Significance
Diabetic foot ulcerations (DFUs) represent a major medical and economic problem with still-unclear pathogenic mechanisms. The Notch pathway plays a pivotal role in cell differentiation, proliferation, and angiogenesis, processes that are profoundly disturbed in diabetic wounds. Notch signalling is activated upon interactions between membrane-bound Notch receptors (Notch 1–4) and ligands (Jagged 1–2 and Delta-like 1, 3, 4). Here, we report that a specific positive Delta-like 4–Notch1 feedback loop is activated by high glucose levels and specifically impairs wound healing in diabetes. Local inhibition of Notch signalling in experimental wounds using chemical and genetic approaches markedly improves healing exclusively in diabetic, but not in nondiabetic, animals, making Notch1 signalling an attractive locally therapeutic target for the treatment of DFUs.
Keywords: diabetic foot ulcer, Notch1, Dll4, wound healing, diabetes
Abstract
Diabetic foot ulcerations (DFUs) represent a major medical, social, and economic problem. Therapeutic options are restricted due to a poor understanding of the pathogenic mechanisms. The Notch pathway plays a pivotal role in cell differentiation, proliferation, and angiogenesis, processes that are profoundly disturbed in diabetic wounds. Notch signaling is activated upon interactions between membrane-bound Notch receptors (Notch 1–4) and ligands (Jagged 1–2 and Delta-like 1, 3, 4), resulting in cell-context-dependent outputs. Here, we report that Notch1 signaling is activated by hyperglycemia in diabetic skin and specifically impairs wound healing in diabetes. Local inhibition of Notch1 signaling in experimental wounds markedly improves healing exclusively in diabetic, but not in nondiabetic, animals. Mechanistically, high glucose levels activate a specific positive Delta-like 4 (Dll4)–Notch1 feedback loop. Using loss-of-function genetic approaches, we demonstrate that Notch1 inactivation in keratinocytes is sufficient to cancel the repressive effects of the Dll4–Notch1 loop on wound healing in diabetes, thus making Notch1 signaling an attractive locally therapeutic target for the treatment of DFUs.
Diabetes affects ∼9.5% of the world population, and the incidence will increase by >50% by 2030 (1, 2). Diabetic foot ulceration (DFU) is a chronic diabetes complication that represents a major medical, social, and economic problem. The lifetime risk of a person with diabetes to develop a foot ulcer is 15%, and it is estimated that every 30 s, a lower limb is lost due to diabetes worldwide (3). Even in the best clinical setting, with broad expertise in a multidisciplinary team (endocrinologist, vascular surgeon, orthopedic surgeon, and podiatrist), the therapeutic options offered to a patient with DFU are restricted due to a poor understanding of the pathogenic mechanisms (4–6).
Wound healing is a complex process that involves extensive coordination among soluble mediators (growth factors, cytokines, and chemokines), the extracellular matrix (ECM), and different cellular players (7). This process can be divided into several stages. The early stage involves homeostasis and inflammation. During the intermediate stage, keratinocyte and fibroblast proliferation and migration, angiogenesis, and matrix deposition are observed. The late stage involves remodeling and reepithelialization (7).
Several mechanisms contribute to the delayed wound healing in DFUs, including decreased angiogenesis, reduced recruitment of bone marrow-derived endothelial progenitor cells (EPCs), and decreased proliferation and migration of fibroblasts and keratinocytes (5). Efforts have been made to stimulate the healing process in DFUs through administration of biological factors or stem cells, but their efficacies have not been sufficient to guarantee adequate DFU healing (7, 8). Given the complexity of the multicellular and multifactorial processes of wound healing, it is conceivable that a therapeutic strategy targeting a signaling pathway that regulates multiple cellular processes important for wound healing would serve as a more efficient or at least complementary solution for DFU treatment.
Notch is an evolutionarily conserved signaling mechanism, and its action is highly pleiotropic. Notch signaling is fundamental for cell-fate determination and also plays a pivotal role in regulating proliferation, apoptosis/survival, and angiogenesis (9, 10), processes that are profoundly disturbed in diabetic wounds. Notch signaling is a cell–cell communication mechanism activated as a result of the interaction between membrane-bound Notch receptors (Notch 1–4) and their ligands (Jagged 1–2 and Delta-like 1, 3, 4) on juxtaposed cells. Binding of the ligand to the receptor triggers two consecutive proteolytic cleavages (metalloproteinase-mediated and subsequent γ-secretase-mediated cleavage) of the Notch receptor that release the Notch intracellular domain (ICD) from the plasma membrane. The Notch ICD translocates into the nucleus, where it forms a complex with the DNA-binding protein CSL [named after CBF1, Su(H), and LAG-1] and its coactivator Mastermind to promote the transcription of Notch target genes, such as Hes and Hey (10, 11).
