Significance
The mechanisms underlying rapid adaptation to changing environments in species with reduced genetic variation, referred to as the “genetic paradox of invasion,” are unknown. We report that transposable elements (TEs) are highly enriched in the gene promoter regions of Capsella rubella compared with its outcrossing sister species Capsella grandiflora. We also show that a number of polymorphic TEs in C. rubella are associated with changes in gene expression. Frequent TE insertions at FLOWERING LOCUS C of C. rubella affect flowering-time variation, an important life history trait correlated with fitness. These results indicate that TE insertions drive rapid phenotypic variation, which could potentially help adapting to novel environments in species with limited genetic variation.
Keywords: Brassicaceae, Capsella rubella, natural variation, rapid phenotypic variation, transposable elements
Abstract
Rapid phenotypic changes in traits of adaptive significance are crucial for organisms to thrive in changing environments. How such phenotypic variation is achieved rapidly, despite limited genetic variation in species that experience a genetic bottleneck is unknown. Capsella rubella, an annual and inbreeding forb (Brassicaceae), is a great system for studying this basic question. Its distribution is wider than those of its congeneric species, despite an extreme genetic bottleneck event that severely diminished its genetic variation. Here, we demonstrate that transposable elements (TEs) are an important source of genetic variation that could account for its high phenotypic diversity. TEs are (i) highly enriched in C. rubella compared with its outcrossing sister species Capsella grandiflora, and (ii) 4.2% of polymorphic TEs in C. rubella are associated with variation in the expression levels of their adjacent genes. Furthermore, we show that frequent TE insertions at FLOWERING LOCUS C (FLC) in natural populations of C. rubella could explain 12.5% of the natural variation in flowering time, a key life history trait correlated with fitness and adaptation. In particular, we show that a recent TE insertion at the 3′ UTR of FLC affects mRNA stability, which results in reducing its steady-state expression levels, to promote the onset of flowering. Our results highlight that TE insertions can drive rapid phenotypic variation, which could potentially help with adaptation to changing environments in a species with limited standing genetic variation.
Rapid phenotypic changes in traits of adaptive significance provide organisms with potentials for local adaptation, which is critical for both survival and range expansion of organisms. This becomes even more crucial in the context of global climate change (1). Species often experience a genetic bottleneck after speciation or introduction into a new area that diminishes their genetic variation, but rapidly adapt and become invasive thereafter, which is referred to as the “genetic paradox of invasion” (2–4). However, the underlying mechanisms mediating these rapid diversification phenotypes, in traits that are correlated with adaptation, are unclear.
Transposable elements (TEs) are among the most variable components of the genome, which can replicate and integrate into new positions. Changes in the environment, such as climate change, can alter both the copy number of TEs and their effects on gene regulation, generating novel genetic and phenotypic variation of potential adaptive significance (5). Therefore, TEs have the potential to quickly create abundant genetic diversity and thus be agents of rapid adaptation (6–10). Despite extensive studies on their phenotypic effects, the extent to which TEs can contribute to the process of rapid adaptation is largely unknown (11–13).
To explore whether TEs could drive rapid phenotypic diversification in species with limited genetic variation, we focused on Capsella rubella (Brassicaceae), an annual and inbreeding forb that experienced a genetic bottleneck during speciation (14, 15). C. rubella provides a typical example of the genetic paradox of invasion with a much wider distribution than that of its outcrossing sister species Capsella grandiflora (16). C. rubella provides an excellent opportunity to unravel the underlying mechanisms, in addition to its transition to selfing that helps with rapid colonization, for the genetic paradox of invasion (14, 15, 17).
In this study, we reasoned that, if TEs are critical for rapid phenotypic variation and adaptation, their distribution in the C. rubella genome might reflect this. Through population genomics analyses, we show that TEs are highly enriched in promoters and downstream regions of genes in C. rubella compared with C. grandiflora. TEs are also highly polymorphic in natural populations of C. rubella, and 4.2% of polymorphic TE insertions are associated with significant changes in expression levels of their adjacent genes. In particular, we demonstrate that frequent TE insertions at the FLOWERING LOCUS C (FLC) locus in natural populations of C. rubella affect its expression and could explain 12.5% of the variation in flowering time, one of the most important life history traits correlated with adaptation. We also reveal that a TE insertion in the 3′ UTR affects mRNA stability. Overall, our results indicate that TEs play a crucial role in rapid phenotypic variation, which could potentially promote adaptation to changing environments in plants and could explain the genetic paradox of invasion.
Results
TEs Are Highly Enriched in C. rubella and Affect the Expression Levels of Its Adjacent Genes.
