Significance
Identifying cell-surface receptors required for viral infection is important for developing antiviral therapies and effective vaccines. Human cytomegalovirus (HCMV) infects epithelial cells during interhost transmission. The pentameric protein complex (PC) was identified as being required for infection of this clinically relevant cell type. The majority of neutralizing antibody responses are directed against the PC. Here we describe an HCMV receptor, OR14I1, which binds to the PC and defines epithelial tropism. Importantly, we identified a peptide representing the N terminus of OR14I1 that blocks HCMV infection of epithelial cells. Moreover, an adenylate cyclase signaling pathway functioning downstream of OR14I1 contributes to HCMV infection and endocytosis. These findings reveal information useful for the development of new anti-HCMV therapies and vaccines.
Keywords: human cytomegalovirus, CRISPR screen, OR14I1, virus receptor, pentameric glycoprotein complex
Abstract
A human cytomegalovirus (HCMV) pentameric glycoprotein complex (PC), gH–gL–UL128–UL130–UL131A, is necessary for viral infection of clinically relevant cell types, including epithelial cells, which are important for interhost transmission and disease. We performed genome-wide CRISPR/Cas9 screens of different cell types in parallel to identify host genes specifically required for HCMV infection of epithelial cells. This effort identified a multipass membrane protein, OR14I1, as a receptor for HCMV infection. This olfactory receptor family member is required for HCMV attachment, entry, and infection of epithelial cells and is dependent on the presence of viral PC. OR14I1 is required for AKT activation and mediates endocytosis entry of HCMV. We further found that HCMV infection of epithelial cells is blocked by a synthetic OR14I1 peptide and inhibitors of adenylate cyclase and protein kinase A (PKA) signaling. Identification of OR14I1 as a PC-dependent HCMV host receptor associated with epithelial tropism and the role of the adenylate cyclase/PKA/AKT–mediated signaling pathway in HCMV infection reveal previously unappreciated targets for the development of vaccines and antiviral therapies.
Human cytomegalovirus (HCMV) is a leading cause of infection-associated birth defects and a major cause of morbidity and mortality in immune-insufficient individuals such as AIDS patients and transplant recipients (1–3). There are no effective vaccines, and antiviral therapies for HCMV infection are limited. The antiviral drugs currently in use are prone to resistance and can be highly toxic to patients. Thus, there remains a need for host-directed therapeutics and vaccines against HCMV. Most viruses exploit a receptor(s) on the cell surface to invade the host cell, initiate replication, and mediate subsequent viral propagation. Receptor targeting often restricts virus infection efficiently and can determine tissue and/or species tropism. HCMV can infect multiple cell types including epithelial and endothelial cells, fibroblasts, cells within the monocytic lineage, smooth muscle cells, neurons, stromal cells, and hepatocytes (4). As a result, HCMV can be found in a variety of tissues. It has been thought there could be multiple host cell receptors mediating virus entry, given this complexity. Although a few cellular receptors have been identified for certain cells (5–9), they remain elusive for the majority of cell types.
HCMV encodes at least 25 membrane glycoproteins that are found in the viral envelope. HCMV infects a wide spectrum of cell types by utilizing different forms of glycoprotein gH–gL to enter different cell types by distinct routes of entry. It is not always clear whether these glycoprotein complexes act directly in membrane fusion or in binding receptors in virus entry. As reported, gB is a fusion protein and gH–gL complexes act upstream of gB to bind receptors, thereby activating gB for fusion (10). HCMV enters human fibroblasts by direct fusion of the viral envelope with the plasma membrane at neutral pH (11). By contrast, HCMV enters into epithelial and endothelial cells through internalization of virions into endosomes and low pH-dependent fusion with endosomal membranes (12). HCMV infection of epithelial cells and fibroblasts is mediated by distinct viral gH–gL envelope glycoprotein complexes (13). A gH–gL–gO trimer complex (TC) is essential for entry into all cell types (14). An HCMV pentamer complex (PC) is composed of three proteins, UL128, UL130, and UL131, that complex with gH–gL. This PC is important for infection of epithelial and endothelial cells and monocytes/macrophages (15–17). So-called low-passage, clinical HCMV isolates express both the PC and TC and have broad tropism, including the ability to infect all of the above-mentioned cell types (18). Omission of any one of the five members of the PC reduces assembly and entry into epithelial/endothelial cells (19). High-passaged laboratory strains of HCMV often lack the PC on virions but retain the TC and gB. For example, the AD169 strain lacks UL128 to UL150 ORFs (12), including UL128 to UL131, which prevent PC formation, resulting in virions expressing only gB and the TC. TC-expressing viruses have restricted tropism and infect fibroblasts. Restoration of functional PC genes in HCMV laboratory strains results in their ability to infect epithelial cells (17).