Notch signaling is sensitive to gene-dosage deviations, and the outcome of Notch signaling differs depending on spatial and temporal cellular contexts (10). Notch signaling plays fundamental roles during development, but accumulating evidence supports its involvement in the regulation of tissue homeostasis in adults as well (10). Moreover, Notch malfunction is associated with a diverse range of human diseases, including hereditary diseases, cancer, CVD, and kidney disease (10). Notch signaling was reported to be modulated in different and opposite ways in diabetes depending on the tissue (12–15).
Notch signaling also plays important roles in the development and postnatal physiology of the skin (16–21). In addition, Notch signaling is involved in normal wound healing through the positive regulation of angiogenesis, cell migration, and inflammation (22, 23). However, it is unknown whether Notch signaling in the skin is involved in impaired wound healing in diabetes.
In this study, we identify that Notch signaling is overactivated in the diabetic skin, and we demonstrate that high glucose levels activate a Delta-like 4 (Dll4)–Notch1 positive-feedback loop. Furthermore, local inhibition of Notch1 signaling in experimental wounds using either chemical or loss-of-function genetic approaches improves healing exclusively in diabetic animals. These results suggest that local Notch1 signaling is an attractive therapeutic target for the treatment of DFUs in addition to the current multidisciplinary standard therapy.
Results
Notch1 Signaling Is Activated in Diabetic Skin.
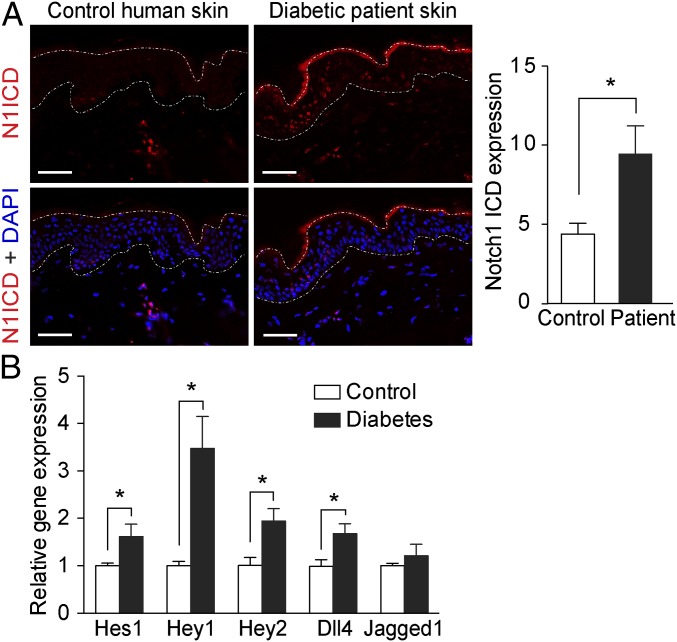
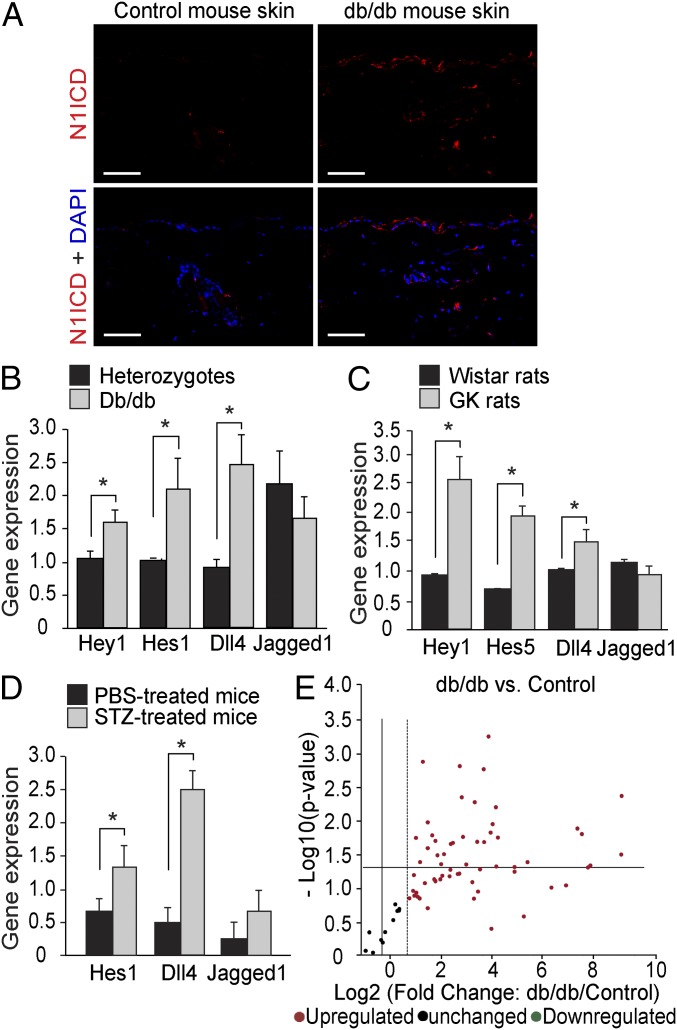
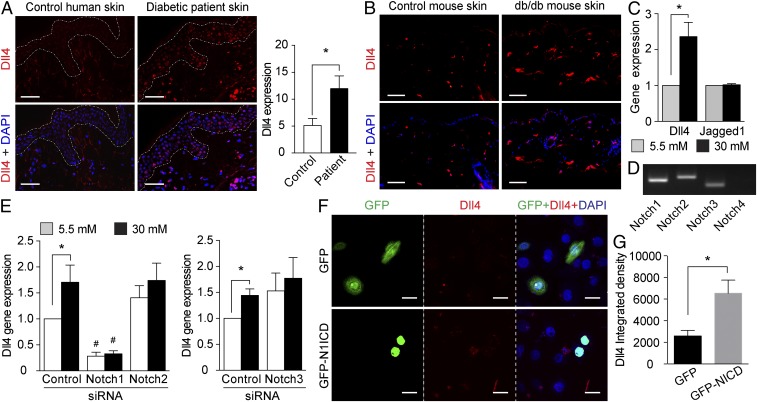
Using a specific antibody for activated Notch1, we detected increased Notch1 ICD levels in the skin from patients with diabetes mainly, but not exclusively, in the epidermis (Fig. 1A). The same distribution of increased Notch1 ICD levels was also identified in the skin from db/db diabetic mice (Fig. 2A). The activation of Notch signaling in diabetic skin was further confirmed by increased mRNA expression of Notch target genes (Hey1, Hey2, Hes1, or Hes5) in the skin from patients with diabetes (Fig. 1B) and from animal models for both type 2 diabetes (db/db mice and GK rats) and type 1 diabetes [streptozotocin (STZ)-induced diabetic mice] (Fig. 2 B–D). Moreover, the expression of the Notch ligand Dll4, but not of Jagged1, was also increased in the skin from patients with diabetes (Fig. 1B) and from diabetic animals (Fig. 2 B–D).
Fig. 1.
Notch1 signaling is activated in the skin of diabetic patients. (A) Immunohistochemistry staining for Notch1 ICD (N1ICD; red) and DAPI (blue) in skin from patients with diabetes and age-matched normoglycemic controls. Dashed lines delineate boundaries of epidermis. (Scale bars: 50 µm.) Quantification of the Notch1 ICD expression in the epidermis are shown in the histogram (n = 5). (B) Relative mRNA expression of Notch target genes Hes1, Hey1, and Hey2 and Notch ligands Dll4 and Jagged1 in the skin from diabetic patients (n = 7–9) and controls (n = 13). *P < 0.05.
Fig. 2.
Notch1 signaling is activated in the skin of diabetic animals. (A) Representative images of immunohistochemistry staining for Notch1 ICD (N1ICD; red) and DAPI (blue) in skin from db/db diabetic mice and control mice. (Scale bars: 50 µm.) (B–D) Relative mRNA expression of Notch target genes Hey1, Hes1, or Hes5 and Notch ligands Dll4 and Jagged1 in the skin of diabetic animals [db/db mice (B; n = 10), GK rats (C; n = 5), and STZ-induced diabetic mice (D; n = 3)] and in their nondiabetic control littermates. *P < 0.05 (analyzed using the Student t test). (E) Mouse Notch Signaling Target RT2 Profiler PCR Array results comparing the expression of key Notch target genes in skin from db/db mice (n = 5) and controls (n = 4).
To further explore the magnitude of change in Notch signaling in diabetic skin, we performed Notch Signaling Target RT2 Profiler PCR Array analysis, in which the gene expression of key genes regulated by the Notch pathway was analyzed in skin from db/db diabetic mice and their controls. As shown in Fig. 2E, most of the tested Notch target genes had higher expression in diabetic skin from db/db mice than controls. These genes are involved in regulating many cellular processes, such as angiogenesis, cell-fate decision, proliferation and apoptosis, inflammation, and development, with potential roles in wound healing (SI Appendix, section 1 and Table S1).
High Glucose Levels Activate Notch1 Signaling, Severely Affecting the Function of Cells Central for Wound Healing.
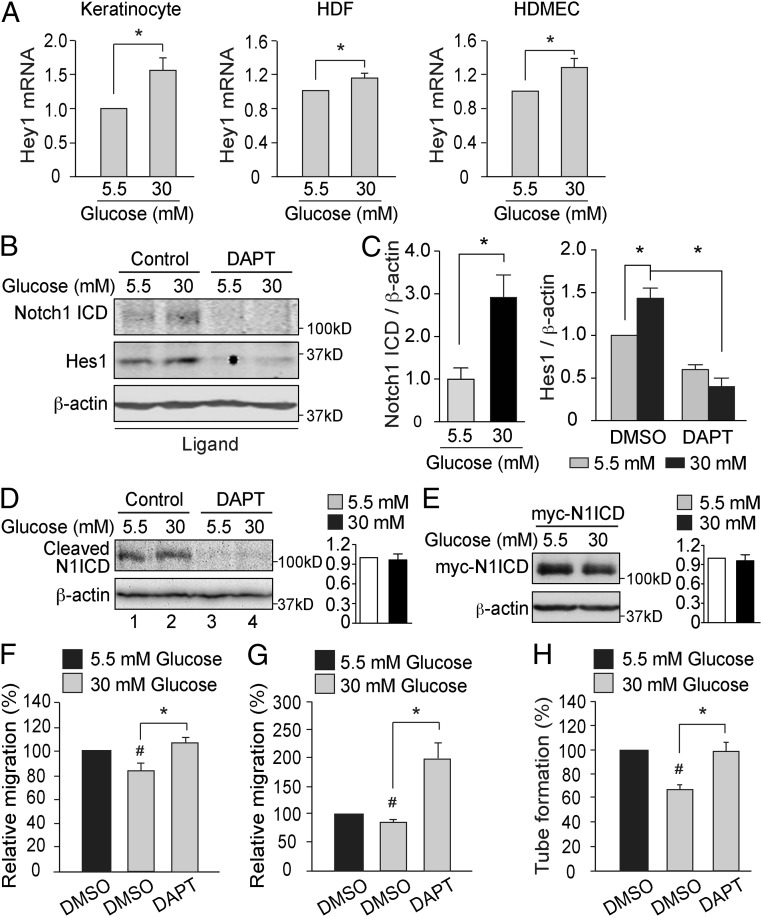
The major cellular components of the epidermis are the keratinocytes, while fibroblasts and endothelial cells are the major components of the dermis. We have therefore evaluated the effects of high glucose levels on Notch signaling in all these three cell types and observed an increase of Hey1 mRNA expression levels in primary cultures of human keratinocytes, human dermal fibroblasts (HDFs), and human dermal microvascular endothelial cells (HDMECs) (Fig. 3A). In addition, exposure to high glucose levels was followed by an increase in Notch1 ICD and Hes1 protein levels (Fig. 3 B and C), but no change in Notch2 ICD or Notch3 ICD expression (SI Appendix, Fig. S1). The specificity for Notch1 receptor was confirmed by specifically silencing Notch1 by siRNA (SI Appendix, Fig. S2). The stimulatory effects of high glucose levels on Notch1 ICD and Hes1 protein levels were abolished by the γ-secretase inhibitor (GSI) DAPT (Fig. 3 B and C). Thus, Notch signaling activation in hyperglycemia is dependent on γ-secretase–mediated cleavage of the Notch receptor.
Fig. 3.