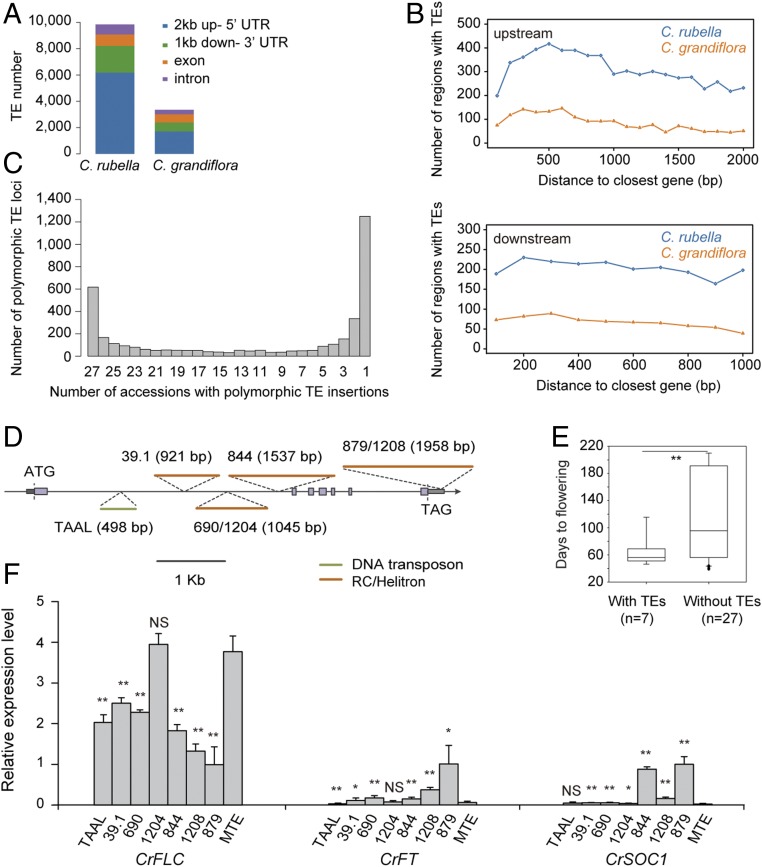
We hypothesized that if TEs are among the key determinants of phenotypic variation, their distribution may differ between C. rubella and its outcrossing congeneric sister species C. grandiflora. To test this, we compared the distribution of TEs in 19,795 orthologous gene pairs between C. rubella and C. grandiflora. TEs are highly enriched in different genic regions of the C. rubella compared with the corresponding regions of C. grandiflora (Fig. 1A). Our analysis with orthologous regions unmasked these divergent distribution patterns compared with the previous study (18), since the genomes of these two species have varied coverages/assemblies. In addition, TEs are highly enriched in the promoter and downstream regions of genes in C. rubella compared with C. grandiflora (Fig. 1B), indicating that TEs could potentially contribute to the diversification of gene expression in C. rubella.
Fig. 1.
Transposable elements cause phenotypic diversification and changes in gene expression in C. rubella. (A) Number of TEs in different genic regions of the C. rubella and C. grandiflora genomes. (B) Enrichment of TEs in 100-bp bins in the regions 2 kb upstream of the start codon and 1 kb downstream of the stop codon between orthologous gene pairs of C. rubella and C. grandiflora. The x axis indicates the distance from TEs to the start codon (stop codon) of the closest gene; the y axis indicates the number of genes with TEs at different distances to the start codon (stop codon). (C) Frequency distribution of polymorphic TEs in the genomes of 27 C. rubella natural accessions with MTE as a reference. The x axis indicates the number of accessions with the same TE insertions. (D) Insertion of TEs in FLC in C. rubella populations. Thick lines in different colors and lengths indicate the classification and length of the inserted TEs. Numbers in parentheses indicate the sequence lengths of inserted TEs; the names before the parentheses indicate the accessions with the inserted TEs; when there are two accessions, accession names are separated by a backslash (/). (E) Flowering-time variation between accessions with or without a TE insertion at the CrFLC locus. (F) Relative expression levels of CrFLC, CrFT, and CrSOC1 in accessions with TE insertions. Four biological replicates, each with three technical replicates, were analyzed. Expression data were normalized with CrTubulin and then to the levels of 879 plants and are shown as means ± SD; *P < 0.05; **P < 0.01; NS, insignificant.
To assess whether TEs are polymorphic in C. rubella, we scanned 28 C. rubella genomes, which included 27 accessions (18, 19) (SI Appendix, Table S1), with MTE accession as a reference (20). A phylogenetic tree based on whole-genome sequences suggested that these 28 C. rubella accessions are closely related (SI Appendix, Fig. S1). We identified 3,808 polymorphic TEs among these genomes (SI Appendix, Fig. S2A). As reported earlier (18), these polymorphic TEs are enriched (89.84%) in intergenic regions (SI Appendix, Fig. S2B). The number of polymorphic TEs in these accessions ranged from 1,024 to 1,688, and 1,731 in the reference MTE (SI Appendix, Fig. S3 and Table S1). The polymorphic TEs that are present in at least two accessions were considered as common. In total, of the 3,808 polymorphic TEs, 2,558 (67.17%) were present in at least two accessions, suggesting a common allelic variation (Fig. 1C).
To assess the functional implications of this variation, we performed transcriptome sequencing for three representative accessions of C. rubella, MTE, 86IT1, and 879 (SI Appendix, Table S2), based on their positions on the phylogenetic tree of 28 C. rubella accessions (SI Appendix, Fig. S1). In these three accessions there are 1,309 polymorphic TEs, of which 5.45% were located in genic regions and 94.55% were in intergenic regions. We found that 55 of 1,309 polymorphic TE loci in these three accessions (4.2%) were associated with significant changes in the expression levels of their adjacent genes [Wilcoxon sum test, false discovery rate (FDR) corrected, P < 0.05, SI Appendix, Table S3]. Among these TEs that are associated with changes in gene expression, retrotransposons are more common (36.37%, SI Appendix, Fig. S4). A Gene Ontology enrichment analysis (21) of the genes that are affected by the TEs using the Arabidopsis orthologous genes suggested that these genes are involved in lateral root formation and defense response (SI Appendix, Fig. S5). Taken together, these findings suggest that polymorphic TEs can diversify gene expression, which has the potential to shape the phenotypic variation in C. rubella as in other plants (8, 22, 23).
