Significance
Human papillomaviruses (HPV) uncouple proliferation from differentiation to enable virus replication in epithelial cells. HPV E7 proteins are well established to promote proliferation by binding to and inactivating retinoblastoma family proteins and other cell cycle inhibitors. However, mechanisms by which high-risk HPV oncoproteins inhibit differentiation have not been defined. This paper identifies a mechanism by which high-risk HPV E7 inhibit keratinocyte differentiation. The inhibition of differentiation requires degradation of the cellular protein PTPN14 by high-risk HPV E7, and this degradation is related to the ability of high-risk HPV oncoproteins to immortalize keratinocytes and to cause cancer.
Keywords: HPV, papillomavirus, differentiation, carcinogenesis, PTPN14
Abstract
High-risk human papillomavirus (HPV) E7 proteins enable oncogenic transformation of HPV-infected cells by inactivating host cellular proteins. High-risk but not low-risk HPV E7 target PTPN14 for proteolytic degradation, suggesting that PTPN14 degradation may be related to their oncogenic activity. HPV infects human keratinocytes but the role of PTPN14 in keratinocytes and the consequences of PTPN14 degradation are unknown. Using an HPV16 E7 variant that can inactivate retinoblastoma tumor suppressor (RB1) but cannot degrade PTPN14, we found that high-risk HPV E7-mediated PTPN14 degradation impairs keratinocyte differentiation. Deletion of PTPN14 from primary human keratinocytes decreased keratinocyte differentiation gene expression. Related to oncogenic transformation, both HPV16 E7-mediated PTPN14 degradation and PTPN14 deletion promoted keratinocyte survival following detachment from a substrate. PTPN14 degradation contributed to high-risk HPV E6/E7-mediated immortalization of primary keratinocytes and HPV+ but not HPV− cancers exhibit a gene-expression signature consistent with PTPN14 inactivation. We find that PTPN14 degradation impairs keratinocyte differentiation and propose that this contributes to high-risk HPV E7-mediated oncogenic activity independent of RB1 inactivation.
Human papillomaviruses (HPVs) are nonenveloped, double-stranded DNA viruses that infect and replicate in the stratified squamous epithelium. HPV initially infects keratinocytes in the basal, proliferative layer of the epithelium, and subsequent steps in the HPV replicative cycle—including viral genome amplification, encapsidation, and egress—are dependent on keratinocyte differentiation (1–3). However, HPV genome amplification also requires components of the cellular machinery for DNA replication that are not expressed in differentiating cells. Thus, productive HPV infection must uncouple proliferation and differentiation in the epithelium. Infection with one of the 13–15 “high-risk” HPVs causes nearly all cervical cancer, some other anogenital cancer, and an increasing proportion of HPV+ head and neck squamous cell carcinomas (HNSCC) (4–6). In total, HPV infection causes ∼5% of cancers worldwide.
The high-risk HPV E7 oncoprotein is able to immortalize human keratinocytes and the efficiency of immortalization is increased by high-risk HPV E6 (7–9). A well-characterized activity of many HPV E7 is to bind and inactivate the retinoblastoma tumor suppressor (RB1) via the LxCxE motif present in HPV E7 conserved region 2 (10–12). In addition, HPV16 E7 can direct the proteasome-mediated degradation of RB1 (13–16). RB1 inactivation releases the inhibition of E2F transcription factors (TF), thus allowing cell cycle progression and acting as a major driver of proliferation. HPV E7 also promotes proliferation by inhibiting the CDK inhibitors p21WAF1/CIP1 and p27KIP1 (17–19). In addition to promoting proliferation, transcriptional studies indicate that human cells harboring high-risk HPV genomes express lower levels of differentiation marker genes and that both high-risk HPV E6 and E7 likely contribute to this repression (20–26). However, a mechanism by which high-risk HPV E6 and E7 inhibit differentiation has not been defined.
RB1 binding by HPV E7 is necessary but insufficient for immortalization and transformation, and several observations highlight the need for other contributors to transformation. First, in multiple assays, the oncogenic activity of high-risk HPV E7 is disrupted by mutations in regions that do not include the LxCxE motif (27–31). Second, low-risk HPV E7 bind RB1 but do not have activity in transformation assays, and other HPV E7, such as HPV1 E7, bind RB1 with high affinity but do not transform (32–34). Finally, bovine papillomavirus (BPV) E7 does not bind to RB1, but in some assays it is required for BPV-mediated transformation (30, 35–37). The idea that RB1 inactivation is insufficient for transformation is additionally supported by studies in mouse models of cervical cancer (38, 39). Overall, updates to the model of transformation by HPV E6 and E7 have been suggested (40) and additional binding partners of HPV E7 have been proposed to mediate transformation independent of RB1 binding (41–43). However, not all of these interactions are conserved among the high-risk HPV E7.
The E3 ubiquitin ligase UBR4 is a conserved interactor of diverse papillomavirus E7 (44). UBR4 is required by both HPV16 E7 and BPV E7 for RB1-independent transformation but for some years the reason for this requirement was unknown (45, 46). Recently, we discovered that the cellular protein PTPN14 binds to HPV E7 proteins from diverse HPV genotypes and that high-risk HPV E7 use UBR4 to direct PTPN14 for proteasome-mediated degradation. Although low-risk HPV E7 also binds UBR4, only high-risk HPV E7 mediates PTPN14 degradation, and HPV E7 binding to PTPN14 and to UBR4 does not require interaction with RB1 (44, 47).
PTPN14 is a nonreceptor protein tyrosine phosphatase that is evolutionarily conserved as a regulator of developmental signaling from Drosophila to humans; however, phenotypes associated with PTPN14 loss vary (48–52). Hereditary variations in human PTPN14 are associated with developmental disorders, including dysregulated angiogenesis, improper lymphatic development, and improper choanal development (48, 51). Mutations in human cancer have implicated PTPN14 as a putative tumor suppressor (53–56). PTPN14 is mutated in cancers, such as colorectal cancer and basal cell carcinoma, and in both cancer types mutations occur along the length of the gene (54, 57). Several potential substrates for dephosphorylation by PTPN14 are related to cell growth control (53, 56, 58). PTPN14 also has phosphatase-independent activities, such as the ability to regulate Hippo signaling through direct interaction with YAP1 or with its upstream regulators LATS1/2 (55, 59–61). These interactions are mediated through central PPxY motifs in PTPN14.
Based upon the observations that the ability of HPV E7 to degrade PTPN14 correlates with HPV E7 oncogenic activity, that the regions of high-risk HPV E7 required for PTPN14 degradation are the same as those that confer RB1-independent transforming activity, and that PTPN14 is a putative tumor suppressor, we hypothesized that PTPN14 degradation could be required for high-risk HPV E7-mediated oncogenic transformation. The biological activities of PTPN14 in keratinocytes have not been studied, and the molecular consequences of PTPN14 degradation by high-risk HPV E7 have not been defined. Here we report that PTPN14 loss impaired the differentiation program in human keratinocytes and that HPV16 E7 could inhibit the expression of differentiation marker genes in response to a stimulus. This inhibition was dependent upon HPV16 E7’s ability to degrade PTPN14 and was retained in the absence of RB1 binding. Moreover, the ability of HPV16 E7 to degrade PTPN14 contributed to the immortalization of primary human keratinocytes by HPV16 E6 and E7. Repression of differentiation is a potentially oncogenic event, and we found that repression of keratinocyte differentiation describes the major gene-expression differences between HPV+ and HPV− HNSCC. Taken together, our results suggest that high-risk HPV E7-mediated PTPN14 degradation impairs keratinocyte differentiation. This is an RB1-independent, and potentially carcinogenic, activity of high-risk HPV E7.
Results
The HPV16 E7 E10K Variant Is Impaired in PTPN14 Degradation but Binds RB1 and Promotes E2F Target Gene Expression.
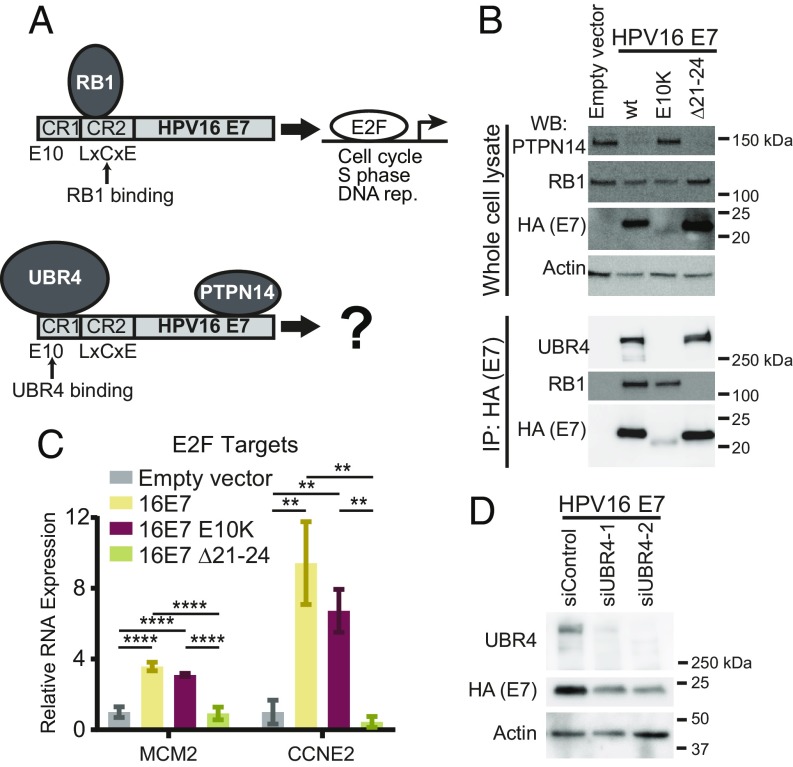
PTPN14 degradation by high-risk HPV E7 requires the E3 ubiquitin ligase UBR4, which interacts with the N terminus of HPV E7. PTPN14 binding maps broadly to the HPV E7 C terminus (Fig. 1A). The recent identification of HPV16 E7 variants from over 5,000 patient samples (62) prompted us to test whether an N-terminal variant might be impaired in the ability to degrade PTPN14. One variant, HPV16 E7 E10K (glutamic acid to lysine change at amino acid 10), is altered in the region that is required for binding to UBR4 (46, 47). To assess the biological activities of this HPV16 E7 variant, we used hTert-immortalized human foreskin keratinocytes (N/Tert-1) (63) to establish cell lines that stably express Flag and HA epitope-tagged versions of the prototypical HPV16 E7 (WT), the HPV16 E7 E10K variant, HPV16 E7 ∆21–24, or an empty vector control. The ∆21–24 deletion eliminates the LxCxE motif that is required for E7 to bind to RB1 (12). HPV16 E7 cells exhibited reduced PTPN14 protein levels and binding to RB1 was not required for this effect (Fig. 1B). However, HPV16 E7 E10K did not promote the reduction in steady-state PTPN14 protein levels. UBR4 did not coimmunoprecipitate with the HPV16 E7 E10K variant (Fig. 1B), suggesting that this variant cannot target PTPN14 for degradation because it is deficient in binding to the required E3 ubiquitin ligase.
