Significance
Cells form structures and perform functions through intricate 3D tissue organizations. However, due to tissue scattering, coherent Raman microscopy—a powerful method complementary to fluorescence imaging—suffers from limited imaging depth in tissues. Here, we develop a volumetric chemical imaging method with greater than 10-fold depth increase. We formulate a Raman-tailored tissue-clearing recipe and combine it with advanced Raman microscopies. Equipped with the toolbox of volumetric chemical imaging and analyses, we elucidate complex 3D structures, chemical compositions, and metabolic dynamics in diverse tissues including lipid synthesis throughout tumor spheroids, 3D networks of axons, vasculatures, and cell bodies in brain regions, as well as heterogeneous tumor structures and tumor metabolism.
Keywords: volumetric imaging, stimulated Raman scattering, tissue clearing, metabolic imaging, cancer metabolism
Abstract
Three-dimensional visualization of tissue structures using optical microscopy facilitates the understanding of biological functions. However, optical microscopy is limited in tissue penetration due to severe light scattering. Recently, a series of tissue-clearing techniques have emerged to allow significant depth-extension for fluorescence imaging. Inspired by these advances, we develop a volumetric chemical imaging technique that couples Raman-tailored tissue-clearing with stimulated Raman scattering (SRS) microscopy. Compared with the standard SRS, the clearing-enhanced SRS achieves greater than 10-times depth increase. Based on the extracted spatial distribution of proteins and lipids, our method reveals intricate 3D organizations of tumor spheroids, mouse brain tissues, and tumor xenografts. We further develop volumetric phasor analysis of multispectral SRS images for chemically specific clustering and segmentation in 3D. Moreover, going beyond the conventional label-free paradigm, we demonstrate metabolic volumetric chemical imaging, which allows us to simultaneously map out metabolic activities of protein and lipid synthesis in glioblastoma. Together, these results support volumetric chemical imaging as a valuable tool for elucidating comprehensive 3D structures, compositions, and functions in diverse biological contexts, complementing the prevailing volumetric fluorescence microscopy.
Three-dimensional structures of biological tissues are closely associated with their functions in both health and disease. Representative examples range from neuroscience and cancer biology, to developmental biology: 3D distributions of neurites determine neuronal wiring of the brain (1–6); 3D interactions between tumor cells and their microenvironment influence tumor growth, invasion, and metastasis (7, 8); and 3D organ morphogenesis reveals key mechanisms of embryonic development (9, 10). Therefore, the ability to visualize structures and functions of tissues in 3D is crucial for enhancing fundamental understanding across biomedical disciplines.
Toward this goal, light microscopy presents an appealing tool to noninvasively probe biological processes with subcellular resolution. In particular, fluorescence microscopy is the method of choice for bio-imaging, offering high sensitivity, molecular specificity, and biocompatibility (11–13). However, its imaging depth is limited to superficial layers of tissues due to inevitable light scattering originating from heterogeneous refractive indices (RIs) within tissues. Typically, fluorescence imaging depth in tissues is limited to ∼500 µm (1 mm in rare cases), achieved by two-photon excited fluorescence microscopy (14). A number of advanced techniques have been developed to extend this depth limit, including adaptive optics, longer excitation wavelengths, and higher-order nonlinear excitation (15–19). Unfortunately, these techniques usually require complicated instrumentations or procedures, and the achievable depth extensions are often within a factor of two or three.
Recently, a series of tissue-clearing–based fluorescence microscopies have emerged and demonstrated remarkable results toward volumetric visualization of whole organs (20, 21). Compared with instrumentation-based approaches, these sample-centered methods adopt a fundamentally different strategy by reducing light scattering with active homogenization of tissue RIs (20, 21). Enabled by tissue clearing, volumetric fluorescence imaging provides a window to peer deep into a variety of tissues (e.g., brain, lung, bones) of different organisms in both physiology and pathology (e.g., Alzheimer’s disease, cancer) (7, 8, 22–25).
Complementary to fluorescence microscopy in many aspects, stimulated Raman scattering (SRS) microscopy allows for imaging of chemical bonds in biological samples with subcellular resolution. By harnessing quantum amplification via stimulated emission, SRS microscopy offers bond-selective vibrational specificity with high sensitivity (down to micromolars), fast acquisition (up to video-rate), and general biocompatibility, making it a powerful technique for biomedicine (26–29). Compared with X-ray tomography (30), magnetic resonance imaging (31), and positron emission tomography (32), SRS offers higher spatial resolution and unique bond-selective specificity. However, the typical imaging depth of SRS has been limited to ∼100 µm inside highly scattering tissues, such as the brain, because of the scattering loss of laser power and overwhelming background (29). In less scattering tissues, the imaging depth is still limited to 300–500 μm. Therefore, extending SRS microscopy to deep volumetric imaging (millimeter range) would provide holistic chemical information in a host of environments relevant to biomedicine.
Inspired by the success of tissue clearing, we devised an SRS-based volumetric chemical imaging method to generate chemical-specific 3D maps deep into tissues. We note that many of tissue-clearing methods developed for volumetric fluorescence imaging alter the tissue composition, and are thus unsuited for comprehensive chemical imaging. Therefore, we identified two Raman-specific criteria, developed a Raman-tailored tissue-clearing strategy, and coupled it with SRS microscopy, including its advanced variations, such as multispectral imaging and deuterium oxide (D2O) probing coupled with SRS (DO-SRS) of metabolic probes (33) (Fig. 1A). Our method achieves a greater than 10-times depth increase compared with the standard SRS. We further developed a volumetric phasor analysis for 3D clustering and segmentation of multispectral SRS images. With this whole toolkit, we reveal complex 3D tissue structures, compositions, and functions in the context of neurobiology and cancer biology.
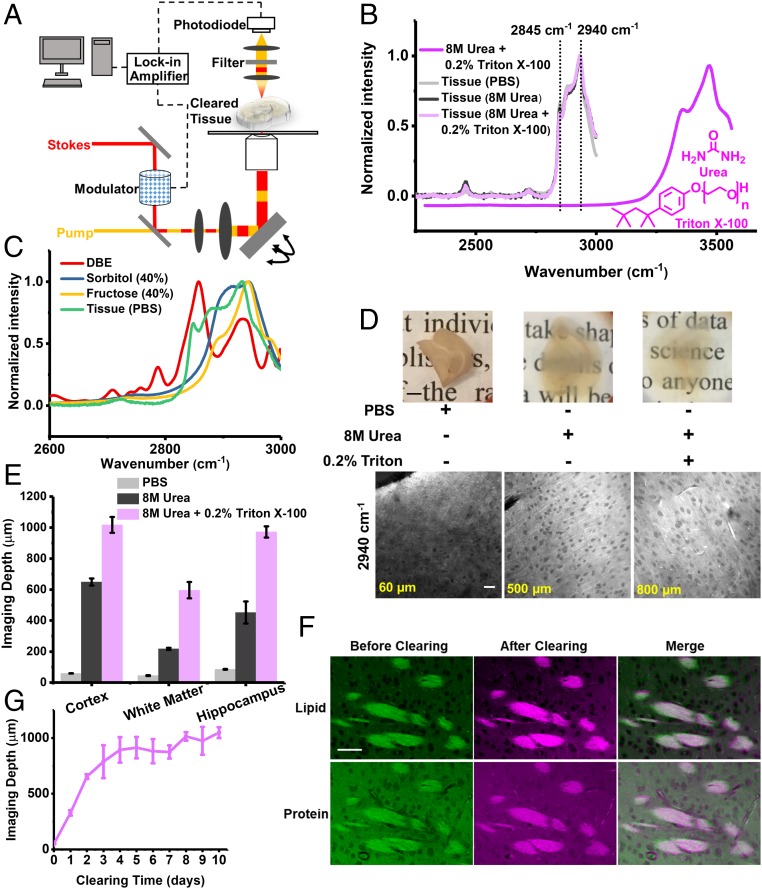
Fig. 1.