HCMV utilizes multiple host cell receptors to bind and infect cells, which may also explain its broad tropism. EGFR is reported to be an HCMV receptor that binds gB (20). While some have challenged the notion that EGFR is important for HCMV entry, EGFR signaling enhances HCMV replication in certain cell types (5, 21, 22). PDGFR-α is another HCMV receptor and binds to gB (9). However, PDGFR-α does not function as an entry receptor for pentamer-expressing HCMV infection of epithelial or endothelial cells (23). Recently, the trimer has been shown to bind directly to PDGFR-α (24), which makes virus binding and entry more complex. Wu et al. further identified the entry of TC viruses via PDGFR-α (25). In addition, integrins have been shown to be coreceptors for HCMV entry of fibroblasts, monocytes, and trophoblasts (7, 8, 26, 27) through their interactions with gB and the PC. The surface molecule THY-1 (CD90) also plays an important role in the initial stage of virus infection. It binds to both gB and gH and promotes entry of both laboratory-adapted and clinical isolates of HCMV (6, 28). It is uncertain whether THY-1 directly binds to these proteins, although THY-1 likely facilitates HCMV entry in many cell types. CD147 promotes entry of pentamer-expressing HCMV into epithelial and endothelial cells. However, soluble CD147 does not block HCMV entry, and the TC and PC do not bind directly to CD147 (29). During the preparation of this manuscript, the single-pass membrane protein NRP2 was shown to function as a receptor for the PC and HCMV infection of epithelial/endothelial cells (30). Epithelial cells are important for interhost transmission and many diseases associated with HCMV, such as retinitis, gastroenteritis, and encephalitis. Therefore, discovery of all relevant cell receptors and signaling pathways on epithelial cells will reveal important targets for preventing HCMV transmission and reducing diseases associated with this virus.
Here, we adapt the CRISPR/Cas9 screening (31–34) technique to identify host factors specifically required for HCMV infection of epithelial cells. These efforts identified an olfactory receptor family member, OR14I1, as being required for HCMV attachment, entry, and infection of epithelial cells. OR14I1 interacts with the HCMV PC, which can be blocked by anti-PC neutralizing antibodies. Exogenous OR14I1 protein in membrane vesicles or a synthetic OR14I1 N-terminal peptide prevents HCMV entry into epithelial cells. Olfactory receptors signal via the generation of cAMP by adenylate cyclase (AC) (35–38). We show that HCMV entry and infection of epithelial cells are blocked by inhibitors of AC and downstream protein kinase A (PKA) signaling when OR14I1 is present. OR14I1 also mediates AKT signaling and endocytosis-mediated entry of HCMV. The identification of OR14I1 as a PC-dependent HCMV host receptor associated with epithelial tropism and the role of an AC-mediated signaling pathway in HCMV infection provide previously unappreciated targets for the development of vaccines and antiviral therapies.
Results
Genetic Screen for Host Receptors Required for Epithelial Infection.
To identify host factors required for HCMV infection and replication, we performed two parallel genome-wide CRISPR screens using either epithelial-tropic TB40E infection of ARPE-19 epithelial cells or fibroblast-tropic AD169 infection of HEL fibroblasts (SI Appendix, Fig. S1). Cells expressing Cas9 and the GeCKO v.2 sgRNA library [19,050 genes targeted with 6 single-guide RNAs (sgRNAs) per gene] were repeatedly exposed to HCMV infection over 3 mo. Cells with sgRNA-induced resistance to HCMV survived this challenge and expanded their numbers. To identify enriched sgRNAs in the resistant cell population, their genomic DNA was subjected to targeted next-generation sequencing (NGS). The reagent redundancy principle (39) was used to select high-confidence candidates that scored with ≥3 unique sgRNAs, each with ≥20 NGS reads. These selection criteria resulted in 312 candidates in the epithelial cell screen. Genetic screens are notorious for generating false positives, so specific hits are suspect until independently confirmed. However, a collective analysis may offer insight. To provide a sense of relevance to this sample set, potential enriched pathways were identified using ConsensusPathDB (SI Appendix, Table S1). Many of these pathways have been previously identified as relevant to HCMV infection and replication by molecular and biochemical approaches. Two pathways, “signaling pathway from G-protein families” and “olfactory transduction,” directed our attention to olfactory receptors as potential receptors for HCMV.