High glucose levels activate Notch1 signaling in vitro. (A) Primary human keratinocytes (n = 6), HDFs (n = 3), and HDMECs (n = 3) were exposed to 5.5 or 30 mM glucose for 24 h. Relative Hey1 mRNA expression levels are shown. *P < 0.05. (B and C) Endogenous Notch1 ICD (n = 4) and Hes1 (n = 3) protein levels in keratinocytes that were exposed to 5.5 or 30 mM glucose and treated with DMSO (control) or DAPT (10 µM) for 24 h. Quantification of Western blots are shown in C. *P < 0.05. (D) HDFs were transiently transfected with Notch1 ΔE, exposed to 5.5 or 30 mM glucose for 2 d, and treated with DMSO (Control) or 10 μM DAPT for 18 h before harvest. The cleaved product Notch1 ICD (N1ICD) was detected by using an antibody raised against activated Notch1. Quantification of N1ICD/β-actin is shown in the histogram (n = 3). (E) HDFs were transiently transfected with myc-tagged Notch1 ICD (myc-N1ICD) and exposed to 5.5 or 30 mM glucose for 2 d. Myc-N1ICD protein levels were detected by using an anti-Myc antibody. Quantification of myc-N1ICD/β-actin is shown in the histogram (n = 3). (F–H) Keratinocytes (F), HDFs (G), and HDMECs (H) were cultured in medium containing 5.5 or 30 mM glucose and exposed to DMSO or 10 μM DAPT for 24 h before the in vitro migration assay (F and G; n = 5) or in vitro angiogenesis assay (H; n = 3). #P < 0.05 (compared with cells exposed to 5.5 mM glucose); *P < 0.05 (compared with the corresponding controls).
To investigate the effect of high glucose levels on γ-secretase activity, we evaluated cleaved Notch1 ICD levels in cells expressing the Notch1 extracellular truncation (NEXT) encoded by the Notch1 ΔE construct. NEXT is the membrane-tethered intermediate of the Notch1 receptor after Notch1 ectodomain shedding, and it is continuously cleaved by γ-secretase in a ligand-independent manner to generate Notch1 ICD (24). Hence, the level of cleaved Notch1 ICD from NEXT reflects γ-secretase activity, as demonstrated by its absence after DAPT treatment (Fig. 3 D, lanes 3 and 4). High glucose did not affect cleaved Notch1 ICD levels in cells expressing NEXT, indicating that γ-secretase activity per se is not modulated by glucose (Fig. 3 D, lanes 1 and 2). Moreover, the levels of ectopically expressed myc-Notch1 ICD were similar in cells exposed to normal or high glucose levels (Fig. 3E), indicating that Notch1 ICD stability is not affected by high glucose levels.
The migration of keratinocytes and fibroblasts together with angiogenesis are central cellular processes during wound healing. We therefore investigated the functional role of the activated Notch signaling in regulating migration and angiogenesis in diabetes. As expected, the migration of keratinocytes and HDFs, as well as the tube formation of the HDMECs, were significantly inhibited by high glucose levels (Fig. 3 F–H). Exposure of the cells to DAPT abrogated the inhibitory effects of hyperglycemia on HDMEC tube formation and cell migration, underscoring the role of Notch signaling for the function of cells essential for wound healing, which are impaired by high glucose levels.
Notch Activation Impairs Wound Healing in Diabetes.
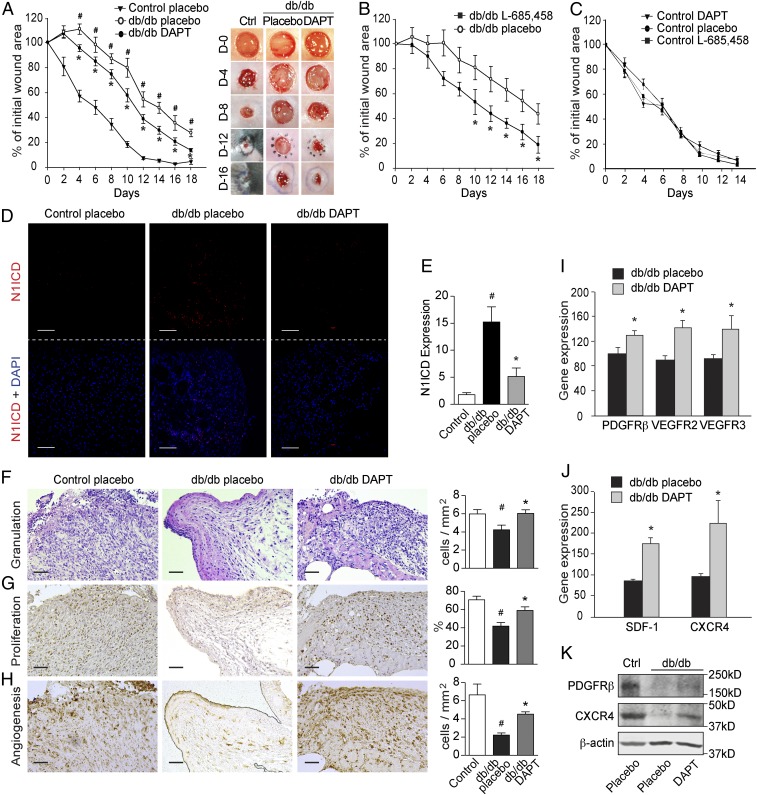
To investigate the potential role of activated Notch signaling in the impaired wound healing in diabetes, we locally applied two GSIs, DAPT or L-685,458, in a wound-healing model in db/db or STZ-induced diabetic mice together with control mice. GSI-mediated abrogation of Notch signaling improved the characteristic delayed wound-healing rate in db/db (Fig. 4 A and B), as well as in STZ-induced diabetic mice (SI Appendix, Fig. S3). Interestingly, Notch signaling inhibition with DAPT and L-685,458 did not affect the wound-healing rate in nondiabetic control mice (Fig. 4C), indicating that this effect was specific to impaired wound healing in diabetes.
Fig. 4.
Inhibition of Notch signaling improves wound healing in diabetic mice. Full-thickness wounds were made on the dorsum of db/db and control mice, and the wounds were treated locally with DMSO (placebo) or GSIs DAPT (100 μM) or L-685,458 (100 μM) every second day. (A–C) The wound-healing rate was measured every second day and is shown as the percentage of the initial wound area (n = 5 per group). (A, Right) Representative wound images at days (D) 0, 4, 8, 12, and 16 during the healing process. (D) Representative images of immunohistochemistry staining for Notch1 ICD (N1ICD; red) and DAPI (blue) in wounds from control and db/db mice treated with placebo or DAPT. (E) Quantification results are shown (n = 9 or 10). (F–H) Levels of granulation, proliferation, and angiogenesis (microvessel density) were evaluated by histological analysis of hematoxylin and eosin staining (F), nuclear PCNA staining (G), and GSL-I isolectin B4 staining (H), respectively. Semiquantitative evaluations are presented in the histograms (n = 3). (I and J) The mRNA expression of PDGFRβ, VEGFR2, VEGFR3, SDF-1, and CXCR4 in diabetic wounds treated with DMSO (placebo) or DAPT (n = 10). (K) Western blotting results showing the expression of PDGFRβ and CXCR4 in control (Ctrl) and db/db mice treated with placebo or DAPT. (Scale bars: 50 µm.) #P < 0.05 (compared with control mice treated with placebo); *P < 0.05 (compared with db/db mice treated with placebo).
Compared with nondiabetic control wounds, Notch1 ICD expression in diabetic wounds was significantly increased and was inhibited by local DAPT treatment, as expected (Fig. 4 D and E). Expression of the ICDs of Notch2, 3, and 4 and Dll4 in db/db diabetic wounds was also inhibited by local DAPT treatment, but there was no change in Jagged1, 2, or Dll1 expression (SI Appendix, Fig. S4).
To investigate the mechanisms underlying the positive effects of GSIs on wound healing in diabetes, we evaluated their effects on granulation, proliferation, and angiogenesis, the pivotal cellular mechanisms for the progression of wound healing. Local treatment with DAPT increased granulation tissue in db/db mice to levels similar to those noted in nondiabetic animals (Fig. 4F). This outcome was at least partially attributed to increased proliferation, as demonstrated by increased nuclear proliferating cell nuclear antigen (PCNA) staining (Fig. 4G). Inhibition of overactive Notch signaling in diabetic wounds was also followed by increased angiogenesis, as assessed by isolectin B4 staining (Fig. 4H), as well as by induction of mRNA and protein expression of other angiogenic markers such as CD31, VEGFR-2 (25), VEGFR-3 (26, 27), and PDGFRβ (28) (Fig. 4 I and K and SI Appendix, Fig. S5 A–C). Moreover, DAPT treatment increased the mRNA and protein expression of chemokines and receptors that play essential roles in the recruitment of EPCs, such as SDF-1 (29) and CXCR4 (30) (Fig. 4 J and K and SI Appendix, Fig. S4D). Together, these results suggest that Notch inhibition in diabetic wounds significantly promotes wound healing by improving granulation, proliferation, and angiogenesis, which are repressed by hyperglycemia.