Fig. 1.
The HPV16 E10K variant is impaired in PTPN14 degradation but binds RB1 and promotes the expression of E2F-regulated genes. (A) Schematic of protein complexes including HPV E7/RB1 and HPV E7/PTPN14/UBR4. (B) N/Tert-1 keratinocytes were transduced with control and HPV16 E7 retroviruses. Total cell lysates were analyzed by SDS/PAGE/Western blotting and probed with antibodies to PTPN14, RB1, HA, and actin (Upper). HPV16 E7-FlagHA was immunoprecipitated with anti-HA from N/Tert lysates and coimmunoprecipitation of UBR4 and RB1 was assessed by SDS/PAGE/Western blotting (Lower). (C) qRT-PCR for E2F-regulated genes in primary HFK transduced with control and HPV16 E7 retroviruses. Bar graphs display the mean ± SD of two (16E7 Δ21–24) or three (empty vector, 16E7 WT, and 16E7 E10K) independent experiments. Statistical significance was determined by ANOVA followed by multiple t tests with the Holm-Šídák family-wise error rate correction (**P < 0.01; ****P < 0.0001). (D) NTert-1 keratinocytes expressing HPV16 E7 WT were treated with control siRNAs or siRNAs targeting UBR4. Protein levels were assessed by Western blot.
HPV16 E7 E10K was comparable to HPV16 E7 WT in its ability to bind RB1 (Fig. 1B). In primary human foreskin keratinocytes (HFK) stably transduced with the same retroviral vectors, both HPV16 E7 WT and HPV16 E7 E10K could induce the expression of E2F target genes CCNE1 and MCM2 (Fig. 1C). This supported the notion that UBR4 binding and PTPN14 degradation by HPV16 E7 is independent of RB1 binding.
In all stable keratinocyte cell lines we have tested, the steady-state levels of HPV16 E7 E10K protein are lower than those of prototypical HPV16 E7 (Fig. 1B). Depletion of UBR4 was previously observed to reduce the steady-state level of HPV18 E7 (47), leading us to speculate that the ability to bind UBR4 is a determinant of HPV E7 protein levels. To test whether HPV16 E7 is expressed at a lower level when it cannot bind to UBR4, we depleted UBR4 transcripts from N/Tert-HPV16 E7 cells using small interfering RNA (siRNA). Indeed, siRNA knockdown of UBR4 resulted in reduced expression of HPV16 E7 WT (Fig. 1D). This suggested that the inability of HPV16 E7 E10K to bind to UBR4 could itself contribute to the reduced expression of HPV16 E7 E10K.
In subsequent experiments, we used HPV16 E7 E10K and HPV16 E7 ∆21–24 together to separate RB1-dependent and RB1-independent activities of HPV16 E7. We emphasize that the HPV16 E7 E10K variant retained the ability to bind RB1 and activate E2F-dependent promoters and that HPV16 E7 ∆21–24 has lost RB1-dependent activities (Fig. 1 B and C). It is possible that even though the reduced levels of HPV16 E7 E10K did not impact the RB1-dependent activity that we assessed, they could impact other activities of HPV16 E7 related to or independent of PTPN14 degradation.
HPV16 E7 Degrades PTPN14 to Inhibit Keratinocyte Differentiation.
To determine whether HPV16 E7 has effects on cellular gene expression that are dependent on its ability to degrade PTPN14, we performed an unbiased analysis of gene expression in keratinocytes expressing HPV16 E7 variants. Duplicate or triplicate primary HFK cell populations were established by transduction with retroviral vectors encoding HPV16 E7 WT, HPV16 E7 E10K, and HPV16 E7 ∆21–24 and selected with puromycin. Total RNA was isolated from independent cell populations, then polyA-selected RNA was subjected to RNA sequencing (RNA-seq). As predicted by our initial validation of the E10K variant, HPV16 E7 E10K behaved like HPV16 E7 WT with respect to the up-regulation of DNA replication genes and had a comparable effect on genes related to RB1 binding (SI Appendix, Fig. S1 and Dataset S1).
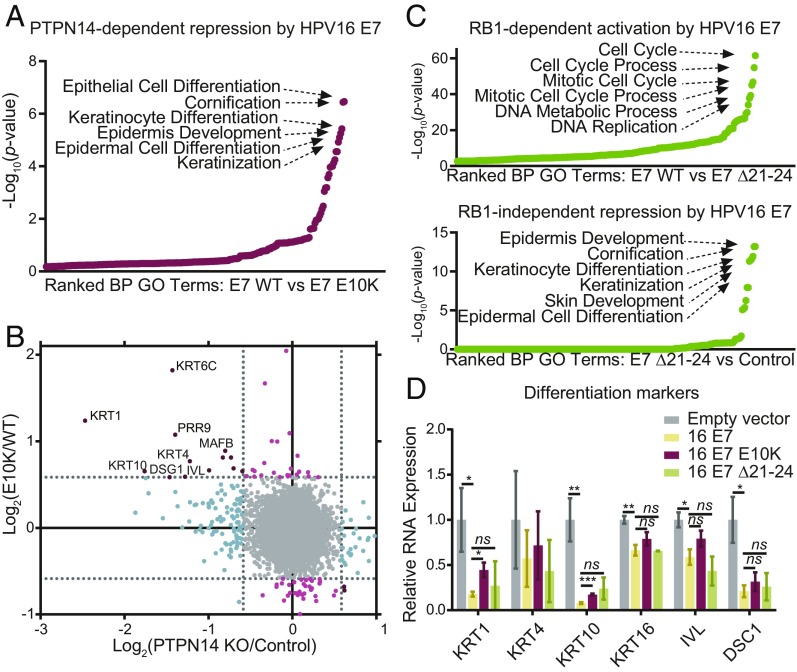
Next, we assessed the differences between HPV16 E7 WT and HPV16 E7 E10K. Seventy-five genes were differentially regulated in HPV16 E7 E10K cells compared with HPV16 E7 WT cells with fold-change ≥1.5 and adjusted P ≤ 0.05. Approximately half of these differentially regulated genes were repressed more by HPV16 E7 WT than by HPV16 E7 E10K. Gene ontology (GO) enrichment analysis showed that the genes repressed by HPV16 E7, dependent on its ability to degrade PTPN14, were described by developmental GO terms that are related to the keratinocyte differentiation program (Fig. 2A). These included epithelial cell differentiation, cornification, keratinocyte differentiation, epidermis development, epidermal cell differentiation, and keratinization. Many of the individual genes that were repressed by HPV16 E7 but not by HPV16 E7 E10K are differentiation markers (Fig. 2B, y axis). In contrast, genes that were activated by HPV16 E7, dependent on its ability to degrade PTPN14, were not significantly enriched for any GO terms (SI Appendix, Fig. S2A).
Fig. 2.
HPV16 E7-mediated degradation of PTPN14 inhibits keratinocyte differentiation. Primary HFK were transduced with retroviruses encoding HPV16 E7, HPV16 E7 E10K, HPV16 E7 Δ21–24, or an empty vector control. PolyA selected RNA was analyzed by RNA-seq. (A) GO enrichment analysis of genes with ≥1.5-fold lower expression in HPV16 E7 WT cells relative to HPV16 E7 E10K cells and P ≤ 0.05. (B) Scatter plot of log2(fold-change) in gene expression compares the gene expression changes of HPV16 E7 E10K relative to HPV16 E7 WT to those of PTPN14 KO relative to control. Colors denote whether genes are altered by PTPN14 KO only (blue), by HPV16 E7 WT more than HPV16 E7 E10K only (light red), or both (dark red). (C) Same analysis as A of (Upper) genes with ≥1.5-fold higher expression in HPV16 E7 WT than HPV16 E7 Δ21–24 cells, and (Lower) genes ≥1.5-fold lower expression in HPV16 E7 Δ21–24 cells relative to empty vector control cells and P ≤ 0.05. (D) Impacts of HPV16 E7 WT, HPV16 E7 E10K, and HPV16 E7 Δ21–24 on gene expression in primary HFK cells were validated by qRT-PCR targeting markers of differentiation. Bar graphs display the mean ± SD of two (16E7 Δ21–24) or three (empty vector, 16E7 WT, and 16E7 E10K) independent experiments. Statistical significance was determined by ANOVA followed by multiple t tests with the Holm-Šídák family-wise error rate correction (*P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant).
To test whether repression of keratinocyte differentiation was related to RB1 inactivation, we used HPV16 E7 Δ21–24 to assess the transcriptional impact of E7 in the absence of RB1 binding. As expected, cell cycle and DNA replication-related GO categories were the most significantly enriched categories among genes differentially regulated by HPV16 E7 WT versus HPV16 E7 Δ21–24 (Fig. 2C, Upper). In contrast, comparing HPV16 E7 Δ21–24 to empty vector control identified the genes that are repressed by HPV16 E7 independent of RB1 binding (Fig. 2C, Lower). GO analysis of these genes identified the same keratinocyte differentiation-related gene sets that were seen in our analysis of PTPN14 degradation-dependent effects of HPV16 E7. Furthermore, individual genes repressed by HPV16 E7 Δ21–24 relative to control (SI Appendix, Fig. S2C, y axis) are similar to those repressed by HPV16 E7 WT, dependent on its ability to degrade PTPN14. We concluded that repression of keratinocyte differentiation through the degradation of PTPN14 was independent of RB1 binding. In the absence of RB1 binding, HPV16 E7 acted mainly as a repressor but retained a modest ability to promote gene expression (SI Appendix, Fig. S2B).
