Significance
The canonical Wnt/β-catenin signaling pathway is tightly regulated and plays a crucial role in a broad range of biological processes. Perturbations in this pathway are associated with multiple pathologies, including different cancers. Efforts to develop Wnt-pathway inhibitors as cancer therapeutics have been met with significant challenges in balancing therapeutic efficacy and toxicity, due to the ubiquitous nature of the pathway. Here we describe the development and characterization of broadly specific frizzled antibodies that block the Wnt ligand–receptor interaction. A combination of structural, biochemical, and cellular studies allowed refining the pharmacokinetics, tolerability, and antitumor profile of multispecific frizzled antibodies. We provide a rationale for structure-based refinement of therapeutic antibody leads and contribute to guiding the development of anti-Wnt/FZD cancer therapeutics.
Keywords: antibody therapeutic, Frizzled receptors, Wnt signaling, X-ray crystallography, protein engineering
Abstract
Aberrant activation of Wnt/β-catenin signaling occurs frequently in cancer. However, therapeutic targeting of this pathway is complicated by the role of Wnt in stem cell maintenance and tissue homeostasis. Here, we evaluated antibodies blocking 6 of the 10 human Wnt/Frizzled (FZD) receptors as potential therapeutics. Crystal structures revealed a common binding site for these monoclonal antibodies (mAbs) on FZD, blocking the interaction with the Wnt palmitoleic acid moiety. However, these mAbs displayed gastrointestinal toxicity or poor plasma exposure in vivo. Structure-guided engineering was used to refine the binding of each mAb for FZD receptors, resulting in antibody variants with improved in vivo tolerability and developability. Importantly, the lead variant mAb significantly inhibited tumor growth in the HPAF-II pancreatic tumor xenograft model. Taken together, our data demonstrate that anti-FZD cancer therapeutic antibodies with broad specificity can be fine-tuned to navigate in vivo exposure and tolerability while driving therapeutic efficacy.
Wnt signaling is an evolutionarily conserved signaling cascade that plays a critical role in diverse biological processes, including embryonic development, tissue differentiation, organogenesis, stem cell maintenance, and normal adult tissue homeostasis (1–3). Activation of the well-characterized β–catenin-dependent canonical pathway is initiated by the binding of secreted Wnt proteins to Frizzled (FZD) receptors and coreceptors such as LRP5 and LRP6 (4–8). To date, 19 human Wnt and 10 FZD receptors have been identified that mediate differential cellular functions (9, 10). FZD receptors interact with Wnt through their N-terminal extracellular cysteine-rich domain (CRD). A Wnt residue, Ser187, (position number according to Xenopus Wnt8) is posttranslationally modified, leading to its palmitoleation, which mediates interactions with the FZD–CRD (site 1); a second site of interaction between FZD and Wnt is located at the opposing end of Wnt (11).
Wnt/FZD signaling is essential for normal cell function, but aberrations in the pathway are frequently found in cancers, fibrosis, and degenerative diseases (12, 13). Abnormal activation of the Wnt pathway is an essential driver of primary tumor formation and metastasis in multiple cancer types (14–18). Inactivating mutations in E3 ubiquitin ligase RNF43 inhibit the down-modulation of FZD expression on the cell surface and sensitize tumor cells to Wnt-dependent growth. These mutations have been identified in pancreatic, biliary duct, and colorectal cancers (19–21). FZD5 expression is up-regulated in renal cell carcinoma (22), prostate cancer (23), and pancreatic tumors (16); aberrant FZD7 expression is observed in hepatocellular carcinoma and colorectal and triple negative breast cancer (14, 24, 25); FZD8 is up-regulated in acute lymphoblastic leukemia and lung cancer (17, 26); and FZD4 is elevated and drives epithelial-to-mesenchymal transition in TMRESS2–ERG fusion prostate cancer (27). Signaling through FZD4 is also essential for normal angiogenesis (28, 29). Mutations in FZD4 and its alternative ligand, Norrin, are primary drivers of retinal hypovascularization in familial exudative vitreoretinopathy (30). Therefore, therapeutic modulation of the Wnt pathway is an attractive approach to treat multiple disease indications (13, 31).
Several approaches have been taken to develop drugs capable of abrogating the Wnt pathway in cancers (13, 31, 32). These include inhibition of Wnt palmitoleation with PORCN (a Wnt-specific acyltransferase) inhibitor LGK974, pan-Wnt neutralization with decoy receptor FZD8-Fc OMP54-28 (31–33), interference in downstream signaling components such as tankyrase inhibitor XAV939, or inhibitors that disrupt β-catenin/cotranscriptional inhibitor interactions (e.g., ICG-001) (34). However, therapeutically targeting the WNT/β-catenin pathway is challenging due to the critical involvement of the pathway in normal tissue homeostasis, and complete abrogation may have severe adverse effects. Gastrointestinal toxicity has been a frequent dose-limiting toxicity observed in clinical trials, among other toxicities (35). Therapeutic antibodies potentially offer advantages in that they can target specific FZD receptors rather than broadly inhibiting the Wnt pathway. For example, OMP-18R5 (vantictumab), a monoclonal antibody (mAb) that interacts with the CRD of FZD1, -2, -5, -7, and -8, has been shown to inhibit tumor growth in several tumor types, including breast, colon, lung, and pancreatic cancer and did not reportedly demonstrate gastrointestinal toxicity (31, 32).
Targeting FZD4 in addition to FZD1, -2, -5, -7, and -8 may have a broader impact on the efficacy of FZD-targeting antibodies. We have recently shown that antibodies specific to these six FZD receptors inhibit endothelial tube formation in vitro, whereas antibodies lacking FZD4 specificity did not (36). Here we expand on these findings and present the structural definition, functional characterization, and refinement of FZD antibodies that target FZD1, -2, -4, -5, -7, and -8 to potently inhibit Wnt signaling, while balancing their tolerability. Indeed, despite similar epitopes, these antibodies showed a wide range of selectivity, potency, tolerability, and developability. Here, we demonstrate how structure-guided activity relationships were critical to fine-tune antibody-binding profiles, which led to the development of a tolerable anti-FZD therapeutic antibody with broad FZD-binding specificity and effective antitumor activity.
