Significance
The formation of multivesicular endosomes (MVEs) enables the sequestration of numerous membrane-associated cargo molecules, including signaling receptors, to downregulate signal transduction pathways initiated by various stimuli. In this work, we demonstrate that the machinery responsible for MVE biogenesis responds to growth factor stimulation by extending its duration of action at endosomes. Specifically, we show that endosomal sorting complex required for transport-I (ESCRT-I) and Vps4 accumulation at MVEs is dramatically prolonged following growth factor stimulation. Additionally, we demonstrate that native ESCRT complexes exhibit an oscillatory behavior on endosomes, which is dependent on the sequential recruitment of downstream components. Our studies strongly suggest that components of the ESCRT machinery act cyclically and in a coordinated manner to enable a rapid response to changing environmental cues.
Keywords: lattice light sheet microscopy, ESCRT microdomain, CRISPR/Cas9, epidermal growth factor receptor, organelle
Abstract
The formation of multivesicular endosomes (MVEs) mediates the turnover of numerous integral membrane proteins and has been implicated in the down-regulation of growth factor signaling, thereby exhibiting properties of a tumor suppressor. The endosomal sorting complex required for transport (ESCRT) machinery plays a key role in MVE biogenesis, enabling cargo selection and intralumenal vesicle (ILV) budding. However, the spatiotemporal pattern of endogenous ESCRT complex assembly and disassembly in mammalian cells remains poorly defined. By combining CRISPR/Cas9-mediated genome editing and live cell imaging using lattice light sheet microscopy (LLSM), we determined the native dynamics of both early- and late-acting ESCRT components at MVEs under multiple growth conditions. Specifically, our data indicate that ESCRT-0 accumulates quickly on endosomes, typically in less than 30 seconds, and its levels oscillate in a manner dependent on the downstream recruitment of ESCRT-I. Similarly, levels of the ESCRT-I complex also fluctuate on endosomes, but its average residency time is more than fivefold shorter compared with ESCRT-0. Vps4 accumulation is the most transient, however, suggesting that the completion of ILV formation occurs rapidly. Upon addition of epidermal growth factor (EGF), both ESCRT-I and Vps4 are retained at endosomes for dramatically extended periods of time, while ESCRT-0 dynamics are only modestly affected. Our findings are consistent with a model in which growth factor stimulation stabilizes late-acting components of the ESCRT machinery at endosomes to accelerate the rate of ILV biogenesis and attenuate signal transduction initiated by receptor activation.
The intracellular trafficking of integral membrane proteins is directed by cytosolic sorting signals, including short peptide motifs and posttranslational modifications, which are recognized by specific adaptors that facilitate the movement of cargoes to their ultimate destinations. Components of the endosomal sorting complex required for transport (ESCRT) machinery have been demonstrated to bind ubiquitin-modified, membrane-associated cargoes for transport into vesicles that bud into the lumen of specialized multivesicular endosomes (MVEs) (1–7). This process is critical for the attenuation of receptor-mediated signaling, and perturbations in ESCRT function have been implicated in various developmental disorders, including cancer and neurodegenerative disease (8–16). Based on a large number of genetic and biochemical studies, a consensus has emerged, indicating that the hierarchical assembly of five protein complexes (ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III, and the Vps4 complex) drives cargo sequestration, membrane bending, and vesicle budding at MVEs (4, 17–23). In particular, the early-acting ESCRT complexes (ESCRT-0, ESCRT-I, and ESCRT-II) promote clustering of ubiquitin-modified cargoes, while ESCRT-III and Vps4 play critical roles in manipulating membrane curvature to facilitate the inward budding and release of intralumenal vesicles (ILVs) (24–30). The ESCRT machinery also plays critical roles in several other membrane scission and resealing events, including cytokinetic abscission, plasma membrane repair, retroviral budding, lysosome repair, and nuclear envelope reassembly, each in a topologically similar orientation (31–39).
In mammalian cells, our understanding of the native distribution of ESCRT complexes has relied mainly on static immunofluorescence- and immunoelectron microscopy-based studies. These approaches have suggested that ESCRT-0 stably associates with early endosomes on microdomains via interaction with phosphatidylinositol 3-phosphate (PI3P), while other ESCRT complexes are more transient in their assembly at MVEs (21, 24, 30, 31, 40–44). More recently, the dynamics of several ESCRT subunits have been analyzed under various conditions following the ectopic overexpression of fluorescently tagged components (45–48). However, interpretation of these data is confounded by inherent cell-to-cell variability of transgene expression, even after isolation of stably transduced cell lines. Moreover, it is unclear how the dynamics of exogenous, tagged subunits are influenced by the continued presence of their untagged, endogenous counterparts. With advances in CRISPR-mediated genome editing, it is no longer necessary to artificially overexpress proteins of interest to analyze their spatiotemporal dynamics in living cells, as long as functional fusion proteins can be generated (49). Notably, numerous studies have documented an increased risk of artifacts following even modest alterations in the concentrations of ESCRT components, further emphasizing the need to examine these factors at native levels of expression (50–52).
Mammalian cells constitutively synthesize MVEs to direct membrane protein turnover. However, the activity of the ESCRT machinery at endosomes is subject to regulation and can be dramatically stimulated in response to ligand-induced activation of growth factor receptors. In particular, the quantity of MVEs in the cytoplasm, their size, and the number of ILVs within them are all elevated in response to epidermal growth factor (EGF) exposure (53, 54). Previous work has demonstrated that both ESCRT-0 subunits (Hrs and STAM) are rapidly phosphorylated by several kinases downstream of EGF receptor (EGFR) signaling to promote receptor degradation (55–59), but little is known about how the dynamics of ESCRT-0 or other components of the ESCRT machinery are altered to facilitate increased MVE biogenesis. Here, we leverage CRISPR/Cas9 technology to functionally tag multiple ESCRT complexes, enabling us to examine their native distributions in live cells under various conditions using lattice light sheet imaging. Surprisingly, we find that ESCRT-0 dynamics are not substantially altered following EGFR stimulation, but downstream ESCRT complexes exhibit dramatically increased stability at MVEs, suggesting they act multiple times to facilitate more rapid ILV production. Additionally, using live cell superresolution imaging, we demonstrate that cargoes delivered to MVEs initially associate with ESCRT-0, but rapidly accumulate at microdomains that are directly juxtaposed to the earliest-acting ESCRT complex. Taken together, our data are consistent with a model in which growth factor stimulation promotes prolonged activity of the late-acting ESCRT machinery to promote rapid cargo deposition into ILVs that form immediately adjacent to ESCRT-0 microdomains on MVEs.
Results
Use of CRISPR-Mediated Genome Editing to Functionally Tag Components of the ESCRT Machinery.
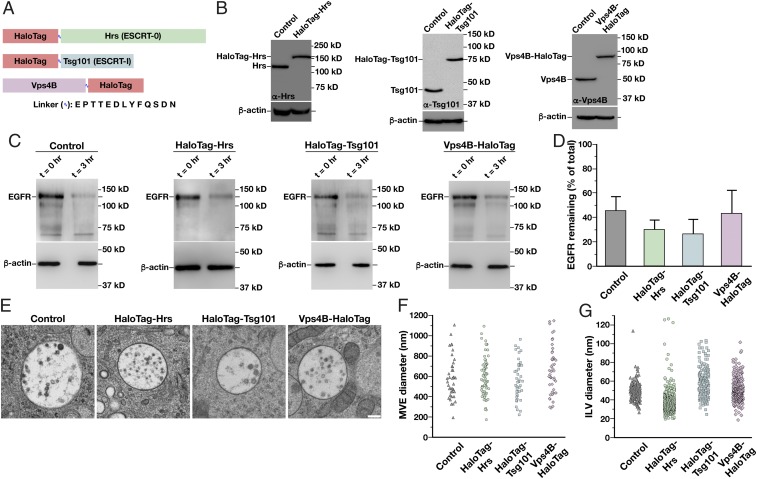
To determine the spatiotemporal distribution of mammalian ESCRT complexes as they assemble and disassemble natively on intracellular membranes, we used CRISPR/Cas9-technology to append the HaloTag onto three core ESCRT subunits, Hrs (ESCRT-0), Tsg101 (ESCRT-I), and Vps4B, at their endogenous loci (Fig. 1A). The monomeric HaloTag was selected based largely on its amenability to rapid and irreversible labeling with bright, fluorogenic dyes (e.g., JF646), which exhibit improved fluorescence properties compared with genetically encoded fluorescent proteins that have been used previously. Following genome editing, human RPE1 cells that had been immortalized by the forced expression of telomerase and maintain a stable diploid karyotype were sorted based on HaloTag expression. Clonal populations were subsequently screened using immunoblot analysis to identify cell lines that exhibited homozygous incorporation of the HaloTag at each locus (Fig. 1B). We isolated several independent clones in each case and subjected them to a series of validation assays to ensure that the functionality of each fusion protein had been preserved.