High Glucose Levels Activate a Positive Dll4–Notch1 Feedback Loop.
Consistent with the increased Dll4 mRNA expression in the skin from patients with diabetes (Fig. 1B) and diabetic animals (Fig. 2 B–D), we detected increased Dll4 protein levels in the skin from both patients with diabetes (Fig. 5A) and db/db diabetic mice (Fig. 5B). Similar to Notch1 ICD, the increased Dll4 expression was distributed mainly in the epidermis (Fig. 5 A and B), in perfect agreement with its functional induction in human primary keratinocytes by high glucose levels (Fig. 5C). In concordance with the in vivo data, Jagged1 expression was not affected by changing the glucose levels (Fig. 5C). Similarly, high glucose levels did not affect the gene expression of other Notch ligands that are expressed in keratinocytes, Dll1, or Jagged2 (SI Appendix, Fig. S6).
Fig. 5.
High glucose levels activate a positive Dll4–Notch1 feedback loop. (A and B) Immunohistochemistry staining for Dll4 (red) and DAPI (blue) in skin from patients with diabetes (A) and db/db diabetic mice (B) and the corresponding controls. Dashed lines delineate boundaries of human epidermis. (Scale bars: 50 µm.) Quantification of Dll4 intensity in human epidermis is shown in the histogram. (C) Relative Dll4 and Jagged1 (Jag1) gene expression in human primary keratinocytes exposed to 5.5 or 30 mM glucose (n = 8). (D) Agarose gel image showing the gene expression of Notch1, 2, 3, and 4 in primary human keratinocytes. (E) Relative Dll4 gene expression in keratinocytes transfected with siRNA for Notch1, Notch2, Notch3, or control siRNA (n = 4 or 5). (F and G) Representative images (F) and quantification (G) of immunocytochemistry of Dll4 (red) in keratinocytes that express GFP (n = 6) or GFP-Notch1 ICD (GFP-N1ICD; n = 8). Cell nuclei were counterstained with DAPI (blue). (Scale bars: 25 µm.) *P < 0.05; #P < 0.05 (compared with the corresponding control siRNA).
We further investigated whether glucose-dependent Dll4 induction is mediated by Notch signaling using specific siRNA silencing. Although several Notch receptors are highly expressed in keratinocytes, e.g., Notch1, Notch2, and Notch3 (Fig. 5D), only Notch1 siRNA, but not Notch2 or Notch3 siRNA, abolished the glucose-induced increase in Dll4 mRNA expression (Fig. 5E and SI Appendix, Figs. S7 and S8), suggesting that high glucose levels induce a positive Dll4–Notch1 feedback loop. This notion is corroborated by the fact that expression of Notch1 ICD in keratinocytes led to elevated Dll4 mRNA expression (Fig. 5F).
The induction of Dll4 gene expression by high glucose levels was also observed in HDMECs (SI Appendix, Fig. S9A), and the Dll4–Notch1 positive feedback loop was confirmed in HDMECs, as silencing of Notch1, but not Notch2 or Notch4, cancelled the effect of high glucose levels on Dll4 expression (SI Appendix, Fig. S9 B–D). Moreover, silencing either Dll4 or Notch1 in HDMECs abolished the inhibitory effect of high glucose levels on the angiogenic capacity of HDMECs (SI Appendix, Fig. S10), suggesting an important functional role of the Dll4–Notch1 loop in wound healing in diabetes.
Ablation of Notch1 in the Skin Improves Diabetic Wound Healing.
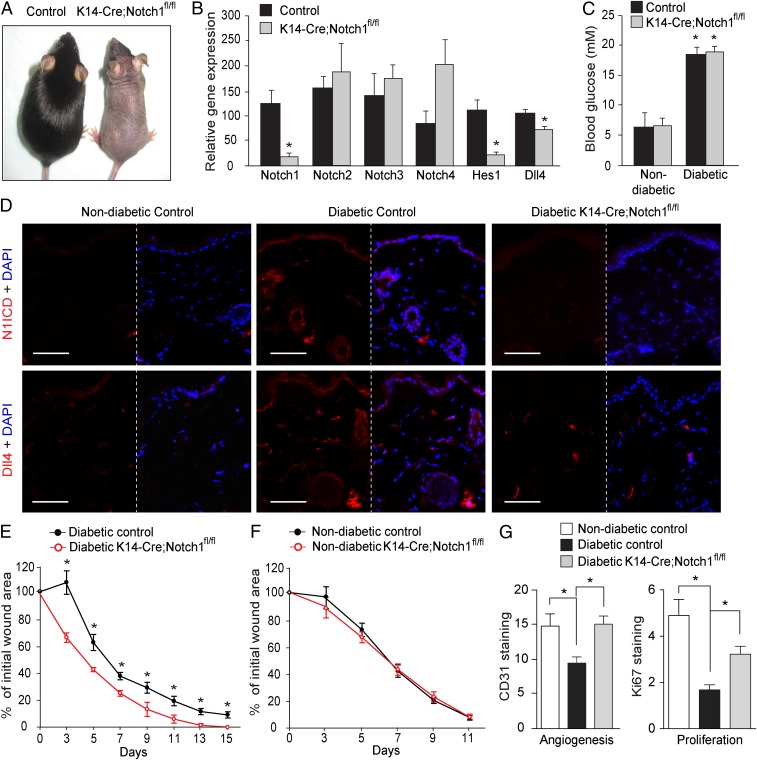
We next wanted to assess whether genetically abrogating the Dll4–Notch1 loop in the epidermis would impact on the glucose-induced phenotype since GSIs block signaling from all four Notch receptors (SI Appendix, Fig. S4). To this end, we used a genetic mouse model, where Notch1 was specifically ablated in the keratinocytes (KRT14–Cre;Notch1fl/fl mice). The KRT14–Cre;Notch1fl/fl mice exhibited a phenotype (less and gray hair) similar to other mice where Notch1 was conditionally ablated in the skin (17–19) (Fig. 6A). The specific Notch1 knockout in the skin of KRT14–Cre;Notch1fl/fl mice was restricted to keratinocytes, as confirmed by significantly decreased Notch1 mRNA expression in epidermis isolated by laser microdissection (SI Appendix, Fig. S11). The specific ablation of Notch1 in the skin of KRT14–Cre;Notch1fl/fl mice did not affect the expression of other Notch receptors (Fig. 6B), but significantly decreased the overall Notch signaling in the skin, as demonstrated by reduced Hes1 and Dll4 mRNA expression (Fig. 6B).
Fig. 6.
Dll4–Notch1 signaling in skin impairs diabetic wound healing. (A) Representative photos of the skin-specific Notch1-knockout KRT14-Cre;Notchfl/fl (K14-Cre;Notchfl/fl) and control mice. (B) The mRNA expression levels of Notch receptors and Notch target gene Hes1 and Dll4 in the skin of KRT14-Cre;Notchfl/fl mice and their controls (n = 5). (C) The blood glucose level in KRT14-Cre;Notchfl/fl mice and their controls before wound induction (n = 5). (D) Representative images of immunohistochemical staining for Notch1 ICD (N1ICD; red; Upper), Dll4 (red; Lower), and DAPI (blue) in skin from nondiabetic control mice, diabetic control mice, and diabetic KRT14-Cre;Notchfl/fl mice. (E) Wound-healing rates in diabetic KRT14-Cre;Notchfl/fl mice and diabetic controls (n = 5). (F) Wound-healing rates in nondiabetic KRT14-Cre;Notchfl/fl mice and their controls (n = 5). (G) Levels of angiogenesis and proliferation in the wounds were evaluated by quantification of the immunohistochemical staining of CD31 (n = 3 or 4) and Ki67 (n = 4–6), respectively. *P < 0.05 (compared with the corresponding control littermates).
Diabetes induced by STZ was followed by the expected induction of both Notch1 ICD and Dll4 in the skin of wild-type mice, but not in KRT14–Cre;Notch1fl/fl mice (Fig. 6D), despite similar blood glucose levels (Fig. 6C). This was paralleled by a more rapid wound-healing rate in diabetic KRT14–Cre;Notch1fl/fl mice than in the diabetic wild-type controls (Fig. 6E). Similar to the effects of DAPT on wounds in db/db mice (Fig. 4), Notch1 gene ablation in the skin improved angiogenesis and proliferation in the granulation tissues of diabetic wounds (Fig. 6G and SI Appendix, Fig. S12). No difference was observed in the wound-healing rate between nondiabetic KRT14–Cre;Notch1fl/fl mice and nondiabetic controls (Fig. 6F). These results corroborate the notion that a Dll4–Notch1 positive loop mediates the repressive effects of hyperglycemia on wound healing in diabetes (Fig. 7).