Results
Unique Binding and Potency Profiles of FZD Antibodies.
In vitro cell-binding experiments were performed to assess the specificity and selectivity of three mAbs generated from a phage display panel (36). mAbs F2.I and F7.B bound FZD1, -2, -4, -5, -7, and -8 on the cell surface, but did not bind FZD3, -6, -9, or -10 at the concentrations tested (Fig. 1A and SI Appendix, Fig. S1). On the other hand, mAb F6 bound FZD1, -2, -5, -7, and -8, but not FZD4, similar to the profile of OMP-18R5. Within the CRD region, human and mouse FZD4 and -5 have a 96.7% and 98.4% homology, respectively (37), and all other relevant FZD receptors are 100% homologous between mouse and human. Importantly, mAbs F6, F2.I, and F7.B showed cross-reactivity to mouse FZD4 and FZD5 (SI Appendix, Fig. S1C).
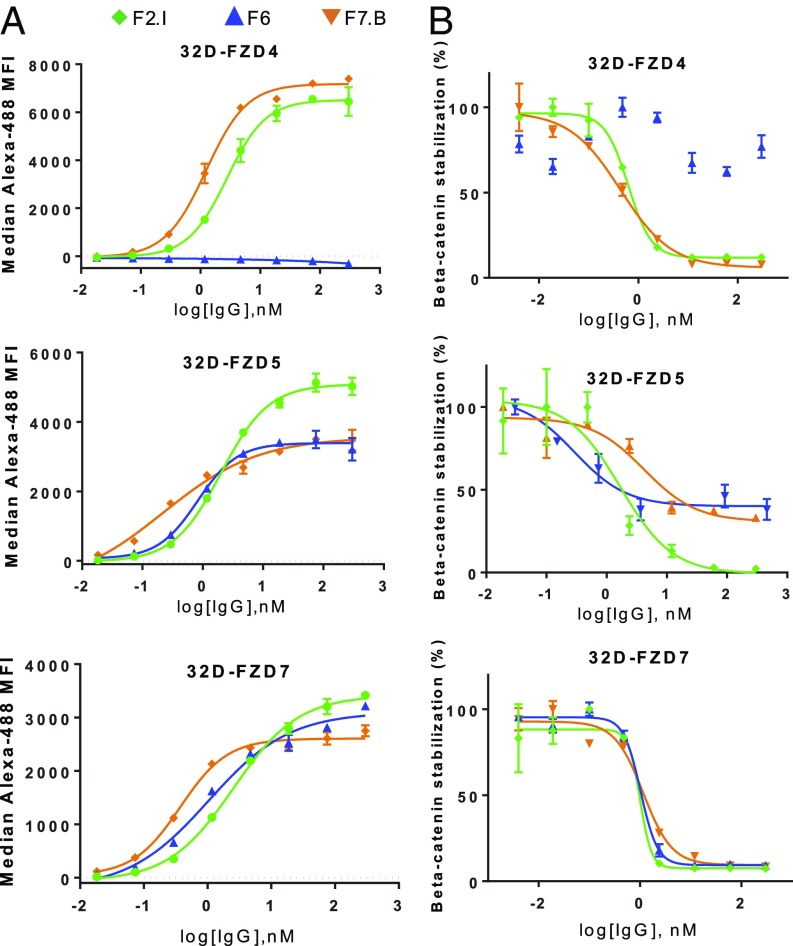
Fig. 1.
Binding and functional characteristics of lead FZD mAbs. (A) Binding curves for mAbs F2.I, F7.B, and F6 to 32D cells overexpressing FZD4, -5, or -7. MFI indicates median fluorescence intensity. (B) Dose–response curves for mAbs F2.I, F7.B, and F6 inhibition of Wnt3a-induced β-catenin stabilization in 32D cells overexpressing FZD4, -5, or -7. As F6 was not found to inhibit FZD4, no trendline could be drawn.
mAbs F6, F2.I, and F7.B functionally inhibited Wnt3a-induced β-catenin stabilization in 32D cells expressing each of their respective FZD-binding partners (Fig. 1B and SI Appendix, Fig. S1). Interestingly, while mAb F2.I fully inhibited β-catenin stabilization in 32D–FZD5 cells, mAbs F6 and F7.B both showed a maximum inhibition of ∼50%, suggesting that they are partial antagonists of FZD5. Correspondingly, mAb F2.I had a higher maximum binding to 32D–FZD5 cells compared to mAbs F6 and F7.B (Fig. 1A). Thus, each mAb had unique binding and potency profiles, with mAb F2.I being the most potent across most FZDs.
Molecular Basis of FZD Antibody Recognition.
Crystal structures of FZD5–CRD with F2.I Fab, FZD7–CRD with F7.B Fab, and FZD7–CRD with F6 Fab were determined at 1.8-Å, 2.1-Å, and 2.5-Å resolutions, respectively (Fig. 2 and SI Appendix, Table S1). The structures revealed that mAbs F2.I, F7.B, and F6 bind to overlapping epitopes on FZD–CRD. This finding is consistent with the antibodies, including OMP-18R5, competing for binding to FZD–CRD (SI Appendix, Fig. S3A). Nonetheless, the three antibodies differ slightly in their angle of approach and in the residues they contact on FZD–CRD (Fig. 2D). Epitope mapping revealed that the antibodies interact predominantly with a hydrophobic cleft on the surface of FZD–CRD (Fig. 2F and SI Appendix, Fig. S2). The contacts also extend to surrounding polar residues, particularly in the case of mAbs F2.I and F6 (SI Appendix, Tables S2–S4). mAbs F2.I and F6 form numerous polar interactions with FZD (mAb F2.I, 12 H bonds and one salt bridge; mAb F6, 8 H bonds and three salt bridges). In contrast, mAb F7.B uses predominantly nonpolar CDR residues to largely mediate hydrophobic interactions with FZD7, with only 8 H bonds and no salt bridges.