Fig. 1.
Generation of functionally tagged ESCRT subunits using CRISPR/Cas9-mediated genome editing. (A) Cartoon highlighting the placement of the HaloTag on three ESCRT subunits to track ESCRT-0 (green), ESCRT-I (blue), and Vps4 (purple) dynamics. The sequence of the flexible linker used in each case, which contains the tobacco etch virus protease recognition sequence, is also shown. (B) Representative immunoblot analyses of control and CRISPR/Cas9-modified cell lines (n = 3 each) using antibodies directed against Hrs (Left), Tsg101 (Center), Vps4B (Right), and β-actin (load control). (C) Control and CRISPR/Cas9-modified clonal cell lines were incubated in the presence or absence of 30 ng/mL EGF for 3 h following serum starvation, and extracts were immunoblotted using antibodies directed against EGFR (Top) and β-actin (Bottom, load control). (D) Quantification of the percentage of EGFR remaining after 3 h of EGF treatment in control and CRISPR/Cas9-modified clonal cell lines. Error bars represent mean ± SEM (n = 4 each). No statistically significant difference was found, as calculated using an ANOVA test. (E) Representative thin-section EM images of MVEs from control and CRISPR/Cas9-modified clonal cell lines (more than 35 MVEs and 200 ILVs examined in each). (Scale bar: 200 nm.) The size distribution of MVEs (F) and ILVs (G) in control and CRISPR/Cas9-modified clonal cell lines is shown. No statistically significant differences were found, as calculated using an ANOVA test.
We first monitored the growth rate of each cell line expressing an ESCRT subunit fused to the HaloTag and found no differences compared with the parental cell line (SI Appendix, Fig. S1A). In contrast, individual depletion of Hrs, Tsg101, or Vps4B using siRNAs validated previously for specificity each resulted in a dramatic decline in cell growth (SI Appendix, Fig. S1 B and C). Notably, depletion of the other Vps4 paralog (Vps4A) in cells expressing Vps4B-HaloTag failed to impact the rate of cell division any more than that observed in control RPE1 cells, and Vps4B-HaloTag accumulated normally at midbodies after mitosis (SI Appendix, Fig. S1 D and E). We additionally examined early and late endosome morphology in immunofluorescence studies using antibodies directed against EEA1 and LAMP1, markers that have each been shown to exhibit partial overlap with components of the ESCRT machinery (21, 40). Again, we found no differences in the size distribution of endosomes in cell lines expressing HaloTag-ESCRT fusion proteins, while depletion of ESCRT components led to the formation of enlarged endosomal compartments (SI Appendix, Fig. S1 F and G). To determine whether cargo down-regulation was impacted, we treated cells with EGF and monitored turnover of its receptor (EGFR) by immunoblot analysis. These studies indicated that the presence of HaloTag on Hrs, Tsg101, and Vps4B did not impair their functions in cargo sorting and degradation (Fig. 1 C and D). Electron microscopy-based analysis of MVEs identified in each cell line further confirmed the functionality of each fusion protein (Fig. 1E). Specifically, the size of MVEs and the ILVs found therein exhibited no statistically significant differences in cell lines expressing HaloTag-ESCRT fusions compared with parental RPE1 cells (Fig. 1 F and G).
Together, our data strongly support the functionality of HaloTag-Hrs, HaloTag-Tsg101, and Vps4B-HaloTag, providing a unique and unprecedented opportunity to study their dynamics when expressed at endogenous levels.
Components of the ESCRT Machinery Exhibit Distinct Patterns of Endosomal Localization.
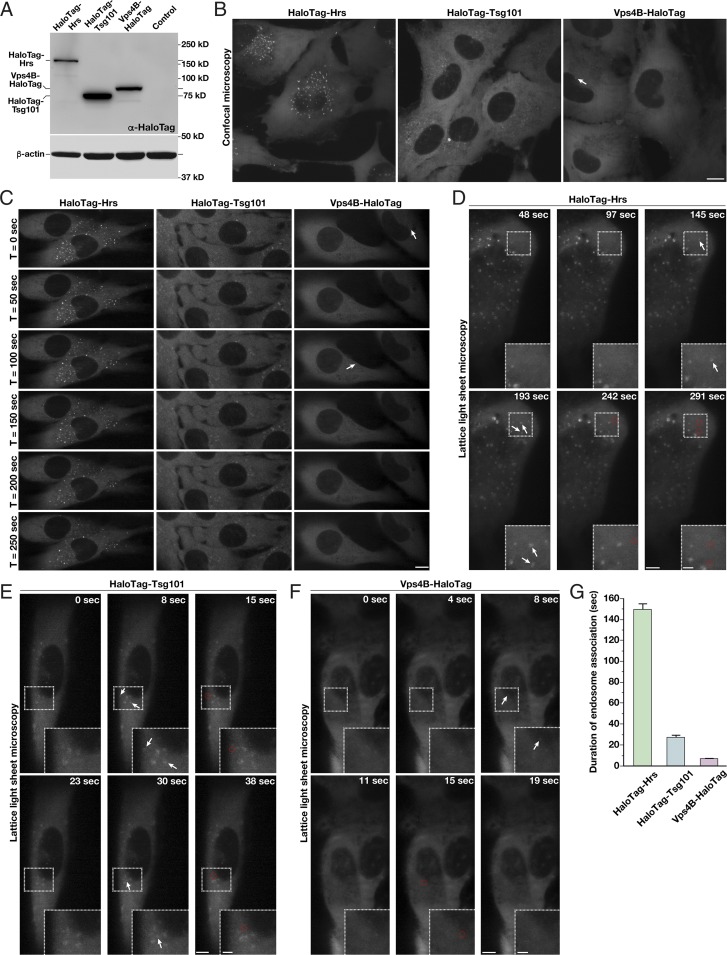
Based on immunoblot analysis using antibodies directed against HaloTag, we found that Hrs, Tsg101, and Vps4B are all expressed within an order of magnitude, enabling us to more easily make direct comparisons of their distributions in cells (Fig. 2A and SI Appendix, Fig. S2A). Using confocal microscopy, we imaged the CRISPR-modified cell lines after a brief period of dye labeling. HaloTag-Hrs localized throughout the cytoplasm, and additionally accumulated at numerous punctate structures that colabeled with a probe for PI3P, a marker of endosomal membranes (Fig. 2B and SI Appendix, Fig. S2B). In contrast, both HaloTag-Tsg101 and Vps4B-HaloTag were largely cytosolic and less frequently associated with PI3P-positive membranes under steady-state conditions (Fig. 2B and SI Appendix, Fig. S2B).
Fig. 2.