Fig. 7.
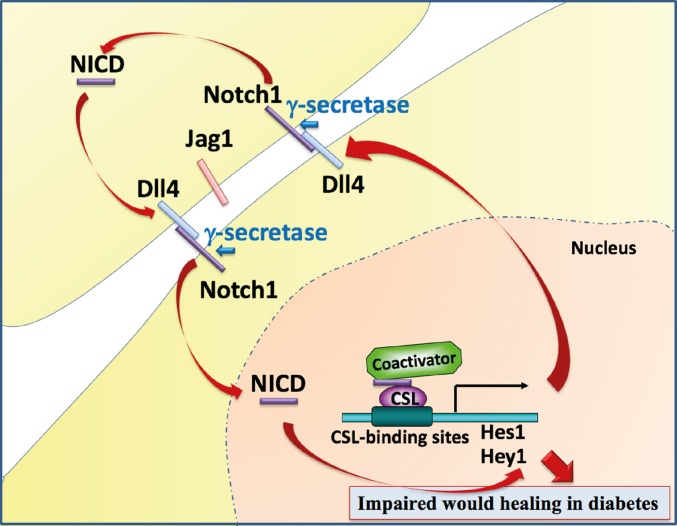
Schematic illustration of the glucose-induced overactive Dll4–Notch1 loop that contributes to impaired wound healing in diabetes. High glucose levels induce a positive Dll4–Notch1 feedback loop. In this loop, Notch1 signaling enhances the expression of the ligand Dll4, which in turn further activates Notch1. This positive Dll4–Notch1 feedback loop is induced and sustained by high glucose levels, thereby resulting in an amplification of the Notch1 activity that contributes to impaired wound healing in diabetes.
Discussion
Notch signaling is crucial for embryonic development, but also for the regulation of postnatal tissue homeostasis. Notch signaling can also be reactivated in pathophysiological conditions, such as atherosclerosis (31, 32), cardiac hypertrophy and failure (33, 34), myocardial infarction (34–36), stroke (37), kidney diseases (38, 39), and cancer (40). The role of Notch in diabetes is more enigmatic, and roles of both activated and reduced Notch signaling have been reported in various tissues (13, 15, 38, 41, 42). Here, we demonstrate that Notch1 signaling is activated in diabetic skin and at least partially mediates the inhibitory effect of diabetes on wound healing. Notch1 signaling blockade via either chemical or genetic approaches improved wound healing in diabetes, suggesting that Notch1 signaling is a potential therapeutic target for diabetic wounds.
Interestingly, this effect is specific for diabetes, where Notch1 is overactivated, and inhibition of Notch signaling using GSIs or skin-specific Notch1 gene ablation did not affect wound healing in nondiabetic control animals. The latter observation receives support from other studies, reporting that in nondiabetic animals, there was either no effect (43) or even delaying (22, 44) of the wound-healing rate when Notch signaling was blocked.
It is probably the difference of the magnitude of Notch signaling in skin that explains the difference in the responses to Notch-signaling blockage. In diabetes, the elevated Notch signaling can be brought back to near-normal levels by Notch inhibition, whereas in a normal, nondiabetes condition, the blockage has no effect, since Notch signaling is faint, if any. The extreme sensitivity of the Notch signaling to dosage has been observed in both normal development and pathology (10, 11). Activation of the Notch signaling was observed in diabetes nephropathy (12), even though it is common for other glomerular diseases as well (39). Whether Notch signaling is activated in other medical conditions in which wound healing is impaired warrants further investigation.
Mechanistically, the inhibition of the overactivated Notch1 signaling reverses the inhibitory effect of hyperglycemia on several mechanisms central for wound healing as the cellular migration, proliferation, and angiogenesis both in vitro and in vivo. These results suggest a central role of Notch1 inhibition on the granulation process and are consistent with previous findings that revealed a hyperproliferative phenotype in the presence of Notch-signaling repression (17, 45) and an enhanced angiogenesis in animals treated with GSIs (46, 47). In other tissues, suppression of Notch signaling has an inhibitory role on the epithelial–mesenchymal transition with potential relevance for diabetic wounds (48). The effect of abrogating Notch1 function appears to be specific for keratinocytes, since deletion of Notch1 signaling in macrophages does not exert a similar effect on wound healing (23). The cell-type-specific diversity in the Notch-signaling output is not restricted to the skin, since in the kidney, Notch signaling has opposite effects in podocytes (49) compared with tubular precursor cells (50). The fact that Notch signaling negatively regulates the expression of SDF-1 (29) and CXCR4 (30), which are important factors for EPC recruitment, may provide a mechanism for how Notch regulates angiogenesis. A reduced EPC incorporation to the wound area secondary to decreased SDF-1 expression is an important defect that contributes to impaired wound healing in diabetes (51). Together, our results indicate that Notch inhibition is a promising therapeutic strategy for DFUs, since it simultaneously improves multiple cellular defects.
Interestingly, increased Dll4 expression was noted in diabetic skin of patients as well as in animal models and in cells exposed to high glucose levels. This induction was Notch1-dependent both in vitro and in vivo. Hence, a hyperglycemia-induced Dll4–Notch1 positive feedback loop was identified that contributes to pathogenic sustained Notch activation in diabetes, and the Dll4–Notch1 loop may further amplify the signaling effect and thus contribute to dosage sensitivity (10, 11). The loop is highly specific, given that Jagged1 expression is not modulated in diabetic skin. This is in concordance with a negative effect of Dll4-dependent Notch1 signaling on angiogenesis (47, 52, 53). It is, however, likely that different ligands are important in different diabetes contexts, as Jagged1 seems to be central for nephropathy (49) and is induced in the retina (42) and myocardium (54). This is another example of the high versatility of Notch signaling that has many well-documented tissue- and cell-type-specific outputs (10, 11).
In addition, our study sheds light on the mechanism underlying hyperglycemia-induced Notch1 signaling activation. Insulin is not a modulator of Notch1 signaling, since Notch1 activation was noticed in the skin of animal models of both type 2 and type 1 diabetes, and insulin had no significant effect on Notch target gene expression in vitro (SI Appendix, Fig. S13). Glucose increased the level of cleaved Notch1 ICD and Notch target gene expression, and the increase in target gene expression was suppressed by GSIs, indicating that it was mediated by canonical Notch1 signaling. Hyperglycemia, however, does not modulate γ-secretase activity per se, given that high glucose levels did not affect the cleavage of ectopically expressed NEXT, which is a ligand-independent substrate for γ-secretase that purely reflects γ-secretase activity (24). Moreover, our results indicate that high glucose levels did not affect Notch1 ICD protein stability. Given that hyperglycemia-induced Notch1 signaling is not a consequence of increased γ-secretase activity or Notch1 ICD stability, but is suppressed by GSIs, it may simply reflect an elevated activation of the Notch1 receptor. Indeed, high glucose levels increased the expression of the ligand Dll4 that was in its turn dependent on the specific activation of Notch1 signaling. Thus, glucose activates a positive feedback loop between the Dll4 ligand and the Notch1 receptor, contributing to triggering the pathologic activation of Notch1 signaling in diabetes (Fig. 7). The pathologic Dll4–Notch1 loop identified here is consistent with previous observations that increased Dll4 expression was sensitive to GSIs in cells cultured with recombinant Notch ligands (31, 55, 56).
The central role of the Notch1 receptor in the glucose-dependent overactivation of Notch signaling was demonstrated by an improved wound-healing rate in diabetic mice, but not in nondiabetic animals, when Notch1 was specifically ablated in the skin. These results confirmed the important role of Notch1 signaling in postnatal skin physiology. Supporting these results, Notch1 activation has been shown to cause keratinocyte growth arrest, whereas postnatal loss of Notch1 in keratinocytes has been shown to result in epidermal hyperplasia, followed by skin tumors in mice (16, 17). However, deletion of Notch2 or Notch3 in the skin does not result in any apparent phenotype (18, 19).
Together, our results demonstrate that glucose activates a Dll4–Notch1 positive feedback loop that contributes to deleterious wound healing in diabetes. These observations can serve as the basis for the future development of new therapeutic approaches for DFUs, in addition to the standard current interventions (improvement of glycemic control, off-loading, treatment of infections, and revascularization, if needed) for a proper wound healing. Even though the specific genetic Notch1 deletion used in our experimental design is not feasible for clinical use, other specific interventions using specific Notch1 inhibitors have already been tested in clinical trials for other clinical applications (57, 58). A specific blockade of Notch1 for the treatment of diabetic wounds would avoid potential risks of pan-Notch inhibition in the skin, such as a systemic lymphoproliferative disease (19, 59).
Materials and Methods
Materials.