To better understand the impacts of PTPN14 degradation on gene expression, we examined individual genes that were significantly lower in HPV16 E7 WT cells than in HPV16 E7 E10K cells. Many of the genes that are repressed by HPV16 E7 WT and HPV16 E7 Δ21–24, but not by HPV16 E7 E10K, are described by epidermis development and more specific GO terms (SI Appendix, Fig. S2D and Dataset S2). Although certain genes were not repressed by either HPV16 E7 E10K or HPV16 E7 Δ21–24, these genes were largely related to other biological processes.
To validate the results obtained from RNA-seq, we used qRT-PCR to confirm the altered expression of several genes related to keratinocyte differentiation in our cell lines. Markers of keratinocyte differentiation, such as keratin 1 (KRT1), keratin 4 (KRT4), keratin 10 (KRT10), keratin 16 (KRT16), involucrin (IVL), and desmocollin 1 (DSC1) were repressed by HPV16 E7 by 1.5- to 12-fold (Fig. 2D). KRT1, KRT4, KRT10, and KRT16 are cytokeratins associated with the suprabasal layers of differentiating keratinocytes. IVL constitutes a major component of the cornified envelope and is expressed at high levels in the upper layers of the epidermis. DSC1 is a component of desmosome complexes associated with keratinization and is expressed at higher levels in the upper spinous layer and granular layer of the epidermis. Comparison of both HPV16 E7 WT and Δ21–24 to the HPV16 E7 E10K variant indicated that the ability of HPV16 E7 to repress expression of these genes was at least partially dependent on its ability to target PTPN14 for degradation, but was not dependent on RB1 binding.
The Ability of HPV E7 to Degrade PTPN14 Correlates with Its Ability to Inhibit the Cellular Response to a Stimulus of Differentiation.
Next, we wanted to determine whether the ability of HPV E7 to alter differentiation-related gene expression in unstimulated cells correlated with changes following a differentiation stimulus. In vivo, detachment from the basement membrane stimulates keratinocyte differentiation, an effect that can be mimicked by growth of cultured cells in suspension (64–66). HPV E7 has been previously shown to protect against cell death following detachment in a UBR4-dependent manner (46, 67). Consequently, we first chose to induce differentiation using detachment as a stimulus. In these experiments we used N/Tert-1 cells engineered to stably express HPV16 E7 WT, HPV16 E7 E10K, HPV6 E7, or an empty vector control. HPV6 is a low-risk HPV encoding an E7 that, like HPV16 E7 E10K, has an intact C-terminal PTPN14 binding domain but does not direct PTPN14 for proteasome-mediated degradation.
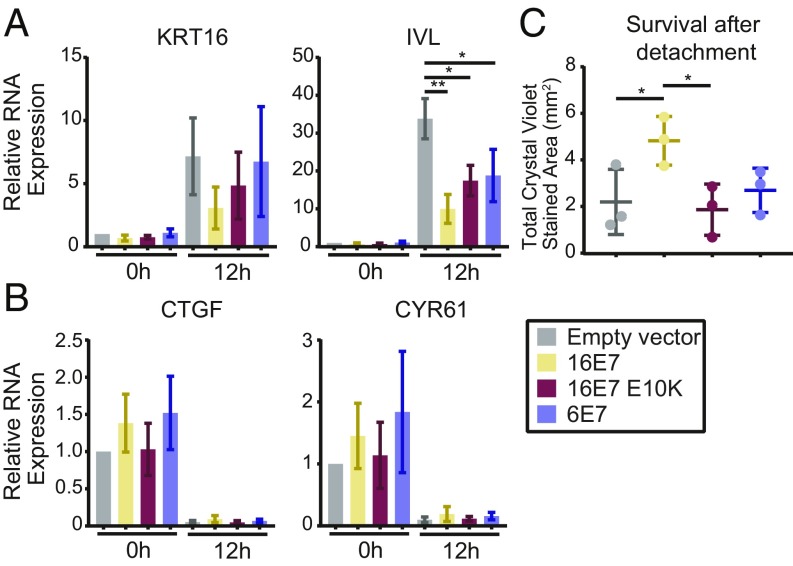
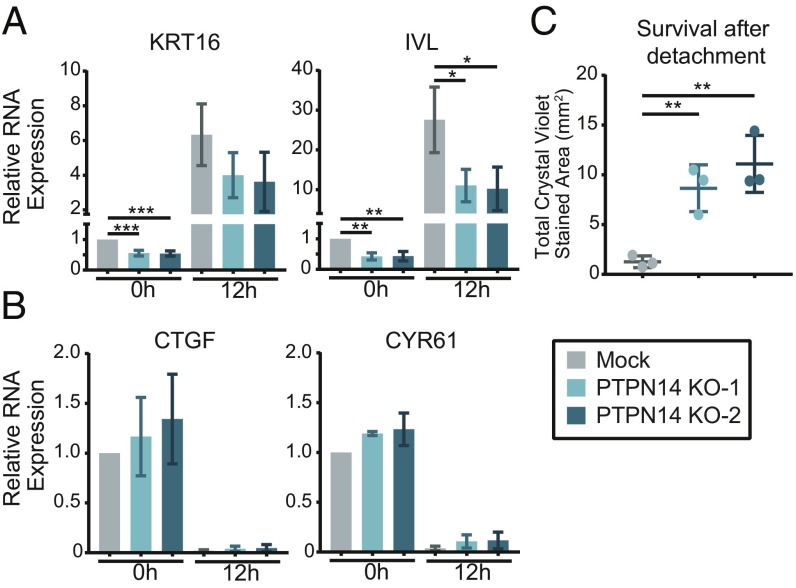
N/Tert-1 cells were harvested directly from adherent culture or subjected to growth in suspension for 12 h to induce differentiation. KRT16 and IVL RNAs were analyzed by qRT-PCR and were induced by detachment in all of the cell lines tested. Detached empty vector cells expressed 7- to 30-fold more of these transcripts compared with adherent cells, and each version of E7 limited the induction of KRT16 and IVL. KRT16 and IVL expression was 2.3- or 3.4-fold lower in N/Tert-HPV16 E7 WT cells compared with the empty vector control and repression of IVL was statistically significant (Fig. 3A). The statistical significance of some other comparisons was limited by the fact that there was a wide range of induction of the differentiation markers following detachment. However, the trend was highly reproducible: in three replicate experiments HPV16 E7 WT always repressed differentiation marker gene expression more than HPV16 E7 E10K and HPV6 E7. This result indicated that PTPN14 degradation is required for maximal repression of detachment-induced differentiation by HPV16 E7.
Fig. 3.
The ability of HPV E7 to degrade PTPN14 correlates with its ability to inhibit differentiation and promote survival upon detachment. N/Tert-1 stably transduced with retroviruses encoding HPV16 E7, HPV16 E7 E10K, HPV6 E7, or an empty vector control were subjected to growth in suspension for 12 h and assayed for markers of differentiation, YAP/TEAD targets, and survival after detachment. (A and B) Gene-expression changes induced by suspension were assayed by qRT-PCR targeting markers of differentiation: KRT16 and IVL (A), and YAP/TEAD targets: CTGF and CYR61 (B). mRNA expression was calculated relative to GAPDH. Bar graphs display the mean ± SD of three independent experiments. (C) Survival after detachment was assayed by replating 1,000 cells from suspension and measuring the surface area covered after 5 d of growth by Crystal violet staining. Three independent experiments are displayed along with mean ± SD. Statistical significance was determined by ANOVA followed by multiple t tests with the Holm-Šídák family-wise error rate correction (*P < 0.05; **P < 0.01).
Several explanations could account for the observation that HPV E7 that does not degrade PTPN14 still partially represses differentiation. Each of the HPV E7 tested here bind and inactivate RB1 and it is possible that some inhibition of differentiation is due to the increased proliferation resulting from RB1 inactivation. Another, not mutually exclusive explanation could be that PTPN14 binding alone is enough to result in some inhibition of differentiation. Our data support this idea, because HPV16 E7 E10K and HPV6 E7 both contain the C-terminal domain that binds PTPN14 and they repressed differentiation to similar levels.
In addition to stimulating differentiation, growth in suspension activates the Hippo signaling pathway (68, 69), which represses the transcription of the well-characterized YAP/TEAD targets CTGF and CYR61. PTPN14 knockdown in MCF10A cells has been shown to induce the transcription of CTGF and CYR61 (59). Because PTPN14 has been characterized as a negative regulator of YAP1 and shown to regulate CTGF and CYR61 in other cell types (55, 59–61), we measured these transcripts to determine whether HPV E7 differentially impacts their expression. Compared with vector controls, none of the HPV E7 cell lines exhibited altered expression of these YAP1/TEAD targets before or after detachment (Fig. 3B).
To further assess cell viability in the detachment experiment, 1,000 cells were taken from suspension culture, replated in coated tissue culture plates, and allowed to grow for 5 d. HPV16 E7 protected against cell death following detachment in a manner that was dependent on the ability of E7 to target PTPN14 for degradation (Fig. 3C). This is consistent with previous reports demonstrating that HPV16 E7 and BPV E7 require UBR4 to protect cells against cell death triggered following detachment from a substrate (46, 67).
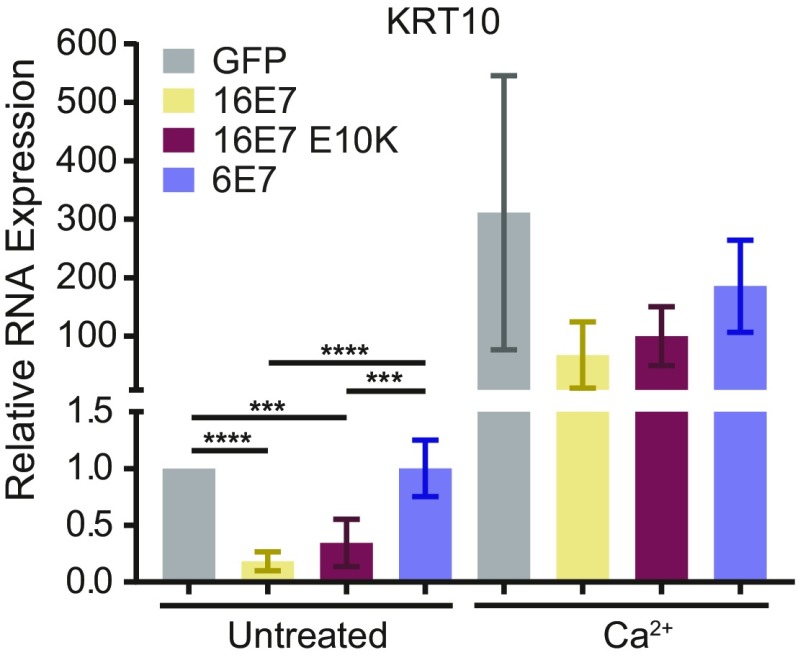
Finally, we used a second stimulus of differentiation to confirm that HPV16 E7 inhibits differentiation dependent on PTPN14 degradation. Treatment of keratinocytes with high calcium induces markers of epithelial differentiation. We treated N/Tert-HPV E7 cells with 1.5-mM calcium for 48 h and measured KRT10 expression by qRT-PCR. In undifferentiated cells, HPV16 E7 WT repressed KRT10 expression to a greater degree than either HPV16 E7 E10K or HPV6 E7 (Fig. 4). Upon calcium treatment, the variability in KRT10 induction was high, but in several independent experiments, HPV16 E7 was maximally able to repress KRT10 in differentiated cells. HPV6 E7 was least able to repress KRT10 and HPV16 E7 E10K exhibited an intermediate effect. Taken together, these data further support the idea that high-risk HPV E7 inhibits keratinocyte differentiation dependent on PTPN14 degradation.