Frequent TE Insertions at CrFLC Affect Natural Variation in Flowering Time.
The geographic distribution of C. grandiflora is much narrower than that of C. rubella, which has colonized most areas of southern and western Europe with varied environmental conditions (24). Flowering time is an important trait for adaptation to changing climates and novel environments (25, 26). Interestingly, we found that FLC, a determinant of natural variation in flowering time of C. rubella (27), has TE insertions in its genic region (27). To assess the frequency of TE insertions at CrFLC, we sequenced the CrFLC genomic region (from 2 kb upstream of the start codon to the end of 3′ UTR) in eight accessions by Sanger sequencing and compared them with 27 other previously published CrFLC genomic sequences (SI Appendix, Table S4). We found five different TE insertions in 7 of 35 accessions (20%). We found a TE insertion in the 3′ UTR in two accessions (879 and 1208) and four additional TE insertions in intron 1 in five accessions (39.1, 690, 1204, 844, and TAAL) (Fig. 1D). The TE insertions ranged from 498 to 1,958 bp, and most were Helitron, except one insertion in the TAAL accession, which was a DNA transposon. All these seven accessions with TE insertions are early flowering compared with the reference accession MTE and accessions without TEs (Fig. 1E and SI Appendix, Table S4). The expression levels of CrFLC are lower (P < 0.05, Student’s t test) than MTE in all of the other six accessions except for 1204 (Fig. 1F). This is also reflected in the inversely correlated expression levels of the CrFLC-downstream genes FT and SOC1 (Fig. 1F). Furthermore, 12.5% of the variation in flowering time could be explained by allelic variation of the TE insertions at CrFLC in C. rubella (Pearson’s correlation coefficient, r2 = 0.125, P < 0.05). These results revealed that TE insertions in CrFLC are associated with early flowering in C. rubella.
Given that each TE insertion is present in only a few C. rubella accessions at different regions of the FLC locus, we concluded that these TE insertions originated recently and independently (SI Appendix, Fig. S1). In the Brassicaceae family, there are frequent TE insertions at FLC (SI Appendix, Fig. S6A). An analysis of a few accessions from C. grandiflora and C. orientalis and 76 accessions of Arabidopsis thaliana (SI Appendix, Table S5) also suggested that the 3′ UTR TE insertion in accession 879 is a recent mutation that is present only in two C. rubella accessions (SI Appendix, Fig. S7). If the TE insertions have any potential to be of adaptive significance, one might expect that these accessions could show some geographic patterns. To assess this, we compared the origin of these accessions. Intriguingly, accessions with TE insertions clustered in the southernmost habitat of this species around the Mediterranean region (SI Appendix, Fig. S6B and Table S4). This region harbors a Mediterranean climate, which is characterized by high precipitation, warm winters, and dry and hot summers (28–31). Plants in the area are usually early flowering to avoid dry and hot summers (28–31). The high TE insertion frequency, associated with early flowering in C. rubella, is consistent with this hypothesis. To assess whether there is significant niche divergence between C. rubella accessions with or without the TE insertion, we performed ecological niche modeling and found significant niche divergence between the two groups (100 random permutations, one-sample t test, P < 0.001, SI Appendix, Fig. S8), consistent with the possibility that TEs in C. rubella show the potential to cause phenotypic variation of adaptive significance.
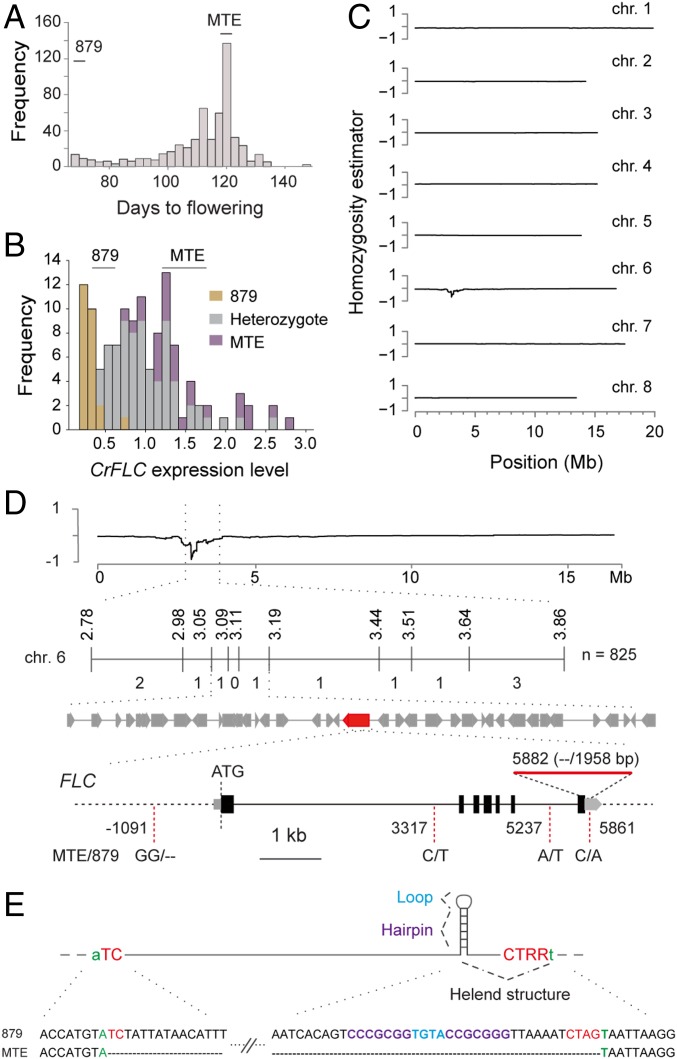
To test whether TE insertions in the 3′ UTR and intron 1 are indeed the causal mutations for early flowering, we performed genetic mapping. First, we analyzed an F2 population (879 × MTE) derived from a cross between the late-flowering, sequenced accession MTE (20) and the early flowering accession 879 that harbors a TE in the 3′ UTR. Flowering time correlates significantly with FLC expression in the Brassicaceae family (32–34). Here, we found that the distribution of flowering time is consistent with the distribution of CrFLC expression in F2 populations (Fig. 2 A and B). The F2 individuals with lower CrFLC expression belong to the 879 genotype (Fig. 2B). Furthermore, the CrFLC expression level was strongly correlated with the TE insertion in the 3′ UTR in 2-wk-old seedlings of the F2 population (Pearson’s correlation coefficient, r2 = 0.462, P < 0.01). These findings suggested that the 3′ UTR TE potentially affected CrFLC expression in this cross.