DAPT (catalog no. 565770) and mitomycin C (catalog no. 475820) were obtained from Calbiochem. L-685,458 (catalog no. L1790), DMSO (catalog no. D4540), and STZ (catalog no. S0120) were obtained from Sigma. Recombinant Jagged-1-Fc (catalog no. 599-JG-100), Dll4-Fc (catalog no. 1506-D4/CF), and IgG-Fc (catalog no. 110-HG-100) were obtained from R&D Systems.
Human Skin Biopsies.
Nine patients with DFUs (age, 69.8 ± 3.2 y old; HbA1c, 60.13 ± 4.45 mmol/mol) and 13 age-matched normoglycemic control subjects (age, 68.1 ± 2.7 y old; HbA1c, 39.62 ± 0.84 mmol/mol) were recruited after agreeing and providing their informed consent. Detailed information of these subjects is presented in SI Appendix, Table S2. The Regional Ethical Review Board in Stockholm approved the study. Skin biopsies were taken by using a 4-mm biopsy punch from the abdomen after local anesthesia.
Animals.
C57BL/KsJm/Leptdb (db/db) mice (stock no. 000662) and their normoglycemic heterozygous littermates were obtained from The Jackson Laboratory. GK rats (strain code 460) and Wistar rats (strain code 003) were obtained from Charles River Laboratories. The animals were housed five per cage in a 12-h light/dark cycle at 22 °C and were provided standard laboratory food and water ad libitum. The animals were caged individually for 1 wk, handled daily, and then wounded as described in Wound Model.
STZ-Induced Diabetic Models.
Diabetes was induced in C57BL/6 mice, KRT14-Cre;Notch1fl/fl mice, and their controls by using STZ, according to the instructions from the Animal Models of Diabetic Complications Consortium. Briefly, the animals were administered 50 mg/kg STZ mixed in sodium citrate buffer (i.p. injection) daily for five consecutive days. All of the treated mice became diabetic 2 wk after the first STZ injection. Animals were maintained in a diabetic state for 3 wk before the start of the wound-healing experiment.
Generation of Skin-Specific Notch1 Conditional-Knockout Mice.
Notch1flox/flox mice (stock no. 007181) possessing LoxP sites on either side of exon 1 of the Notch1 gene and transgenic male mice (KRT14-Cre; stock number 004782) expressing Cre recombinase in the skin and hair under the control of a human keratin 14 (KRT14) promoter were obtained from The Jackson Laboratory. Skin-specific Notch1 conditional-knockout mice were generated as follows: Notch1flox/flox mice were crossed with KRT14-Cre mice, and the F1 KRT14-Cre;Notch1flox/+ male offspring were back-crossed to unrelated Notch1flox/flox females. The F2 KRT14Cre;Notch1flox/flox (KRT14Cre;Notch1fl/fl) mice displayed typical hair phenotypes, but not all of the other offspring exhibited this phenotype. Cre-negative Notch1flox/flox mice were used as controls. All of the animals were genotyped by PCR.
Wound Model.
The wound model was deployed as described (60). Briefly, following blood glucose control and general anesthesia, two full-thickness wounds extending through the panniculus carnosus were made on the dorsum on each side of midline by using a 6-mm biopsy punch. Freshly prepared DAPT (100 μM), L-685,458 (100 μM), or DMSO control was applied topically every second day. Digital photographs were obtained on the day of surgery and every other day after wounding. A circular reference was placed nearby to allow for correction of the distance between the camera and the animals. The wound area was calculated by using ImageJ software (Version 1.32; NIH), corrected for the area of the reference circle, and expressed as a percentage of the original area. The data analysis was blinded. In the experiments aiming to analyze histology and mRNA expression during the wound-healing process, the animals were killed on day 7 after surgery when the wounds were ∼50% closed. The wounds were harvested. One wound was snap-frozen in liquid nitrogen, and the other was fixed in 4% paraformaldehyde (PFA) in PBS.
Histology and Immunohistology.
Histological analysis was performed on formalin-fixed, paraffin-embedded (FFPE) sections (5 µm). The slides were stained with hematoxylin and eosin after deparaffinization and rehydration and were examined by two independent observers, who were blinded to the identity of the biopsy. Image analysis and quantification was done by using the smart segmentation feature on Image-Pro Premier software (Version 9.2; Media Cybernetics). At least three images from each tissue were evaluated, and each condition had three to five slides. Granulation was measured as the ratio of the number of cells to the total area of the granulation layer in an image. Microvessel density was presented as the density of GSL-I isolectin B4 bound to microvascular structures after immunohistochemistry staining with biotinylated GSL-I isolectin B4 (catalog no. B-1205; Vector Laboratories). Proliferation in the wound was analyzed by evaluating the percentage of cells positive for PCNA nuclear staining (catalog no. NCL-L-PCNA, Novocastra; Vision Biosystems).
Fluorescent Immunohistochemistry.
The frozen tissue sections were fixed sequentially with 50% acetone for 30 s and 100% acetone for 5 min. For FFPE sections, the sections were deparaffinized and rehydrated, and antigen retrieval was performed in a microwave (800 W for 20 min) by using citrate buffer. The slides were then washed with PBS-T (0.1% Tween) three times for 3 min each. The sections were blocked with goat serum or 5% BSA in PBS for 30 min at room temperature (RT), then incubated with primary antibodies overnight at 4 °C. After three times washing with PBS-T for 3 min each, the sections were incubated with fluochrome-conjugated secondary antibody for 1 h at RT in the dark. After washing, the slides were treated with 1 μg/mL DAPI (Life Technologies) in PBS for 5 min at RT. The FFPE sections were treated with 0.1% Sudan Black-B solution for 10 min to quench autofluorescence. The sections were then mounted and stored at 4 °C. The fluorescent images were acquired by using a Leica DM3000 LED fluorescence microscope, an LSM Meta 510 confocal microscope (Zeiss), or a Leica TCS SP5 confocal microscope (Leica Microsystems). Image analysis was performed by using Image-Pro Premier (Version 9.2) and ImageJ (Version 1.47) software. Antibody information is provided in SI Appendix, section 3.
Cell Culture.
Primary human keratinocytes and HDMECs were obtained from Promocell and cultured in their special medium provided by Promocell. Primary HDFs (Promocell) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; 5.5 mM glucose) supplemented with 100 IU/mL penicillin and streptomycin and 10% heat-inactivated FBS (Invitrogen). All of the cells were maintained in a humidified atmosphere with 5% CO2 at 37 °C in a cell-culture incubator, and passages 4–9 were used for experiments.
Notch-Signaling Activation Using Recombinant Ligands.
Cell-culture plates were first coated overnight with 50 μg/mL Protein G (Invitrogen) in PBS at RT and were then washed twice with PBS and blocked with 3% BSA in PBS for 2 h at RT. The plates were washed again twice with PBS and incubated with mixture of recombinant Jagged1-Fc (1.5 μg/mL) and Dll4-Fc (0.57 μg/mL) or IgG-Fc for 2–4 h at RT. After two washes with PBS, the cells were immediately plated and grown in culture medium.
Plasmids and Transfection.
Plasmids encoding Notch1 ΔE, myc-tagged Notch1 ICD, and GFP were described (61, 62). The cells were transfected with Lipofectamine or Lipofectamine 3000 Reagent (Life Technologies) according to the manufacturer’s instructions. pFLAG-GFP/Notch1 ICD was constructed by generating a PCR fragment of Notch1 ICD from pCMX/Notch1 ICD-myc (61) using primer pairs carrying restriction sites for EcoRI in the 5′ end and the stop codon/BamHI in the 3′ end. The PCR fragment were inserted in frame into the vector from EcoRI–BamHI-digested pFLAG-GFP/mHIF-1α (559–573) (63).
RNA Interference.
siRNA oligonucleotides against human Notch1 (catalog nos. SASI_Hs02_00350287 and SASI_Hs01_00052328), Notch2 (catalog no. SASI_Hs01_00068801), Notch3 (catalog no. SASI_Hs01_00101286), Notch4 (catalog no. SASI_Hs01_00052678), and Dll4 (catalog no. SASI_Hs02_00352665) were obtained from Sigma, and AllStars negative control siRNA from Ambion was used as a control. The final concentration of each individual siRNA oligo in the cell-culture medium was 20 nM. In keratinocytes, 5 nM Silencer Select siRNA for Notch2 (catalog no. s9637; ThermoFisher Scientific) and Notch3 (catalog nos. s9640 and s9642; ThermoFisher Scientific) and corresponding Silencer Select Negative Control No. 2 siRNA (catalog no. 4390847; ThermoFisher Scientific) were also used. Keratinocytes and HDMECs were transfected with siRNAs by using Lipofectamine RNAiMAX Transfection Reagent (ThermoFisher Scientific) or HiPerFect Transfection Reagent (Qiagen), according to the supplier’s protocol. Twenty-four hours after transfection, the cells were exposed to 5.5 or 30 mM glucose for 24 h before harvest.