Fig. 4.
The ability of HPV E7 to degrade PTPN14 correlates with its ability to inhibit differentiation induced by Ca2+. N/Tert-1 stably transduced with retroviruses encoding HPV16 E7, HPV16 E7 E10K, HPV6 E7, or an empty vector control were treated with 1.5 mM Ca2+ and assayed for KRT10 as a marker of differentiation. Bar graph displays the mean ± SD of four independent experiments. Statistical significance was determined by ANOVA followed by multiple t tests with the Holm-Šídák family-wise error rate correction (***P < 0.001; ****P < 0.0001).
PTPN14 Knockout Limits Differentiation Gene Expression in Primary Human Keratinocytes.
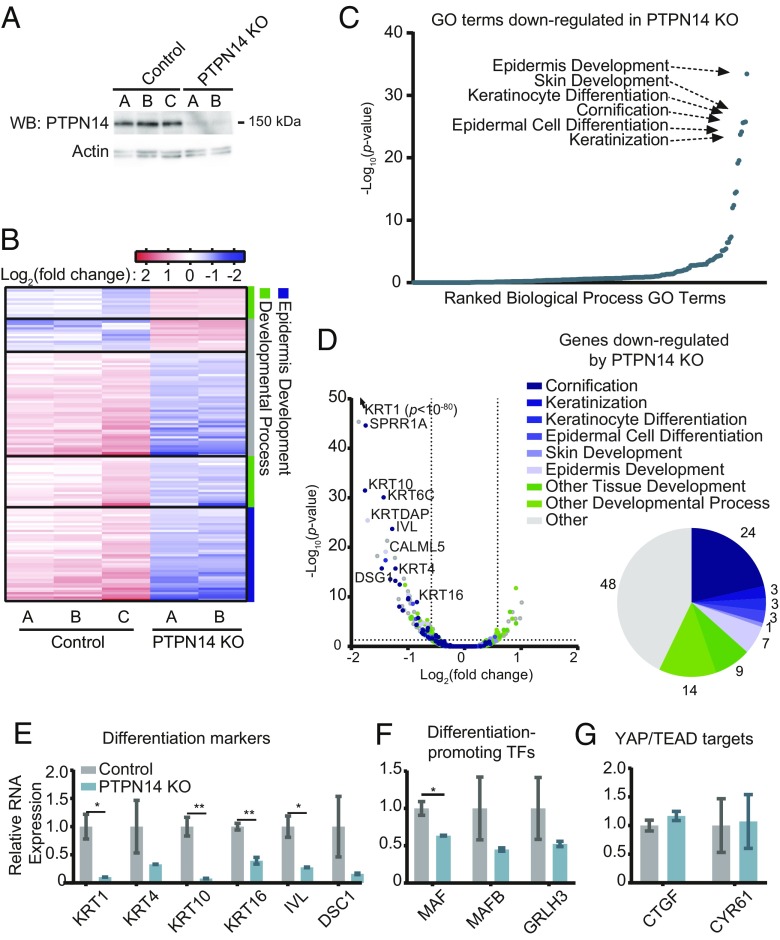
To test what cellular processes are affected when PTPN14 levels are reduced in human keratinocytes, we performed an unbiased analysis of gene expression in the presence and absence of PTPN14. Primary HFK were transduced with lentiviral vectors encoding SpCas9 plus a single-guide RNA (sgRNA) targeting PTPN14 (sgPTPN14-3) or a nontargeting control sgRNA (sgNT-2), then selected with puromycin to generate control (HFK-control) and PTPN14-deleted [HFK-PTPN14 knockout (KO)] cell lines (Fig. 5A). It should be noted that PTPN14 KO cells exhibit a nearly complete loss of PTPN14 protein. In contrast, the ability of high-risk HPV E7 to degrade PTPN14 is conserved, but can result in residual PTPN14 protein levels that vary by HPV genotype (47). Total RNA was isolated from two or three independent isolates of HFK-control and HFK-PTPN14 KO, then polyA-selected RNA was subjected to RNA-seq. In cells that did not express PTPN14, 141 genes were differentially regulated with fold-change ≥1.5 and adjusted P ≤ 0.05. Of these, 29 genes were up-regulated and 112 were down-regulated in the absence of PTPN14 (Fig. 5B and Dataset S3). Thus, PTPN14 appeared to act largely to promote, rather than to repress, gene expression. As in the analysis of the HPV16 E7 variants, keratinocyte differentiation-related GO terms were down-regulated in PTPN14 KO cells (Figs. 2 A and C and 5C). More than half of the down-regulated genes were in epidermis development-related or other developmental process-related GO categories (Fig. 5D). There was not a corresponding enrichment in differentiation-related GO terms among the up-regulated genes; however, one GO category, inflammatory response, was significantly enriched in this analysis (SI Appendix, Fig. S3).
Fig. 5.
PTPN14 depletion impairs differentiation-related gene expression in primary human keratinocytes. Primary HFK were transduced with LentiCRISPRv2 lentiviral vectors encoding SpCas9 and nontargeting or PTPN14-directed sgRNAs and analyzed for changes in gene expression. (A) Cell lysates were subjected to SDS/PAGE/Western analysis and probed with anti-PTPN14 and antiactin antibodies. (B) PolyA selected RNA was analyzed by RNA-seq. Genes differentially expressed by ≥1.5-fold with P value ≤ 0.05 are displayed in the heat map. Color coding on the right side denotes whether genes are related to epidermis development (blue), other developmental processes (green), or neither (gray). (C) GO enrichment analysis of genes down-regulated in HFK-PTPN14 KO compared with HFK-control. (D) Volcano plot of gene-expression changes in HFK-control vs. HFK-PTPN14 KO. Dots colored by GO terms. Pie chart displays the fraction of genes down-regulated in the absence of PTPN14 that fall into enriched GO Terms. (E–G) Transcript abundance for selected genes in HFK-control and HFK-PTPN14 KO was measured by qRT-PCR detecting differentiation markers (E), differentiation promoting TFs (F), and YAP/TEAD targets (G). Bar graphs display the mean ± SD of two or three independent experiments. Statistical significance was determined by Welch’s t tests (*P < 0.05; **P < 0.01).
We hypothesized that individual genes might be similarly regulated by PTPN14 KO and HPV E7-mediated PTPN14 degradation. Indeed, the genes that were both down-regulated by PTPN14 loss and down-regulated by HPV16 E7 WT in a PTPN14 degradation-dependent manner are involved in keratinocyte differentiation (Fig. 2B). Furthermore, gene-expression changes induced by HPV16 E7 Δ21–24 are positively correlated with those resulting from PTPN14 KO (SI Appendix, Fig. S2C). Taken together, gene-expression analysis of HFK-PTPN14 KO and cells expressing HPV16 E7 variants is consistent with degradation of PTPN14 by HPV16 E7 acting to inhibit keratinocyte differentiation. Our data suggest that PTPN14 degradation mediates the predominant RB-independent effect of HPV16 E7 on gene expression.
We selected a subset of genes for validation by qRT-PCR. In agreement with the RNA-seq results, markers of keratinocyte differentiation, such as KRT1, KRT4, KRT10, KRT16, IVL, and DSC1, were expressed at 3- to 12-fold lower levels in the absence of PTPN14 (Fig. 5E). TFs such as MAF, MAFB, and GRLH3 that are transcriptionally regulated during progression of the keratinocyte differentiation program (70–74), exhibited lower expression in the absence of PTPN14 (Fig. 5F). Unlike the published effects in other cell types, we found that PTPN14 loss did not impact the expression of the well-characterized YAP/TEAD targets CTGF and CYR61 (Fig. 5G). These data support the idea that PTPN14 loss impairs the regulation of keratinocyte differentiation but does not affect expression of canonical Hippo-regulated genes in primary HFK. They also suggest that PTPN14 KO itself phenocopied much of the gene repression that was enabled by HPV16 E7 ∆21–24 (does not bind RB1). Even though the expression of the HPV16 E7 E10K variant protein was low, taken together these data support the hypothesis that PTPN14 loss represses keratinocyte differentiation and that PTPN14 inactivation is a major RB1-independent activity of HPV16 E7.
PTPN14 Contributes to the Up-Regulation of Differentiation Markers upon Detachment.
Having determined that PTPN14 loss reduces the basal expression of keratinocyte differentiation-related genes, we next tested whether PTPN14 loss alters the cellular response to a differentiation stimulus. We used CRISPR-Cas9 gene editing in N/Tert-1 cells to engineer control (N/Tert-mock) or PTPN14-deleted (N/Tert-PTPN14 KO) pooled stable cell lines. Again, we stimulated these cells to differentiate through growth in low adherence plates for 12 h. Consistent with the effect in primary cells, PTPN14 KO reduced the expression of KRT16 and IVL in adherent cells. We further found that PTPN14 loss also impaired the expression of KRT16 and IVL upon the induction of differentiation (Fig. 6A), mirroring the results observed in our N/Tert-E7 cells. As we observed in the N/Tert-E7 cells as well as the primary HFK-PTPN14 KO cells, N/Tert-PTPN14 KO cells did not express significantly more CTGF or CYR61 than mock controls in either the adherent condition or following growth in suspension (Fig. 6B).
Fig. 6.