Development of clearing-enhanced SRS microscopy. (A) Set-up for clearing-enhanced SRS microscopy. (B) Spontaneous Raman spectra of 8 M urea and 0.2% Triton X-100 (dark magenta), mouse brain slices in PBS (light gray), slices cleared with 8 M urea (dark gray), and slices cleared with 8 M urea solution with 0.2% Triton X-100 (light magenta). (C) Spontaneous Raman spectra of dibenzyl ether (red), 40% sorbitol solution (blue), 40% fructose solution (yellow), and mouse brain slices in PBS (green, the same as B). (D) Photos and the corresponding SRS images (2,940 cm−1) close to depth limit (60, 500, and 800 µm) of 1-mm-thick brain slices in PBS, cleared with 8 M urea, and with 8 M urea and 0.2% Triton X-100, respectively. [Scale bars: 2 mm in the photos (Upper), 50 µm in the SRS images (Lower).] (E) Quantification of label-free imaging depths for different brain regions in PBS, cleared with 8 M urea, and cleared with 8 M urea and 0.2% Triton X-100. Error bars indicate SEM (n = 4, 6, and 4 for cortex, white matter, and hippocampus in PBS. n = 4, 3, and 3 for cortex, white matter, and hippocampus in 8 M urea. n = 3 for each brain region in 8 M urea and 0.2% Triton X-100). The imaging depth limit was defined as the depth with signal-to-background ratio of 0.5. (F) SRS images of regions in the same white matter region before and after tissue clearing (8 M urea with 0.2% Triton X-100). (Scale bars, 50 µm.) (G) Quantification of imaging depth with clearing time (8 M urea with 0.2% Triton X-100). Error bars indicate SD (n = 3 for each data point).
Results
Development of Raman-Tailored Reagents for Clearing-Enhanced SRS Imaging.
To develop a clearing recipe tailored for SRS microscopy, we identified two Raman-specific criteria that differ from those for volumetric fluorescence imaging. First, the tissue-clearing chemicals should achieve certain “Raman transparency,” defined as the ability to generate optically transparent tissues without introducing interfering Raman signatures in the designated vibrational windows. While a general method to create transparency across the entire Raman spectrum, including the fingerprint region, would be rather challenging, herein we focus on two important spectral windows: the high-wavenumber C-H region (2,800–3,000 cm−1) and the cell-silent region (1,800–2,600 cm−1). The vibrational signatures for lipids and proteins are in the high-wavenumber C-H region, represented by CH2 (2,845 cm−1) and CH3 (2,940 cm−1) bands, respectively (34, 35) (Fig. 1B). The interplay between proteins and lipids has important implications in cancer (36, 37) and Alzheimer’s disease (38, 39). For the cell-silent window, many emerging vibrational tags reside in this region (40–44), allowing imaging of metabolic dynamics of small metabolites and also super multiplexed imaging (45, 46). Hence, our first criterion requires the clearing reagents to make tissues optically transparent without introducing Raman peaks in the CH region or the cell-silent region. Second, the native chemical structures of examined tissues should be largely preserved after tissue clearing. Here the clearing process should maintain the integrity of both proteins and lipids. This is a rather challenging requirement because many clearing methods [including almost all organic-solvent and hydrogel-embedding–based methods (20, 21)] are optimized for imaging specific proteins by removing lipids to reduce the heterogeneity of RIs.
Based on the two Raman-specific criteria, we developed a clearing recipe for Raman microscopy. We examined existing optical-clearing reagents and identified urea as a promising candidate. In fact, urea is the central component in Scale/ScaleS (2, 47). In terms of our first criterion about Raman transparency, urea shows no Raman peaks in a broad window from 1,800 to 3,000 cm−1 (SI Appendix, Fig. S1A) by virtue of its unique structure containing only N-H bonds, but no C-H bonds. In contrast, several common clearing reagents—dibenzyl ether [3DISCO (22)], sorbitol [ScaleS (47)], and fructose [SeeDB (5) and FRUIT (48)]—show strong Raman peaks in the CH stretching region (2,800–3000 cm−1) (Fig. 1C), causing severe background issues. To examine the second criterion about maintenance of tissue integrity, we adopted the spontaneous Raman spectrum as a benchmark because it is highly sensitive to chemical structures. We observed minimum change of characteristic Raman peaks in the CH stretching region for tissues incubated in 8 M urea for 4 d (Fig. 1B, dark gray) compared with those in PBS (Fig. 1B, light gray), indicating preservation of proteins and lipids. Our results agree well with a previous report that urea can preserve the integrity of both protein and lipid ultrastructures at the resolution of electron microscopy (47).
After validating the two Raman-specific clearing criteria, we verified that a high concentration of urea (8 M) was capable of rendering tissues substantially less opaque than the control (Fig. 1D). This effect is likely due to the small size of urea that allows it to penetrate, partially denature, and hydrate the hydrophobic core of high-refractive index proteins (2, 20, 21, 47). We then performed label-free SRS imaging on brain tissues cleared with urea. Indeed, we did not observe background interference from high-concentration urea in either band (2,845 cm−1 or 2940 cm−1). We defined the SRS imaging depth limit as the depth at which the signal-to-background ratio of SRS images at 2940 cm−1 reaches 0.5, below which imaging contrast became too low. By this measure, we found that urea extended the SRS imaging depth for ∼5–10 times at different brain regions (Fig. 1E). Representative SRS images (2,940 cm−1) of 1-mm-thick brain slices are shown in Fig. 1D at depths close to the defined imaging depth limits: 60 µm for the control slice in PBS and 500 µm for the slice treated with 8 M urea. This clearing solution of 8 M urea is well suited for volumetric chemical imaging of multicellular spheroids (Fig. 2).
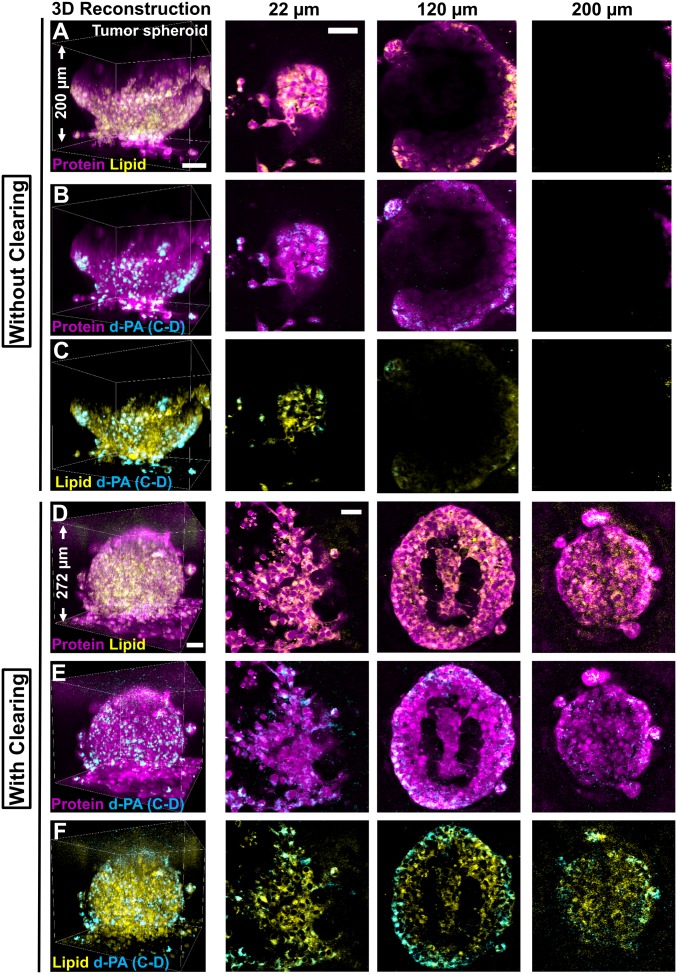
Fig. 2.
Clearing-enhanced volumetric chemical imaging of 3D tumor spheroids. (A–C) Three-dimensional reconstructions and representative 2D images (at z = 22 µm, 120 µm, and 200 µm) of a tumor spheroid of MCF-10AHras breast cells without tissue clearing. (D–F) Three-dimensional reconstructions and representative 2D images (at z = 22 µm, 120 µm, and 200 µm) of a tumor spheroid of MCF-10AHras breast cells with tissue clearing. Protein (magenta) and lipid (green) channels are overlaid in A and D. Protein (magenta) and the d-PA (cyan) channels are overlaid in B and E. Lipid (green) and d-PA (cyan) channels are overlaid in C and F. (Scale bars, 50 µm.)
Improving Raman-Tailored Clearing for Volumetric SRS Imaging.
To improve Raman-tailored clearing for imaging deep tissues, we asked whether adding auxiliary reagents could further improve clearing performance. We found addition of 0.2% Triton X-100, a typical detergent, allows substantial depth extension with well-preserved features (axon fibers and cell bodies) in brain tissues (Fig. 1D). This is perhaps due to increased membrane permeability effected by Triton X-100. Note that 0.2% Triton X-100 alone has only marginal effect on imaging depth (SI Appendix, Fig. S2).