RNA Purification and Quantitative RT-PCR.
Total RNA was extracted from cells by using an RNeasy RNA-extraction kit (Qiagen) and from skin by using an RNeasy Fibrous Tissue Mini Kit (Qiagen) or an miRNeasy Mini kit (Qiagen). cDNA was obtained by reverse-transcribing total RNA with SuperScript III and first-strand synthesis Supermix for quantitative RT-PCR (Invitrogen) or a High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). Quantitative RT-PCR was performed on a 7300 or 7900 Real-Time PCR System (Applied Biosystems) using SYBR Green Master Mix (ThermoFisher Scientific). The primer sequences are presented in SI Appendix, section 2. The internal controls were PBGD, or the average mRNA expression of PBGD and B2M for mice and rats and the average of PBGD, GNBL2, and RLP32 for humans. Human Dll4 and PBGD gene expressions were detected by using Taqman Gene Expression Assays (catalog nos. Hs00184092_m1 and Hs00609296_g1) and Taqman Universal Master Mix (ThermoFisher Scientific).
RT2 Profiler PCR Array.
cDNA were synthesized by using the RT2 First Strand Kit (Qiagen) from 400 ng of total RNA that was extracted from the skin of db/db and control mice, according to the kit protocol. Quantitative PCR was performed by using the RT2 Profiler PCR array for Mouse Notch Signaling Targets (catalog no. PAMM-259ZR; Qiagen), and the data were analyzed according to the manufacturer’s instructions. Data under the detection limit of the methods were excluded.
Protein Extraction and Western Blotting.
To detect endogenous Notch1 ICD and Hes1 levels, keratinocytes were plated in Jagged1-Fc– and Dll4-Fc–coated plates in medium containing normal (5.5 mM) or high (30 mM) glucose levels for 24 h and exposed to DMSO or 10 μM DAPT for 18 h before the cells were collected. To detect ectopically expressed Notch1 ICD, HDFs were transfected as desired. Transfected cells were then exposed to 5.5 or 30 mM glucose for 2 d and were treated with 10 μM DAPT or the same amount of DMSO for 18 h before harvest. Protein extraction and Western blotting were performed as described (64). Antibody information is provided in SI Appendix, section 3.
Immunocytochemistry for Dll4 Expression in Keratinocytes.
Keratinocytes cultured on chambered cell-culture slides (BD Biosciences) were fixed in 4% PFA at RT for 10 min. After three washes with PBS (5 min each wash), the cells were permeabilized with 0.1% Triton X-100 at RT for 10 min, blocked with 1% BSA for 1 h, and then incubated with anti-Dll4 antibody (catalog no. NB600-892; 1:100; Novus Biologicals) in PBS containing 1% BSA and 0.1% Tween 20 at RT overnight. After three washes with PBS containing 0.1% Tween 20, the cells were incubated with goat anti-rabbit IgG and Alexa Fluor 594 (catalog no. A-11037; 1:500; ThermoFisher Scientific) for 1 h. The slides were then mounted with medium containing DAPI (Prolong Gold Antifade Mountant with DAPI; ThermoFisher Scientific) and observed by using a Leica DM3000 LED fluorescence microscope. The integrated intensity of Dll4 in GFP-positive or GFP-Notch1 ICD-positive cells was evaluated by using Image-Pro Premier (Version 9.2).
In Vitro Angiogenesis Assay.
HDMECs were seeded at a density of 1 × 104 cells per well in 150 μL of culture medium in a 96-well plate precoated with 50 μL of EC-Matrigel per well (catalog no. ECM625; Chemicon). Tube formation was quantified 12 h after various treatments or siRNA transfections by scoring (0–5) the sprouting tube-like structures in five randomly selected fields under an inverted phase contrast microscope at 40× magnification.
In Vitro Migration Assay.
HDF and keratinocyte migration was studied by using the scratch assay as described (65). Briefly, the cells were plated in cell-culture plates that were precoated with collagen (50 μg/mL) and blocked with 3% BSA in PBS. After achieving confluence, cells were serum-starved overnight. A scratch was generated beside a reference marker with a micropipette tip on the following day. After rinsing with PBS, the cells were incubated for an additional 16 h with a GSI (10 μM DAPT) or with control (DMSO) dissolved in DMEM supplemented with 0.2% FBS and various glucose concentrations (5.5 or 30 mM). Mitomycin C (10 μg/mL) was included in the medium to prevent cell proliferation. Pictures were obtained immediately after scratching (basal level) and after 16 h by using a digital camera coupled to an inverted phase-contrast microscope. The reference marker was used to make sure the images taken at 0 and 16 h were from the same scratch edge. The images were quantified by using ImageJ software (Version 1.32). For each image, distances between two sides of the scratch were measured at certain intervals by using ImageJ, and the mean of the distance was calculated. Migration rate was calculated as the difference between the mean of distance at 0 h (Distance_0 h) and at 16 h (Distance_16 h) divided by the mean of distance measured at 0 h [(Distance_0 h − Distance_16 h)/Distance_0 h]. For each experiment in Fig. 3 F and G, the migration rate of control condition where cells were cultured in 5.5 mM glucose in the presence of DMSO was considered as 100, and the migration rates of other samples were calculated as a percentage to the control condition and presented as relative migration in the figure.
Statistics.
Differences between groups were computed by using Student’s t test or one-way analysis of variance followed by Bonferroni’s post hoc test. P < 0.05 was considered statistically significant. All of the in vitro experiments were performed at least thrice. All data are presented as the mean ± SEM.
Study Approval.
The experimental procedure in animals was approved by the North Stockholm Ethical Committee for the Care and Use of Laboratory Animals. The study using human material was reviewed and approved by the Regional Ethical Review Board in Stockholm. Written informed consent was received from participants before inclusion in the study. The study was conducted according to the Declaration of Helsinki’s principles.
Supplementary Material
Acknowledgments
We thank Stina Lindberg for her excellent technical assistance. This work was supported by grants from the Swedish Research Council, the Family Erling-Persson Foundation, the Stockholm County Research Council, Stockholm Regional Research Foundation (ALF), the Bert von Kantzows Foundation, the Swedish Society of Medicine, the Karolinska Institute’s Research Foundations, the Integrated CardioMetabolic Center, the Swedish Cancer Society, and the Strategic Research Programme in Diabetes and in Stem Cells/Regenerative Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900351116/-/DCSupplemental.