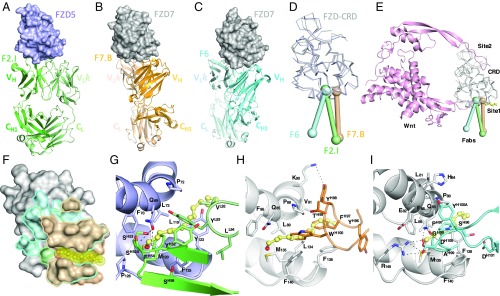
Fig. 2.
Structural insights into antibody recognition of FZD–CRD and mechanism of Wnt-signaling inhibition. Cocrystal structures of (A) F2.I Fab in complex with FZD5, (B) F7.B Fab in complex with FZD7, and (C) F6 Fab in complex with FZD7. (D) Angle of approach of three antibodies against the same subsite on FZDs. (E) mAbs F2.I (green), F7.B (wheat), and F6 (cyan) block the Wnt–FZD interaction at site 1. The modeled Wnt (pink) is represented as cartoon and the FZD–CRDs (gray) are shown as ribbons. The palmitoyl group is shown in yellow as ball and stick. (F) Outline of the epitope for mAbs F2.I (green), F7.B (wheat), and F6 (cyan) traced onto the surface of FZD7 show overlapping sites of interaction and the lipid-binding pocket with the Wnt palmitoyl group modeled in yellow. Molecular basis of lipid blocking by mAbs (G) F2.I (green), (H) F7.B (wheat), and (I) F6 (cyan). FZD5 is shown in purple and FZD7 in gray.
The nature of the interaction of the antibodies with FZD is reflective of their binding affinities. F2.I Fab binds with fast on rates and slow off rates across the three FZD–CRD tested and is the highest affinity binder to FZD5 (1.7 nM) and FZD7 (0.3 nM) (SI Appendix, Fig. S3B). F7.B Fab has much faster off rates and 25 times and 17 times weaker binding to FZD5 (40.8 nM) and FZD7 (5.2 nM) compared to F2.I Fab, respectively; however, F7.B Fab bound to FZD4 with the highest affinity (1.5 nM). F6 Fab bound with nanomolar affinities to FZD5 and FZD7 but, as expected from cellular experiments, did not show any appreciable binding to FZD4 in the concentration range tested, similar to the profile of OMP-18R5.
Residue conservation in the hydrophobic pocket of FZD explains the broad binding specificity of our antibodies (SI Appendix, Fig. S4A). This is particularly true for FZD4, which shares a sequence identity of only 38% with FZD5 and 37% with FZD7 in the CRD domain. The inability of mAb F6 to recognize FZD4 primarily resides in three residue differences with FZD7. Glu77, Gln85, and Arg145 in FZD7 are Thr, Thr, and Ser residues at these corresponding positions in FZD4, respectively; thus the shorter side chains in FZD4 prevent the formation of key H bonds (SI Appendix, Fig. S4B). These insights reveal the basis for the specificities required for broad FZD recognition by therapeutic antibody candidates.
Mechanism of Inhibition of Wnt Signaling.
Our cocrystal structures revealed that the three antibodies interact with FZD–CRD around site 1, thereby blocking a critical component of the Wnt–FZD interaction. Competitive binding experiments between our antibodies and Norrin—a natural ligand that only interacts with FZD4 at a site distant from the lipid-binding site (38)—did not reveal any steric occlusions, as would be expected from inspection of the structures (SI Appendix, Fig. S5). mAbs F2.I and F6 interact with the entrance of the lipid-binding cleft in FZD, whereas mAb F7.B CDRs are instead localized near the center of the lipid-binding cleft. All mAbs utilize hydrophobic side chains to position themselves in the path where the palmitoyl moiety of Wnt would interact with FZD residues (mAb F7.B, HCDR3 residues Phe97 and Trp100; mAb F2.I, HCDR2 residue Phe54 and LCDR3 residue Tyr93; and mAb F6, HCDR3 residues Pro97 and Gly98; Fig. 2 G–I). While the Wnt site 1–FZD–CRD interface is 580 Å2, the total buried surface area between the FZD–CRD and the antibodies is much greater (939 Å2, 800 Å2, and 1,088 Å2 for mAbs F2.I, F7.B, and F6, respectively), which reveals the expanse of antibody interactions that extends beyond the ligand-binding pocket. Our structural characterization of antibody–FZD complexes reveals potent epitopes located around the lipid-binding pocket of the natural ligand on the FZD–CRD and the mechanism of action of the antibodies that utilize slightly different contact residues for inhibition.
Low Tolerability and Exposure of FZD Antibodies.
C57BL/6 mice were dosed every 3–4 d with 30 mg/kg of either mAbs F2.I, F7.B, or F6 (Fig. 3A). Mice dosed with mAb F2.I began losing weight after just one dose, with a drop of 7% body weight overnight. By day 6, treatment-related death was observed in 6 out of 20 mice, and all surviving mice treated with mAb F2.I became moribund and were killed, as many mice lost >20% of their initial body weight. In comparison, both mAbs F7.B and F6 were well tolerated after 2 wk, with no decrease in body weight or physical signs of discomfort or toxicity.