We next validated the fulfillment of two Raman-clearing criteria with 8 M urea and 0.2% Triton X-100. First, 0.2% Triton X-100 introduced no extra Raman background at the CH imaging-window (Fig. 1B, dark magenta). Second, we found minimum changes of characteristic Raman peaks in the CH region (Fig. 1B, light magenta). We further confirmed the distributions of proteins and lipids in tissues were preserved after clearing, using SRS imaging at different brain regions. The SRS images of the same regions aligned well with each other at the subcellular level before and after clearing (Fig. 1F and SI Appendix, Fig. S3). We found a less than 7% increase in protein/lipid ratio comparing the same regions before and after clearing (SI Appendix). As a negative control, treating tissues with a high concentration (15%) of Triton X-100 or the 3DISCO method clearly disrupted the lipid structure (4, 22), as indicated by the attenuation of the lipid peak at 2,845 cm−1 (SI Appendix, Figs. S4A and S5A) and the disappearance of lipid-rich structures (nerve tracts and axon fibers) (SI Appendix, Figs. S4B and S5B). While this recipe of 8 M urea and 0.2% Triton X-100 preserved proteins and lipids in tissues, it removed lipids from multicellular spheroids (SI Appendix, Fig. S6), possibly because compact tissue structures are more resistant to Triton X-100. Therefore, the basic recipe of 8 M urea is the method of choice for volumetric chemical imaging of spheroids (Fig. 2), while the improved recipe of 8 M urea and 0.2% Triton X-100 was used for tissue samples.
Clearing-Enhanced SRS Imaging with Greater than 10 Times Depth Extension.
We then confirmed the optimal clearing time of the improved recipe (8 M urea and 0.2% Triton X-100) on 1-mm-thick coronal brain slices (Fig. 1G and SI Appendix, Fig. S7). The clearing effect plateaued after a 4-d incubation (Fig. 1G). As reported previously (2), we observed a slight volume expansion of cleared brain tissues with an average expansion factor of 1.2 ± 0.1 (SI Appendix). Such a degree of expansion would allow moderately higher imaging resolution, as is harnessed in expansion microscopy (49).
We next determined the depth extension of clearing-enhanced SRS imaging. Remarkably, our improved Raman-tailored clearing recipe increased the depth limit of SRS imaging by 17.2-fold (13.8-fold with correction for expansion) in the cerebral cortex, 13.2-fold (10.6-fold with correction) in the white matter, and 11.4-fold (9.1-fold with correction) in the hippocampus (Fig. 1E). The different depth extensions likely originated from varied chemical compositions among these brain regions. For example, the white matter has more lipids than the cortex and the hippocampus, leading to higher RI heterogeneity, severer scattering, and thus a shallower imaging depth. In summary, we have achieved label-free, dual-color volumetric chemical imaging approximately 1-mm deep in highly scattering tissues.
Clearing-Enhanced SRS Imaging of Whole Tumor Spheroids.
With the recipe of 8 M urea, we aimed to achieve volumetric imaging of multicellular tumor spheroids. Tumor spheroids are widely used in vitro models of solid tumors. Compared with 2D cultures, tumor spheroids are considered to better mimic the 3D tumor microenvironment and are thus increasingly used to study tumor cell physiology and response of tumors to radiotherapy and chemotherapy in a 3D multicellular context (50, 51). In addition to the two-color label-free imaging, we further introduced a third color for bioorthogonal chemical imaging (40, 41). This method allowed imaging not only the spatial distributions of preexisting proteins and lipids, but also the metabolic activity of newly synthesized lipids by incubating spheroids with deuterated palmitic acid (d-PA) (SI Appendix, Fig. S1B) (2,109 cm−1).
Without tissue clearing, SRS imaging of spheroids—prepared from Ras-transformed MCF-10A breast epithelial cells—captured the structure of no more than half of a spheroid (∼120 µm). The spherical shape of multicellular aggregates with tight cellular packing introduced severe light scattering (Fig. 2 A–C). In contrast, with tissue clearing using 8 M urea, the distributions of proteins, preexisting lipids, and newly synthesized lipids were visualized at subcellular resolution throughout the entire spheroid (Fig. 2 D–F). The intricate inner architecture of the spheroid was captured showing the necrotic core (Fig. 2 D–F). Interestingly, while preexisting lipids were distributed relatively evenly throughout the whole 3D structure (Fig. 2 D–F, yellow), d-PA was preferentially incorporated into membrane structures of the cells at the invasion front or the edge of the spheroid (Fig. 2 D–F, cyan). We speculate that such differential enrichment might be correlated with higher metabolic demand of the cells near the invasion front for cell division (52). Without tissue clearing, we could have mistakenly concluded that these tumor spheroids had a hollow structure without any preferential enrichment of newly synthesized lipids (Fig. 2 A–C). Hence, we demonstrated that volumetric metabolic imaging is informative for probing the heterogeneity of tumor activity in 3D spherical models during invasion.
Clearing-Enhanced SRS Imaging of the Mouse Brain.
The development of volumetric imaging has been driven by a strong interest in understanding 3D structure–function relationships in complex brain networks. We thus conducted two-color clearing-enhanced SRS imaging across a variety of mouse brain regions. Pure protein and lipid images were obtained with a linear combination algorithm (SI Appendix). As shown in Fig. 3A, this volumetric chemical imaging method is capable of imaging from the hippocampus all the way to the white matter with a total depth of ∼1.1 mm. In the hippocampus, a key region involved in learning and memory, the distribution of proteins highlights blood vessels and cell bodies while the distribution of lipids delineates axon fibers (Fig. 3A and SI Appendix, Fig. S8 A, I, and M). Because of the effective clearing, the representative 2D images at increasing depths maintained high-resolution contrast for axon fibers, cell bodies, and even individual nucleoli in the dentate gyrus (Fig. 3 B–E and SI Appendix, Fig. S8 B–D and J–L). Compared with the hippocampus, the white matter contains many lipid- and protein-rich nerve tracts (Fig. 3 A and F and SI Appendix, Fig. S8 E–G, arrows) with complex topology, likely related to its role in relaying information between different brain regions.
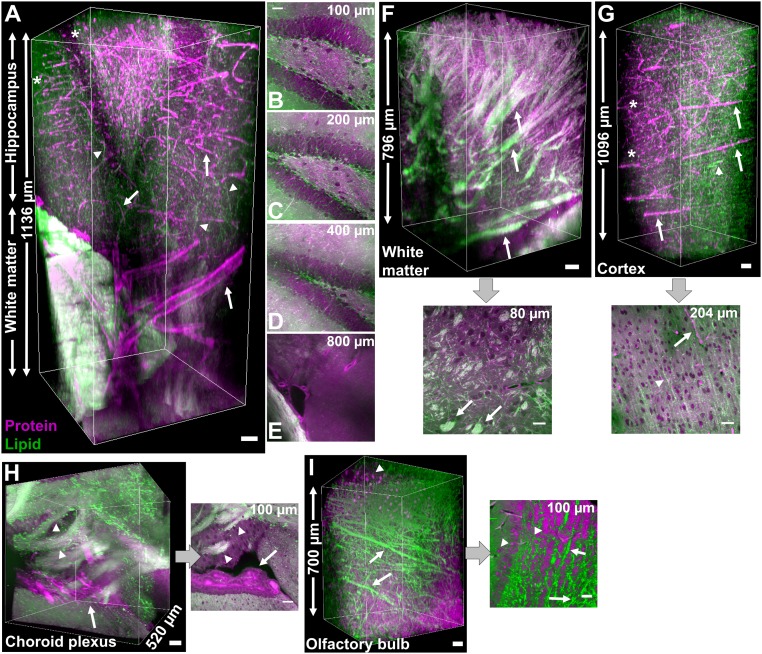
Fig. 3.
Clearing-enhanced volumetric chemical imaging of various regions in mouse brain tissues. (A) Three-dimensional reconstruction of the hippocampus and white matter. Arrows indicate vasculatures, arrowheads indicate axons, and stars indicate cell bodies. (B–E) Representative 2D images at indicated depths in A. (F) Three-dimensional reconstruction and a representative 2D image of the white matter. Arrows indicate nerve tracts. (G) Three-dimensional reconstruction and a representative 2D image of the cerebral cortex. Arrows indicate vasculatures, arrowheads indicate axons, and stars indicate cell bodies. (H) Three-dimensional reconstruction and a representative 2D image of the choroid plexus. Arrows indicate the choroid plexus and arrowheads indicate nerve tracts. (I) Three-dimensional reconstruction and a representative 2D image of the olfactory bulb. Arrows indicate axon fibers and arrowheads indicate cell bodies. Proteins are in magenta and lipids are in green. (Scale bars, 50 µm.)
