDC20-M–low glioma cells and HA cells (Fig. 5 B and C and SI Appendix, Fig. S18 B and C).
To test the effect of MLN8237 in vivo, we established an intracranial PDX glioma model in BALB/c nude mice. RNA-sequencing data showed a ratio of CDC20-M/CREBRF-M at 2.08 ± 0.12 (n = 4), suggesting a CDC20-M–high signature. Mice carrying the PDX glioma were treated with MLN8237 (at 25 mg/kg body weight, two applications every other day, for 4 wk or with the vehicle control). MLN8237 significantly inhibited the growth of intracranial PDX glioma and prolonged the survival of the mice (Fig. 5 D and E). A striking reduction in the p-AURKA and p-H2A.X staining intensity was found in the MLN8237-treated PDX, compared with the vehicle-treated PDX samples (SI Appendix, Fig. S19). Taken together, MLN8237 potently suppressed the growth of CDC20-M–high glioma cells both in vitro and in vivo. These findings suggest that gliomas with high CDC20-M signature may be vulnerable toward treatment targeting core members of CDC20-M.
Discussion
GI is a major causative factor for the progression and treatment resistance of gliomas and other cancers. Here, we have identified a conserved CDC20-M in glioma transcriptomes with anchorage in the NPC pool and the early stages of the human brain development. Members of CDC20-M are involved in the maintenance of genome stability. CDC20-M signature is a robust and independent predictive marker of poor prognosis and rapid progression in both low- and high-grade adult diffuse gliomas. We have developed the ratio between CDC20-M and CREBRF-M expression as a surrogate marker of both CIN and mutation burden, which permits the assignment of individual gliomas into two major categories: with high or low severity of GI and hence distinct prognoses. Furthermore, in vitro and in vivo experiments suggest that core members of CDC20-M may provide potential therapeutic targets for gliomas. Thus, the severities of GI in glioma, reported by the elevated CDC20-M signature, could support patient risk stratification and potentially also guide treatment decisions.
Gliomas with severe GI evolve rapidly and thus require more aggressive treatment. However, the identification of gliomas with severe GI requires detection of CIN and mutation burden, which can be costly and technically challenging in a clinical setting (10). The CDC20-M spans a broad spectrum of activities involved in cell proliferation and genome maintenance, and their concerted actions are likely required for the fidelity of DNA replication and chromosome segregation. Dysregulated activities of CDC20-M members, by inappropriate expression or due to amplifications/mutations, could be the cause of GI. SCNA/mutations in the CDC20-M members such as BRCA1, CHEK1, FANCI, and TOP2A are implicated in replication stress and can increase mutation burden (30–34, 64); and, in the CDC20-M, members such as CDC20, AURKA, BUB1, RAE1, and CENPE can induce CIN (36–39). Complemented by its anticorrelated CREBRF-M signature, our studies developed CDC20-M as a surrogate marker for GI. Whereas previously reported CIN70 signature (25) and FGFR-TACC fusion (5) indicate only severity of CIN, the ratio of CDC20-M/CREBRF-M expression predicts both CIN and mutation burden in concordance. FGFR-TACC fusion (5) and genomic alterations in cohesion complex members (23, 24) can apply to only a minor fraction of gliomas; the ratio of CDC20-M/CREBRF-M expression, however, is applicable to nearly all adult diffuse gliomas irrespective of their morphological subtypes or grades.
In addition to the superior capacity in predicting poor prognosis, the identification of CDC20-M and the capacity to assign individual glioma samples into distinct GI subtypes can provide important support to therapy development. Gliomas with high CDC20-M signature may carry early developmental features compared with gliomas with high CREBRF-M signature. Our findings show that cell proliferation and GI occur hand-in-hand. The CDC20-M contains checkpoint members of both DNA replication and chromosome segregation. Concordant severities of CIN and mutation burden identified in over 600 TCGA glioma samples, ongoing DDR, and inactivation of the TP53 pathway together suggest that, although mutations in genes implicated in chromosome segregation are altered in a sizable proportion of gliomas (23), replication stress is a more likely driver of GI in the bulk of gliomas (55, 65). As indicated by the inhibition of CDC20 and AURKA in the current report, members of CDC20 may contain previously inadequately recognized therapeutic target(s) for glioma. The severe CIN and mutation burden in gliomas expressing high CDC20-M suggest that this subset of gliomas could be particularly vulnerable to therapies that would further induce GI. Because of the high mutation burden, these gliomas might also be more suitable for mutation-related neoantigen vaccine (66).
Finally, as the CDC20-M signature is conserved across organ development, the CDC20-M signature-based risk stratification and treatment decision may find its application in other cancers.
Materials and Methods
Details regarding the integrated analyses of transcriptomic, genomic, and clinical data and in vitro and in vivo analyses of patient-derived glioma cell lines are provided in SI Appendix. The use of glioma samples in Beijing was approved by the Ethics Committee of Beijing Tiantan Hospital, and written informed consent was obtained from all patients. The animal experiment was approved by the Animal Ethical and Welfare Committee of Tianjin Medical University.
Supplementary Material
Acknowledgments
This study was supported by National Natural Science Foundation of China Grants 81773015, 81472348, and 31711530156; The National Key Research and Development Plan Grant 2016YFC0902500; the Beijing Cooperative Construction Project; and the Erik Philip-Sörensen Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814060116/-/DCSupplemental.