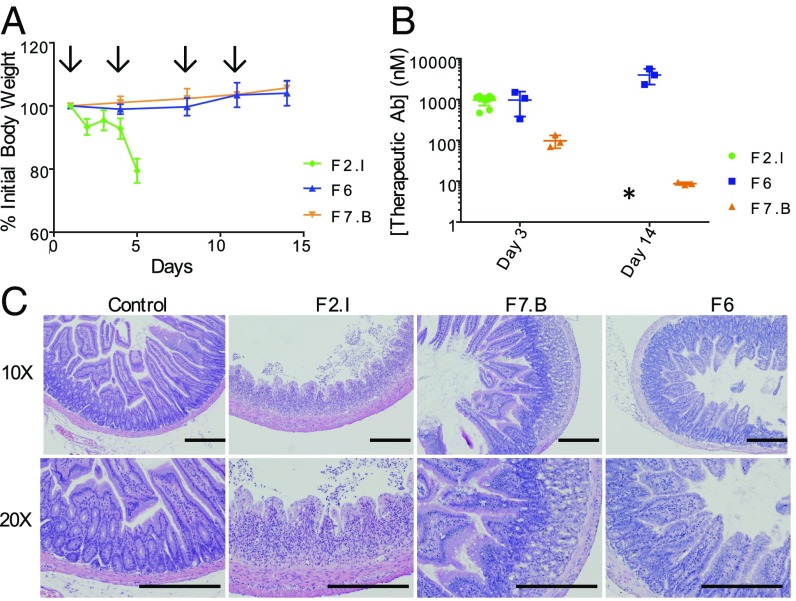
Fig. 3.
Lead FZD mAbs display poor exposure or gut toxicity in vivo. (A) Body weights of mice treated with 30 mg/kg of mAbs F2.I (green), F7.B (orange), or F6 (blue). Arrows indicate when mice were dosed. n = 3 for mAbs F6 and F7.B; n = 10 for mAb F2.I. (B) mAb plasma exposures corresponding to C, presented as mean ± SD. *Mice treated with mAb F2.I were killed on day 5 due to dramatic body weight loss. (C) Histological cross-sections stained with H&E of the duodenum of mice treated with 30 mg/kg of either control IgG, mAbs F2.I, F7.B, or F6. Shown are 10× and 20× magnifications. (Scale bars, 500 μm.)
We also measured the plasma exposure of these mAbs from treated mice 72 h after the first dose (day 3) and 72 h after the fourth dose (day 14) (Fig. 3B). Mice treated with mAb F6 showed good exposure on day 3 (1 µM), with antibody accumulation (4 µM) evident after four doses. Mice treated with mAb F2.I showed good exposure (1 µM) after 3 d but accumulation could not be determined as all mice were killed by day 6. In contrast, mice treated with mAb F7.B had less than 100 nM of mAb in their plasma on day 3, and less than 10 nM 72 h after the fourth dose. This low exposure rendered this molecule unattractive as a drug candidate.
To determine the reason for poor tolerability of mAb F2.I in vivo, histology sections of essential tissues including heart, lung, kidney, brain, liver, and gastrointestinal tract including duodenum, jejunum, and ileum were analyzed. In contrast to most essential tissues appearing to be morphologically normal, the intestinal tract of mice treated with mAb F2.I showed extensive necrosis and structural damage (Fig. 3C). Wnt-pathway inhibition has previously been reported to cause gut toxicity, including severe necrosis and inflammation of the small intestine (39). However, the intestinal sections from mAbs F7.B- and F6-treated mice showed little to no necrosis and no change to the structure of the intestinal crypts. These data suggested that differences in antibody selectivity and potency may account for the differential mAb intestinal toxicity. Unlike F2.I and F7.B, F6 behaved well in the in vivo studies. However, F6 does not bind FZD4, making it unsuitable for FZD4-related indications. Overall, none of the three antibodies had the desired properties for a lead therapeutic candidate, i.e., good exposure, tolerability, and broad specificity that includes FZD4.
Structure-Based Refinement of FZD Antibodies.
mAb F7.B showed poor plasma exposure in vivo, likely due to the presence of several nonpolar residues in its CDRs that render the antibody hydrophobic. An isoelectric point (pI) close to the physiological range further exacerbates the hydrophobic nature of mAb F7.B. To circumvent the high hydrophobicity, near-neutral pI and low exposure of mAb F7.B, we applied structure-based engineering to design resurfaced mutants. A variant (F7.Bv2) was identified with five mutations across CDR-H1, CDR-H2, and CDR-L3 and exhibited an increased pI and shorter retention time in hydrophobic interaction chromatography (SI Appendix, Fig. S6). F7.Bv2 displayed a dramatically improved exposure profile in C57BL/6 mice compared to the parental F7.B (SI Appendix, Fig. S6). However, the CDR mutations led to a reduction in binding affinity across the FZD receptors, and when tested in the HPAF-II xenograft model, F7.Bv2 had no effect on tumor growth compared to the vehicle control (SI Appendix, Fig. S6). We also attempted to engineer F6 specificity toward FZD4 based on our structural understanding of its interaction with FZD7; however, our efforts were not successful.
Next, we used structure-based design in an attempt to resolve the poor tolerability of mAb F2.I. We suspected that the high binding affinity of mAb F2.I across multiple FZDs, compared to mAbs F7.B and F6 which were both tolerable in mice, may be associated with its poor tolerability. Antibody residues involved in interacting with FZD5 were mutated to dampen the affinity for this FZD (Fig. 4A and SI Appendix, Fig. S7A), which we hypothesized could mitigate toxicity. A total of 20 mutants (11 heavy chain and two light chain mutants, and some combinations of heavy and light chain mutants) were generated. While some mutants retained a similar high binding affinity to FZD5 compared to parent mAb F2.I, others showed poor or a complete loss of binding to FZD5, highlighting the spectrum of designed disruptions at the antibody–antigen interface (Fig. 4B and SI Appendix, Fig. S7 and Tables S5 and S6). Approximately a third of the mutants showed an intermediate loss in binding to FZD4, FZD5, and FZD7.