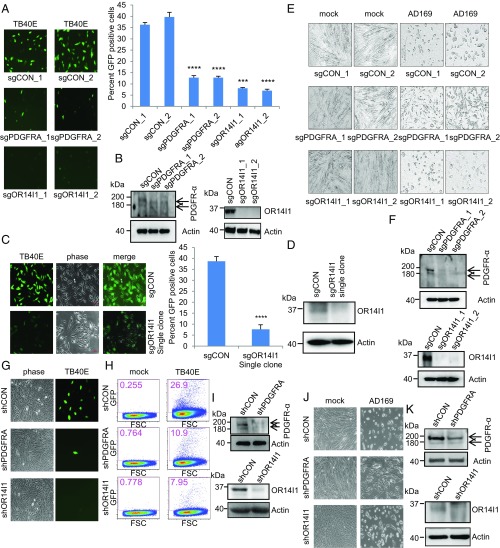
A review of candidates found solely in the epithelial cell screen revealed OR14I1, which encodes an olfactory receptor family membrane protein. In the initial evaluation, ARPE-19 epithelial cells expressing sgRNAs against either OR14I1 or PDGFRA were infected with a PC-positive (PC+) TB40E virus expressing green fluorescent protein (TB40E-GFP) and assessed for GFP expression 2 d post infection (dpi). The results demonstrate a marked reduction of HCMV infection in either OR14I1-deficient or PDGFR-α–deficient cells (Fig. 1 A and B). The OR14I1 observation was confirmed using a clonally derived population of sgRNA-OR14I1 cells (Fig. 1 C and D). The roles of OR14I1 and PDGFR-α were next evaluated in TC-only AD169 virus infections of HEL fibroblasts. HEL cells expressing sgRNAs targeting either OR14I1 or PDGFRA were infected with AD169 virus, and the cultures were monitored for cytopathic effect (Fig. 1 E and F). Consistent with the screen results, AD169 virus infection of HEL fibroblasts was inhibited by the loss of PDGFR-α but not in the absence of OR14I1, suggesting that OR14I1 is not required for AD169 infection of fibroblasts. As expected, reduced virus infection was observed with TB40E-GFP infection of ARPE-19 epithelial cells stably expressing shRNAs against either OR14I1 or PDGFRA, given that this strain contains both the TC and PC (Fig. 1 G–I). In contrast, AD169 virus infection of HEL fibroblasts stably expressing shRNAs against either OR14I1 or PDGFRA was inhibited by reduction of PDGFR-α but not by depletion of OR14I1, as AD169 only expresses the TC (Fig. 1 J and K). Together, these data confirm that PDGFR-α is needed for HCMV infection of fibroblasts and epithelial cells. In contrast, OR14I1 is required for PC+ HCMV infection of epithelial cells.
Fig. 1.
OR14I1 and PDGRFA are required for HCMV infection of epithelial cells. (A) ARPE-19 cells stably expressing the indicated sgRNAs were infected with TB40E-GFP virus (MOI 3.0). Cells were imaged (10×) for GFP expression (green) as an indicator of viral infection at 2 dpi, and the percent GFP-positive cells was quantified. CON, negative control. (B) Immunoblots of lysates from cells in A. (C) Clonally derived sgOR14I1-expressing ARPE-19 cells were infected with TB40E-GFP virus (MOI 3.0). Cells were imaged (10×) for GFP expression (green) as an indicator of viral infection at 2 dpi, and the GFP-positive cells were quantified. (D) Immunoblots of lysates from cells in C. (E) HEL fibroblasts stably expressing the indicated sgRNAs were infected with AD169 virus (MOI 3.0). Infectivity was determined by cytopathic effect at 2 dpi. (F) Immunoblots of lysates from cells in E. (G) ARPE-19 cells stably expressing the indicated shRNAs were infected with TB40E-GFP virus (MOI 2.0) and then imaged (10×) as in A. (H) ARPE-19 cells stably expressing the indicated shRNAs were infected with TB40E-GFP virus (MOI 2.0). GFP expression was determined at 2 dpi using flow cytometry. The plot depicts GFP versus forward scatter (FSC). (I) Immunoblots of lysates from cells in G. (J) HEL fibroblasts stably expressing the indicated shRNAs were infected with AD169 virus (MOI 2.0). Infectivity was determined by cytopathic effect at 2 dpi. (K) Immunoblots of lysates from cells in J. Actin serves as a loading control throughout. Representative images of three independent experiments are shown. Data represent the mean of n = 3 experiments ±SD. ***P < 0.001, ****P < 0.0001.