References
- 1.Stupp R, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103:1139–1143. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartkova J, et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. 2010;29:5095–5102. doi: 10.1038/onc.2010.249. [DOI] [PubMed] [Google Scholar]
- 4.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 5.Singh D, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godek KM, et al. Chromosomal instability affects the tumorigenicity of glioblastoma tumor-initiating cells. Cancer Discov. 2016;6:532–545. doi: 10.1158/2159-8290.CD-15-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piccirillo SG, et al. Genetic and functional diversity of propagating cells in glioblastoma. Stem Cell Rep. 2015;4:7–15. doi: 10.1016/j.stemcr.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee AJX, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–1870. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrell RA, Swanton C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol. 2014;8:1095–1111. doi: 10.1016/j.molonc.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: Therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–538. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sottoriva A, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greaves M. Evolutionary determinants of cancer. Cancer Discov. 2015;5:806–820. doi: 10.1158/2159-8290.CD-15-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: An evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 14.Swanton C, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci USA. 2009;106:8671–8676. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alcantara Llaguno SR, Parada LF. Cell of origin of glioma: Biological and clinical implications. Br J Cancer. 2016;115:1445–1450. doi: 10.1038/bjc.2016.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKinnon PJ. Maintaining genome stability in the nervous system. Nat Neurosci. 2013;16:1523–1529. doi: 10.1038/nn.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Z, et al. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang YT, Wang CL, Van Aelst L. DOCK7 interacts with TACC3 to regulate interkinetic nuclear migration and cortical neurogenesis. Nat Neurosci. 2012;15:1201–1210. doi: 10.1038/nn.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Waters AM, et al. The kinetochore protein, CENPF, is mutated in human ciliopathy and microcephaly phenotypes. J Med Genet. 2015;52:147–156. doi: 10.1136/jmedgenet-2014-102691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fish JL, Kosodo Y, Enard W, Pääbo S, Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci USA. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceccarelli M, et al. TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennan CW, et al. TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 26.Taylor AM, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33:676–689.e3. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du J, et al. The mitotic checkpoint kinase NEK2A regulates kinetochore microtubule attachment stability. Oncogene. 2008;27:4107–4114. doi: 10.1038/onc.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeLuca JG, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- 29.Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell. 2011;19:701–714. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chavali PL, Putz M, Gergely F. Small organelle, big responsibility: The role of centrosomes in development and disease. Philos Trans R Soc Lond B Biol Sci. 2014;369:pii: 20130468. doi: 10.1098/rstb.2013.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heijink AM, Krajewska M, van Vugt MATM. The DNA damage response during mitosis. Mutat Res. 2013;750:45–55. doi: 10.1016/j.mrfmmm.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Hosoya N, Miyagawa K. Targeting DNA damage response in cancer therapy. Cancer Sci. 2014;105:370–388. doi: 10.1111/cas.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malik M, Nitiss KC, Enriquez-Rios V, Nitiss JL. Roles of nonhomologous end-joining pathways in surviving topoisomerase II-mediated DNA damage. Mol Cancer Ther. 2006;5:1405–1414. doi: 10.1158/1535-7163.MCT-05-0263. [DOI] [PubMed] [Google Scholar]
- 34.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diaz-Rodríguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci USA. 2008;105:16719–16724. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver BA, Cleveland DW. The role of aneuploidy in promoting and suppressing tumors. J Cell Biol. 2009;185:935–937. doi: 10.1083/jcb.200905098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol. 2009;185:983–994. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–1064. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, et al. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–7158. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- 40.Audas TE, Li Y, Liang G, Lu R. A novel protein, Luman/CREB3 recruitment factor, inhibits luman activation of the unfolded protein response. Mol Cell Biol. 2008;28:3952–3966. doi: 10.1128/MCB.01439-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Florio M, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 43.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novershtern N, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Graaf CA, et al. Haemopedia: An expression atlas of murine hematopoietic cells. Stem Cell Reports. 2016;7:571–582. doi: 10.1016/j.stemcr.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim SK, et al. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- 47.Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- 48.Brat DJ, et al. Cancer Genome Atlas Research Network Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Louis DN, et al. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 50.Sun Y, et al. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA. Proc Natl Acad Sci USA. 2014;111:3538–3543. doi: 10.1073/pnas.1313814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Z, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Claus EB, et al. Survival and low-grade glioma: The emergence of genetic information. Neurosurg Focus. 2015;38:E6. doi: 10.3171/2014.10.FOCUS12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor AM, et al. Cancer Genome Atlas Research Network Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33:676–689.e3. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burrell RA, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 57.Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng X, et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18:382–395. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen QJ, et al. Targeting megakaryocytic-induced fibrosis in myeloproliferative neoplasms by AURKA inhibition. Nat Med. 2015;21:1473–1480. doi: 10.1038/nm.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Brocklyn JR, et al. Aurora-A inhibition offers a novel therapy effective against intracranial glioblastoma. Cancer Res. 2014;74:5364–5370. doi: 10.1158/0008-5472.CAN-14-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kurokawa C, et al. Alisertib demonstrates significant antitumor activity in bevacizumab resistant, patient derived orthotopic models of glioblastoma. J Neurooncol. 2017;131:41–48. doi: 10.1007/s11060-016-2285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cervantes A, et al. Phase I pharmacokinetic/pharmacodynamic study of MLN8237, an investigational, oral, selective aurora a kinase inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2012;18:4764–4774. doi: 10.1158/1078-0432.CCR-12-0571. [DOI] [PubMed] [Google Scholar]
- 63.Hong X, et al. The selective Aurora-A kinase inhibitor MLN8237 (alisertib) potently inhibits proliferation of glioblastoma neurosphere tumor stem-like cells and potentiates the effects of temozolomide and ionizing radiation. Cancer Chemother Pharmacol. 2014;73:983–990. doi: 10.1007/s00280-014-2430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rasmussen RD, et al. BRCA1-regulated RRM2 expression protects glioblastoma cells from endogenous replication stress and promotes tumorigenicity. Nat Commun. 2016;7:13398. doi: 10.1038/ncomms13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 66.Ott PA, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.