Fig. 2.
QTL mapping analysis of flowering time. (A) Distribution of flowering time of the 557 individuals of the 879 × MTE F2 population. Averages and ranges of flowering times for the two parental lines are shown above. (B) Histogram of CrFLC expression levels variations among F2 population (n = 122) with TE insertion (879 genotype, shown in yellow), without TE insertion (MTE genotype, purple), and heterozygote (gray) at its 3′ UTR. Averages and ranges of CrFLC expression levels for the two parental lines are shown above. Expression data were normalized against CrTubulin. (C) SHOREmap analysis of flowering time. The homozygosity estimator is 0 for even allele frequencies for both parents, 1 when homozygous for the late-flowering accession MTE, and −1 when homozygous for the early-flowering accession 879. (D) Fine mapping by a genetic-linkage analysis and sequence variation in the candidate gene CrFLC. The number of recombinants between the markers and the causal locus is indicated below the linkage map. CrFLC is marked in red. Dashed red lines indicate sequence variation between MTE and 879; red line indicates deleted sequence. (E) Sequence variation in the CrFLC 3′ UTR and the inserted Helitron structure. The Helitron structure is illustrated at the top, composed of the 5′ TC and 3′ CTRR termini (shown in red), loop and two short palindromic sequences close to the 3′ terminus that could form a 20-bp hairpin (shown in blue and purple) and are inserted within a host dinucleotide AT (shown in green). The end sequences of the CrFLC 3′ UTR Helitrons are shown at the bottom in the same color.
Sequencing of a pool of early flowering plants from the F2 population mapped the causal loci to an ∼863-kb region on chromosome 6 (from position 2.78–3.64 Mb) (Fig. 2C). We further used 825 individuals from the BC2F2 population and narrowed the causal region down to a 130-kb interval (SI Appendix, Table S6). This mapping interval contained 39 ORFs, including CrFLC (Fig. 2D). Sanger sequencing revealed that CrFLC has one deletion of 2 bp in the promoter, two SNPs in its intron, one SNP, and a 1,958-bp Helitron TE insertion in the 3′ UTR that differentiated MTE from 879 (Fig. 2 D and E). The CrFLC expression level in 879 was a quarter of that in MTE (Fig. 1F), making it a prime candidate for the observed flowering-time variation.
To assess whether the TE insertion in intron 1 of CrFLC is also the causal mutation for early flowering, we constructed an F2 population of 844 × MTE. Using a similar sequencing approach, we identified an ∼650-kb region on chromosome 6 based on 501 F2 individuals (SI Appendix, Fig. S9 A and B). We narrowed down the mapping interval to a 210-kb region using 561 more F2 individuals and additional markers, which included the CrFLC locus (SI Appendix, Fig. S9 C and D and Table S7). The location of TEs in intron 1 was roughly similar to the position of Ler in A. thaliana, in which a TE insertion regulates histone modification via siRNA, thereby regulating flowering time (35). Therefore, it is likely that a similar mechanism may mediate the onset of early flowering in 844.