Both OR14I1 and PDGFR-α Contribute to HCMV Binding to ARPE-19 Epithelial Cells.
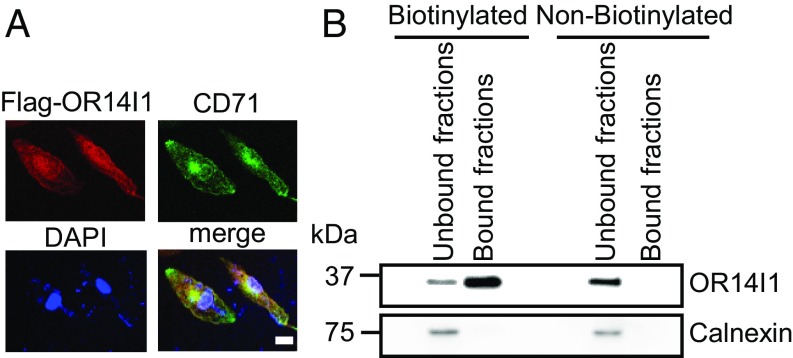
To establish the cellular localization of OR14I1, ARPE-19 cells were transiently transfected with a vector expressing Flag-tagged OR14I1 (Flag-OR14I1). OR14I1 was found to reside at the plasma membrane and other membrane-associated intracellular compartments (Fig. 2A). To verify that endogenous OR14I1 is located on the cell surface, proteins exposed to the extracellular environment were biotinylated and enriched over a streptavidin column. Both enriched and flow-through fractions were analyzed by immunoblot for OR14I1. As shown in Fig. 2B, OR14I1 was detected primarily in the biotinylated cell-surface fraction while an intracellular protein, calnexin, was observed only in the pooled unbound fractions. To determine if OR14I1 is necessary for HCMV binding to epithelial cells, shRNA-OR14I1 or shRNA-control ARPE-19 cells were chilled and then incubated on ice with chilled PC+ TB40E-GFP virus. After incubation and washing of unbound virus, viral DNA associated with the cell surface was quantified by qPCR. These studies detected a 79.3 ± 4.3% reduction of TB40E virus binding to OR14I1-depleted epithelial cells (Fig. 3 A and B). As expected, similar observations were made using shPDGFR-α ARPE-19 cells, given that TB40E virus contains viral TC as well as PC. Optimal binding of HCMV to OR14I1-deficient cells could be recovered in cells ectopically expressing an sgRNA-resistant OR14I1 cDNA at physiological and supraphysiological levels (Fig. 3 C and D and SI Appendix, Fig. S2), indicating that OR14I1 is required for optimal binding to epithelial cells. These results are consistent with the noted requirements for PC+ HCMV infection, in that both OR14I1 and PDGFR-α are necessary for HCMV binding to epithelial cells.
Fig. 2.
OR14I1 localizes to the plasma and additional cellular membranes. (A) Flag-tagged OR14I1 plasmid was transiently transfected into ARPE-19 cells. Thirty-six hours after transfection, cells were fixed, permeabilized, and immunostained with anti-Flag (red) and anti-transferrin receptor, CD71 (green) antibodies, and then imaged by confocal microscopy. CD71 served as a surrogate for membrane proteins. DAPI staining was used to define nuclei. (Scale bar, 20 µm.) (B) Immunoblot analysis for OR14I1 in the cell surface-bound fractions of biotinylated proteins and the flow-through (unbound fraction representing proteins that did not bind to the EZ-Link Sulfo-NHS-SS-Biotin column). Calnexin immunodetection was included to define the intracellular protein pool.