ESCRT-0 associates more stably with endosomes compared with downstream ESCRT complexes under normal growth conditions. (A) Representative immunoblot analysis (n = 3) of control and CRISPR/Cas9-modified cell lines using antibodies directed against HaloTag (Top) and β-actin (Bottom, load control). (B and C) Representative CRISPR/Cas9-modified cells imaged live using SFC optics following dye labeling with the JF549-HaloTag ligand (more than 15 different cells each, more than three biological replicates each). Maximum intensity projections of z-stacks (B) or individual confocal sections over time (C) are shown. Arrows highlight transient Vps4-positive endosomes. (Scale bars: 10 μm.) (D–F) Representative CRISPR/Cas9-modified cells imaged live using LLSM following dye labeling using the JF646-HaloTag ligand (more than 10 cells each, more than three biological replicates each). Projected z-stacks are shown for each time point. Arrows highlight the appearance of ESCRT-positive endosomes, and red circles denote their disappearance. (Scale bars: 5 μm; Insets, 2 μm.) (G) Quantification of the average duration of each HaloTag-ESCRT fusion protein on endosomes. Error bars represent mean ± SEM (more than 600 endosomes analyzed for each cell line).
Time-lapse confocal imaging suggested that each component of the ESCRT machinery associates with endosomes for a period of time, but ultimately disappears (Fig. 2C). However, it was unclear whether the disappearance correlated with the dissociation of the ESCRT component from endosomes or with movement of endosomes into a different focal plane. To circumvent this issue, we took advantage of lattice light sheet microscopy (LLSM), which is capable of imaging full cellular volumes with high temporal resolution (60). Using this approach, we imaged each cell line continuously over the course of 10 min and analyzed all endosomal membranes that were positive for components of the ESCRT machinery (Fig. 2 D–F and Movies S1–S3). This systematic strategy demonstrated that endogenous Hrs remains associated with endosomes for a longer period of time compared with Tsg101 or Vps4B. On average, we found that Hrs resides on endosomes for 149.4 (±5.7) s, while Tsg101 remains for only 27.2 (±2.1) s and Vps4B appears transiently for 6.8 (±1.4) s (Fig. 2G). In all cases, the duration of endosomal association varied with endosome size. Based on diffraction-limited fluorescence measurements, the majority of endosomes exhibited a diameter of 350–950 nm, and a clear Gaussian distribution was apparent following statistical analysis of all structures visualized (SI Appendix, Fig. S2 C and D). The smallest endosomes (potentially endosomal microdomains) exhibited the shortest durations of ESCRT complex assembly, while larger structures generally exhibited more prolonged accumulation of the ESCRT machinery (SI Appendix, Fig. S2E).
Although relatively uncommon, the appearance of large endosomes as determined by LLSM (greater than 75th percentile in size) could correspond to natural variability in the diameters of individual MVEs or a preponderance of closely juxtaposed endosomes that cannot be easily distinguished by diffraction-limited imaging. To address this issue, we tracked all ESCRT-labeled structures during each time-lapse imaging series and plotted their trajectories (SI Appendix, Fig. S2F). Endosome displacement was modest in the perinuclear region (11.0 ± 0.8-μm track length, 1.18 ± 0.05-μm track displacement), while peripheral endosomes generally exhibited more long-range movements (47.9 ± 7.6-μm track length, 7.94 ± 0.84-μm track displacement). We largely failed to observe MVE fission events, but in a minority of cases (less than 4% overall), we observed Hrs-positive endosomes coming into direct contact with one another. The duration of these associations ranged from a few seconds to several minutes, at which point we concluded that a homotypic fusion event had likely occurred (SI Appendix, Fig. S2 G–I). Overall, our fluorescence data, together with our electron microscopy-based analysis, suggest that the vast majority of ESCRT-positive endosomes in human RPE1 cells are individual, nonclustered structures that do not undergo continual fission/fusion events under steady-state conditions.
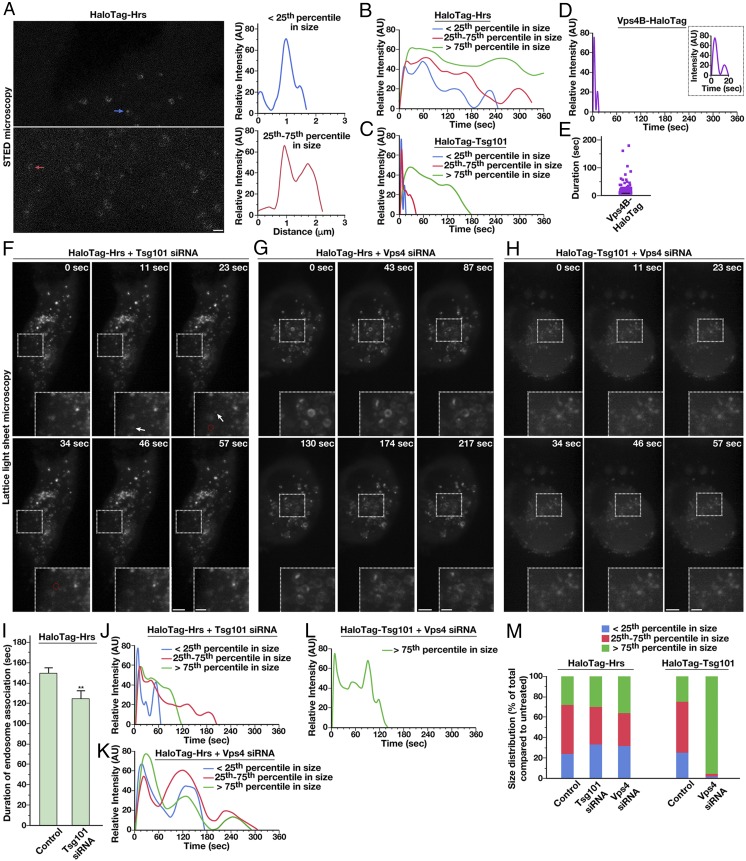
At the opposite end of the size spectrum, we identified a number of ESCRT-positive structures that were diffraction-limited, potentially highlighting a class of small MVEs. However, our electron microscopy-based studies failed to identify such a population. Instead, we considered the possibility that the diffraction-limited structures correspond to individual ESCRT microdomains present on endosomes. To investigate this possibility, we carried out live stimulated emission depletion (STED) microscopy, which is capable of achieving 50- to 60-nm lateral resolution in cells (61, 62). Using this approach, we consistently found Hrs in small patches on endosomes (Fig. 3A), similar to previous findings in other human cell lines and Caenorhabditis elegans using immunogold electron microscopy (24, 42, 43). Although many larger endosomes harbored several distinct microdomains, suggesting that ILV formation occurs simultaneously at multiple sites, we also found numerous examples of smaller endosomes on which only a single ESCRT-positive endosomal microdomain was visible (Fig. 3A).
Fig. 3.
ESCRT subunit dynamics are regulated by the action of downstream ESCRT complexes. (A, Left) Representative HaloTag-Hrs–expressing cells imaged live using STED microscopy following dye labeling using the SiR-HaloTag ligand (more than 10 cells imaged, more than three biological replicates). Arrows highlight endosomes of two different size classes (blue, less than 25th percentile; red, 25th–75th percentile). (Scale bar: 1 μm.) (A, Right) Fluorescence intensity based on line-scan analysis around the circumference of each is shown, reflecting the distribution of Hrs. (B–D) Averaged fluorescence intensity profiles of labeled endosomes classified by compartment size (more than 95% of Vps4B-HaloTag–labeled compartments were diffraction-limited). Only compartments that acquire and lose fluorescence during the imaging series were used (more than 350 endosomes per condition in more than 10 cells each, more than three biological replicates). (E) Individual residency times of Vps4B-HaloTag at endosomes are plotted (more than 140 endosomes, at least 10 different cells in more than three biological replicates). A black line indicates the average. (F–H) Representative CRISPR/Cas9-modified cells imaged live using LLSM following dye labeling with the JF646-HaloTag ligand and treatment with siRNA targeting either Tsg101 or both Vps4 isoforms (more than 10 cells each, more than three biological replicates each). Projected z-stacks are shown for each time point. Arrows highlight the appearance of ESCRT-positive endosomes, and red circles denote their disappearance. (Scale bars: 5 μm; Insets, 2 μm.) (I) Quantification of the average duration of HaloTag-Hrs on endosomes in control and Tsg101-depleted cells. Error bars represent mean ± SEM (more than 400 endosomes analyzed for each cell line, more than 10 cells imaged per condition in more than three biological replicates). **P < 0.01, as calculated using a Student’s t test. (J–L) Averaged fluorescence intensity profiles of labeled endosomes classified by compartment size under the conditions shown (relative to sizes determined under normal conditions). Only compartments that acquire and lose fluorescence during the imaging series were used (more than 400 endosomes per condition, more than 10 cells imaged per condition in more than three biological replicates). (M) Size distributions of ESCRT-positive endosomes under the conditions indicated (more 400 endosomes per condition, more than 10 different cells each in more than three biological replicates).