References
- 1.Danaei G, et al. Global Burden of Metabolic Risk Factors of Chronic Diseases Collaborating Group (Blood Glucose) National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: Systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2·7 million participants. Lancet. 2011;378:31–40. doi: 10.1016/S0140-6736(11)60679-X. [DOI] [PubMed] [Google Scholar]
- 2.Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract. 2010;87:4–14. doi: 10.1016/j.diabres.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Boulton AJ, Vileikyte L, Ragnarson-Tennvall G, Apelqvist J. The global burden of diabetic foot disease. Lancet. 2005;366:1719–1724. doi: 10.1016/S0140-6736(05)67698-2. [DOI] [PubMed] [Google Scholar]
- 4.Hinchliffe RJ, et al. A systematic review of the effectiveness of interventions to enhance the healing of chronic ulcers of the foot in diabetes. Diabetes Metab Res Rev. 2008;24(Suppl 1):S119–S144. doi: 10.1002/dmrr.825. [DOI] [PubMed] [Google Scholar]
- 5.Brem H, Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J Clin Invest. 2007;117:1219–1222. doi: 10.1172/JCI32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaper NC, Havekes B. Diabetes: Impaired damage control. Diabetologia. 2012;55:18–20. doi: 10.1007/s00125-011-2368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: Mechanisms, signaling, and translation. Sci Transl Med. 2014;6:265sr6. doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markakis K, Bowling FL, Boulton AJ. The diabetic foot in 2015: An overview. Diabetes Metab Res Rev. 2016;32:169–178. doi: 10.1002/dmrr.2740. [DOI] [PubMed] [Google Scholar]
- 9.Phng LK, Gerhardt H. Angiogenesis: A team effort coordinated by Notch. Dev Cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 10.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- 11.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: Recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sweetwyne MT, et al. Notch1 and Notch2 in podocytes play differential roles during diabetic nephropathy development. Diabetes. 2015;64:4099–4111. doi: 10.2337/db15-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hachisuga M, et al. Hyperglycemia impairs left-right axis formation and thereby disturbs heart morphogenesis in mouse embryos. Proc Natl Acad Sci USA. 2015;112:E5300–E5307. doi: 10.1073/pnas.1504529112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min XH, et al. Abnormal differentiation of intestinal epithelium and intestinal barrier dysfunction in diabetic mice associated with depressed Notch/NICD transduction in Notch/Hes1 signal pathway. Cell Biol Int. 2014;38:1194–1204. doi: 10.1002/cbin.10323. [DOI] [PubMed] [Google Scholar]
- 15.Hall E, et al. The effects of high glucose exposure on global gene expression and DNA methylation in human pancreatic islets. Mol Cell Endocrinol. 2018;472:57–67. doi: 10.1016/j.mce.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Rangarajan A, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicolas M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 18.Pan Y, et al. Gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev Cell. 2004;7:731–743. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Dumortier A, et al. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PLoS One. 2010;5:e9258. doi: 10.1371/journal.pone.0009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melnik BC. Does therapeutic intervention in atopic dermatitis normalize epidermal Notch deficiency? Exp Dermatol. 2014;23:696–700. doi: 10.1111/exd.12460. [DOI] [PubMed] [Google Scholar]
- 21.Ali N, et al. Regulatory T cells in skin facilitate epithelial stem cell differentiation. Cell. 2017;169:1119–1129.e11. doi: 10.1016/j.cell.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chigurupati S, et al. Involvement of Notch signaling in wound healing. PLoS One. 2007;2:e1167. doi: 10.1371/journal.pone.0001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimball AS, et al. Notch regulates macrophage-mediated inflammation in diabetic wound healing. Front Immunol. 2017;8:635. doi: 10.3389/fimmu.2017.00635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: Importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerhardt H, et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tammela T, et al. Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature. 2008;454:656–660. doi: 10.1038/nature07083. [DOI] [PubMed] [Google Scholar]
- 27.Benedito R, et al. Notch-dependent VEGFR3 upregulation allows angiogenesis without VEGF-VEGFR2 signalling. Nature. 2012;484:110–114. doi: 10.1038/nature10908. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, et al. Differential roles of PDGFR-alpha and PDGFR-beta in angiogenesis and vessel stability. FASEB J. 2009;23:153–163. doi: 10.1096/fj.08-113860. [DOI] [PubMed] [Google Scholar]
- 29.Ceradini DJ, et al. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10:858–864. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi J, et al. Stromal cell-derived factor-1 effects on ex vivo expanded endothelial progenitor cell recruitment for ischemic neovascularization. Circulation. 2003;107:1322–1328. doi: 10.1161/01.cir.0000055313.77510.22. [DOI] [PubMed] [Google Scholar]
- 31.Fung E, et al. Delta-like 4 induces Notch signaling in macrophages: Implications for inflammation. Circulation. 2007;115:2948–2956. doi: 10.1161/CIRCULATIONAHA.106.675462. [DOI] [PubMed] [Google Scholar]
- 32.Aoyama T, et al. Gamma-Secretase inhibitor reduces diet-induced atherosclerosis in apolipoprotein E-deficient mice. Biochem Biophys Res Commun. 2009;383:216–221. doi: 10.1016/j.bbrc.2009.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Croquelois A, et al. Control of the adaptive response of the heart to stress via the Notch1 receptor pathway. J Exp Med. 2008;205:3173–3185. doi: 10.1084/jem.20081427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Øie E, et al. Activation of Notch signaling in cardiomyocytes during post-infarction remodeling. Scand Cardiovasc J. 2010;44:359–366. doi: 10.3109/14017431.2010.511256. [DOI] [PubMed] [Google Scholar]
- 35.Gude NA, et al. Activation of Notch-mediated protective signaling in the myocardium. Circ Res. 2008;102:1025–1035. doi: 10.1161/CIRCRESAHA.107.164749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kratsios P, et al. Distinct roles for cell-autonomous Notch signaling in cardiomyocytes of the embryonic and adult heart. Circ Res. 2010;106:559–572. doi: 10.1161/CIRCRESAHA.109.203034. [DOI] [PubMed] [Google Scholar]
- 37.Arumugam TV, et al. Gamma secretase-mediated Notch signaling worsens brain damage and functional outcome in ischemic stroke. Nat Med. 2006;12:621–623. doi: 10.1038/nm1403. [DOI] [PubMed] [Google Scholar]
- 38.Lin CL, et al. Modulation of Notch-1 signaling alleviates vascular endothelial growth factor-mediated diabetic nephropathy. Diabetes. 2010;59:1915–1925. doi: 10.2337/db09-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niranjan T, et al. The Notch pathway in podocytes plays a role in the development of glomerular disease. Nat Med. 2008;14:290–298. doi: 10.1038/nm1731. [DOI] [PubMed] [Google Scholar]
- 40.Aster JC, Pear WS, Blacklow SC. The varied roles of Notch in cancer. Annu Rev Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S, et al. Down regulation of Jag-1 in VSMCs contributes to impaired angiogenesis under high glucose condition: Experimental study using aortic rings of rats. Clin Hemorheol Microcirc. 2015;61:497–511. doi: 10.3233/CH-141915. [DOI] [PubMed] [Google Scholar]
- 42.Yoon CH, et al. Diabetes-induced Jagged1 overexpression in endothelial cells causes retinal capillary regression in a murine model of diabetes mellitus: Insights into diabetic retinopathy. Circulation. 2016;134:233–247. doi: 10.1161/CIRCULATIONAHA.116.014411. [DOI] [PubMed] [Google Scholar]
- 43.Vagnozzi AN, Reiter JF, Wong SY. Hair follicle and interfollicular epidermal stem cells make varying contributions to wound regeneration. Cell Cycle. 2015;14:3408–3417. doi: 10.1080/15384101.2015.1090062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel J, Baz B, Wong HY, Lee JS, Khosrotehrani K. Accelerated endothelial to mesenchymal transition increased fibrosis via deleting Notch signalling in wound vasculature. J Invest Dermatol. 2017;138:1166–1175. doi: 10.1016/j.jid.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009;16:55–66. doi: 10.1016/j.ccr.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao L, et al. Modulating Notch signaling to enhance neovascularization and reperfusion in diabetic mice. Biomaterials. 2010;31:9048–9056. doi: 10.1016/j.biomaterials.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hellström M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 48.Chen X, et al. MicroRNA-26a and -26b inhibit lens fibrosis and cataract by negatively regulating Jagged-1/Notch signaling pathway. Cell Death Differ. 2017;24:1431–1442, and erratum (2017) 24:1990. doi: 10.1038/cdd.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majumder S, et al. Shifts in podocyte histone H3K27me3 regulate mouse and human glomerular disease. J Clin Invest. 2018;128:483–499. doi: 10.1172/JCI95946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Q, Wang Y, Zhang T, Zuo W. Notch-mediated Sox9+ cell activation contributes to kidney repair after partial nephrectomy. Life Sci. 2018;193:104–109. doi: 10.1016/j.lfs.2017.11.041. [DOI] [PubMed] [Google Scholar]
- 51.Gallagher KA, et al. Diabetic impairments in NO-mediated endothelial progenitor cell mobilization and homing are reversed by hyperoxia and SDF-1 alpha. J Clin Invest. 2007;117:1249–1259. doi: 10.1172/JCI29710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lobov IB, et al. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci USA. 2007;104:3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suchting S, et al. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc Natl Acad Sci USA. 2007;104:3225–3230. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu F, Yu B, Zhang X, Zhang Y. Cardioprotective effect of Notch signaling on the development of myocardial infarction complicated by diabetes mellitus. Exp Ther Med. 2017;14:3447–3454. doi: 10.3892/etm.2017.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ridgway J, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444:1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 56.Benedito R, et al. The Notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 57.Wu Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 58.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling–Are we there yet? Nat Rev Drug Discov. 2014;13:357–378. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 59.Demehri S, et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6:e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Botusan IR, et al. Stabilization of HIF-1alpha is critical to improve wound healing in diabetic mice. Proc Natl Acad Sci USA. 2008;105:19426–19431. doi: 10.1073/pnas.0805230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gustafsson MV, et al. Hypoxia requires Notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 62.Kallio PJ, et al. Signal transduction in hypoxic cells: Inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 1998;17:6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng X, et al. Cell-type-specific regulation of degradation of hypoxia-inducible factor 1 alpha: Role of subcellular compartmentalization. Mol Cell Biol. 2006;26:4628–4641. doi: 10.1128/MCB.02236-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gu HF, et al. Impact of the hypoxia-inducible factor-1 α (HIF1A) Pro582Ser polymorphism on diabetes nephropathy. Diabetes Care. 2013;36:415–421. doi: 10.2337/dc12-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W, et al. Signals that initiate, augment, and provide directionality for human keratinocyte motility. J Invest Dermatol. 2004;123:622–633. doi: 10.1111/j.0022-202X.2004.23416.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.