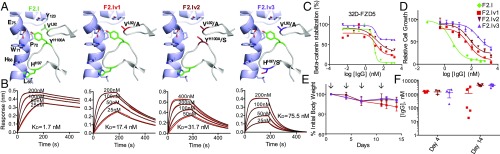
Fig. 4.
Structure-based design of mAb F2.I variants reduces in vivo toxicity. (A) Mutants designed to reduce the affinity toward FZD5. (B) The corresponding binding curves for F2.I Fab and Fab variants, F2.Iv1 (V92A, light chain), F2.Iv2 (Y100AS, heavy chain/V92A, light chain) and F2.Iv3 (H97S, heavy chain/V92A, light chain) are shown. The data (red) were fit using a 1:1 model (black). (C) β-Catenin stabilization assays in 32D–FZD5 cells for mAb F2.I and its variants, mAbs F2.Iv1 (red), F2.Iv2 (brown), and F2.Iv3 (purple). (D) HPAF-II cell proliferation assay for mAb F2.I and its variants. (E) Body weights of mice treated with 30 mg/kg of mAbs F2.I, F2.Iv1, F2.Iv2, and F2.Iv3. Arrows indicate when mice were dosed; n = 5 per group. (F) IgG plasma exposure corresponding to E, presented as mean ± SD. Note: one of five mice from mAb F2.Iv1, two of five mice from mAb F2.Iv2, and one of five mice from mAb F2.Iv3 had undetectable plasma IgG exposure on day 4, indicating that these mice likely did not receive their first dose (technical error).
Binding results correlated well with 32D–FZD cell-based reporter assays (Fig. 4C and SI Appendix, Fig. S7C and Table S7) and with the HPAF-II proliferation assay (Fig. 4D), an RNF43-mutant cell line shown to be highly dependent on FZD5 for viability (16). Specifically, similar to the parental molecule (mAb F2.I), mAb F2.Iv1 inhibited HPAF-II proliferation nearly 100%, whereas mAbs F2.Iv2 and F2.Iv3 inhibited proliferation to a maximum of 82% and 66%, respectively.
In C57BL/6 mice, our engineered mAb F2.I variants F2.Iv1, F2.Iv2, and F2.Iv3 showed improved tolerability at a high dose (30 mg/kg; Fig. 4E). Noticeably, while mAb F2.Iv1 was initially well tolerated for three doses at 30 mg/kg, body weights in these mice began to drop after the fourth dose. One of five mice was killed due to >20% initial body weight loss. For mice treated with mAbs F2.Iv2 and F2.Iv3, no significant loss of body weight was observed out to 14 d, which correlated well with their lower binding affinity toward multiple FZD receptors, including FZD5 (SI Appendix, Table S7). Additionally, mAb F2.Iv1 was found to show variable, but overall low plasma exposure in three of five mice on day 14 (Fig. 4F). On the other hand, mAbs F2.Iv2 and F2.Iv3 both showed high plasma exposures in these mice.
To further differentiate between mAbs F2.Iv2 and F2.Iv3, we compared their functional activity in vitro. In the 32D β-catenin stabilization assay, mAb F2.Iv3 showed less activity on both FZD5 and FZD7 (39% and 73% maximum inhibition, respectively) compared to mAb F2.Iv2 (66% and 88% maximum inhibition, respectively) (Fig. 4C and SI Appendix, Fig. S7 and Table S7). In addition, mAb F2.Iv3 was found to only inhibit HPAF-II proliferation by a maximum of 66% (compared to 82% for mAb F2.Iv2) (Fig. 4D and SI Appendix, Table S7). Correspondingly, F2.Iv3 showed a 100-fold shift in IC50 in this assay from parental mAb F2.I (3.3 nM to 327.1 nM), compared to a 20-fold shift in IC50 observed for mAb F2.Iv2 (69.9 nM). Based on these profiles of in vitro potency, activity on HPAF-II cell proliferation and in vivo tolerability and exposure, mAb F2.Iv2 was chosen as our lead antibody candidate.
mAb F2.Iv2 Inhibits Tumor Proliferation and Growth in Vivo.
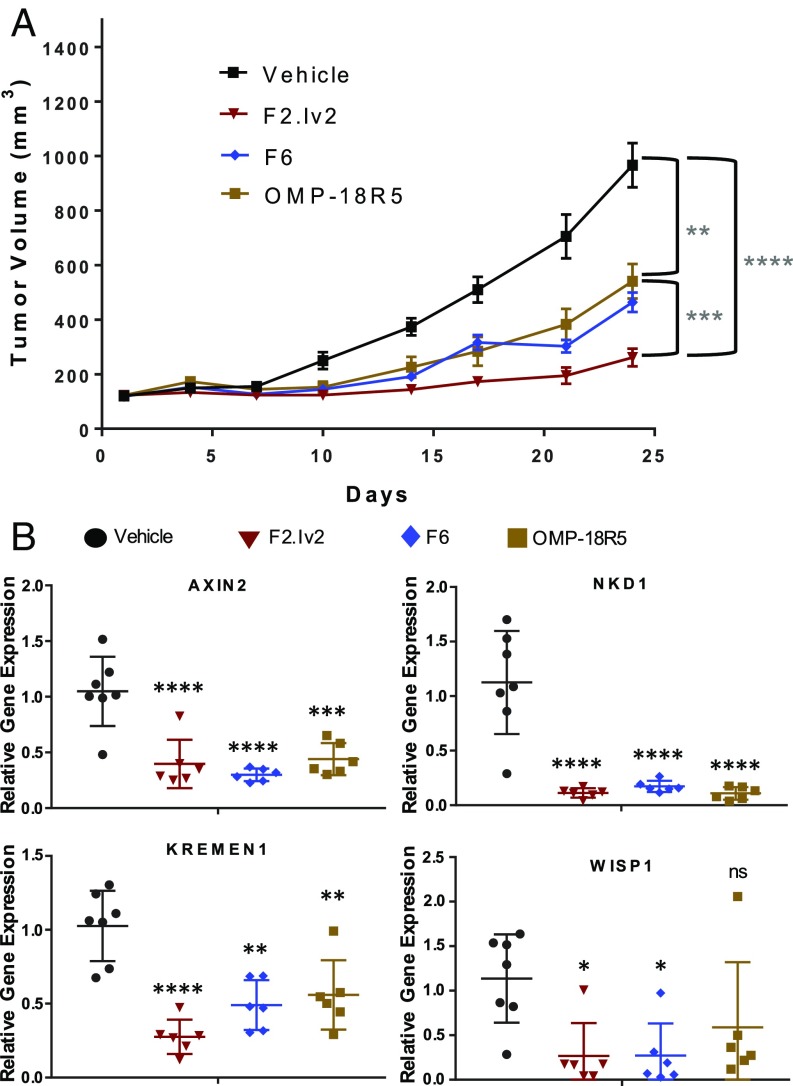