Several studies have examined the kinetics of ESCRT complex assembly and disassembly in mammalian cells, but, to our knowledge, they all use ectopic overexpression of tagged subunits, which can lead to artifacts, especially in the continued presence of endogenous, untagged counterparts (45–50). Using our CRISPR/Cas9-modified cell lines, we examined the native dynamics of early- and late-acting components of the ESCRT machinery. Under normal growth conditions, we monitored the fluorescence intensity of HaloTag-Hrs as it accumulated and dispersed from endosomes of various size. The small endosomes (less than 25th percentile in size), potentially reflecting the assembly of individual microdomains, exhibited a rapid rate of Hrs accumulation, peaking in intensity within 14.0 (±0.1) s after detection over background cytosolic fluorescence (Fig. 3B). Fluorescence intensity fluctuated in a sinusoidal-like pattern, with each peak persisting for 29.9 (±1.1) s on average. Full disassembly occurred with similar kinetics compared with the assembly phase [19.7 (±0.6) s], and the average total residency time of Hrs on small endosomes lasted 44.7 ± 5.9 s (Fig. 3B and SI Appendix, Fig. S2E). Larger Hrs-positive structures (between 25th and 75th percentiles in size) also exhibited multiple peaks of fluorescence intensity, suggestive of distinct assembly and disassembly events occurring on individual endosomes (Fig. 3B). Strikingly, the average duration of each peak was similar to that observed on small endosomes, lasting 29.2 (±1.2) s, with peak intensity occurring within 19.2 (±0.2) s after initial detection over background. In contrast, the largest endosomes (greater than 75th percentile in size), which also exhibited the longest continual duration of Hrs fluorescence, showed broader fluctuations in intensity, suggesting the possible presence of multiple microdomains on individual endosomes, each with unique dynamics that may average one another out over a large sample size. Even in these cases, however, initial peak intensity was achieved quickly, within 31.9 (±0.6) s (Fig. 3B).
Analysis of cells expressing HaloTag-Tsg101 indicated that ESCRT-I reaches peak intensity over 3.5 (±0.1) s on small endosomes (less than 25th percentile in size), substantially more rapidly compared with Hrs (Fig. 3C). Moreover, we found that Tsg101 arrival occurs in multiple waves (Fig. 3C), ultimately followed by rapid disassembly within 3.0 (±0.1) s. On average, the duration of each peak in fluorescence intensity was brief, only 3.9 (±0.1) s. Larger Tsg101-positive structures (between 25th and 75th percentiles in size) exhibited similarly rapid assembly kinetics, with peak intensity occurring within 4.7 (±0.1) s after initial detection over background. Disassembly occurred over 5.8 (±0.2) s, following multiple oscillations in intensity (Fig. 3C). The largest Tsg101-positive structures (greater than 75th percentile in size) exhibited several broad peaks in fluorescence intensity, as was demonstrated in the case of Hrs, suggesting multiple assembly and disassembly events on endosomes (Fig. 3C). Together, our data suggest that ESCRT-0 only briefly recruits ESCRT-I (for less than 4 s) before ESCRT-I is at least partially returned to the cytoplasm, but ESCRT-0 can act repetitively in this manner during the lifetime of an ESCRT microdomain. In contrast to the early-acting ESCRT complexes, Vps4B-HaloTag dynamics at endosomes were extremely transient, although the longest-lived structures exhibited multiple waves of accumulation (Fig. 3D). Nonetheless, in the majority of cases, Vps4B was present for less than 4 s (Fig. 3E), consistent with its proposed function in promoting ILV scission (29, 30, 63).
The Sequential Action of ESCRT Components Regulates Their Assembly and Disassembly Rates.
Previous studies have demonstrated that the ESCRT complexes function in a sequential manner at MVEs, and inhibition of one complex blocks the recruitment of downstream machinery (21, 22). However, it remains unclear how the dynamics of early-acting components respond to inhibition of later acting factors. To address this question, we used validated siRNAs targeting Tsg101 and Vps4 and imaged native ESCRT-0 and ESCRT-I using LLSM. Consistent with previous work, inhibition of Tsg101 or Vps4 had a dramatic impact on endosome morphology and movement (64–66) (Fig. 3 F–H and SI Appendix, Fig. S3 A and B). However, despite the appearance of abnormal compartments, we failed to identify significant changes in the frequency of endosome/endosome contacts compared with control cells during live imaging studies (SI Appendix, Figs. S2I and S3C). Depletion of Vps4 resulted in an increased residency time of Hrs at all endosomes imaged, suggesting that cargo deposition into ILVs may be a prerequisite for ESCRT-0 disassembly (SI Appendix, Fig. S3D). These data are consistent with our previous immunogold electron microscopy-based studies demonstrating that inhibition of ESCRT-III causes ESCRT-0 microdomains on endosomes to expand in surface area (24). Similarly, the residency time of Tsg101 on endosomes, irrespective of their size, increased significantly in the absence of Vps4 (SI Appendix, Fig. S3E).
The kinetics of Hrs and Tsg101 assembly and disassembly were also impacted by the inhibition of downstream ESCRT components. In particular, we found that the average duration of Hrs assembly on endosomes was reduced in the absence of Tsg101 [to 124.0 (±7.8) s] and the oscillatory fluctuations in Hrs intensity were shortened [12.2 (±0.3) s], suggesting that the recruitment of ESCRT-I plays an important role in maintaining ESCRT-0 microdomain stability (Fig. 3I). In contrast, the rate at which Hrs initially accumulated on endosomes was not affected by depletion of downstream ESCRT machinery (Fig. 3 J and K). Similarly, the kinetics of initial ESCRT-I recruitment to endosomes were not affected by inhibition of Vps4 (Fig. 3L). However, inhibition of Vps4 led to longer Hrs and Tsg101 residency times on endosomes [175.1 (±7.1) s and 140.5 (±4.4) s, respectively] and slower disassembly kinetics, requiring 42.5 (±1.3) s on average (Fig. 3 K and L and SI Appendix, Fig. S3E). Additionally, Vps4 depletion increased the size of Tsg101-positive structures dramatically compared with control cells, while the overall size distribution of Hrs-positive endosomes exhibited a more modest change (Fig. 3M). Together, these data are consistent with a model in which the entire ESCRT machinery acts cooperatively to enable cargo down-regulation.
Increased Substrate Load Prolongs the Retention of ESCRT Machinery at Endosomes.