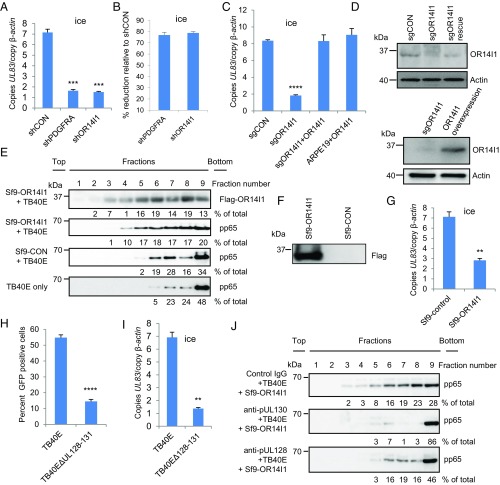
Fig. 3.
OR14I1 is necessary for PC+ HCMV binding to and infection of epithelial cells. (A) Virus binding assay. ARPE-19 cells expressing the indicated shRNAs were incubated with TB40E-GFP virus on ice (MOI 3.0). After washing, cell surface-bound viral DNA (UL83) was quantified using qPCR and normalized to cellular DNA (β-actin). (B) The results in A are presented as the relative reduction of viral DNA in the knockdown cell lines relative to shCON. (C) Binding assays as in A using ARPE-19 cells expressing the indicated sgRNAs and/or cDNAs: sgCON, clonal sgOR14I1 cells, sgOR14I1 cells expressing sgRNA-resistant OR14I1, or WT cells overexpressing OR14I1 (MOI 3.0). (D) Immunoblots of cells in C. (E) Membrane flotation assay. TB40E-GFP virus was incubated with membrane vesicles from control Sf9 cells (Sf9-CON) or Sf9 cells expressing human Flag-OR14I1 (Sf9-OR14I1). After centrifugation, fractions underwent immunoblotting to determine the levels of TB40E-GFP virus (virion protein pp65) and location of membrane vesicles (Flag-OR14I1). (F) Immunoblot of cells in E. (G) TB40E-GFP virus was preincubated with Sf9-control or Sf9-Flag-OR14I1 membrane vesicles before being used in a virus binding assay with ARPE-19 cells (MOI 3.0). Viral (UL83) and cellular (β-actin) DNA levels were quantified by qPCR. (H) ARPE-19 cells infected with TB40E-GFP or a PC-deleted TB40E-GFP (TB40E∆UL128–131; MOI 3.0). Cells were fixed at 2 dpi and assessed for GFP (green)-positive cells. (I) ARPE-19 cells were incubated with either TB40E-GFP or TB40E∆UL128–131 virus (MOI 3.0) on ice. After washing, cell surface-bound viral DNA (UL83) was quantified using qPCR and normalized to cellular DNA (β-actin). (J) Control IgG antibody, anti-pUL128, or anti-pUL130 was preincubated with purified TB40E-GFP virus in a membrane flotation assay. Data represent the mean of n = 3 experiments ±SD. **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine whether HCMV interacts with OR14I1, Sf9 insect cells were transduced with a baculovirus expressing Flag-tagged human OR14I1 or control. Using a membrane flotation assay, membrane vesicles generated from the transduced Sf9 cells were incubated with PC+ TB40E-GFP virions, followed by fractionation of the resultant suspension (40, 41) (Fig. 3 E and F). Individual fractions were assessed by immunoblotting for their respective levels of the HCMV virion protein pp65 and Flag-OR14I1. These assays demonstrated a concordance in the fractional levels of pp65 and OR14I1. Preincubation of TB40E-GFP virions with Sf9-OR14I1 membrane vesicles reduced viral binding to ARPE-19 cells, demonstrating that exogenous OR14I1 competes with HCMV binding to epithelial cells (Fig. 3G). To determine whether TB40E-GFP binding to OR14I1 is dependent on the PC, ARPE-19 epithelial cells were infected with either wild-type TB40E-GFP or TB40E-GFP lacking the PC (TB40E∆UL128–131). Consistent with published results, loss of the viral PC dramatically decreased epithelial cell infection (Fig. 3H and SI Appendix, Fig. S3), and this correlated with a comparable loss of virus binding to the cell surface (Fig. 3I). A requirement for the PC to efficiently infect epithelial cells was found to be independent of the HCMV strain used (SI Appendix, Fig. S4). However, loss of the viral PC did not affect HCMV infection of fibroblasts (SI Appendix, Fig. S5).
To determine if the PC is needed for HCMV binding to OR14I1, PC+ TB40E-GFP virus was preincubated with neutralizing antibodies (19) against either of two PC subunits (pUL130 or pUL128) or a negative control antibody; Sf9-OR14I1 membrane vesicles were then added, and membrane flotation assays were performed (Fig. 3J). These assays showed that viral binding to OR14I1 was diminished by the presence of either anti-PC antibody. Together, these results demonstrate that the virion-associated PC is required for interaction of HCMV with OR14I1.