We also acquired volumetric images of other brain regions: the cerebral cortex (Fig. 3G), the choroid plexus (Fig. 3H and SI Appendix, Fig. S8H), and the olfactory bulb (Fig. 3I). In the cerebral cortex, we observed partially aligned axon fibers (Fig. 3G, arrowheads) together with several thick capillaries toward the same direction (Fig. 3G, arrows). These results suggest an interplay between axon guidance and angiogenesis (53), which would be difficult to capture without volumetric imaging. Punctate protein signals indicated cell bodies in the cortex (Fig. 3G, stars). In addition, the choroid plexus was observed to be a protein-rich plexus of cells surrounded by many lipid-rich nerve tracts (Fig. 3H and SI Appendix, Fig. S8H), likely related to its function of secreting cerebrospinal fluid into ventricles. Interestingly, we observed high-density axon fibers (Fig. 3I, arrows) and cell bodies (Fig. 3I, arrowheads) in the olfactory bulb. These results demonstrate two-color label-free volumetric chemical imaging as a valuable method for providing 3D distributions of blood vessels, axon fibers, and cell bodies in brain tissues with chemical information.
Multispectral SRS Imaging with Volumetric Phasor Analysis.
Next we extended our technique to multispectral SRS imaging in which a broad spectrum was acquired for every pixel, thus offering rich spectroscopic information. To achieve robust 3D image segmentation for analysis and quantification, we developed a spectral phasor method—sensitive to small spectral variations of multispectral images—for volumetric chemical analysis in tissues. Different from the phasor analysis performed on single-plane images (54), our volumetric phasor analysis transforms all voxels in the entire imaging volume to phasor points on a single phasor plot (Fig. 4A). Therefore, clustering and segmentation is performed collectively for the entire imaging volume, resulting in systematic and holistic segmentation of tissue components.
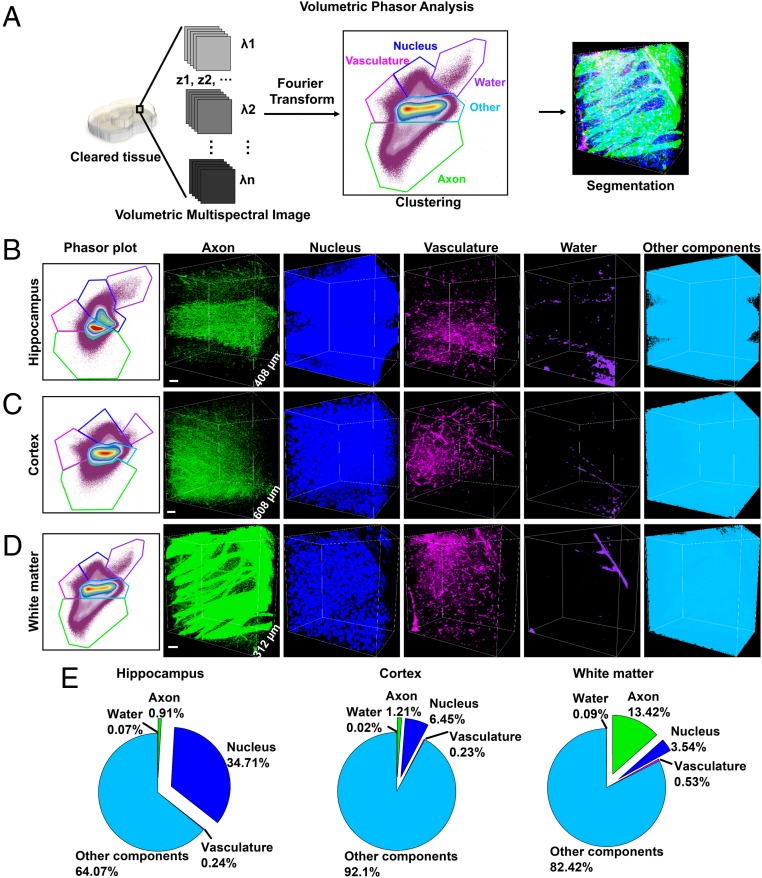
Fig. 4.
Volumetric phasor analysis. (A) Schematic process of volumetric phasor analysis. Volumetric phasor analysis provides segmentation of various regions in mouse brains: the hippocampus (B), the cerebral cortex (C), and the white matter (D). For B–D, the phasor plot is shown (Left) and followed by volumetric images of segmented species. “Other components” include extracellular matrix and cellular structures that are not resolved by SRS. (E) Volume fraction of tissue components in the hippocampus (B), the cerebral cortex (C), and the white matter (D). (Scale bars, 50 µm.)
We demonstrated volumetric phasor analysis on mouse brain tissues. To implement this analysis, we first acquired volumetric multispectral images by taking z-stack images across 13 different wavenumbers covering 2,810–3,090 cm−1 (SI Appendix). Second, we generated a reference phasor plot by selectively cropping out individual regions of axons, vasculature, nuclei, or extracellular matrix and transforming these regions onto a phasor plot. Indeed, each structure was identified as a separable cluster on the reference phasor plot (SI Appendix, Fig. S9). Third, we transformed the whole stack of volumetric multispectral images onto a phasor plot (Fig. 4 B–D) and clustered this phasor plot according to the reference phasor plot. Fourth, we projected these clusters back to the volumetric image and evaluated the clustering results. As shown in Fig. 4 B–D and SI Appendix, Fig. S10, we reliably segmented axons, nuclei, vasculature, water, and other components (including extracellular matrix and cellular structures that were not resolved by SRS) in the cerebral cortex, the hippocampus, and the white matter. The volume fractions of tissue components showed that the hippocampus contained more nuclei densely packed in the dentate gyrus than the cortex and the white matter, while the white matter contained more axons as nerve tracts than the other two brain regions (Fig. 4E). These results indicate volumetric phasor analysis achieves rigorous, chemically specific segmentation for a large number of species in 3D.
Clearing-Enhanced SRS Imaging of Tumors with Volumetric Phasor Analysis.
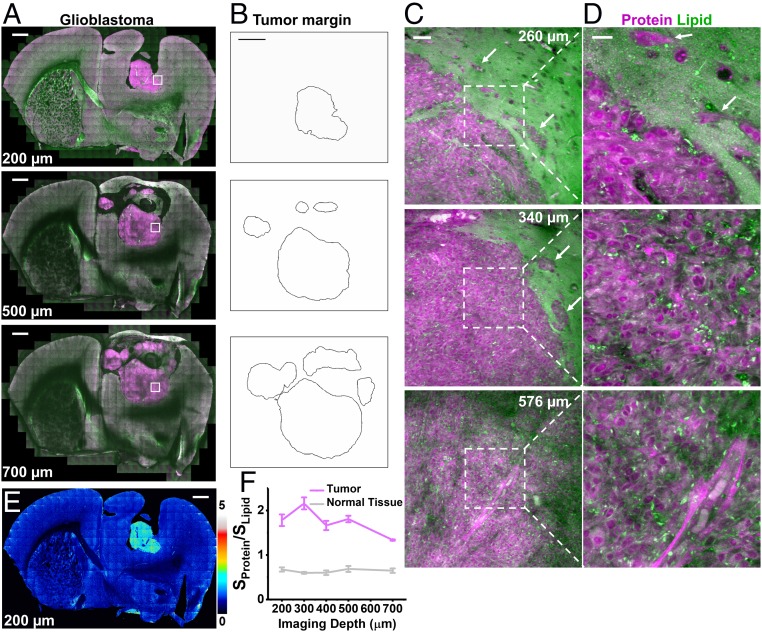
We applied the two-color label-free volumetric imaging to capture 3D chemical information in tumor xenografts. Glioblastoma multiforme is the most aggressive brain cancer with a median survival time of ∼15 mo from diagnosis (55). To investigate its 3D structure, we performed large-area mosaic imaging up to 700 µm into the glioblastoma xenograft in mouse brain (Fig. 5A). The high-protein, low-lipid distribution of the tumor allowed delineation of tumor margins (Fig. 5B) (36, 37). Single-plane images at a series of depths through the same tumor (Fig. 5 A, C, and D and SI Appendix, Fig. S11) revealed distinct features of normal tissues, infiltrating glioma, and dense glioma (36, 37). Dense glioma has a high level of proteins and a low level of lipids. In contrast, normal brain tissues are lipid-rich, containing axon fibers and low density of cell bodies. Infiltrating glioma at the tumor margins (Fig. 5 C and D, arrows, and SI Appendix, Fig. S11 B and C) show lipid-rich areas of brain parenchyma with clusters of infiltrating glioblastoma cells. We validated this method of tumor demarcation by immunolabeling with a human-specific vimentin antibody, which specifically stained human-sourced tumor cells but not mouse cells (SI Appendix, Fig. S12). The z-stack results indicate the margins of the tumor differ at different imaging depths, as the size of the tumor gradually expands with more complex structures from 200 to 700 µm (Fig. 5 A–D and SI Appendix, Fig. S11). We mapped out the protein/lipid ratios (Sprotein/Slipid, the signal of the protein channel over that of the lipid channel) of the whole coronal slices as a function of depth (Fig. 5 E and F and SI Appendix, Fig. S13). This single parameter indicates chemical heterogeneity, both between tumor and normal tissues and within tumor tissues. These results render volumetric chemical imaging a capable tool for capturing the complex geometry and composition of the tumor, because 2D images at a single shallow depth could miss critical information.