PTPN14 loss reduces the expression of differentiation markers after detachment. Adherent N/Tert-mock and -PTPN14 KO cells were detached by trypsinization and replated in ultralow adherence plates before harvesting 12-h postdetachment, then assayed for markers of differentiation, YAP/TEAD targets, and survival after detachment. (A and B) Gene-expression changes induced by suspension were assayed by qRT-PCR targeting markers of differentiation: KRT16 and IVL (A), and YAP/TEAD targets: CTGF and CYR61 (B). mRNA expression was calculated relative to GAPDH. Bar graphs display the mean ± SD of three independent experiments. (C) Survival after detachment was assessed by replating 1,000 cells from suspension, allowing cells to grow for 5 d, and measuring total viable cell area by Crystal violet staining. Three independent experiments are displayed along with mean ± SD. Statistical significance was determined by ANOVA followed by multiple t tests with the Holm-Šídák family-wise error rate correction (*P < 0.05; **P < 0.01; ***P < 0.001).
Finally, we used cell growth after replating as a measure of viability after detachment. The N/Tert-PTPN14 KO cells exhibited improved survival and colony formation after detachment compared with control cells (Fig. 6C). This is consistent with the result that HPV E7 expression improved survival after suspension in a PTPN14 degradation-dependent manner (Fig. 3C) and indicates that loss of PTPN14 is sufficient to improve survival of keratinocytes after detachment.
PTPN14 Degradation Contributes to High-Risk HPV E6/E7 Immortalization of Primary Human Keratinocytes.
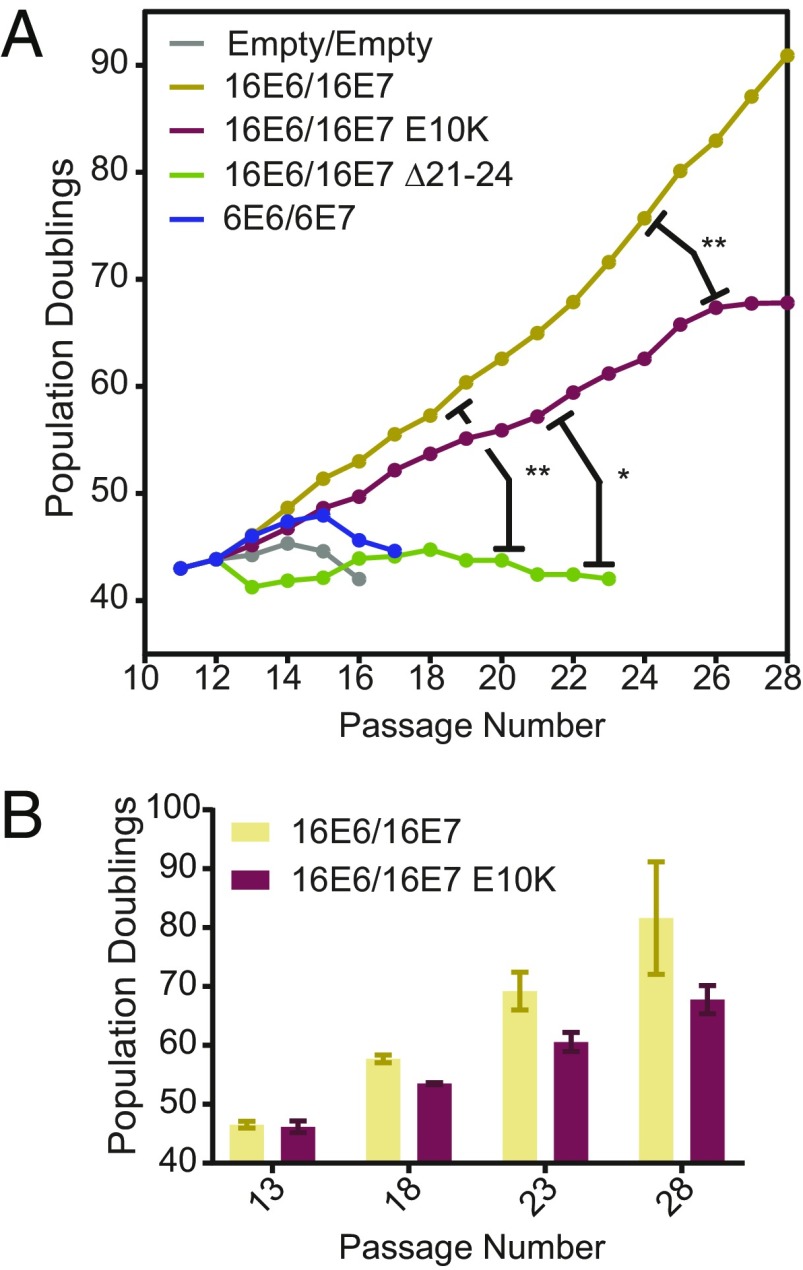
Coexpression of HPV16 E6 and E7 can efficiently immortalize primary keratinocytes in cell culture. To determine whether PTPN14 degradation is required for immortalization by HPV16 oncoproteins, primary HFK were transduced with pairs of HPV E6/E7-encoding retroviruses, selected with puromycin and blasticidin, and monitored for cell growth over the next 17 passages, equivalent to 75 d for WT HPV16 E6/E7 cells (Fig. 7).
Fig. 7.
PTPN14 degradation contributes to high-risk HPV E6/E7 immortalization of primary human keratinocytes. Primary HFK cells were transduced with pairs of retroviruses encoding various HPV E6 and E7 and passaged for up to 75 d. (A) Growth curves from representative immortalization experiment. Population doublings were calculated based upon the number of cells harvested at each passage. Statistical significance was determined from three independent experiments by repeated measures two-way ANOVA. Displayed P values represent the column (cell line) factor (*P < 0.05; **P < 0.01). (B) Bar chart shows mean ± SD of selected passages from growth curves in A.
Primary HFK transduced with HPV6 E6/E7 or with empty vector controls rapidly senesced, while cells transduced with HPV16 E6/E7 were immortalized in three of three replicate experiments. The cells transduced with HPV16 E6/E7 ∆21–24 retroviruses were severely growth-impaired and were not immortalized but exhibited a small degree of lifespan extension, perhaps due to sporadic epigenetic inactivation of RB1. Cells transduced with HPV16 E6/E7 E10K-expressing vectors retained some proliferative capacity, but their growth was reproducibly impaired compared with that of HPV16 E6/E7 WT cells. We hypothesize that these cells are not fully immortalized and that both RB1 inactivation and PTPN14 degradation are required for immortalization of primary HFK by HPV E6 and E7.
Keratinocyte Differentiation Gene Expression Describes the Major Differences Between HPV+ and HPV− HNSCC.
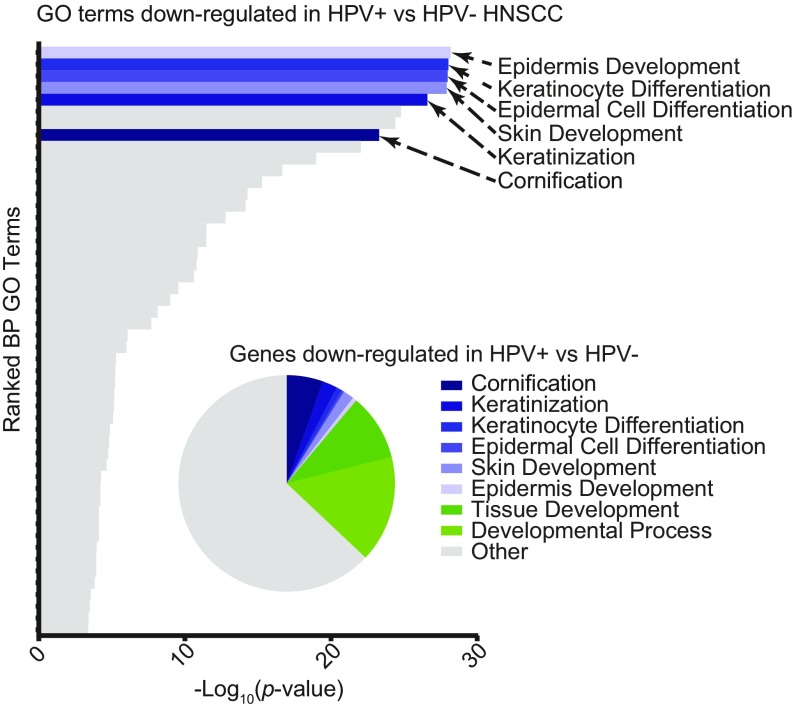
The changes in differentiation-related gene expression in HPV E7-expressing cell lines appeared to be dependent on the ability of HPV E7 to degrade PTPN14 and to reflect the same changes that result from PTPN14 loss in primary HFK. The ability of HPV E7 to degrade PTPN14 also correlates with its ability to immortalize primary HFK. We wished to determine whether E7- or PTPN14-dependent changes in differentiation-related gene expression are reflected in HPV-associated cancers. Using RNA-seq data from the Cancer Genome Atlas (TCGA), we examined gene-expression signatures in 508 HNSCC samples, 60 of which are HPV+ and 448 HPV− (75–77). Genes that were differentially expressed by threefold or more in HPV+ vs. HPV− samples were selected for further analysis.
Strikingly, the most enriched GO terms among genes down-regulated in HPV+ cancers relative to HPV− cancers were epidermis development, keratinocyte differentiation, and epidermal cell differentiation (Fig. 8). As in the PTPN14 KO cells and in the presence of HPV16 E7, down-regulated genes reflected a keratinocyte differentiation signature. Furthermore, many of the other highly enriched GO terms were related to more general developmental processes. In total, epidermis development and other developmental processes accounted for about one-third of the differentially regulated genes in HPV+ vs. HPV− HNSCC. In contrast, GO enrichment identified no clear gene sets enriched among genes up-regulated in HPV+ compared with HPV− HNSCC (SI Appendix, Fig. S4). The down-regulation of differentiation-related genes in HPV+ relative to HPV− cancers is consistent with the changes in gene expression induced by the high-risk HPV E7-mediated degradation of PTPN14.
Fig. 8.
Keratinocyte differentiation gene signature describes the major differences between HPV+ and HPV− HNSCC. Data from HNSCC samples on the TCGA database were determined to be HPV+ or HPV− and analyzed for differences in gene expression. Bar chart portrays the ranked −log10(P values) of enriched GO terms among genes down-regulated in HPV+ HNSCC. Pie chart displays fraction of total down-regulated genes that fall into selected GO categories.