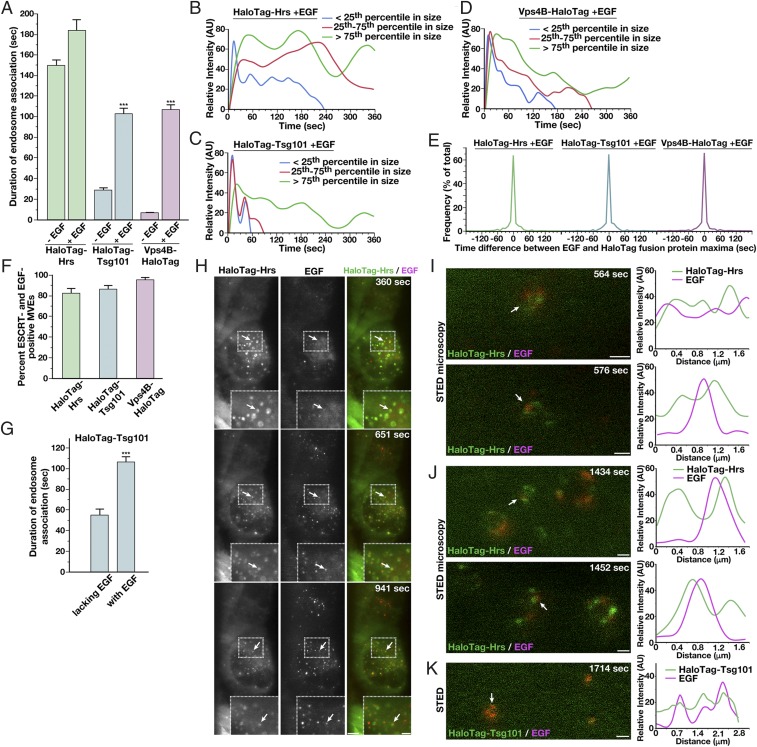
Elevated growth factor signaling has been previously implicated in altering ESCRT function to promote MVE biogenesis (53, 54). To understand how this activity influences the dynamics of the ESCRT machinery, we treated cells with a low concentration of EGF for 2 min following a period of growth factor deprivation and imaged HaloTag-ESCRT fusion proteins using LLSM. During serum starvation, the ESCRT complexes and Vps4B continued to assemble onto membranes (SI Appendix, Fig. S3F). However, following EGF stimulation, the duration of ESCRT machinery association with endosomes was significantly altered. Specifically, ESCRT-0 remained on endosomes for an average of 183.3 ± 7.5 s, ESCRT-I for 102.4 ± 7.4 s, and Vps4B for 106.6 ± 5.8 s, all significant increases relative to steady-state growth conditions, but especially dramatic in the cases of Tsg101 and Vps4B, which were present for a fourfold and 15-fold longer period of time, respectively (Fig. 4A). Analysis of initial ESCRT assembly kinetics following EGF stimulation indicated minimal changes relative to those determined at steady state, and we continued to observe periodic oscillations in Hrs and Tsg101 levels, irrespective of endosome size or the duration of their association with endosomes (Fig. 4 B and C, and SI Appendix, Fig. S3G, and Movies S4 and S5). Effects on Hrs dynamics at endosomes were surprisingly modest, even though previous studies indicate that it is a downstream target of EGFR signaling (55–59) (Fig. 4B). In contrast, analysis of Vps4B levels following EGF stimulation revealed periodic oscillations, similar to those observed for Tsg101 (Fig. 4D, SI Appendix, Fig. S3G, and Movie S6). These data suggest that Vps4 acts repeatedly at endosomes under these conditions, potentially driving multiple ILV budding events on individual endosomes to facilitate the down-regulation of EGFR and other cargoes. Consistent with this idea, electron tomography-based analysis of MVEs indicated the formation of ILV clusters near the limiting membrane specifically following EGF stimulation (SI Appendix, Fig. S3H and Movies S7 and S8).
Fig. 4.
Growth factor stimulation alters ESCRT dynamics at endosomes. (A) Quantification of the average duration of each HaloTag-ESCRT fusion protein on endosomes in the presence or absence of EGF stimulation. Error bars represent mean ± SEM (more than 600 endosomes analyzed for each cell line, more than 10 cells each in more than three biological replicates). ***P < 0.005, as calculated using an ANOVA test. (B–D) Averaged fluorescence intensity profiles of labeled endosomes classified by compartment size following treatment with 30 ng/mL EGF. Only compartments that acquire and lose fluorescence during the imaging series were used (more than 350 endosomes per condition, more than 10 cells each in more than three biological replicates). (E) Time differences between Alexa Fluor 555-EGF and JF646-labeled HaloTag-ESCRT achieving maximum fluorescence intensity at endosomes are plotted (more than 400 endosomes analyzed for each cell line, more than 10 cells each in more than three biological replicates). (F) Percentages of ESCRT-labeled endosomes that exhibited EGF accumulation at any point during time-lapse imaging were calculated for each Halo-Tag ESCRT fusion. Error bars represent mean ± SEM (endosomes from more than 10 cells each in more than three biological replicates). (G) Quantification of the average duration of HaloTag-Tsg101 on endosomes, either EGF-positive or EGF-negative, following EGF stimulation. Error bars represent mean ± SEM (more than 500 endosomes analyzed, more than 10 cells each in more than three biological replicates). ***P < 0.005, as calculated using a t test. (H) Representative HaloTag-Hrs–expressing cells imaged live using LLSM following dye labeling using the JF646-HaloTag ligand and incubation with Alexa Fluor 555-EGF for 2 min, followed by washout (more 10 cells each, more than three biological replicates each). Projected z-stacks are shown for each time point. Arrows highlight HaloTag-Hrs–positive endosomes that also show accumulation of EGF. (Scale bar, 5 μm; Insets, 2 μm.) Representative HaloTag-Hrs–expressing cells (I and J) or HaloTag-Tsg101–expressing cells (K) imaged live using STED microscopy following dye labeling using the SiR-HaloTag ligand and incubation with Alexa Fluor 594-EGF (Left; more than 10 cells in more than three biological replicates) are shown. Fluorescence intensity based on line-scan analysis around the circumference of representative endosomes (Right, indicated by arrows on Left) is also shown, reflecting the relative distributions of each ESCRT complex and EGF. (Scale bars: I and J, 500 nm; K, 1 μm.)
Using fluorescently labeled EGF, we were also able to monitor the distribution of cargo relative to components of the ESCRT machinery. Strikingly, we found a direct correlation between cargo and ESCRT accumulation at endosomes. Specifically, in the majority of cases, maximum fluorescence intensity of EGF at endosomes peaked simultaneously with each of the ESCRT components examined, strongly suggesting a direct relationship between the presence of cargo and ESCRT function at endosomes (Fig. 4E). Further analysis revealed that only a subset (82 ± 5%) of Hrs-positive endosomes accumulated EGF (Fig. 4F). These findings are in line with prior work suggesting that EGF stimulation promotes the inward budding on some, but not all, MVEs (53). The majority of Tsg101-positive membranes (86 ± 4%) also showed EGF labeling, and nearly all Vps4B-positive endosomes (95 ± 2%) harbored EGF, suggesting that local growth factor signaling promotes sustained ESCRT activity at MVEs. In agreement with this idea, we found that EGF-positive endosomes, but not endosomes lacking EGF, exhibited dramatically increased residency times for Tsg101 (Fig. 4G). These data argue against a global cellular effect for EGF signaling in driving MVE formation.
Additionally, using STED microscopy, we examined the distribution of EGF relative to HaloTag-Hrs specifically (Fig. 4H). Within 6–10 min after addition of EGF to cells, we found it had accumulated at Hrs-positive endosomes (Fig. 4I). At this time point, we found that EGF colocalized with Hrs, but the distribution of Hrs at the endosome-limiting membrane was relatively broad. With additional time, however, Hrs became highly concentrated into discrete microdomains, but, surprisingly, EGF was distributed immediately adjacent to these regions (Fig. 4J and SI Appendix, Fig. S3I), colocalizing instead with Tsg101 (Fig. 4K). These data strongly suggest that ESCRT-0 only temporarily associates with cargoes before they are transferred to a juxtaposed microdomain and likely incorporated into budding ILVs via the action of the downstream ESCRT machinery.