Fig. 5.
Clearing-enhanced volumetric chemical imaging of glioblastoma in mouse brain. (A) Representative 2D images of the tumor in the whole coronal slice at increasing imaging depths. (B) Tumor margins outlined from A. (C and D) Zoomed-in 2D images of the white-boxed region in A at increasing imaging depths. (E) Protein/lipid ratiometric image at 200-µm depth. (F) Plots of protein/lipid signal ratios as a function of imaging depths in tumor and normal tissues. Error bars indicate SD. (n = 3 for each data point) Proteins are in magenta and lipids are in green. Infiltrating glioblastoma cells are indicated by arrows in C and D. (Scale bars: 1 mm in A, B, and E; 50 µm in C; 20 µm in D.)
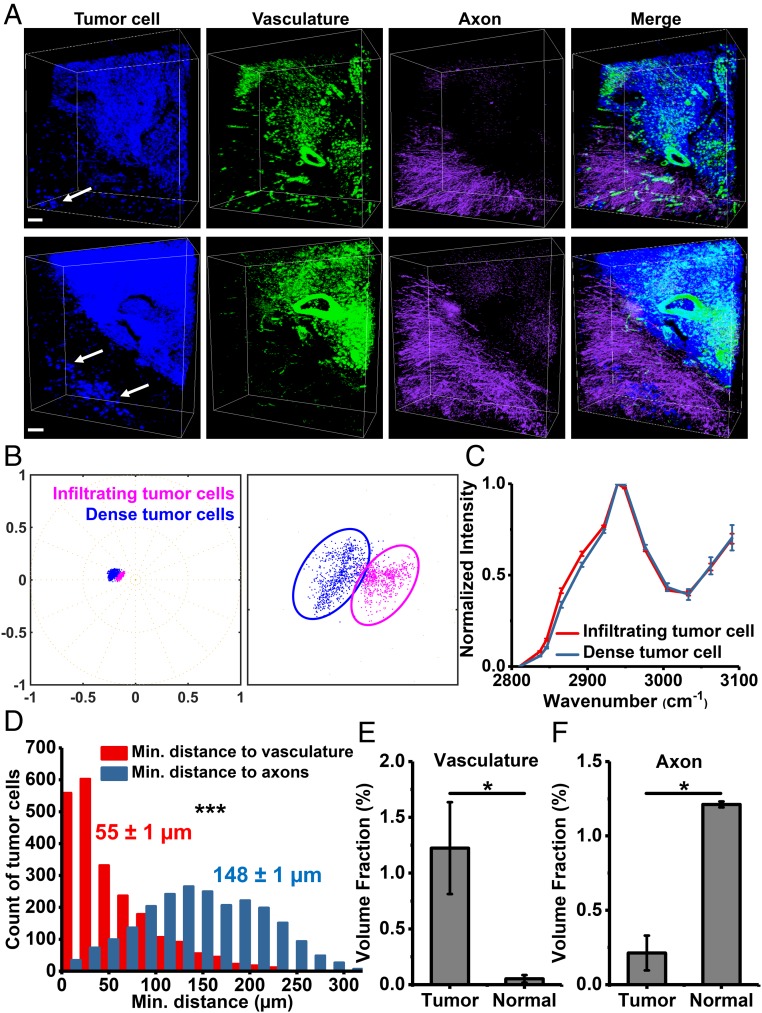
Next we quantitatively analyzed tissue structures of brain glioblastoma using multispectral SRS imaging together with volumetric phasor analysis. The reference phasor plot showed five clusters representing axons, vasculature, tumor cells, extracellular matrix of normal brain tissue, and normal cell nuclei (SI Appendix, Fig. S14A). We segmented axons, vasculature, and tumor cells for further analysis (Fig. 6A and SI Appendix, Fig. S14B). The distribution of tumor cells showed the boundary between normal and dense tumor tissues (Fig. 6A). With volumetric phasor analysis, infiltrating tumor cells were readily identified to spread beyond tumor margins and disperse in normal tissues (Fig. 6A, arrows). We validated volumetric phasor analysis for identifying tumor margins and infiltrating tumor cells by immunolabeling (SI Appendix, Fig. S15) and stimulated Raman histology (56) (SI Appendix, Fig. S16). Infiltrating tumor cells are of clinical importance, because these cells are often left behind after surgical resection and give rise to recurrence and poor prognosis (36, 37). We hypothesized that these properties of infiltrating tumor cells might be associated with their unique chemical profile. To test this hypothesis, we selected individual infiltrating tumor cells and dense tumor cells and transformed the multispectral data of these cells onto a phasor plot (Fig. 6B). Indeed, these two types of tumor cells formed two clusters on the phasor plot, indicating their differences in chemical composition (Fig. 6B). We then compared averaged single-cell SRS spectrum of infiltrating tumor cells to that of dense tumor cells (Fig. 6C). The spectrum of infiltrating tumor cells showed a slightly higher shoulder peak, from 2,837 cm−1 to 2,921 cm−1, than that of dense tumor cells (Fig. 6C). These spectral differences indicate a higher lipid-to-protein ratio for the infiltrating tumor cells, which was not known before and implies that glioblastoma exhibit cellular heterogeneity in chemical composition. The altered composition of infiltrating tumor cells might derive from their distinct metabolism (57, 58) and enhance cell motility and invasion (59).
Fig. 6.
Segmentation and quantification of brain glioblastoma tissues using volumetric phasor analysis. (A) Volumetric images of segmented tumor cells, vasculature, and axons in two different regions of brain glioblastoma. Arrows indicate infiltrating tumor cells. (Scale bars, 50 µm.) (B) Phasor plots of infiltrating tumor cells (n = 6 cells) and dense tumor cells (n = 6 cells). The plot (Right) is a zoom-in of the Left. (C) SRS spectra of infiltrating tumor cells (n = 6 cells) and dense tumor cells (n = 6 cells). Error bars are SD. (D) Histograms of the minimal distances from individual tumor cells (n = 2,265) to vasculature and axons. Mean ± SEM are shown near the histograms. (E) Volume fraction of vasculature in tumor (n = 5) and normal (n = 3) tissues. Error bars are SEM. (F) Volume fraction of axons in tumor (n = 3) and normal (n = 3) tissues. Error bars are SEM. Statistical significance was determined by pair-sample two-tailed t test. *P < 0.05 and ***P < 0.001.
In addition, we found tumor tissues were associated with more blood vessels and fewer axons than neighboring normal brain tissues (Fig. 6A). Hence, we quantified the minimal distances from individual tumor cells to vasculature and axons (Fig. 6D). Indeed, the average minimal distance from tumor cells to axons was almost three times of that to vasculature (Fig. 6D). We also found a much higher volume fraction of vasculature but a much lower volume fraction of axons in tumor tissues than those in normal tissues (Fig. 6 E and F). The differential distribution of axons agreed with previous reports (36, 37). Furthermore, the vasculature in tumor tissues contained very thick blood vessels and showed extravasation of blood cells (Fig. 6A), which agreed with the abnormal structure of dilated and leaky vasculature in the tumor (60). The high volume fraction and abnormal morphology of tumor-associated vasculature suggest the dysregulated, sustained process of angiogenesis, one of the hallmarks of cancer (52). These heterogeneous information of multiple species—revealing morphology and distributions inside tissues in a label-free and quantitative manner—would be difficult to acquire without volumetric phasor analysis on clearing-enhanced multispectral SRS images.
Metabolic Volumetric Chemical Imaging of Tumors in the Cell-Silent Window.
To further study tumor metabolism and angiogenesis, we conducted metabolic imaging using a recently developed method of DO-SRS (33). Going beyond structural imaging, DO-SRS—coupling deuterium oxide probing with SRS microscopy—could reveal metabolic dynamics of animals in situ (33). The metabolic incorporation of deuterium from deuterium oxide into macromolecules produces carbon–deuterium bonds that vibrate in the cell-silent spectral window. DO-SRS microscopy enables simultaneous visualization of both lipid and protein synthesis through spectral unmixing, allowing the study of tumor metabolism in an integrated manner (33).