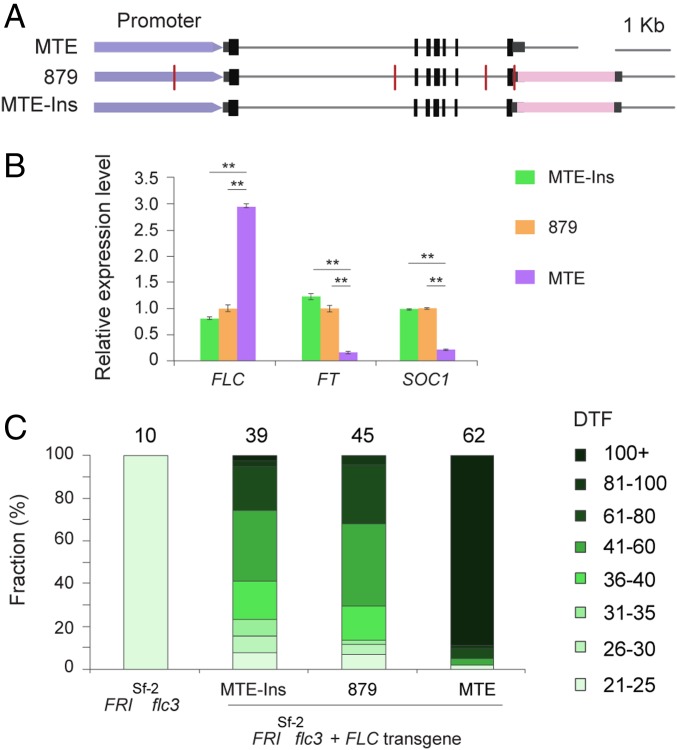
However, the TE insertion at the 3′ UTR in accession 879 is unique (SI Appendix, Fig. S6). To confirm whether the TE insertion in the 3′ UTR modulates flowering time, we generated constructs with genomic CrFLC from MTE and 879 and an additional construct of MTE-CrFLC in which the Helitron TE from 879 was introduced (MTE-Ins) and compared their ability to complement FRIsf2 flc-3 A. thaliana plants (Fig. 3A). CrFLC expression was higher in transgenic plants harboring the MTE construct than in those with 879 or MTE-Ins constructs (Fig. 3B; see SI Appendix, Fig. S10, for 10 randomly selected independent transgenic lines). Consistent with these findings, levels of the CrFLC downstream genes CrFT and CrSOC1 were inversely correlated with the CrFLC expression level (Fig. 3B). Flowering time was correlated with CrFLC expression. Transgenic plants with the MTE genomic sequence were late flowering, while transgenic plants with either 879 or MTE with the TE insertion were earlier flowering (all P < 0.001, Student’s t test; Fig. 3C). Taken together, these results suggested that the allelic variation caused by the TE insertion at the 3′ UTR of CrFLC was responsible for the flowering-time difference between 879 and MTE accessions of C. rubella.
Fig. 3.
TE insertion at CrFLC explains variation in flowering time. (A) Diagrams of constructs. Red vertical lines indicate variation between MTE and 879. Pink rectangles indicate the TE insertion. (B) Expression of CrFLC and two downstream flowering regulators (FT, SOC1) in A. thaliana transgenic plants, as assayed using a pool of 25 independent transgenic lines. Expression data were normalized against AtTubulin and then to the levels of transgenic plants with 879-CrFLC and are shown as means ± SD; **P < 0.01. (C) Flowering time for transgenic plants. Flowering times for T1 transgenic lines with FRIsf2 and flc-3 alleles in the A. thaliana Col-0 background are shown; the number of independent transgenic lines scored for each construct is given above the bar graph. DTF, days to flowering.
TE in 3′ UTR of CrFLC Causes Alternative Polyadenylation and Affects Its mRNA Stability.
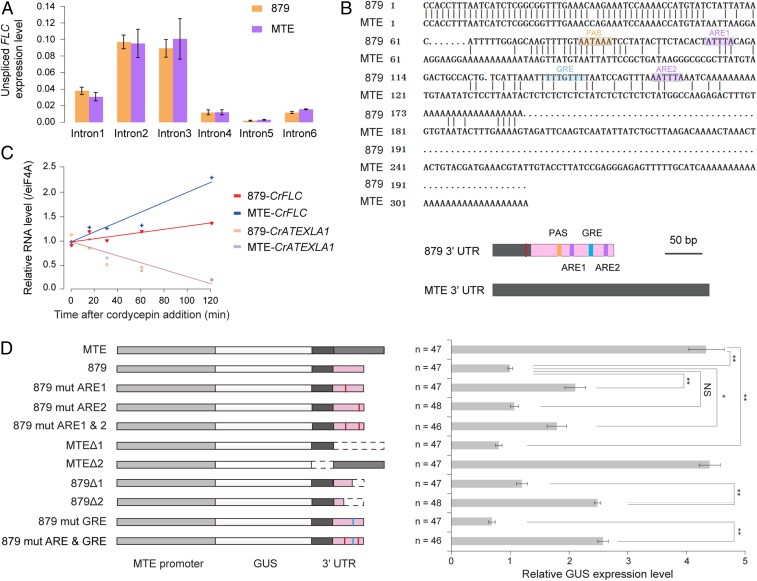
TE insertions can change gene expression by altering transcription and/or by modulating mRNA stability (8). To assess whether the differential CrFLC expression between MTE and 879 is caused by transcriptional differences, we quantified transcription rates by measuring unspliced mRNA abundance (18, 36). The abundance of unspliced CrFLC nascent transcript is similar between MTE and 879 (Fig. 4A). Consistent with this, in both accessions, low levels of DNA methylation were detected around the CrFLC transcription start site and the end of intron 1, two regions important for the regulation of FLC expression (37) (SI Appendix, Fig. S11). These findings suggest that transcriptional differences do not account for the observed changes in gene expression.
Fig. 4.