Discussion
Our previous finding that PTPN14 is targeted for degradation by high-risk HPV E7 but not by low-risk HPV E7 suggested that PTPN14 loss might be related to the biology of the high-risk HPV (47). PTPN14 is a candidate tumor suppressor based on the observation that it is mutated in some cancers (54, 57, 78–81). The targeted degradation of PTPN14 by high-risk HPV E7 requires the E3 ubiquitin ligase UBR4 and the interaction of UBR4 with papillomavirus E7 is required for E7 to transform cells (45, 46). Thus, PTPN14 degradation could be analogous to the well-established ability of high-risk HPV E6 but not low-risk HPV E6 to target p53 for proteasome-mediated degradation using the E3 ubiquitin ligase UBE3A (82, 83). However, neither our previous studies nor those from another group provided insight regarding the downstream effects of HPV E7-mediated PTPN14 degradation in human keratinocytes (47, 84).
PTPN14 has been implicated as a negative regulator of YAP1, a transcriptional coactivator that is regulated by the Hippo signaling pathway (59, 61, 85). An appealing hypothesis was that HPV E7-mediated PTPN14 degradation would activate YAP1 and promote the expression of proproliferative YAP target genes, such as CTGF and CYR61. However, we have not identified any cell type in which high-risk HPV E7 expression causes an increase in CTGF or CYR61 RNA. In addition, we found that depletion or KO of PTPN14 in human keratinocytes did not cause CTGF or CYR61 up-regulation (Figs. 5 and 6). However, our cell-detachment experiments suggested that these genes are indeed regulated by Hippo signaling in keratinocytes (Figs. 3 and 6). Thus, our results suggest that PTPN14 may not regulate canonical Hippo-YAP signaling in keratinocytes.
In the absence of support for this initial hypothesis, we took an unbiased approach to determine the effect of high-risk HPV E7-mediated PTPN14 degradation in keratinocytes. By using an HPV16 E7 variant that cannot degrade PTPN14 (Figs. 1 and 2) and by directly testing the effect of PTPN14 KO in primary HFK (Fig. 5), we determined that PTPN14 loss results in a down-regulation of several markers of epidermal cell differentiation. Consistent with this idea, PTPN14 appears to be a target of regulation by p53 in mouse cells but is likely a p63 target in human cells (79, 86–88). p63 is a master regulator of epidermal development (89). The link between PTPN14 and differentiation directly connected PTPN14 degradation to HPV biology.
To further test how high-risk HPV E7-mediated PTPN14 degradation affects processes related to epidermal cell differentiation, we used a keratinocyte detachment and replating assay (Fig. 3). Our studies indicated that high-risk HPV E7 inhibit the expression of differentiation markers following cell detachment in a PTPN14 degradation-dependent manner. The same inhibition of differentiation markers occurred in detached PTPN14 KO keratinocytes (Fig. 6). Anoikis is cell death triggered by detachment from a substrate, and the ability to survive anoikis and proliferate in the absence of contact with the basement membrane is a hallmark of cancer cells. The HPV E7 proteins that inhibited differentiation marker gene expression promoted cell survival following detachment and this correlated with the ability to degrade PTPN14 (Fig. 3).
In support of the notion that HPV E7-mediated PTPN14 degradation contributes to oncogenic transformation, our subsequent experiments indicated that PTPN14 degradation by high-risk HPV contributes to keratinocyte immortalization. Primary keratinocytes were fully immortalized by HPV16 E6/E7 but not by HPV16 E6/E7 E10K (Fig. 7). In transcriptional profiles of human head and neck cancer samples, changes in gene expression consistent with PTPN14 loss were reflected in HPV+ but not HPV− cancers (Fig. 8). Strikingly, we found that the GO terms related to keratinocyte differentiation and epidermis development described both the PTPN14-dependent differential gene expression in primary cells and the most significant differences between HPV+ and HPV− head and neck carcinomas. We also observed that in previously published data, these same GO terms were down-regulated by the coexpression of HPV16 E6 and E7 in primary HFKs (90). These findings are consistent with the effect of PTPN14 loss being maintained throughout HPV-mediated carcinogenesis. Notably, the HPV16 E7 E10K variant that cannot bind UBR4 or degrade PTPN14 (Fig. 1) was identified in a CIN3 lesion (62). We hypothesize that other patient-specific genetic or epigenetic changes may have compensated for the inability of HPV16 E7 to degrade PTPN14 in this lesion; however, only viral sequence information was collected from the patient samples in this study. Alternatively, this mutation may have impaired the progression of this lesion from CIN3 to a malignant cancer.
Some previous studies suggested that differentiation inhibition by HPV E7 could be RB/E2F-dependent. E2F TFs have been shown to limit keratinocyte differentiation (91) and both the RB1-binding domain and the N terminus of HPV16 E7 contributed to HPV-mediated differentiation inhibition in one study (20). However, our unbiased transcriptional analysis clearly showed that much of the HPV16 E7-mediated repression of differentiation is independent of RB1 binding. Here we have focused on the genes repressed by HPV16 E7, which includes many markers of keratinocyte differentiation that were also down-regulated upon PTPN14 KO (Figs. 2 and 5). Using the HPV16 E7 ∆21–24 mutant, we have found that RB1 binding is not required for the repression of most of these genes (Fig. 2). RB1 binding did allow for the repression of some differentiation genes by HPV16 E7 (SI Appendix, Fig. S2D) and certain genes up-regulated by HPV16 E7 but not by HPV16 E7 ∆21–24 were related to keratinocyte differentiation. Nonetheless, differentiation-related GO terms comprised a minor part of the HPV16 E7 gene induction signature, whereas they were the most significant terms repressed by HPV16 E7 in the presence or absence of RB1 binding.
All HPVs, not only the high-risk types, likely manipulate differentiation to replicate. PTPN14 is a conserved interactor of HPV E7, suggesting an evolutionary pressure to maintain this interaction regardless of the ability to direct it for proteasomal degradation. Future studies could address whether low-risk HPV E7 impact keratinocyte differentiation via their ability to bind to (but not degrade) PTPN14. Our study supports this hypothesis as the HPV16 E7 E10K variant and the low-risk HPV6 E7 proteins both inhibited keratinocyte differentiation to similar levels after stimulation and both contain a C-terminal domain that binds PTPN14 (Figs. 3 and 4). More broadly, genus β HPV E6 proteins bind MAML1 to inhibit Notch signaling, resulting in impaired keratinocyte differentiation and a cellular environment more conducive to virus replication (92–94). The genus α HPVs, which include all of the high-risk and low-risk HPV discussed here, do not engage MAML1 in the same way. It is interesting to speculate that all HPV promote proliferation but that pathogenesis is related to their ability to further impair differentiation: via the HPV E7-PTPN14 interaction in the case of mucosal, genus α HPV and via the E6-MAML interaction in the case of cutaneous, genus β HPV.
Our results suggest that PTPN14 does not regulate canonical Hippo-YAP signaling. Instead, it is possible that PTPN14 regulates differentiation by modulating other cellular signaling pathways that have identified roles in epithelial development, such as transforming growth factor β (TGFβ) or protein kinase C (PKC) signaling. Both TGFβ and PKC have been proposed to be regulated by PTPN14 in other systems (50, 53). It also remains possible that PTPN14 regulates YAP1 independently of Hippo signaling.
The binding and degradation of RB1 is a major component of high-risk HPV E7-mediated transformation. However, many observations have suggested that there must be RB1-independent contributions to E7-mediated transformation. We propose that PTPN14 degradation is a critical contributor to the oncogenic activity of high-risk HPV E7 and that PTPN14 inactivation impairs keratinocyte differentiation. PTPN14 degradation is conserved across the high-risk HPV E7 that we have tested so far, and it is not dependent on the ability of E7 to bind or inactivate RB1. We have not yet established whether PTPN14 binding is sufficient to impair differentiation or whether PTPN14 degradation has additional oncogenic effects. In either case, the identification of a mechanism by which HPV E7 controls differentiation is significant. The potential of differentiation therapy has been validated by the highly successful use of all-trans retinoic acid to treat acute promyelocytic leukemia (95). It is tantalizing to speculate that inhibiting PTPN14 inactivation could similarly restore the cellular differentiation program in HPV+ cancer cells and have therapeutic potential. Future work could aim to elucidate the mechanism of PTPN14 signal transduction in keratinocytes and further characterize the role of PTPN14 degradation in HPV replication and in HPV-associated cancers.
Materials and Methods
Cells.
Primary HFK (G5-Ep isolate, gift of James Rheinwald, Harvard Medical School, Boston) and hTert-immortalized HFK (63) were cultured as previously described (96). PTPN14 KO or nontargeting control primary HFK were established by transduction with LentiCRISPR v2 vectors (Dataset S4) followed by puromycin selection. N/Tert-Cas9 cells were generated by transduction with pXPR_111 (Addgene #59702) and blasticidin selection. PTPN14 KO or mock control N/Tert cell lines were established by transfection of N/Tert-Cas9 with sgRNA targeting PTPN14 (Synthego) (Dataset S4). Retroviruses and lentiviruses were generated as previously described (96).
siRNA Transfection.
NTert-1 cells expressing HPV16 E7 WT were transfected with a control siRNA (siControl) or siRNAs targeting UBR4 (Dharmacon) (Dataset S4) using Dharmafect 1 transfection reagent (Dharmacon). siRNA treated cells were harvested for Western blot 48-h posttransfection.
Keratinocyte Differentiation Assays.
To assess keratinocyte survival and changes in gene expression following detachment from a substrate, N/Tert-mock or N/Tert-sgPTPN14, or N/Tert-1 cells engineered to stably express HPV16 E7 WT, HPV16 E7 E10K, HPV6 E7, or an empty vector control were harvested by trypsinization and replated in ultralow attachment plates (Sigma-Aldrich CLS3471). After 0 or 12 h of culture in suspension, cells were harvested for RNA analysis or 1,000 cells were replated in standard six-well tissue culture plates. Replated cells were stained with Crystal violet 5-d postreplating.
To assess keratinocyte differentiation induced by calcium, NTert-1 cells engineered to stably express HPV16 E7 WT, HPV16 E7 E10K, HPV6 E7, or an empty vector control were treated with 1.5 mM CaCl2 in KBM media (Lonza) without supplements. At 48-h posttreatment, cells were harvested for RNA analysis.
Keratinocyte Immortalization Assay.