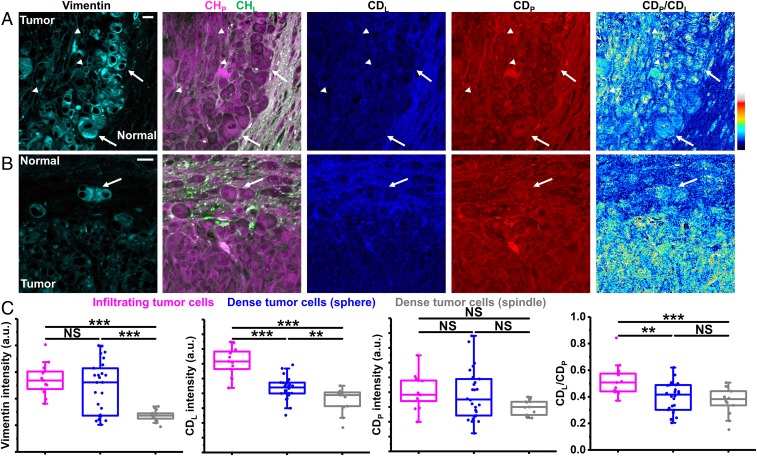
Because infiltrating tumor cells had higher lipid-to-protein ratio revealed by label-free imaging earlier (Fig. 6C), we hypothesized that infiltrating tumor cells had reprogrammed metabolism compared with dense tumor cells. To test this hypothesis, we performed correlative DO-SRS and immunofluorescence on glioblastoma. Immunolabeling of vimentin not only confirmed the presence of infiltrating tumor cells but also provided a marker for epithelial-mesenchymal transition (EMT), which has emerged as a key regulator of tumor invasion (61) and malignancy (62). Based on vimentin expression and cell morphology, we found dense tumor cells could be categorized into two groups: one is high vimentin-expression sphere-shaped cells that are located close to tumor margins (arrows in Fig. 7A); the other is low vimentin-expression spindle-shaped cells that are narrow and elongated and located far away from tumor margins (arrowheads in Fig. 7A). Infiltrating tumor cells were also sphere-shaped with high vimentin expression similar to sphere-shaped dense tumor cells (Fig. 7 B and C). These results contradict with canonical phenotypes of mesenchymal cells—high vimentin expression and spindle-cell morphology, indicating that EMT of these tumor cells is a gradual process with multiple intermediate or hybrid states instead of an “all-or-none” process (52, 63). Detailed quantification with DO-SRS (Fig. 7C) revealed: (i) infiltrating tumor cells exhibited higher activity of lipid synthesis (indicated by CDL) than both sphere-shaped and spindle-shaped dense tumor cells; (ii) sphere-shaped dense tumor cells showed higher activity of lipid synthesis than spindle-shaped ones; (iii) infiltrating tumor cells and two types of dense tumor cells had similar levels of newly synthesized proteins (indicated by CDP); and (iv) infiltrating tumor cells showed higher CDL/CDP ratio than both sphere-shaped and spindle-shaped dense tumor cells, which likely contributes to higher lipid-to-protein ratio of infiltrating tumor cells observed earlier (Fig. 6C). These results indicate enhanced lipid synthesis is central to the metabolic reprogramming of infiltrating tumor cells, which agrees with an emerging role of lipid metabolism in glioblastoma pathology (57–59). Infiltrating tumor cells were more similar—in cellular morphology, vimentin expression, and metabolic activities—to sphere-shaped dense tumor cells than to spindle-shaped dense tumor cells (Fig. 7C), suggesting an infiltration process that dense tumor cells acquire the capacity to invade normal tissues through EMT and enhanced lipid synthesis.
Fig. 7.
Correlative DO-SRS and immunofluorescence of glioblastoma. (A) Correlative images of immunolabeled vimentin, label-free SRS, and DO-SRS show two types of dense tumor cells: arrows indicate sphere-shaped dense tumor cells and arrowheads indicate spindle-shaped dense tumor cells. (Scale bar, 20 µm.) (B) Correlative images of immunolabeled vimentin, label-free SRS, and DO-SRS show infiltrating tumor cells (arrow). (Scale bar, 20 µm.) (C) Intensities of vimention, CDL, CDP, and CDL/CDP of infiltrating tumor cells (n = 13 cells), sphere-shaped dense tumor cells (n = 26 cells), and spindle-shaped dense tumor cells (n = 13 cells), from one mouse. Statistical significance was determined by one-way ANOVA with the Holm–Bonferroni method. **P < 0.01 and ***P < 0.001. NS, not significant.
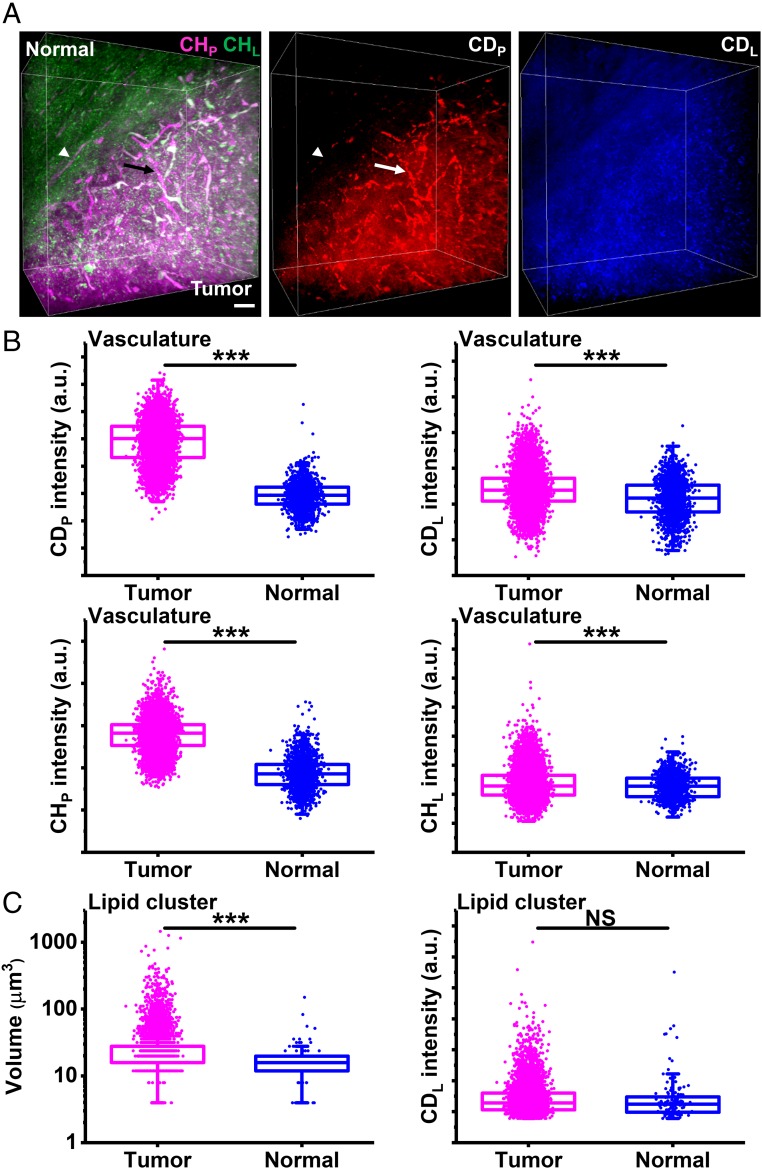
Next we studied the metabolic basis of angiogenesis in tumor. Based on the high-volume fraction of vasculature in tumor (Fig. 6E), we hypothesized that tumor-associated vasculature had high activities of protein and lipid synthesis for sustained angiogenesis. To test this hypothesis, we conducted volumetric DO-SRS imaging on glioblastoma with Raman-tailored tissue clearing. As previously reported (33), tumor tissues exhibited higher activity of protein synthesis (CDP) than normal tissues (Fig. 8A). What’s more, tumor vasculature was highlighted in CDP channel, indicating highly active protein synthesis in tumor vasculature (Fig. 8A). We then segmented tumor and normal vasculature (SI Appendix) and compared their metabolic activities and chemical composition. As shown in Fig. 8B, tumor vasculature had much higher signals in CDP and CHP and slightly higher signals in CDL and CHL than normal blood vessels. These data indicate: (i) active angiogenesis takes place in the tumor, (ii) metabolic reprogramming for tumor angiogenesis involves a major increase in protein synthesis and a minor increase in lipid synthesis, and (iii) tumor vasculature has a different chemical composition from normal vasculature. Based on high metabolic activity and distinct chemical composition of tumor vasculature (Fig. 8B) and its close association with tumor cells (Fig. 6 D and E), tumor vasculature could possibly derive from differentiation of glioblastoma stem cells into endothelial cells or pericytes, a phenomenon termed vascular mimicry (64–66).