To assess in vivo efficacy, we tested our new lead mAb F2.Iv2 against F6 and OMP-18R5 (both of which do not bind FZD4) in the RNF43-mutant HPAF-II pancreatic adenocarcinoma tumor xenograft model (Fig. 5A). Female Ath/nu mice were implanted with 4 × 106 HPAF-II cells, and once tumors were palpable, mice were dosed i.p. with either vehicle (PBS) control, mAb F2.Iv2 (30 mg/kg), mAb F6 (30 mg/kg), or OMP-18R5 (30 mg/kg) twice per week (SI Appendix, Fig. S8). Mice in the three antibody-treated groups showed significant tumor growth inhibition (TGI) compared to vehicle-treated mice (P value of 7.4 × 10−6 for F2.Iv2, 8.9 × 10−5 for F6, and 1.4 × 10−2 for OMP-18R5; Fig. 5A). mAb F2.Iv2 treatment resulted in a significantly reduced tumor growth compared to both mAb F6 (77% vs. 56% TGI, P value 3.7 × 10−3) and OMP-18R5 (49% TGI, P value 6.6 × 10−3). While treated mice showed minor body weight loss, there was no difference in weight loss between mAb F6-, mAb OMP-18R5-, and mAb F2.Iv2-treated mice (SI Appendix, Fig. S8D). Importantly, all antibodies showed excellent plasma exposures throughout the study (SI Appendix, Fig. S8E).
Fig. 5.
In vivo efficacy of engineered FZD antibody variant shows significant tumor growth inhibition. (A) HPAF-II xenograft study showing tumor volume for mice treated with 30 mg/kg of mAbs OMP-18R5, F6, F2.Iv2, or vehicle i.p. twice per week (n = 12 per group). (B) Tumor gene expression for mice treated with mAbs OMP-18R5, F6, F2.Iv2, or vehicle showing representative genes for Wnt-pathway gene modulation at study endpoint (29 d after HPAF-II cell injection) (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). ns, not significant.
To confirm on-target Wnt-pathway inhibition, tumors were collected at the endpoint of the study (day 29) and gene expression analysis was conducted on tumor cDNA. Axin2, Nkd1, Kremen1, and Wisp1 are well-characterized Wnt-pathway target genes known to be down-regulated upon Wnt-pathway inhibition (40), especially in RNF43-mutant cells (16). These genes were found to be significantly down-regulated in tumors collected from almost all anti-FZD mAb treatment groups compared to the vehicle control group (Fig. 5B).
Discussion
Inhibition of the Wnt signaling pathway has emerged as a therapeutic strategy for numerous disease indications, including cancer (41). However, targeting the Wnt pathway is associated with significant challenges due to its pervasive roles in normal tissue homeostasis. Hence it is imperative to develop therapeutic approaches that appropriately navigate specificity, efficacy, and toxicity. For this purpose, monoclonal antibodies can offer key advantages by providing fine specificity and selective inhibition by blocking defined receptor–ligand interactions. OMP-18R5, a mAb that specifically recognizes five FZD receptors (FZD1, -2, -5, -7, and -8), was previously shown to inhibit tumor growth in different tumor types (31, 32). Here, we characterized and refined anti-FZD therapeutic antibodies that bind FZD4 in addition to FZD1, -2, -5, -7, and -8, to expand on the therapeutic potential of anti-FZD mAbs such as OMP-18R5 to inhibit Wnt/FZD-driven tumor growth.
Our structural characterization of three previously reported multispecific FZD antibodies (36) revealed that they have overlapping epitopes that map to the lipid-binding cleft of FZD. The lipid-binding site on FZD receptors is indispensable for Wnt–FZD interaction and activation of β-catenin signaling and thus represents an attractive target site for therapeutic intervention. However, targeting a hydrophobic patch on FZD was found to be associated with developability challenges. To recognize this site, the mAb F7.B paratope is highly hydrophobic and displayed limited bioavailability in vivo. mAb F2.I overcame this hurdle with a more hydrophilic paratope, which resulted in CDR residues that interact extensively with polar regions that surround the FZD–CRD hydrophobic cleft. These sizable interactions led to mAb F2.I having the highest binding affinity to FZD5 (1.7 nM) and FZD7 (0.3 nM), which we propose resulted in its toxicity in vivo. On the other hand, mAb F6 lacked critical residues required for its interaction with FZD4. These data highlight the importance of extensively characterizing antibody leads, even within the same epitope bin, when targeting a broad family of highly related members, as is the case for human FZD receptors.