To assess the ability of HPV16 E7 variants to support keratinocyte immortalization, primary HFK were transduced with one MSCV-based retroviral vector encoding conferring puromycin resistance (HPV16 or HPV6 E6 or an empty vector control) and one retroviral vector conferring blasticidin resistance (HPV16 or HPV6 E7 or an empty vector control) (Dataset S4). Cells were selected in puromycin and blasticidin and passaged for ∼110 d. Population doublings were calculated based upon the number of cells collected and replated at each passage.
Plasmids and Cloning.
LentiCRISPR v2 vectors were cloned according to standard protocols using sgRNA sequences as contained in the Broad Institute Brunello library (97). The E10K mutation was introduced by site-directed mutagenesis into pDONR-Kozak-16E7 and recombined into MSCV-IP N-FlagHA GAW, as previously described (96). Additional HPV E6 and E7 retroviral vectors used in the study are listed in Dataset S4.
Western Blotting.
Western blots were performed as previously described (47) using Mini-PROTEAN or Criterion (Bio-Rad) SDS/PAGE gels and transfered to PVDF. Membranes were blocked in 5% nonfat dried milk in TBS-T [Tris buffered saline (pH 7.4) with 0.05% Tween-20], then incubated with primary antibodies as follows: RB1 (Calbiochem/EMD), actin (Millipore), PTPN14 (R&D Systems), and UBR4 [gift of Yoshihiro Nakatani, Dana-Farber Cancer Institute, Boston (98)]. Membranes were washed in TBS-T and incubated with horseradish peroxidase-coupled anti-mouse or anti-rabbit antibodies and detected using an Amersham 600 chemiluminescent imaging system. HA-tagged proteins were detected using an HA antibody conjugated to HRP (Roche) and visualized in the same way. For anti-HA immunoprecipitations, HA-tagged proteins were immunoprecipitated and processed for Western blot, as previously described (47).
RNA-Seq.
Total RNA was isolated from two to three independent isolates of HFK-control, HFK-PTPN14 KO, HFK-empty vector control, or HFK E7 cells using the RNeasy mini kit (Qiagen). PolyA selection, reverse transcription, library construction, sequencing, and initial analysis were performed by Novogene. Differentially expressed genes were selected based on a 1.5-fold change and adjusted P ≤ 0.05 cut-off and were analyzed for enriched biological processes using the GO enrichment analysis tool of the PANTHER classification system (99). All GO terms in enrichment analyses are displayed in rank order by adjusted P value. RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus with accession number GSE121906 (100).
qRT-PCR.
Total RNA was isolated from N/Tert cells using the NucleoSpin RNA extraction kit (Macherey-Nagel). RNA was then reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). cDNAs were assayed by qPCR using Fast SYBR Green Master Mix (Applied Biosystems) using a QuantStudio 3 96-Well, 0.2-mL Block instrument (ThermoFisher). All gene qRT-PCR data were normalized to GAPDH or to G6PD. qRT-PCR primer sequences are listed in Dataset S4.
Supplementary Material
Acknowledgments
We thank the members of our laboratories for helpful discussions and suggestions and Yoshihiro Nakatani for the anti-UBR4 antibody. This work was supported by American Cancer Society Grant 131661-RSG-18-048-01-MPC (to E.A.W.) and by National Institutes of Health R01 CA066980 (to K.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data depostion: The RNA-sequencing data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE121906).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819534116/-/DCSupplemental.
References
- 1.Doorbar J, Egawa N, Griffin H, Kranjec C, Murakami I. Human papillomavirus molecular biology and disease association. Rev Med Virol. 2015;25:2–23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham SV. The human papillomavirus replication cycle, and its links to cancer progression: A comprehensive review. Clin Sci (Lond) 2017;131:2201–2221. doi: 10.1042/CS20160786. [DOI] [PubMed] [Google Scholar]
- 3.McBride AA, Münger K. Expert views on HPV infection. Viruses. 2018;10:E94. doi: 10.3390/v10020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol. 2015;33:3235–3242. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muñoz N, et al. International Agency for Research on Cancer Multicenter Cervical Cancer Study Group Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 6.Schiffman M, et al. The carcinogenicity of human papillomavirus types reflects viral evolution. Virology. 2005;337:76–84. doi: 10.1016/j.virol.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Halbert CL, Demers GW, Galloway DA. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Münger K, Phelps WC, Bubb V, Howley PM, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyson N, Guida P, Münger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson N, Howley PM, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 12.Münger K, et al. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 14.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez SL, Stremlau M, He X, Basile JR, Münger K. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J Virol. 2001;75:7583–7591. doi: 10.1128/JVI.75.16.7583-7591.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones DL, Münger K. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J Virol. 1997;71:2905–2912. doi: 10.1128/jvi.71.4.2905-2912.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funk JO, et al. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zerfass-Thome K, et al. Inactivation of the cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein. Oncogene. 1996;13:2323–2330. [PubMed] [Google Scholar]
- 19.Jones DL, Alani RM, Münger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins AS, Nakahara T, Do A, Lambert PF. Interactions with pocket proteins contribute to the role of human papillomavirus type 16 E7 in the papillomavirus life cycle. J Virol. 2005;79:14769–14780. doi: 10.1128/JVI.79.23.14769-14780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duffy CL, Phillips SL, Klingelhutz AJ. Microarray analysis identifies differentiation-associated genes regulated by human papillomavirus type 16 E6. Virology. 2003;314:196–205. doi: 10.1016/s0042-6822(03)00390-8. [DOI] [PubMed] [Google Scholar]
- 22.Garner-Hamrick PA, et al. Global effects of human papillomavirus type 18 E6/E7 in an organotypic keratinocyte culture system. J Virol. 2004;78:9041–9050. doi: 10.1128/JVI.78.17.9041-9050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hudson JB, Bedell MA, McCance DJ, Laiminis LA. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. J Virol. 1990;64:519–526. doi: 10.1128/jvi.64.2.519-526.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCance DJ, Kopan R, Fuchs E, Laimins LA. Human papillomavirus type 16 alters human epithelial cell differentiation in vitro. Proc Natl Acad Sci USA. 1988;85:7169–7173. doi: 10.1073/pnas.85.19.7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nees M, et al. Human papillomavirus type 16 E6 and E7 proteins inhibit differentiation-dependent expression of transforming growth factor-beta2 in cervical keratinocytes. Cancer Res. 2000;60:4289–4298. [PubMed] [Google Scholar]
- 26.Zehbe I, et al. Human papillomavirus 16 E6 variants differ in their dysregulation of human keratinocyte differentiation and apoptosis. Virology. 2009;383:69–77. doi: 10.1016/j.virol.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helt AM, Galloway DA. Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J Virol. 2001;75:6737–6747. doi: 10.1128/JVI.75.15.6737-6747.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jewers RJ, Hildebrandt P, Ludlow JW, Kell B, McCance DJ. Regions of human papillomavirus type 16 E7 oncoprotein required for immortalization of human keratinocytes. J Virol. 1992;66:1329–1335. doi: 10.1128/jvi.66.3.1329-1335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIntyre MC, Frattini MG, Grossman SR, Laimins LA. Human papillomavirus type 18 E7 protein requires intact Cys-X-X-Cys motifs for zinc binding, dimerization, and transformation but not for Rb binding. J Virol. 1993;67:3142–3150. doi: 10.1128/jvi.67.6.3142-3150.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White EA, et al. Papillomavirus E7 oncoproteins share functions with polyomavirus small T antigens. J Virol. 2015;89:2857–2865. doi: 10.1128/JVI.03282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciccolini F, Di Pasquale G, Carlotti F, Crawford L, Tommasino M. Functional studies of E7 proteins from different HPV types. Oncogene. 1994;9:2633–2638. [PubMed] [Google Scholar]
- 33.Ibaraki T, Satake M, Kurai N, Ichijo M, Ito Y. Transacting activities of the E7 genes of several types of human papillomavirus. Virus Genes. 1993;7:187–196. doi: 10.1007/BF01702398. [DOI] [PubMed] [Google Scholar]
- 34.Storey A, Osborn K, Crawford L. Co-transformation by human papillomavirus types 6 and 11. J Gen Virol. 1990;71:165–171. doi: 10.1099/0022-1317-71-1-165. [DOI] [PubMed] [Google Scholar]
- 35.Neary K, DiMaio D. Open reading frames E6 and E7 of bovine papillomavirus type 1 are both required for full transformation of mouse C127 cells. J Virol. 1989;63:259–266. doi: 10.1128/jvi.63.1.259-266.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarver N, Rabson MS, Yang YC, Byrne JC, Howley PM. Localization and analysis of bovine papillomavirus type 1 transforming functions. J Virol. 1984;52:377–388. doi: 10.1128/jvi.52.2.377-388.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vande Pol SB, Brown MC, Turner CE. Association of bovine papillomavirus type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif. Oncogene. 1998;16:43–52. doi: 10.1038/sj.onc.1201504. [DOI] [PubMed] [Google Scholar]