A Peptide Representing the N Terminus of OR14I1 Blocks HCMV Binding and Infection of Epithelial Cells.
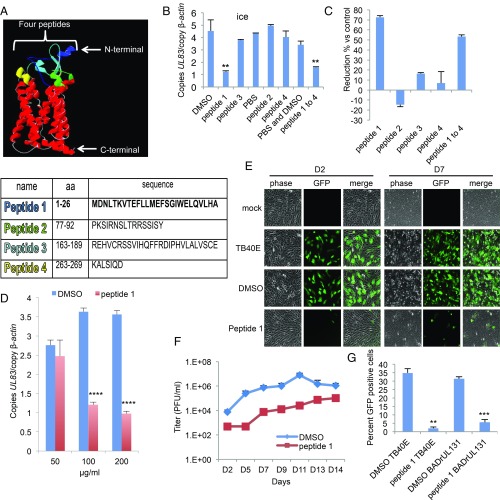
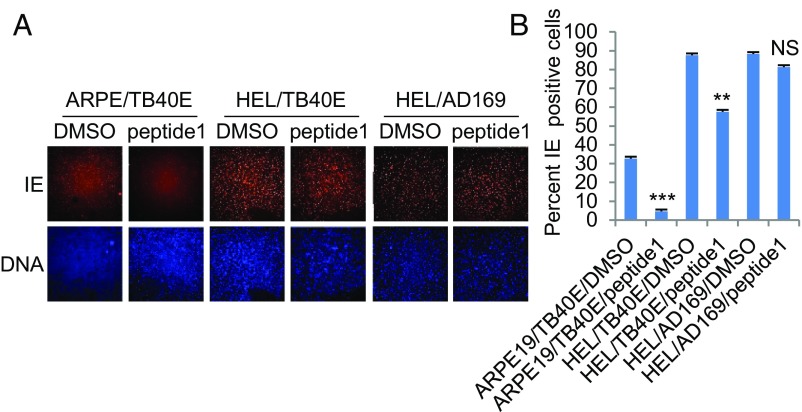
A structural model of OR14I1 inferred from the crystal structure of another G protein-coupled receptor, 4yay.1.A (soluble cytochrome b562, type 1 angiotensin II receptor), predicts four regions of OR14I1 that are exposed to the cell surface (Fig. 4A). Peptides were generated that represent each of these regions, and their effects on TB40E-GFP virus binding to, and infection of, ARPE-19 epithelial cells were determined. These assays revealed that only the most N-terminal peptide (peptide 1) of OR14I1 prevented TB40E-GFP binding and infection of ARPE-19 cells (Fig. 4 B and C) in dose-dependent and sustained manners (Fig. 4 D and E). The initial treatment with peptide 1 also limited the long-term replication of HCMV (Fig. 4F). The antiviral activity of peptide 1 was independent of viral strain (Fig. 4G and SI Appendix, Figs. S6 and S7), but was dependent on the presence of the PC (Figs. 4 and 5). Peptide 1 had a modest effect on PC+ HCMV infection of fibroblasts but prevented PC+ virus infection of ARPE-19 cells (Fig. 5) and several other epithelial cell lines (SI Appendix, Fig. S8). Together, these results show that a peptide representing the N-terminal sequence of OR14I1 can limit the binding, infection, and replication of PC+ HCMV in epithelial cells.
Fig. 4.
OR14I1 peptide inhibits HCMV binding and infection of epithelial cells. (A) OR14I1 model depicting four extracellular domains (blue, green, aqua, yellow) with their amino acid (aa) sequences listed below. (B) Binding assay. TB40E-GFP virus was preincubated with the indicated OR14I1 peptides (100 μg/mL) or relevant solvent followed by incubation of the virus with ARPE-19 cells on ice (MOI 2.0). After washing, cell surface-bound viral DNA (UL83) was quantified using qPCR and normalized to cellular DNA (β-actin). (C) The results in B are presented as the relative reduction in cell-bound viral DNA by peptide treatment relative to the relevant control. (D) Cell binding assay using the indicated concentrations of peptide 1 (MOI 2.0). (E) TB40E-GFP virus was preincubated with peptide 1 (100 μg/mL) or DMSO, followed by infection of ARPE-19 cells (MOI 2.0). Cells were imaged (10×) for GFP expression at 2 dpi (D2) and 7 dpi (D7). (F) Culture media supernatants from E were harvested on the indicated dpi and assayed for infectious virus by plaque assay. (G) TB40E-GFP or BADrUL131-GFP virus, both expressing the PC, was preincubated with peptide 1 (100 μg/mL) or DMSO before infection of ARPE-19 cells (MOI 2.0). Cells were fixed and assessed for GFP (green)-positive cells at 2 dpi. Data represent the mean of n = 3 experiments ±SD. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Fig. 5.