Structure-based design provides a means for optimizing the affinity, specificity, and pharmacokinetic attributes of lead molecules (42, 43). In most antibody development cases reported to date, this strategy has been utilized to enhance the affinity of antibody–antigen interactions (44, 45). Here, structure-based antibody design was instead employed to reduce the binding affinity of mAb F2.I toward FZD, with the aim to improve its in vivo tolerability, while keeping its target specificity. Our antibody–antigen crystal structure allowed accurate selection of paratope residues to design mAb F2.I variants that possess a wide spectrum of binding affinities to FZDs. The antibody variant that showed an ideal in vivo combination of effective tumor growth inhibition and tolerability has binding affinities that range between 15.5 and 250.7 nM against FZD1, -2, -4, -5, -7, and -8 (SI Appendix, Table S7), indicating that for Wnt inhibition, higher affinity is not necessarily a direct indicator of a better therapeutic. Instead a balance between affinity, tolerability, and efficacy needs to be achieved. We also note that the slightly reduced binding affinities of our lead variant mAb F2.Iv2, compared to parental mAb F2.I, are primarily a result of faster off rates, which in turn are associated with incomplete FZD mediated β-catenin stabilization. Our results provide a concrete example for the rational development of a lead therapeutic antibody and a unique structure–function dataset from which principles can be derived for the inhibition of other signaling pathways that seek to block receptor–ligand interactions.
Our lead variant, mAb F2.Iv2, was found to induce tumor stasis in the RNF43-mutant HPAF-II tumor xenograft model and was significantly more potent than F6 and OMP-18R5, both of which lack FZD4 reactivity. Gene expression analyses of tumors extracted after antibody treatment revealed on-target specificity of these mAbs, as indicated by the down-modulation of Wnt pathway-related genes. As HPAF-II tumors have been shown to be primarily driven by FZD5 signaling (16), future studies will determine the efficacy of mAb F2.Iv2 in additional pancreatic ductal adenocarcinoma tumor models, including those driven by aberrant FZD4 signaling or FZD4-driven angiogenesis. Additional studies with our lead mAb F2.Iv2 will reveal its long-term tolerability and additional effects associated with Wnt-pathway inhibition, such as impaired bone mineralization (46).
mAb OMP-18R5 has been tested in three phase 1b clinical trials in combination with chemotherapy: one in HER2-negative breast cancer, one in non-small-cell lung carcinoma, and one in advanced pancreatic cancer. We propose that mAb F2.Iv2, with its refined anti-FZD activity profile that includes FZD4, is the most multispecific FZD monoclonal antibody therapeutic yet described that possesses desirable in vivo exposure, tolerability, and efficacy and adds to the molecular arsenal of Wnt-pathway inhibitors for clinical development.
Materials and Methods
FZD antibody variants were generated by site-directed mutagenesis. Functional inhibition was assessed by β-catenin stabilization assay quantified using a β-Catenin ELISA Kit. Total IgG4 protein concentrations were determined using the Human Therapeutic IgG4 ELISA Kit. Recombinant human FZD–CRDs and Fabs were transiently expressed in HEK 293 cells. Crystals of FZD5–F2.I, FZD7–F7.B, and FZD7–F6 complexes were obtained and subjected to X-ray diffraction experiments. Structures of these complexes were solved by molecular replacement. Binding kinetics and competition assays were performed by biolayer interferometry. The antitumor effectiveness of variant mAbs was tested in HPAF-II proliferation experiments and in a HPAF-II xenograft tumor model. Real-time PCR was performed to analyze gene expression. Animal work was performed according to the guidelines of the University of Toronto Animal Care Committee (UACC) under AUP# 5565.2 or at Charles River Discovery Services North Carolina, accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Details are included in SI Appendix.
Supplementary Material
Acknowledgments
Part of this work was funded by Mitacs Accelerate Grant IT06689 and funding from the Canada Research Chairs program (to J.-P.J.). X-ray diffraction experiments were performed using beamline 08ID-1 at the Canadian Light Source, which is supported by the Canada Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, the University of Saskatchewan, the Government of Saskatchewan, Western Economic Diversification Canada, the National Research Council Canada, and the Canadian Institutes of Health Research. X-ray diffraction experiments were also performed at the General Medical Sciences and Cancer Institutes Structural Biology Facility at the Advanced Photon Source, which has been funded in whole or in part with federal funds from the National Cancer Institute (Grant ACB-12002) and the National Institute of General Medical Sciences (Grant AGM-12006). The Eiger 16M detector was funded by an NIH–Office of Research Infrastructure Programs, High-End Instrumentation Grant (1S10OD012289-01A1). This research used resources of the Advanced Photon Source, a DOE Office of Science user facility, operated for the DOE Office of Science by the Argonne National Laboratory under Contract DE-AC02-06CH11357. The Structural & Biophysical Core Facility (The Hospital for Sick Children) provided access to the Biolayer Interferometry (BLI) instrument.
Footnotes
Conflict of interest statement: The authors have filed a patent application for the antibodies described in this work.
This article is a PNAS Direct Submission.
Data deposition: Structural data have been deposited in the Protein Data Bank (PDB), www.wwpdb.org (PDB ID codes 6O39, 6O3A, and 6O3B).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817246116/-/DCSupplemental.