- 38.Shin MK, Sage J, Lambert PF. Inactivating all three rb family pocket proteins is insufficient to initiate cervical cancer. Cancer Res. 2012;72:5418–5427. doi: 10.1158/0008-5472.CAN-12-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strati K, Lambert PF. Role of Rb-dependent and Rb-independent functions of papillomavirus E7 oncogene in head and neck cancer. Cancer Res. 2007;67:11585–11593. doi: 10.1158/0008-5472.CAN-07-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munger K, Jones DL. Human papillomavirus carcinogenesis: An identity crisis in the retinoblastoma tumor suppressor pathway. J Virol. 2015;89:4708–4711. doi: 10.1128/JVI.03486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Todorovic B, et al. The human papillomavirus E7 proteins associate with p190RhoGAP and alter its function. J Virol. 2014;88:3653–3663. doi: 10.1128/JVI.03263-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLaughlin-Drubin ME, Crum CP, Münger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA. 2011;108:2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McLaughlin-Drubin ME, Park D, Munger K. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc Natl Acad Sci USA. 2013;110:16175–16180. doi: 10.1073/pnas.1310432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White EA, et al. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc Natl Acad Sci USA. 2012;109:E260–E267. doi: 10.1073/pnas.1116776109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeMasi J, Huh KW, Nakatani Y, Münger K, Howley PM. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc Natl Acad Sci USA. 2005;102:11486–11491. doi: 10.1073/pnas.0505322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huh KW, et al. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc Natl Acad Sci USA. 2005;102:11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White EA, Münger K, Howley PM. High-risk human papillomavirus E7 proteins target PTPN14 for degradation. MBio. 2016;7:e01530-16. doi: 10.1128/mBio.01530-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Au AC, et al. Protein tyrosine phosphatase PTPN14 is a regulator of lymphatic function and choanal development in humans. Am J Hum Genet. 2010;87:436–444. doi: 10.1016/j.ajhg.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poernbacher I, Baumgartner R, Marada SK, Edwards K, Stocker H. Drosophila Pez acts in Hippo signaling to restrict intestinal stem cell proliferation. Curr Biol. 2012;22:389–396. doi: 10.1016/j.cub.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 50.Wyatt L, Wadham C, Crocker LA, Lardelli M, Khew-Goodall Y. The protein tyrosine phosphatase Pez regulates TGFbeta, epithelial-mesenchymal transition, and organ development. J Cell Biol. 2007;178:1223–1235. doi: 10.1083/jcb.200705035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benzinou M, et al. Mouse and human strategies identify PTPN14 as a modifier of angiogenesis and hereditary haemorrhagic telangiectasia. Nat Commun. 2012;3:616. doi: 10.1038/ncomms1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith AL, et al. Pez: A novel human cDNA encoding protein tyrosine phosphatase- and ezrin-like domains. Biochem Biophys Res Commun. 1995;209:959–965. doi: 10.1006/bbrc.1995.1591. [DOI] [PubMed] [Google Scholar]
- 53.Belle L, et al. The tyrosine phosphatase PTPN14 (Pez) inhibits metastasis by altering protein trafficking. Sci Signal. 2015;8:ra18. doi: 10.1126/scisignal.2005547. [DOI] [PubMed] [Google Scholar]
- 54.Bonilla X, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48:398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 55.Wilson KE, et al. PTPN14 forms a complex with Kibra and LATS1 proteins and negatively regulates the YAP oncogenic function. J Biol Chem. 2014;289:23693–23700. doi: 10.1074/jbc.M113.534701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang P, et al. Identification and functional characterization of p130Cas as a substrate of protein tyrosine phosphatase nonreceptor 14. Oncogene. 2013;32:2087–2095. doi: 10.1038/onc.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Z, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 58.Wadham C, Gamble JR, Vadas MA, Khew-Goodall Y. The protein tyrosine phosphatase Pez is a major phosphatase of adherens junctions and dephosphorylates beta-catenin. Mol Biol Cell. 2003;14:2520–2529. doi: 10.1091/mbc.E02-09-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X, et al. PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene. 2013;32:1266–1273. doi: 10.1038/onc.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michaloglou C, et al. The tyrosine phosphatase PTPN14 is a negative regulator of YAP activity. PLoS One. 2013;8:e61916. doi: 10.1371/journal.pone.0061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W, et al. PTPN14 is required for the density-dependent control of YAP1. Genes Dev. 2012;26:1959–1971. doi: 10.1101/gad.192955.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mirabello L, et al. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell. 2017;170:1164–1174.e6. doi: 10.1016/j.cell.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dickson MA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adams JC, Watt FM. Fibronectin inhibits the terminal differentiation of human keratinocytes. Nature. 1989;340:307–309. doi: 10.1038/340307a0. [DOI] [PubMed] [Google Scholar]
- 65.Banno T, Blumenberg M. Keratinocyte detachment-differentiation connection revisited, or anoikis-pityriasi nexus redux. PLoS One. 2014;9:e100279. doi: 10.1371/journal.pone.0100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Green H. Terminal differentiation of cultured human epidermal cells. Cell. 1977;11:405–416. doi: 10.1016/0092-8674(77)90058-7. [DOI] [PubMed] [Google Scholar]
- 67.DeMasi J, Chao MC, Kumar AS, Howley PM. Bovine papillomavirus E7 oncoprotein inhibits anoikis. J Virol. 2007;81:9419–9425. doi: 10.1128/JVI.00422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Totaro A, et al. YAP/TAZ link cell mechanics to Notch signalling to control epidermal stem cell fate. Nat Commun. 2017;8:15206. doi: 10.1038/ncomms15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao B, et al. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klein RH, et al. GRHL3 binding and enhancers rearrange as epidermal keratinocytes transition between functional states. PLoS Genet. 2017;13:e1006745. doi: 10.1371/journal.pgen.1006745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lopez-Pajares V, et al. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell. 2015;32:693–706. doi: 10.1016/j.devcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miyai M, et al. Transcription factor MafB coordinates epidermal keratinocyte differentiation. J Invest Dermatol. 2016;136:1848–1857. doi: 10.1016/j.jid.2016.05.088. [DOI] [PubMed] [Google Scholar]
- 73.Miyai M, et al. Ectopic expression of the transcription factor MafB in basal keratinocytes induces hyperproliferation and perturbs epidermal homeostasis. Exp Dermatol. 2017;26:1039–1045. doi: 10.1111/exd.13364. [DOI] [PubMed] [Google Scholar]
- 74.Yu Z, et al. The Grainyhead-like epithelial transactivator Get-1/Grhl3 regulates epidermal terminal differentiation and interacts functionally with LMO4. Dev Biol. 2006;299:122–136. doi: 10.1016/j.ydbio.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 75.Nulton TJ, Olex AL, Dozmorov M, Morgan IM, Windle B. Analysis of the Cancer Genome Atlas sequencing data reveals novel properties of the human papillomavirus 16 genome in head and neck squamous cell carcinoma. Oncotarget. 2017;8:17684–17699. doi: 10.18632/oncotarget.15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evans MR, et al. An oral keratinocyte life cycle model identifies novel host genome regulation by human papillomavirus 16 relevant to HPV positive head and neck cancer. Oncotarget. 2017;8:81892–81909. doi: 10.18632/oncotarget.18328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Evans MR, et al. 2018. Human papillomavirus 16 E2 regulates keratinocyte gene expression relevant to cancer and the viral life cycle. bioRxiv, 10.1101/461715. Preprint, posted November 5, 2018.
- 78.Mehra R, et al. Biallelic alteration and dysregulation of the Hippo pathway in mucinous tubular and spindle cell carcinoma of the kidney. Cancer Discov. 2016;6:1258–1266. doi: 10.1158/2159-8290.CD-16-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mello SS, et al. A p53 super-tumor suppressor reveals a tumor suppressive p53-Ptpn14-Yap axis in pancreatic cancer. Cancer Cell. 2017;32:460–473.e6. doi: 10.1016/j.ccell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pellegrini C, et al. Understanding the molecular genetics of basal cell carcinoma. Int J Mol Sci. 2017;18:E2485. doi: 10.3390/ijms18112485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schramm A, et al. Mutational dynamics between primary and relapse neuroblastomas. Nat Genet. 2015;47:872–877. doi: 10.1038/ng.3349. [DOI] [PubMed] [Google Scholar]
- 82.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 83.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 84.Szalmás A, et al. The PTPN14 tumor suppressor is a degradation target of human papillomavirus E7. J Virol. 2017;91:e00057-17. doi: 10.1128/JVI.00057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang JM, et al. YAP modifies cancer cell sensitivity to EGFR and survivin inhibitors and is negatively regulated by the non-receptor type protein tyrosine phosphatase 14. Oncogene. 2013;32:2220–2229. doi: 10.1038/onc.2012.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Perez CA, Ott J, Mays DJ, Pietenpol JA. p63 consensus DNA-binding site: Identification, analysis and application into a p63MH algorithm. Oncogene. 2007;26:7363–7370. doi: 10.1038/sj.onc.1210561. [DOI] [PubMed] [Google Scholar]
- 87.Armstrong SR, et al. The regulation of tumor suppressor p63 by the ubiquitin-proteasome system. Int J Mol Sci. 2016;17:E2041. doi: 10.3390/ijms17122041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sethi I, Gluck C, Zhou H, Buck MJ, Sinha S. Evolutionary re-wiring of p63 and the epigenomic regulatory landscape in keratinocytes and its potential implications on species-specific gene expression and phenotypes. Nucleic Acids Res. 2017;45:8208–8224. doi: 10.1093/nar/gkx416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soares E, Zhou H. Master regulatory role of p63 in epidermal development and disease. Cell Mol Life Sci. 2017;75:1179–1190. doi: 10.1007/s00018-017-2701-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Harden ME, Prasad N, Griffiths A, Munger K. Modulation of microRNA-mRNA target pairs by human papillomavirus 16 oncoproteins. MBio. 2017;8:e02170-16. doi: 10.1128/mBio.02170-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wong CF, et al. E2F modulates keratinocyte squamous differentiation: Implications for E2F inhibition in squamous cell carcinoma. J Biol Chem. 2003;278:28516–28522. doi: 10.1074/jbc.M301246200. [DOI] [PubMed] [Google Scholar]
- 92.Brimer N, Lyons C, Wallberg AE, Vande Pol SB. Cutaneous papillomavirus E6 oncoproteins associate with MAML1 to repress transactivation and NOTCH signaling. Oncogene. 2012;31:4639–4646. doi: 10.1038/onc.2011.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Meyers JM, Spangle JM, Munger K. The human papillomavirus type 8 E6 protein interferes with NOTCH activation during keratinocyte differentiation. J Virol. 2013;87:4762–4767. doi: 10.1128/JVI.02527-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tan MJ, et al. Cutaneous β-human papillomavirus E6 proteins bind Mastermind-like coactivators and repress Notch signaling. Proc Natl Acad Sci USA. 2012;109:E1473–E1480. doi: 10.1073/pnas.1205991109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ablain J, de Thé H. Retinoic acid signaling in cancer: The parable of acute promyelocytic leukemia. Int J Cancer. 2014;135:2262–2272. doi: 10.1002/ijc.29081. [DOI] [PubMed] [Google Scholar]
- 96.White EA, et al. Comprehensive analysis of host cellular interactions with human papillomavirus E6 proteins identifies new E6 binding partners and reflects viral diversity. J Virol. 2012;86:13174–13186. doi: 10.1128/JVI.02172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nakatani Y, et al. p600, a unique protein required for membrane morphogenesis and cell survival. Proc Natl Acad Sci USA. 2005;102:15093–15098. doi: 10.1073/pnas.0507458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mi H, et al. PANTHER version 11: Expanded annotation data from gene ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.White EA. 2019 Analysis of gene expression in primary human foreskin keratinocytes +/- HPV16 E7 or PTPN14 knockout. NCBI Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE121906. Deposited October 29, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.