Discussion
Until recently, our ability to define the timing and duration of vesicle formation events that occur on intracellular membranes has lagged behind our understanding of the protein machineries that function in cargo selection and lipid bilayer remodeling. With advances in live cell imaging approaches and genome editing technologies, barriers to studying native vesicle-mediated transport processes in mammalian cells are beginning to diminish. The dynamics of clathrin-mediated endocytosis are arguably the best understood, at least in part due to the availability of genome-edited cell lines and facile access to total internal reflection fluorescence microscopy, which is capable of illuminating a narrow, ∼200-nm window at the surface of cells (67–71). Light sheet imaging with diffraction-limited optics now affords us the opportunity of studying vesicle formation anywhere within mammalian cells, including MVEs that accumulate in the perinuclear region and facilitate the turnover of many integral membrane proteins (60, 72). Here, we generate a series of CRISPR/Cas9-modified human cell lines and define the native spatiotemporal recruitment patterns of early- and late-acting ESCRT complexes under normal growth conditions and following growth factor stimulation. Our findings unexpectedly demonstrate that the residency times of ESCRT-I and Vps4 at MVEs become dramatically prolonged upon EGFR activation, while ESCRT-0, a known downstream target of EGFR signaling (55–59), remains at MVEs for a similar length of time both before and after treatment with EGF. These data suggest that the increased rates of MVE and ILV formation that occur after growth factor stimulation are driven by elevations in ESCRT-I recruitment/retention that facilitate the coupling of cargo capture at MVEs to internalization by ESCRT-III/Vps4.
Although numerous studies have examined the dynamics of mammalian ESCRT components at various membrane compartments in cells, most have tracked ectopically overexpressed subunits, which may not reflect the behavior of their endogenous counterparts (45–50). In contrast, recent studies in yeast have defined the assembly and disassembly rates of a native ESCRT-III subunit (Vps24), as well as the ATPase Vps4, demonstrating that these factors associate only transiently with MVEs (3–45 s), with the majority of recruitment events lasting less than 20 s (30). Approximately 80% of these occurrences were suggested to be productive in generating ∼25-nm ILVs. Although ILVs in metazoan organisms are approximately twice this size, the findings using yeast are in agreement with our data collected from mammalian cells grown under normal conditions, suggesting that ILV scission typically requires only a fraction of a minute, irrespective of the size of the budding vesicles. Importantly, the increased duration of Vps4B at MVEs following EGF stimulation implies that multiple ILVs are likely generated during these extended phases of recruitment. Consistent with this idea, electron tomography of MVEs produced in several systems demonstrates that ILVs are generated repetitively from single endosomal microdomains (24, 73). Without the need to reassemble ESCRT machinery de novo for each ILV budding event, the rate at which ubiquitin-modified cargoes are sequestered from the cytoplasm can be accelerated, more rapidly attenuating signal transduction pathways initiated at the cell surface.
Using confocal imaging, another study also recently examined the kinetics of ESCRT machinery assembly and disassembly at endosomes following EGF stimulation using HeLa cells stably overexpressing tagged subunits (45). Although some of our findings are in broad agreement, including the average total duration of ESCRT-0 and Vps4B recruitment to MVEs, many of our results differ significantly, potentially due to our distinct strategies to express tagged ESCRT components and/or the different imaging approaches we each employed. In particular, the prior work suggested that ESCRT-0 and ESCRT-I require ∼2 min to reach peak intensity on MVEs (45), while our data indicate that they are recruited significantly faster. Moreover, the earlier study showed that disassembly of the late-acting ESCRT machinery required more than 1 min (45), which contrasts with our findings that Vps4B dissociates from endosomes on the order of seconds. The implications of these differences are important to consider. The previous study suggested that during the prolonged phase of ESCRT-0 and ESCRT-I complex recruitment, cargo is sequestered and membrane deformation to a U-shaped topology occurs (45). However, there is little evidence supporting the idea that the early-acting ESCRT machinery is capable of membrane bending. Instead, recent studies have largely implicated the ESCRT-III complex, potentially in coordination with Vps4, in the membrane remodeling process (24, 30, 52, 63, 74). Using LLSM, our imaging studies revealed that native ESCRT-0 and ESCRT-I are recruited rapidly onto endosomes, but their levels at the limiting membrane subsequently oscillate, perhaps indicative of association/dissociation events that lead to the formation of a discrete ESCRT microdomain. Our live STED imaging data are consistent with this idea, showing that the initial distribution of ESCRT-0 at MVEs is broad but, upon cargo influx, coalesces into specific regions of the membrane.
Although we have not yet been able to generate cell lines that express tagged ESCRT-III subunits in a homozygous manner, potentially due to their intolerance of large protein tags (30, 52, 75–77), our analysis of native Vps4B dynamics contrasts with the previous interpretation of its slow dissociation kinetics following EGF stimulation. Under normal growth conditions, Vps4B assembles and disassembles rapidly from endosomes, suggesting that the process of ILV bud neck constriction and scission is rapid. Although every recruitment event observed may not correlate to ILV budding, the extended duration of Vps4B at MVEs subsequent to growth factor stimulation is inconsistent with the generation of a single ILV, as previously suggested (45). Instead, we find that the levels of Vps4B undergo oscillations after reaching peak intensity, similar to those observed for other components of the ESCRT machinery, suggesting repetitive recruitment and disassembly events, likely producing multiple ILVs. Together, these data support a model in which the increased number of MVEs and ILVs that form upon growth factor stimulation is a result of prolonged recruitment of the late-acting ESCRT complexes to endosomal membranes.
Materials and Methods
CRISPR-Mediated Genome Editing and siRNA Treatments.
Guide RNA (gRNA) target sequences directed at the amino terminus of Hrs (5′-TTTGGGCTGGAGGTCGCCAT-3′), the amino terminus of Tsg101 (5′-GAGTCGCCAGGCGGCCGTCA-3′), and the carboxyl-terminus of Vps4B (5′-ACAGAAGATTTTGGTCAAGA-3′) were selected based on recommendations made by multiple gRNA design tools. Plasmid-based homology directed repair (HDR) templates included sequences encoding the HaloTag, a linker, and ∼800 bp of flanking genomic DNA, both upstream and downstream of the Cas9 cut site. Human hTERT-immortalized RPE1 cells were electroporated with plasmids encoding Cas9-GFP, a specific gRNA, and an HDR template, and single cells were isolated using fluorescence-activated cell sorting based on GFP expression. Homozygous genome-edited clones were confirmed using immunoblot analysis and Sanger sequencing. For depletion studies, RPE1 cells were transfected using previously validated siRNAs targeting Hrs (54), Tsg101 (78), Vps4A (79), and Vps4B (79) using Lipofectamine RNAiMAX (Invitrogen). For immunoblotting studies, antibodies directed against the following proteins were used (1 μg/mL): Hrs (GTX101718; GeneTex), Tsg101 (GTX70255; GeneTex), Vps4A (ab180581; Abcam), Vps4B (sc-377162; Santa Cruz Biotechnology), β-actin (A1978; Sigma–Aldrich), EGFR (PA1-1110; Thermo Fisher Scientific) and HaloTag (G9211; Promega).
Fluorescence Imaging Studies.