References
- 1.Dravid G, et al. Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells. 2005;23:1489–1501. doi: 10.1634/stemcells.2005-0034. [DOI] [PubMed] [Google Scholar]
- 2.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 3.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 4.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenov MV, Habas R, Macdonald BT, He X. SnapShot: Noncanonical Wnt signaling pathways. Cell. 2007;131:1378. doi: 10.1016/j.cell.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- 8.He X, Semenov M, Tamai K, Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: Arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 9.Huang HC, Klein PS. The Frizzled family: Receptors for multiple signal transduction pathways. Genome Biol. 2004;5:234. doi: 10.1186/gb-2004-5-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dijksterhuis JP, et al. Systematic mapping of WNT-FZD protein interactions reveals functional selectivity by distinct WNT-FZD pairs. J Biol Chem. 2015;290:6789–6798. doi: 10.1074/jbc.M114.612648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 13.Kahn M. Can we safely target the WNT pathway? Nat Rev Drug Discov. 2014;13:513–532. doi: 10.1038/nrd4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merle P, et al. Functional consequences of frizzled-7 receptor overexpression in human hepatocellular carcinoma. Gastroenterology. 2004;127:1110–1122. doi: 10.1053/j.gastro.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Phesse T, Flanagan D, Vincan E. Frizzled7: A promising achilles’ heel for targeting the Wnt receptor complex to treat cancer. Cancers (Basel) 2016;8:50. doi: 10.3390/cancers8050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinhart Z, et al. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat Med. 2017;23:60–68. doi: 10.1038/nm.4219. [DOI] [PubMed] [Google Scholar]
- 17.Wang HQ, Xu ML, Ma J, Zhang Y, Xie CH. Frizzled-8 as a putative therapeutic target in human lung cancer. Biochem Biophys Res Commun. 2012;417:62–66. doi: 10.1016/j.bbrc.2011.11.055. [DOI] [PubMed] [Google Scholar]
- 18.Ueno K, Hirata H, Hinoda Y, Dahiya R. Frizzled homolog proteins, microRNAs and Wnt signaling in cancer. Int J Cancer. 2013;132:1731–1740. doi: 10.1002/ijc.27746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koo BK, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 20.Jiang X, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA. 2013;110:12649–12654. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fennell LJ, et al. RNF43 is mutated less frequently in Lynch syndrome compared with sporadic microsatellite unstable colorectal cancers. Fam Cancer. 2018;17:63–69. doi: 10.1007/s10689-017-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssens N, Andries L, Janicot M, Perera T, Bakker A. Alteration of frizzled expression in renal cell carcinoma. Tumour Biol. 2004;25:161–171. doi: 10.1159/000081098. [DOI] [PubMed] [Google Scholar]
- 23.Thiele S, et al. Expression profile of WNT molecules in prostate cancer and its regulation by aminobisphosphonates. J Cell Biochem. 2011;112:1593–1600. doi: 10.1002/jcb.23070. [DOI] [PubMed] [Google Scholar]
- 24.Vincan E, et al. Frizzled-7 dictates three-dimensional organization of colorectal cancer cell carcinoids. Oncogene. 2007;26:2340–2352. doi: 10.1038/sj.onc.1210026. [DOI] [PubMed] [Google Scholar]
- 25.Ueno K, et al. Frizzled-7 as a potential therapeutic target in colorectal cancer. Neoplasia. 2008;10:697–705. doi: 10.1593/neo.08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan NI, Bradstock KF, Bendall LJ. Activation of Wnt/beta-catenin pathway mediates growth and survival in B-cell progenitor acute lymphoblastic leukaemia. Br J Haematol. 2007;138:338–348. doi: 10.1111/j.1365-2141.2007.06667.x. [DOI] [PubMed] [Google Scholar]
- 27.Gupta S, et al. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70:6735–6745. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- 28.Ye X, et al. Norrin, frizzled-4, and Lrp5 signaling in endothelial cells controls a genetic program for retinal vascularization. Cell. 2009;139:285–298. doi: 10.1016/j.cell.2009.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Descamps B, et al. Frizzled 4 regulates arterial network organization through noncanonical Wnt/planar cell polarity signaling. Circ Res. 2012;110:47–58. doi: 10.1161/CIRCRESAHA.111.250936. [DOI] [PubMed] [Google Scholar]
- 30.Gilmour DF. Familial exudative vitreoretinopathy and related retinopathies. Eye (Lond) 2015;29:1–14. doi: 10.1038/eye.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le PN, McDermott JD, Jimeno A. Targeting the Wnt pathway in human cancers: Therapeutic targeting with a focus on OMP-54F28. Pharmacol Ther. 2015;146:1–11. doi: 10.1016/j.pharmthera.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurney A, et al. Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci USA. 2012;109:11717–11722. doi: 10.1073/pnas.1120068109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu D, et al. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc Natl Acad Sci USA. 2011;108:13253–13257. doi: 10.1073/pnas.1110431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emami KH, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected] Proc Natl Acad Sci USA. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madan B, Virshup DM. Targeting Wnts at the source–New mechanisms, new biomarkers, new drugs. Mol Cancer Ther. 2015;14:1087–1094. doi: 10.1158/1535-7163.MCT-14-1038. [DOI] [PubMed] [Google Scholar]
- 36.Pavlovic Z, et al. A synthetic anti-Frizzled antibody engineered for broadened specificity exhibits enhanced anti-tumor properties. MAbs. 2018;10:1157–1167. doi: 10.1080/19420862.2018.1515565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aken BL, et al. The Ensembl gene annotation system. Database (Oxford) 2016;2016:baw093. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang TH, et al. Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan. eLife. 2015;4:1–27. doi: 10.7554/eLife.06554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau T, et al. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res. 2013;73:3132–3144. doi: 10.1158/0008-5472.CAN-12-4562. [DOI] [PubMed] [Google Scholar]
- 40.Liu J, et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci USA. 2013;110:20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50–60. doi: 10.1016/j.ctrv.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiyoshi M, et al. Affinity improvement of a therapeutic antibody by structure-based computational design: Generation of electrostatic interactions in the transition state stabilizes the antibody-antigen complex. PLoS One. 2014;9:e87099. doi: 10.1371/journal.pone.0087099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Julian MC, Li L, Garde S, Wilen R, Tessier PM. Efficient affinity maturation of antibody variable domains requires co-selection of compensatory mutations to maintain thermodynamic stability. Sci Rep. 2017;7:45259. doi: 10.1038/srep45259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yanaka S, Moriwaki Y, Tsumoto K, Sugase K. Elucidation of potential sites for antibody engineering by fluctuation editing. Sci Rep. 2017;7:9597. doi: 10.1038/s41598-017-10246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diskin R, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Funck-Brentano T, et al. Porcupine inhibitors impair trabecular and cortical bone mass and strength in mice. J Endocrinol. 2018;238:13–23. doi: 10.1530/JOE-18-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.