Synthetic N-terminal peptide of OR14I1 blocks HCMV infection of ARPE-19 epithelial cells and is dependent on the presence of viral PC. (A) TB40E-GFP encoding the viral PC or AD169 lacking the PC was preincubated with peptide 1 (100 μg/mL) and then infected on ARPE-19 epithelial cells or HEL fibroblasts (MOI 2.0). Cells were fixed, permeabilized, stained for DNA (blue), immunostained for viral immediate-early protein (IE), and imaged (4×). (B) Quantitation of data in A indicating the percent IE-positive cells. Data represent the mean of n = 3 experiments ±SD. **P < 0.01, ***P < 0.001; NS, not significant.
AC/PKA/AKT Signaling Is Required for HCMV Entry and Infection of Epithelial Cells.
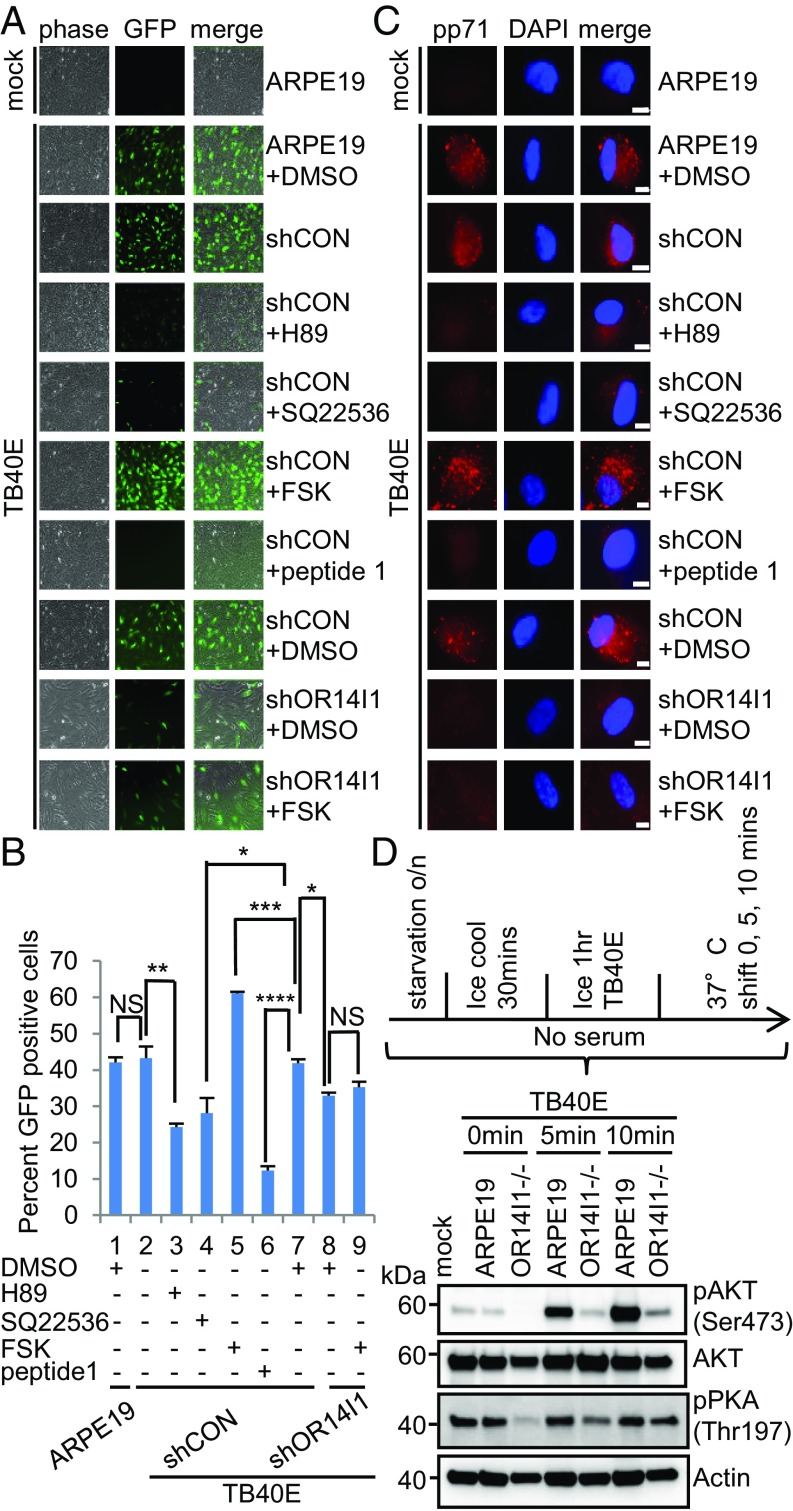
OR14I1 belongs to the family of G protein-coupled receptors (GPCRs) that initiate a cascade of cellular signaling events. Downstream signaling by olfactory receptors is mediated by adenylate cyclase and protein kinase A activities (38). Given that OR14I1 is required for PC-mediated HCMV attachment and infection of epithelial cells, a role for AC and PKA in HCMV replication was accessed. ARPE-19 epithelial cells expressing either a control shRNA, or an shRNA against OR14I1 expression, were pretreated with the following: the AC antagonist SQ22536, AC agonist forskolin (FSK), PKA inhibitor H-89, or OR14I1 peptide 1. The signaling inhibitors H-89, SQ22536, as well as peptide 1 significantly reduced infectivity (Fig. 6 A and B). In contrast, the AC agonist FSK enhanced infection of ARPE-19 epithelial cells, but only in cells expressing OR14I1, suggesting that the combination of OR14I1 engagement and AC activity is required for HCMV infection. These observations were further explored by synchronizing infections by temperature shift and virus entry measured at 2 hpi by intracellular staining of the virion pp71 protein (Fig. 6C). The patterns were similar to those in Fig. 6A, where antagonists reduced entry and the agonist enhanced entry in the presence of OR14I1, thereby reinforcing the conclusion that the combination of OR14I1 engagement