TE insertion-induced alternative polyadenylation regulates CrFLC expression by modulating RNA stability. (A) Expression levels of unspliced CrFLC. Five biological replicates, each with three technical replicates, were analyzed. Expression data were normalized against CrTubulin and are shown as means ± SD. (B) Alternative polyadenylation at the 3′ UTR between 879 and MTE and a schematic model. The MTE 3′ UTR is shown in dark gray. Pink rectangles indicate part of Helitron. The ARE (purple rectangles) and GRE (blue rectangles) sites and the polyadenylation signal (PAS, orange rectangles) domain are shown. Red line indicates variation. (C) RNA stability of CrFLC. CrATEXLA1 was used as a control, representing an unstable transcript. Three biological replicates, each with three technical replicates, were analyzed. (D) Various deletion constructs with different 3′ UTRs and their effects on gene expression in transgenic lines. The MTE promoter is shown in light gray; GUS genes are in white; the same 3′ UTR region is shown in dark gray, and the latter half of the MTE 3′ UTR is shown in gray; the deleted regions are shown in dashed lines. The vertical red line indicates the mutation of the ARE motif, and the vertical blue line indicates the mutation of the GRE motif. The pooled number of independent transgenic lines is given next to the bar graphs. Expression data were normalized against AtTubulin and are shown as means ± SD; *P < 0.05; **P < 0.01; NS, insignificant.
TE insertions at the 3′ UTR can give rise to alternative polyadenylation (APA), which in turn could modulate mRNA stability (38, 39). The Helitron insertion in 879 introduces a canonical poly(A) signal (AAUAAA). The 3′ RACE assays revealed that there is APA in 879-CrFLC (Fig. 4B). To assess whether APA could fully account for the observed phenotypic differences, we generated an additional 879-CrFLC construct in FRIsf2 flc-3 A. thaliana plants, in which the poly(A) signal was mutated (879-PAS-mut) (SI Appendix, Fig. S12). These plants were also early flowering like transgenic plants of 879-CrFLC compared with transgenic plants of MTE-CrFLC, which suggests that, while the TE insertion at the 3′ UTR causes APA, other mechanisms also contribute to the reduction in gene expression.
To assess mRNA stability, we measured CrFLC expression in the presence of cordycepin, which inhibits transcription (40), in parental lines. These experiments revealed that 879-CrFLC mRNA degraded faster than MTE-CrFLC mRNA (Fig. 4C). To further confirm the effect of the 3′ UTR, we generated GUS-reporter constructs in FRIsf2 flc-3 A. thaliana plants driven by the CrFLC promoter from MTE with either MTE or 879 3′ UTRs (Fig. 4D). We found that the MTE 3′ UTR led to an almost threefold increase in expression compared with that of 879. These results are consistent with findings from Arabidopsis, indicating that the 3′ UTR could affect the stability of FLC mRNA (36).
MTE and 879 also differ in AU-rich elements (AREs) and GU-rich elements (GREs) before their poly(A) tails (Fig. 4B). These elements, which are present only in 879, have the potential to affect RNA stability (41). To assess whether AREs or GREs could mediate changes in gene expression, we compared expression levels of GUS after specifically mutagenizing these elements in transgenic lines (Fig. 4D). While the loss of the first ARE had a significant impact on gene expression, the loss of the GRE and the second ARE did not, suggesting that the first ARE at the 3′ UTR of 879 is critical for modulating mRNA expression in these transgenic lines. An analysis of transgenic lines harboring reporter constructs with partial UTRs from MTE suggested that the latter part of the MTE 3′ UTR is more critical as the deletion of the first half of the MTE 3′ UTR (MTEΔ2) resulted in the same expression level as that of MTE, but the deletion of the latter half (MTEΔ1) significantly reduced the expression level (Fig. 4D). These results indicate that the presence of AREs and the absence of the latter part of the MTE 3′ UTR in 879-CrFLC might reduce CrFLC expression probably by affecting mRNA stability (Fig. 4D).
Discussion
Invasive species with limited genetic variation often rapidly acquire diversification of phenotypes to adapt to changing environments and become successful in a process referred to as the genetic paradox of invasion (2). How this process occurs is a fundamental question in biology. C. rubella experienced an extreme genetic bottleneck but has a wide distribution and is therefore a good model system to address this question. TEs have been suggested as an agent of rapid adaptation because they can produce abundant genetic variation in a limited time (5–9) and can affect phenotypic variation (11–13). For example, in peppered moths, a TE insertion within intron 1 of the cortex gene increases its expression level and induces melanization (13). However, the extent to which TEs contribute to rapid phenotypic diversification and the mechanisms by which they influence phenotypes are unknown.
Our results show that TEs play a crucial role in rapid phenotypic variation and suggest that they can potentially aid in adaptation to changing environments. Previous studies in other systems have showed that TEs in 3′ UTRs can alter gene expression via the regulation of methylation in Arabidopsis (42) and can change the translation efficiency in rice (43). We have found that a TE insertion in the 3′ UTR affects mRNA stability. This mechanism also has the potential to affect gene expression and phenotypes. While our study hints at a correlation with potential adaptive significance, future studies should explore whether such phenotypic diversification ultimately confers an adaptive advantage and whether TEs can drive adaptation.