Fig. 8.
Metabolic volumetric chemical imaging of glioblastoma. (A) Three-dimensional reconstruction of label-free SRS and DO-SRS images. The arrow indicates a blood vessel in the tumor. The arrowhead indicates a blood vessel in the normal tissue. (Scale bar, 50 µm.) (B) Intensities of CDP, CDL, CHP, and CHL of tumor vasculature (n = 7,588) and normal vasculature (n = 1,605) from one mouse. (C) Volume and CDL intensities of clusters of newly synthesized lipids in the tumor (n = 6,894) and those in the normal tissue (n = 136) from one mouse. Statistical significance was determined by two-sample two-tailed t test. ***P < 0.001. NS, not significant.
Besides, we observed that in CDL channel, newly synthesized lipids formed large clusters mainly in the tumor (Fig. 8A). We then segmented these clusters (SI Appendix) and compared the volume and metabolic activities of these clusters in tumor and normal tissues. As shown in Fig. 8C, there are far more clusters in the tumor than in the normal tissue: the clusters in the tumor had larger volume than those in the normal tissue, while their CDL intensities were not significantly different. These results indicate that tumor tissues exhibited more and larger localized hotspots of lipid synthesis than normal tissues.
Multimodal Imaging of Tumors.
Using brain and subcutaneous glioblastoma as models, we further extended our method to multimodal imaging. Correlative two-photon fluorescence and SRS imaging showed the network of vasculature in the autofluorescence channel and the boundaries between tumor and normal tissues determined by the SRS channel (SI Appendix, Fig. S17). Correlative second-harmonic generation and SRS imaging illustrated collagen fibers from second-harmonic generation and heterogeneous distributions of proteins and lipids from SRS (SI Appendix, Fig. S18). These results demonstrate that our technique can be readily combined with fluorescence imaging and other nonlinear deep-tissue imaging methods in synergy for the characterization of complex biological tissues.
Discussion
We have developed a tissue-clearing–enhanced volumetric chemical imaging technique capable of extending the current SRS imaging depth greater than 10 times. We designed Raman-tailored tissue-clearing strategies for the high-wavenumber C-H region and cell-silent window. Our method achieved clearing-enhanced SRS microscopy with a greater than 10-fold depth extension to ∼1 mm in tissues, although several reports demonstrated clearing-enhanced Raman spectroscopy (67, 68). In addition, we developed a robust volumetric phasor analysis method to analyze and quantify multispectral images of tissues. Furthermore, we demonstrated metabolic volumetric chemical imaging by employing DO-SRS, a newly developed method for visualizing metabolic dynamics (33).
With the advantages of tissue-clearing–enhanced volumetric chemical imaging and analyses, we applied this toolbox to 3D contexts of neurobiology and cancer biology. First, we found increased lipid synthesis at the outer surface of tumor spheroids, indicating rapid cell division and proliferation at the invasion front. Second, we quantitatively determined the distributions of blood vessels, axon fibers, and cell bodies in various brain regions. Third, we delineated tumor margins and unveiled complex 3D structures of glioblastoma xenografts. Fourth, we found infiltrating glioblastoma cells had high lipid-to-protein ratio, high vimentin expression, and enhanced lipid synthesis compared with dense tumor cells. Fifth, we found close association between tumor cells and vasculature and tumor vasculature had higher metabolic activities and higher protein and lipid content than normal vasculature.
Our clearing-enhanced SRS microscopy is complementary to volumetric fluorescence imaging with several distinct advantages. First, unlike fluorescence, SRS is capable of providing comprehensive chemical information of sample composition, identifying and distinguishing molecules, such as lipids, proteins, DNA, water, sugars, cholesterol, and phosphates (29). The spatial distributions, chemical subtypes (e.g., saturated vs. unsaturated lipids), and detailed conformations (e.g., α-helices vs. β-sheets of proteins) of these species are crucial for their functions. Second, label-free SRS enables probing chemical structures in tissues without exogenous labels, free from the difficulties of probe delivery for fluorescence microscopy. Third, the emerging bioorthogonal chemical imaging platform with vibrational tags (such as DO-SRS) allows visualization of metabolic dynamics of biomolecules, such as proteins, lipids, and nucleic acids, inside tissues and organisms (33, 40, 41). Therefore, volumetric chemical imaging by clearing-enhanced SRS microscopy can be a valuable tool for characterizing 3D structures, compositions, dynamics, and functions in diverse biological contexts.
Our technique could be further optimized on several fronts. In terms of next-generation clearing reagents, more chemicals could be screened to achieve increased depth extension. Instrumentation-wise, SRS microscopes can be constructed for simultaneous multichannel imaging (69–72), hence allowing high-throughput volumetric chemical imaging. In addition, the clearing-enhanced SRS could be combined with Bessel-beam SRS tomography for fast 3D imaging (73). Regarding applications, the Raman-clearing strategies could be tailored for specialized imaging purposes. For example, by combining SRS with hydrogel-based clearing [e.g., CLARITY (3)], chemical imaging of both protein distributions and protein secondary structures in whole organs may be achievable. Taken together, clearing-enhanced SRS will find broad use for comprehensive understanding of structure–function relationships in 3D tissue context.
Methods
Samples were immersed in the clearing solution (8 M urea or 8 M urea supplemented with 0.2% Triton X-100) for more than 4 d at room temperature and were then mounted in the clearing solution for SRS microscopy. The animal experimental protocol (AC-AAAQ0496) was approved by the Institutional Animal Care and Use Committee at Columbia University. Full details are given in SI Appendix.
Supplementary Material
Acknowledgments
We thank E. Silveira for technical assistance; Y. Yang and X. Qu for discussion; and T. Swayne and the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University for help with 3D image analysis, supported by NIH Grant P30 CA013696. W.M. acknowledges support of R01EB020892 from the NIH and the Camille and Henry Dreyfus Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.L.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813044116/-/DCSupplemental.