Confocal imaging was conducted using a Nikon Eclipse TiE swept-field confocal (SFC) microscope using a 60× oil immersion objection and a CoolSNAP HQ2 CCD camera. Cells were grown in glass-bottomed dishes and maintained at 37 °C and 5% CO2 during imaging. STED microscopy was performed on a Leica TCS SP8 3× imaging system. Data were collected using a 775-nm depletion laser. The full width at half maximum (FWHM) of the STED point-spread function was calculated to be 90 nm, based on the imaging of fluorescent beads. LLSM was carried out as described previously (60), using cells grown in media lacking phenol red. Briefly, samples were illuminated by a 2D optical lattice generated by a spatial light modulator (Fourth Dimension Displays), using a 560-nm or 641-nm diode laser (MPB Communications) at 100% acousto-optic tunable filter (AOTF) transmittance and an excitation objective [0.65 N.A., 3.74-mm working distance (WD); Special Optics]. Fluorescent emission was collected on a sCMOS camera (Hamamatsu Orca Flash 4.0 v2) using a detection objective (CFI Apo LWD 25XW, 1.1 N.A.; Nikon). Point-spread functions for deconvolution were experimentally measured using 200-nm tetraspeck beads adhered to 5-mm glass coverslips (Invitrogen) for each excitation wavelength. For all live cell imaging, dye labeling (30-min incubation followed by washout into growth media) was performed using the following HaloTag ligands: SiR (STED), JF646 (LLSM), JF549 (SFC), or TMR (SFC). EGF labeled with Alexa Fluor 555 (LLSM) or Alexa Fluor 594 (STED) was used in cargo-sorting studies (2-min incubation, followed by washout; 30 ng/mL EGF). For immunofluorescence studies, cells were fixed using 4% paraformaldehyde, permeabilized in PBS + 0.5% Triton X-100, blocked using 3% BSA for 1 h, and incubated overnight with primary antibodies at 4 °C directed against EEA1 (610457; BD Transduction Laboratories) or LAMP1 (H4A3; Developmental Studies Hybridoma Bank). Coverslips were subsequently incubated with appropriate secondary antibodies labeled with either Alexa Fluor 488 or Cy3, washed, and mounted on slides using Vectashield.
Image Analysis and Statistics.
Data acquired by LLSM were deskewed as described previously (60). Images were analyzed using Imaris Bitplane software. Surfaces, spots, and tracking algorithms were used to analyze the dynamics of fluorescently labeled structures. Specifically, size and normalized relative intensity statistics were reported from the surfaces and spots algorithms, and tracking algorithms were used to assess contact events, duration, displacement, and length of run. Only tracks that were initiated and concluded during the 10-min imaging window were included in intensity analysis curves. The durations of fluorescence intensity peaks were calculated at FWHM. Visualization and statistical testing were carried out using SAS JMP software.
Electron Microscopy Studies.
Cells grown on sapphire disks were loaded into 100-μm-deep aluminum sample holders, cryoprotected with 1-hexadecene, and frozen using a Balzers HPM 010 high-pressure freezer. Samples were freeze-substituted in acetone containing 2% OsO4 and 1% H2O using a Leica Microsystems AFS instrument over a period of 4 d (−90 °C for 72 h, followed by a warming period of 4 °C per hour until the samples reached 4 °C), embedded through a graded series of Epon EMbed 812 (Electron Microscopy Sciences), and polymerized for 24 h at 60 °C. Micrographs of 80-nm sections were collected on a Phillips CM120 80 kV transmission electron microscope equipped with an AMT BioSprint 12 series digital camera or a MegaView III digital camera (Olympus Soft Imaging Solutions). For tomography (300-nm sections), single-axis tilt series were collected using SerialEM1 software on a 300-kV Tecnai TF-30 transmission electron microscope equipped with a Gatan 2k × 2k Ultrascan camera. Tilt series were captured at magnifications of 12,000 × (0.94 nm per pixel) to 23,000 × (0.467 nm per pixel) at 1° increments from −60° to 60°. Reconstruction and modeling of tilt series were carried out using IMOD software (80, 81).
Supplementary Material
Acknowledgments
We thank Sarah Tanjung for initial assistance with immunoblotting studies and members of the A.A. laboratory for critically reading this manuscript. Lattice light sheet imaging was conducted at the Advanced Imaging Center at the Janelia/Howard Hughes Medical Institute (HHMI), which is a jointly funded venture of the Gordon and Betty Moore Foundation and the HHMI, and STED microscopy was performed at the University of Wisconsin-Madison Optical Imaging Core facility. This work was supported, in part, by NIH Grant GM088151 (to A.A.), National Science Foundation Grant DMS1661900 (to A.A.), and University of Wisconsin Carbone Cancer Center Grant P30 CA014520. K.B.Q. and E.B.F. received support from Grant T32 GM007215, and P.L. currently receives support from Grant T32 GM008688.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817898116/-/DCSupplemental.
References
- 1.Bache KG, Raiborg C, Mehlum A, Stenmark H. STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes. J Biol Chem. 2003;278:12513–12521. doi: 10.1074/jbc.M210843200. [DOI] [PubMed] [Google Scholar]
- 2.Mayers JR, et al. ESCRT-0 assembles as a heterotetrameric complex on membranes and binds multiple ubiquitinylated cargoes simultaneously. J Biol Chem. 2011;286:9636–9645. doi: 10.1074/jbc.M110.185363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayers JR, et al. Regulation of ubiquitin-dependent cargo sorting by multiple endocytic adaptors at the plasma membrane. Proc Natl Acad Sci USA. 2013;110:11857–11862. doi: 10.1073/pnas.1302918110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 5.Slagsvold T, et al. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem. 2005;280:19600–19606. doi: 10.1074/jbc.M501510200. [DOI] [PubMed] [Google Scholar]
- 6.Huang F, et al. Lysine 63-linked polyubiquitination is required for EGF receptor degradation. Proc Natl Acad Sci USA. 2013;110:15722–15727. doi: 10.1073/pnas.1308014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frankel EB, Audhya A. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin Cell Dev Biol. 2018;74:4–10. doi: 10.1016/j.semcdb.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Q, Hope LW, Brasch M, Reinhard C, Cohen SN. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci USA. 2003;100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malerød L, Stuffers S, Brech A, Stenmark H. Vps22/EAP30 in ESCRT-II mediates endosomal sorting of growth factor and chemokine receptors destined for lysosomal degradation. Traffic. 2007;8:1617–1629. doi: 10.1111/j.1600-0854.2007.00630.x. [DOI] [PubMed] [Google Scholar]
- 10.Vaccari T, Bilder D. The Drosophila tumor suppressor vps25 prevents nonautonomous overproliferation by regulating notch trafficking. Dev Cell. 2005;9:687–698. doi: 10.1016/j.devcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Lee MP, Feinberg AP. Aberrant splicing but not mutations of TSG101 in human breast cancer. Cancer Res. 1997;57:3131–3134. [PubMed] [Google Scholar]
- 12.Thompson BJ, et al. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 13.Zivony-Elboum Y, et al. A founder mutation in Vps37A causes autosomal recessive complex hereditary spastic paraparesis. J Med Genet. 2012;49:462–472. doi: 10.1136/jmedgenet-2012-100742. [DOI] [PubMed] [Google Scholar]
- 14.Ghazi-Noori S, et al. Progressive neuronal inclusion formation and axonal degeneration in CHMP2B mutant transgenic mice. Brain. 2012;135:819–832. doi: 10.1093/brain/aws006. [DOI] [PubMed] [Google Scholar]
- 15.Sadler JBA, et al. A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability. Proc Natl Acad Sci USA. 2018;115:E8900–E8908. doi: 10.1073/pnas.1805504115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horner DS, et al. ESCRT genes and regulation of developmental signaling. Semin Cell Dev Biol. 2018;74:29–39. doi: 10.1016/j.semcdb.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 17.Schuh AL, Audhya A. The ESCRT machinery: From the plasma membrane to endosomes and back again. Crit Rev Biochem Mol Biol. 2014;49:242–261. doi: 10.3109/10409238.2014.881777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teo H, et al. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell. 2006;125:99–111. doi: 10.1016/j.cell.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 19.Teo H, Perisic O, González B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: Crystal structure and interactions with ESCRT-III and membranes. Dev Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Fyfe I, Schuh AL, Edwardson JM, Audhya A. Association of the endosomal sorting complex ESCRT-II with the Vps20 subunit of ESCRT-III generates a curvature-sensitive complex capable of nucleating ESCRT-III filaments. J Biol Chem. 2011;286:34262–34270. doi: 10.1074/jbc.M111.266411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bache KG, Brech A, Mehlum A, Stenmark H. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol. 2003;162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katzmann DJ, Stefan CJ, Babst M, Emr SD. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J Cell Biol. 2003;162:413–423. doi: 10.1083/jcb.200302136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frankel EB, et al. Ist1 regulates ESCRT-III assembly and function during multivesicular endosome biogenesis in Caenorhabditis elegans embryos. Nat Commun. 