Materials and Methods
More detailed information on the materials and methods used in this study is provided in SI Appendix, Supplementary Information Text. The C. rubella accessions were described in our previous study (14, 27, 32) or this study (SI Appendix, Table S4). Flowering time was assayed as days to flowering after planting. TEs in C. grandiflora (20) and C. rubella (20) were annotated using RepeatMasker v.4.0.6 (www.repeatmasker.org). The polymorphic TEs were identified using TEPID (44) based on the raw reads of 27 resequenced genomes of C. rubella accessions (18, 19) (SI Appendix, Table S1) with the MTE genome as the reference. A neighbor-joining tree of 28 C. rubella accessions was constructed using PHYLIP (version 3.696) (45). Ecological niche modeling was performed to measure niche similarity using ENMTools 1.3 (46). Pooled leaves of early-flowering 879 × MTE (844 × MTE) F2 plants were sequenced to map the causal loci using SHOREmap (47). For the complementation assay and the poly(A) signal mutation assay of CrFLC in A. thaliana, the fragments with (or without) mutations were cloned into the pCAMBIA1300 vector using the KpnI and SalI sites and introduced into A. thaliana with FRIsf2 and flc-3 alleles in the Col-0 background. For the GUS-reporter assay, the promoter was amplified from MTE and cloned into the pBI121 vector using the SbfI and XmaI sites, and the 3′ UTR was cloned using the SacI and EcoRI sites.
RNA-seq was performed on total RNA samples extracted from flower bud tissues with three biological repeats for each accession (SI Appendix, Table S2). Expression levels of genes were estimated by fragments per kilobase of exon per million reads mapped. RT-qPCR was performed to estimate RNA expression levels. RNA stability was measured using RT-qPCR to quantify gene expression levels at different time points after incubation of 3-deoxyadenosine (cordycepin). DNA after bisulfite conversion was sequenced and analyzed to measure DNA methylation levels with the online tool Kismeth (48). All primers are listed in SI Appendix, Table S8. All statistical analyses were performed in R (www.r-project.org/).
The data described in this paper have been deposited in the NCBI Sequence Read Archive under accession nos. PRJNA392709, PRJNA392711, and PRJNA511520 and in GenBank under accession nos. MF422379–MF422413, MF422414–MF422449, MF422450–MF422525, MF142149–MF142156, and MG492014–MG492020.
Supplementary Material
Acknowledgments
We thank Detlef Weigel, John Bowman, Xiaofeng Cao, and Wolfgang Busch for valuable suggestions and discussions; Li Yang and Mao-Fu Li for assistance with the experiments; and especially the anonymous reviewers for their help in improving the manuscript. This study was funded by National Natural Science Foundation of China Grant 91731306 (to Y.-L.G.); Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB27010305 (to Y.-L.G.); the Innovative Academy of Seed Design, Chinese Academy of Sciences (Y.-L.G.); Chinese Academy of Sciences President’s International Fellowship for Visiting Scientists (S.B.); and Monash University Larkins Fellowship (to S.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data described in the paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive under accession nos. PRJNA392709, PRJNA392711, and PRJNA511520 and in GenBank under accession nos. MF422379–MF422413, MF422414–MF422449, MF422450–MF422525, MF142149–MF142156, and MG492014–MG492020.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811498116/-/DCSupplemental.
References
- 1.Scheffers BR, et al. The broad footprint of climate change from genes to biomes to people. Science. 2016;354:719. doi: 10.1126/science.aaf7671. [DOI] [PubMed] [Google Scholar]
- 2.Estoup A, et al. Is there a genetic paradox of biological invasion? Annu Rev Ecol Evol Syst. 2016;47:51–72. [Google Scholar]
- 3.Kolbe JJ, et al. Genetic variation increases during biological invasion by a Cuban lizard. Nature. 2004;431:177–181. doi: 10.1038/nature02807. [DOI] [PubMed] [Google Scholar]
- 4.Frankham R. Resolving the genetic paradox in invasive species. Heredity (Edinb) 2005;94:385. doi: 10.1038/sj.hdy.6800634. [DOI] [PubMed] [Google Scholar]
- 5.Stapley J, Santure AW, Dennis SR. Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol Ecol. 2015;24:2241–2252. doi: 10.1111/mec.13089. [DOI] [PubMed] [Google Scholar]
- 6.Schrader L, et al. Transposable element islands facilitate adaptation to novel environments in an invasive species. Nat Commun. 2014;5:5495. doi: 10.1038/ncomms6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casacuberta E, González J. The impact of transposable elements in environmental adaptation. Mol Ecol. 2013;22:1503–1517. doi: 10.1111/mec.12170. [DOI] [PubMed] [Google Scholar]
- 8.Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: From conflicts to benefits. Nat Rev Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrón MG, Fiston-Lavier AS, Petrov DA, González J. Population genomics of transposable elements in Drosophila. Annu Rev Genet. 2014;48:561–581. doi: 10.1146/annurev-genet-120213-092359. [DOI] [PubMed] [Google Scholar]
- 10.Li ZW, et al. Transposable elements contribute to the adaptation of Arabidopsis thaliana. Genome Biol Evol. 2018;10:2140–2150. doi: 10.1093/gbe/evy171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin A, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–1138. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 13.Van’t Hof AE, et al. The industrial melanism mutation in British peppered moths is a transposable element. Nature. 2016;534:102–105. doi: 10.1038/nature17951. [DOI] [PubMed] [Google Scholar]
- 14.Guo YL, et al. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci USA. 2009;106:5246–5251. doi: 10.1073/pnas.0808012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foxe JP, et al. Recent speciation associated with the evolution of selfing in Capsella. Proc Natl Acad Sci USA. 2009;106:5241–5245. doi: 10.1073/pnas.0807679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurka H, Neuffer B. Evolutionary processes in the genus Capsella (Brassicaceae) Plant Syst Evol. 1997;206:295–316. [Google Scholar]