References
- 1.Murray E, et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 2015;163:1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hama H, et al. Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 3.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Susaki EA, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 5.Ke MT, Fujimoto S, Imai T. SeeDB: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 6.Pan C, et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat Methods. 2016;13:859–867. doi: 10.1038/nmeth.3964. [DOI] [PubMed] [Google Scholar]
- 7.Kubota SI, et al. Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep. 2017;20:236–250. doi: 10.1016/j.celrep.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka N, et al. Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. Nat Biomed Eng. 2017;1:796–806. doi: 10.1038/s41551-017-0139-0. [DOI] [PubMed] [Google Scholar]
- 9.Belle M, et al. Tridimensional visualization and analysis of early human development. Cell. 2017;169:161–173.e12. doi: 10.1016/j.cell.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Renier N, et al. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 12.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 13.Mertz J. Optical sectioning microscopy with planar or structured illumination. Nat Methods. 2011;8:811–819. doi: 10.1038/nmeth.1709. [DOI] [PubMed] [Google Scholar]
- 14.Yang W, Yuste R. In vivo imaging of neural activity. Nat Methods. 2017;14:349–359. doi: 10.1038/nmeth.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji N. Adaptive optical fluorescence microscopy. Nat Methods. 2017;14:374–380. doi: 10.1038/nmeth.4218. [DOI] [PubMed] [Google Scholar]
- 16.Shu X, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouzounov DG, et al. In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat Methods. 2017;14:388–390. doi: 10.1038/nmeth.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu XX, Kao YT, Min W. Molecular-switch-mediated multiphoton fluorescence microscopy with high-order nonlinearity. J Phys Chem Lett. 2012;3:2082–2086. [Google Scholar]
- 19.Chen Z, Wei L, Zhu X, Min W. Extending the fundamental imaging-depth limit of multi-photon microscopy by imaging with photo-activatable fluorophores. Opt Express. 2012;20:18525–18536. doi: 10.1364/OE.20.018525. [DOI] [PubMed] [Google Scholar]
- 20.Richardson DS, Lichtman JW. Clarifying tissue clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tainaka K, Kuno A, Kubota SI, Murakami T, Ueda HR. Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu Rev Cell Dev Biol. 2016;32:713–741. doi: 10.1146/annurev-cellbio-111315-125001. [DOI] [PubMed] [Google Scholar]
- 22.Ertürk A, et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- 23.Greenbaum A, et al. Bone CLARITY: Clearing, imaging, and computational analysis of osteoprogenitors within intact bone marrow. Sci Transl Med. 2017;9:eaah6518. doi: 10.1126/scitranslmed.aah6518. [DOI] [PubMed] [Google Scholar]
- 24.Liebmann T, et al. Three-dimensional study of Alzheimer’s disease hallmarks using the iDISCO clearing method. Cell Rep. 2016;16:1138–1152. doi: 10.1016/j.celrep.2016.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nojima S, et al. CUBIC pathology: Three-dimensional imaging for pathological diagnosis. Sci Rep. 2017;7:9269. doi: 10.1038/s41598-017-09117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freudiger CW, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saar BG, et al. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science. 2010;330:1368–1370. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min W, Freudiger CW, Lu S, Xie XS. Coherent nonlinear optical imaging: Beyond fluorescence microscopy. Annu Rev Phys Chem. 2011;62:507–530. doi: 10.1146/annurev.physchem.012809.103512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng JX, Xie XS. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science. 2015;350:aaa8870. doi: 10.1126/science.aaa8870. [DOI] [PubMed] [Google Scholar]
- 30.Bouxsein ML, et al. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 31.Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- 32.Shokeen M, Anderson CJ. Molecular imaging of cancer with copper-64 radiopharmaceuticals and positron emission tomography (PET) Acc Chem Res. 2009;42:832–841. doi: 10.1021/ar800255q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi L, et al. Optical imaging of metabolic dynamics in animals. Nat Commun. 2018;9:2995. doi: 10.1038/s41467-018-05401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu ZL, et al. Label-free chemical imaging in vivo: Three-dimensional non-invasive microscopic observation of amphioxus notochord through stimulated Raman scattering (SRS) Chem Sci. 2012;3:2646–2654. [Google Scholar]
- 35.Lu FK, et al. Multicolor stimulated Raman scattering (SRS) microscopy. Mol Phys. 2012;110:1927–1932. doi: 10.1080/00268976.2012.695028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji M, et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci Transl Med. 2013;5:201ra119. doi: 10.1126/scitranslmed.3005954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji M, et al. Detection of human brain tumor infiltration with quantitative stimulated Raman scattering microscopy. Sci Transl Med. 2015;7:309ra163. doi: 10.1126/scitranslmed.aab0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiskis J, et al. Plaque-associated lipids in Alzheimer’s diseased brain tissue visualized by nonlinear microscopy. Sci Rep. 2015;5:13489. doi: 10.1038/srep13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JH, Kim DH, Song WK, Oh MK, Ko DK. Label-free imaging and quantitative chemical analysis of Alzheimer’s disease brain samples with multimodal multiphoton nonlinear optical microspectroscopy. J Biomed Opt. 2015;20:56013. doi: 10.1117/1.JBO.20.5.056013. [DOI] [PubMed] [Google Scholar]
- 40.Wei L, et al. Live-cell bioorthogonal chemical imaging: Stimulated Raman scattering microscopy of vibrational probes. Acc Chem Res. 2016;49:1494–1502. doi: 10.1021/acs.accounts.6b00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Z, Shen Y, Hu F, Min W. Applications of vibrational tags in biological imaging by Raman microscopy. Analyst. 2017;142:4018–4029. doi: 10.1039/c7an01001j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HJ, et al. Assessing cholesterol storage in live cells and C. elegans by stimulated Raman scattering imaging of phenyl-diyne cholesterol. Sci Rep. 2015;5:7930. doi: 10.1038/srep07930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tipping WJ, Lee M, Serrels A, Brunton VG, Hulme AN. Imaging drug uptake by bioorthogonal stimulated Raman scattering microscopy. Chem Sci. 2017;8:5606–5615. doi: 10.1039/c7sc01837a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang J, et al. Small unnatural amino acid carried Raman tag for molecular imaging of genetically targeted proteins. J Phys Chem Lett. 2018;9:4679–4685. doi: 10.1021/acs.jpclett.8b01991. [DOI] [PubMed] [Google Scholar]
- 45.Hu F, et al. Supermultiplexed optical imaging and barcoding with engineered polyynes. Nat Methods. 2018;15:194–200. doi: 10.1038/nmeth.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei L, et al. Super-multiplex vibrational imaging. Nature. 2017;544:465–470. doi: 10.1038/nature22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hama H, et al. ScaleS: An optical clearing palette for biological imaging. Nat Neurosci. 2015;18:1518–1529. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- 48.Hou B, et al. Scalable and DiI-compatible optical clearance of the mammalian brain. Front Neuroanat. 2015;9:19. doi: 10.3389/fnana.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen F, Tillberg PW, Boyden ES. Optical imaging. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thoma CR, Zimmermann M, Agarkova I, Kelm JM, Krek W. 3D cell culture systems modeling tumor growth determinants in cancer target discovery. Adv Drug Deliv Rev. 2014;69–70:29–41. doi: 10.1016/j.addr.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 51.Guzman A, Sánchez Alemany V, Nguyen Y, Zhang CR, Kaufman LJ. A novel 3D in vitro metastasis model elucidates differential invasive strategies during and after breaching basement membrane. Biomaterials. 2017;115:19–29. doi: 10.1016/j.biomaterials.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 52.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Adams RH, Eichmann A. Axon guidance molecules in vascular patterning. Cold Spring Harb Perspect Biol. 2010;2:a001875. doi: 10.1101/cshperspect.a001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu D, Xie XS. Reliable cell segmentation based on spectral phasor analysis of hyperspectral stimulated Raman scattering imaging data. Anal Chem. 2014;86:4115–4119. doi: 10.1021/ac500014b. [DOI] [PubMed] [Google Scholar]
- 55.Stupp R, et al. European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups; National Cancer Institute of Canada Clinical Trials Group Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 56.Orringer DA, et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng. 2017;1:0027. doi: 10.1038/s41551-016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agnihotri S, Zadeh G. Metabolic reprogramming in glioblastoma: The influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol. 2016;18:160–172. doi: 10.1093/neuonc/nov125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun P, et al. Lipid metabolism enzyme ACSVL3 supports glioblastoma stem cell maintenance and tumorigenicity. BMC Cancer. 2014;14:401. doi: 10.1186/1471-2407-14-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo D, Bell EH, Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncol. 2013;2:289–299. doi: 10.2217/cns.13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jain RK. Normalization of tumor vasculature: An emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 61.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iser IC, Pereira MB, Lenz G, Wink MR. The epithelial-to-mesenchymal transition-like process in glioblastoma: An updated systematic review and in silico investigation. Med Res Rev. 2017;37:271–313. doi: 10.1002/med.21408. [DOI] [PubMed] [Google Scholar]
- 63.Burger GA, Danen EHJ, Beltman JB. Deciphering epithelial-mesenchymal transition regulatory networks in cancer through computational approaches. Front Oncol. 2017;7:162. doi: 10.3389/fonc.2017.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ricci-Vitiani L, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 65.Wang R, et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468:829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- 66.Cheng L, et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schulmerich MV, et al. Optical clearing in transcutaneous Raman spectroscopy of murine cortical bone tissue. J Biomed Opt. 2008;13:021108. doi: 10.1117/1.2892687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sdobnov AY, Tuchin VV, Lademann J, Darvin ME. Confocal Raman microscopy supported by optical clearing treatment of the skin-influence on collagen hydration. J Phys D Appl Phys. 2017;50:285401. [Google Scholar]
- 69.Liao CS, et al. Microsecond scale vibrational spectroscopic imaging by multiplex stimulated Raman scattering microscopy. Light Sci Appl. 2015;4:e265. doi: 10.1038/lsa.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ozeki Y, et al. High-speed molecular spectral imaging of tissue with stimulated Raman scattering. Nat Photonics. 2012;6:845–851. [Google Scholar]
- 71.Fu D, Holtom G, Freudiger C, Zhang X, Xie XS. Hyperspectral imaging with stimulated Raman scattering by chirped femtosecond lasers. J Phys Chem B. 2013;117:4634–4640. doi: 10.1021/jp308938t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He R, et al. Stimulated Raman scattering microscopy and spectroscopy with a rapid scanning optical delay line. Opt Lett. 2017;42:659–662. doi: 10.1364/OL.42.000659. [DOI] [PubMed] [Google Scholar]
- 73.Chen X, et al. Volumetric chemical imaging by stimulated Raman projection microscopy and tomography. Nat Commun. 2017;8:15117. doi: 10.1038/ncomms15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.