2017;8:1439. doi: 10.1038/s41467-017-01636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shields SB, et al. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J Cell Biol. 2009;185:213–224. doi: 10.1083/jcb.200811130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sundquist WI, et al. Ubiquitin recognition by the human TSG101 protein. Mol Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 27.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 28.Adell MA, et al. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J Cell Biol. 2014;205:33–49. doi: 10.1083/jcb.201310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schöneberg J, Lee IH, Iwasa JH, Hurley JH. Reverse-topology membrane scission by the ESCRT proteins. Nat Rev Mol Cell Biol. 2017;18:5–17. doi: 10.1038/nrm.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adell MAY, et al. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife. 2017;6:e31652. doi: 10.7554/eLife.31652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 33.Jimenez AJ, et al. ESCRT machinery is required for plasma membrane repair. Science. 2014;343:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- 34.Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003;100:12414–12419, and erratum (2003) 100:152845. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 36.Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522:236–239. doi: 10.1038/nature14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vietri M, et al. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 2015;522:231–235. doi: 10.1038/nature14408. [DOI] [PubMed] [Google Scholar]
- 38.Skowyra ML, Schlesinger PH, Naismith TV, Hanson PI. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science. 2018;260:eaar5078. doi: 10.1126/science.aar5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radulovic M, et al. ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 2018;37:e99753. doi: 10.15252/embj.201899753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raiborg C, et al. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci. 2001;114:2255–2263. doi: 10.1242/jcs.114.12.2255. [DOI] [PubMed] [Google Scholar]
- 41.Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA. High-affinity binding of a FYVE domain to phosphatidylinositol 3-phosphate requires intact phospholipid but not FYVE domain oligomerization. Biochemistry. 2001;40:8581–8587. doi: 10.1021/bi010425d. [DOI] [PubMed] [Google Scholar]
- 42.Raiborg C, et al. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat Cell Biol. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 43.Sachse M, Urbé S, Oorschot V, Strous GJ, Klumperman J. Bilayered clathrin coats on endosomal vacuoles are involved in protein sorting toward lysosomes. Mol Biol Cell. 2002;13:1313–1328. doi: 10.1091/mbc.01-10-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raiborg C, Malerød L, Pedersen NM, Stenmark H. Differential functions of Hrs and ESCRT proteins in endocytic membrane trafficking. Exp Cell Res. 2008;314:801–813. doi: 10.1016/j.yexcr.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 45.Wenzel EM, et al. Concerted ESCRT and clathrin recruitment waves define the timing and morphology of intraluminal vesicle formation. Nat Commun. 2018;9:2932. doi: 10.1038/s41467-018-05345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bleck M, et al. Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. Proc Natl Acad Sci USA. 2014;111:12211–12216. doi: 10.1073/pnas.1321655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mierzwa BE, et al. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat Cell Biol. 2017;19:787–798. doi: 10.1038/ncb3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koch B, et al. Generation and validation of homozygous fluorescent knock-in cells using CRISPR-Cas9 genome editing. Nat Protoc. 2018;13:1465–1487. doi: 10.1038/nprot.2018.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- 51.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White IJ, Bailey LM, Aghakhani MR, Moss SE, Futter CE. EGF stimulates annexin 1-dependent inward vesiculation in a multivesicular endosome subpopulation. EMBO J. 2006;25:1–12. doi: 10.1038/sj.emboj.7600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Razi M, Futter CE. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol Biol Cell. 2006;17:3469–3483. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komada M, Kitamura N. Growth factor-induced tyrosine phosphorylation of Hrs, a novel 115-kilodalton protein with a structurally conserved putative zinc finger domain. Mol Cell Biol. 1995;15:6213–6221. doi: 10.1128/mcb.15.11.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeshita T, et al. Cloning of a novel signal-transducing adaptor molecule containing an SH3 domain and ITAM. Biochem Biophys Res Commun. 1996;225:1035–1039. doi: 10.1006/bbrc.1996.1290. [DOI] [PubMed] [Google Scholar]
- 57.Stern KA, et al. Epidermal growth factor receptor fate is controlled by Hrs tyrosine phosphorylation sites that regulate Hrs degradation. Mol Cell Biol. 2007;27:888–898. doi: 10.1128/MCB.02356-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Row PE, Clague MJ, Urbé S. Growth factors induce differential phosphorylation profiles of the Hrs-STAM complex: A common node in signalling networks with signal-specific properties. Biochem J. 2005;389:629–636. doi: 10.1042/BJ20050067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bache KG, Raiborg C, Mehlum A, Madshus IH, Stenmark H. Phosphorylation of Hrs downstream of the epidermal growth factor receptor. Eur J Biochem. 2002;269:3881–3887. doi: 10.1046/j.1432-1033.2002.03046.x. [DOI] [PubMed] [Google Scholar]
- 60.Chen BC, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: Stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 62.Klar TA, Hell SW. Subdiffraction resolution in far-field fluorescence microscopy. Opt Lett. 1999;24:954–956. doi: 10.1364/ol.24.000954. [DOI] [PubMed] [Google Scholar]
- 63.Shen QT, et al. Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J Cell Biol. 2014;206:763–777. doi: 10.1083/jcb.201403108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doyotte A, Russell MRG, Hopkins CR, Woodman PG. Depletion of TSG101 forms a mammalian “Class E” compartment: A multicisternal early endosome with multiple sorting defects. J Cell Sci. 2005;118:3003–3017. doi: 10.1242/jcs.02421. [DOI] [PubMed] [Google Scholar]
- 65.Du X, Kazim AS, Dawes IW, Brown AJ, Yang H. The AAA ATPase VPS4/SKD1 regulates endosomal cholesterol trafficking independently of ESCRT-III. Traffic. 2013;14:107–119. doi: 10.1111/tra.12015. [DOI] [PubMed] [Google Scholar]
- 66.Bache KG, et al. The ESCRT-III subunit hVps24 is required for degradation but not silencing of the epidermal growth factor receptor. Mol Biol Cell. 2006;17:2513–2523. doi: 10.1091/mbc.E05-10-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scott BL, et al. Membrane bending occurs at all stages of clathrin-coat assembly and defines endocytic dynamics. Nat Commun. 2018;9:419. doi: 10.1038/s41467-018-02818-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cocucci E, Gaudin R, Kirchhausen T. Dynamin recruitment and membrane scission at the neck of a clathrin-coated pit. Mol Biol Cell. 2014;25:3595–3609. doi: 10.1091/mbc.E14-07-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cocucci E, Aguet F, Boulant S, Kirchhausen T. The first five seconds in the life of a clathrin-coated pit. Cell. 2012;150:495–507. doi: 10.1016/j.cell.2012.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doyon JB, et al. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat Cell Biol. 2011;13:331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dambournet D, et al. Genome-edited human stem cells expressing fluorescently labeled endocytic markers allow quantitative analysis of clathrin-mediated endocytosis during differentiation. J Cell Biol. 2018;217:3301–3311. doi: 10.1083/jcb.201710084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li TL, et al. Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science. 2018;360:eaaq1392. doi: 10.1126/science.aaq1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buono RA, et al. ESCRT-mediated vesicle concatenation in plant endosomes. J Cell Biol. 2017;216:2167–2177. doi: 10.1083/jcb.201612040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chiaruttini N, et al. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell. 2015;163:866–879. doi: 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buchkovich NJ, Henne WM, Tang S, Emr SD. Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev Cell. 2013;27:201–214. doi: 10.1016/j.devcel.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 76.Sweeney NT, Brenman JE, Jan YN, Gao FB. The coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr Biol. 2006;16:1006–1011. doi: 10.1016/j.cub.2006.03.067. [DOI] [PubMed] [Google Scholar]
- 77.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 78.Morita E, et al. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morita E, et al. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci USA. 2010;107:12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 81.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.