- 17.Tang C, et al. The evolution of selfing in Arabidopsis thaliana. Science. 2007;317:1070–1072. doi: 10.1126/science.1143153. [DOI] [PubMed] [Google Scholar]
- 18.Ågren JA, et al. Mating system shifts and transposable element evolution in the plant genus Capsella. BMC Genomics. 2014;15:602. doi: 10.1186/1471-2164-15-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Q, et al. Long-term balancing selection contributes to adaptation in Arabidopsis and its relatives. Genome Biol. 2017;18:217. doi: 10.1186/s13059-017-1342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slotte T, et al. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet. 2013;45:831–835. doi: 10.1038/ng.2669. [DOI] [PubMed] [Google Scholar]
- 21.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22.Hollister JD, et al. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci USA. 2011;108:2322–2327. doi: 10.1073/pnas.1018222108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei L, Cao X. The effect of transposable elements on phenotypic variation: Insights from plants to humans. Sci China Life Sci. 2016;59:24–37. doi: 10.1007/s11427-015-4993-2. [DOI] [PubMed] [Google Scholar]
- 24.Hurka H, Friesen N, German DA, Franzke A, Neuffer B. ‘Missing link’ species Capsella orientalis and Capsella thracica elucidate evolution of model plant genus Capsella (Brassicaceae) Mol Ecol. 2012;21:1223–1238. doi: 10.1111/j.1365-294X.2012.05460.x. [DOI] [PubMed] [Google Scholar]
- 25.Weigel D. Natural variation in Arabidopsis: From molecular genetics to ecological genomics. Plant Physiol. 2012;158:2–22. doi: 10.1104/pp.111.189845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson JT, Inouye DW, McKinney AM, Colautti RI, Mitchell-Olds T. Phenotypic plasticity and adaptive evolution contribute to advancing flowering phenology in response to climate change. Proc Biol Sci. 2012;279:3843–3852. doi: 10.1098/rspb.2012.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, et al. Parallel evolution of common allelic variants confers flowering diversity in Capsella rubella. Plant Cell. 2018;30:1322–1336. doi: 10.1105/tpc.18.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kigel J, et al. Relationships between flowering time and rainfall gradients across Mediterranean-desert Transects. Isr J Ecol Evol. 2011;57:91–109. [Google Scholar]
- 29.Wolfe MD, Tonsor SJ. Adaptation to spring heat and drought in northeastern Spanish Arabidopsis thaliana. New Phytol. 2014;201:323–334. doi: 10.1111/nph.12485. [DOI] [PubMed] [Google Scholar]
- 30.Shavrukov Y, et al. Early flowering as a drought escape mechanism in plants: How can it aid wheat production? Front Plant Sci. 2017;8:1950. doi: 10.3389/fpls.2017.01950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Exposito-Alonso M, et al. Genomic basis and evolutionary potential for extreme drought adaptation in Arabidopsis thaliana. Nat Ecol Evol. 2018;2:352–358. doi: 10.1038/s41559-017-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo YL, Todesco M, Hagmann J, Das S, Weigel D. Independent FLC mutations as causes of flowering-time variation in Arabidopsis thaliana and Capsella rubella. Genetics. 2012;192:729–739. doi: 10.1534/genetics.112.143958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slotte T, Holm K, McIntyre LM, Lagercrantz U, Lascoux M. Differential expression of genes important for adaptation in Capsella bursa-pastoris (Brassicaceae) Plant Physiol. 2007;145:160–173. doi: 10.1104/pp.107.102632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lempe J, et al. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 2005;1:109–118. doi: 10.1371/journal.pgen.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, He Y, Amasino R, Chen X. siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev. 2004;18:2873–2878. doi: 10.1101/gad.1217304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Csorba T, Questa JI, Sun Q, Dean C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA. 2014;111:16160–16165. doi: 10.1073/pnas.1419030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheldon CC, Conn AB, Dennis ES, Peacock WJ. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell. 2002;14:2527–2537. doi: 10.1105/tpc.004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golisz A, Sikorski PJ, Kruszka K, Kufel J. Arabidopsis thaliana LSM proteins function in mRNA splicing and degradation. Nucleic Acids Res. 2013;41:6232–6249. doi: 10.1093/nar/gkt296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Ahmadi W, Al-Ghamdi M, Al-Haj L, Al-Saif M, Khabar KS. Alternative polyadenylation variants of the RNA binding protein, HuR: Abundance, role of AU-rich elements and auto-regulation. Nucleic Acids Res. 2009;37:3612–3624. doi: 10.1093/nar/gkp223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saze H, Kakutani T. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 2007;26:3641–3652. doi: 10.1038/sj.emboj.7601788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen J, et al. Translational repression by a miniature inverted-repeat transposable element in the 3′ untranslated region. Nat Commun. 2017;8:14651. doi: 10.1038/ncomms14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stuart T, et al. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. eLife. 2016;5:e20777. doi: 10.7554/eLife.20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feisenstein J. PHYLIP-phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 46.Warren DL, Glor RE, Turelli M. ENMTools: A toolbox for comparative studies of environmental niche models. Ecography. 2010;33:607–611. [Google Scholar]
- 47.Schneeberger K, et al. SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- 48.Gruntman E, et al. Kismeth: Analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics. 2008;9:371. doi: 10.1186/1471-2105-9-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.