and AC activity is required for HCMV entry. To determine whether OR14I1 engagement activated the signaling cascade, we monitored the AKT phosphorylation state and found it was rapidly activated when virus and cells were synchronously released from a chilled incubation state by a shift to 37 °C (Fig. 6D). Interestingly, AKT and PKA activation is reduced when OR14I1 is depleted (Fig. 6D), which indicates that OR14I1 is required for stimulating the AC/PKA/AKT signaling pathway.
Fig. 6.
AC/PKA/AKT signaling is required for HCMV entry and infection of epithelial cells. (A) ARPE-19 cells expressing the indicated shRNAs were pretreated with PKA inhibitor H-89 (20 μM), adenylate cyclase antagonist SQ22536 (150 μM), AC agonist forskolin (20 μM), or DMSO solvent for 2 h before TB40E-GFP infection (MOI 2.0). Peptide 1 was preincubated with TB40E for 2 h at 37 °C before addition to cells. Cells were imaged (10×) for GFP at 2 dpi. (B) Quantification of data in A after cell fixation and DNA staining. Results are presented as the percent GFP-positive cells. Data represent the mean of n = 3 experiments ±SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (C) ARPE-19 cells expressing the indicated shRNAs were pretreated with the indicated drug for 2 h before TB40E-GFP infection (MOI 2.0); peptide 1 was preincubated with TB40E for 2 h at 37 °C. Cells were cooled on ice for 30 min and then infected with cold TB40E (MOI 2.0) containing the noted small molecules for 1 h on ice. Cells were then transferred to 37 °C. At 2 hpi, cells were washed and treated briefly with trypsin to remove surface-bound virus. They were then fixed, permeabilized, stained with anti-pp71, and imaged (63×). (Scale bars, 10 µm.) Representative images of three independent experiments are shown. (D) ARPE-19 and OR14I1−/− cells were infected with cold TB40E (MOI 2.0) for 1 h on ice. Cells were transferred to 37 °C for 0, 5, and 10 min. Levels of p-AKT (S473), total AKT, p-PKA (T197), and actin were detected by immunoblotting from whole-cell lysates. o/n, overnight.
OR14I1 Mediates Endocytosis of HCMV.
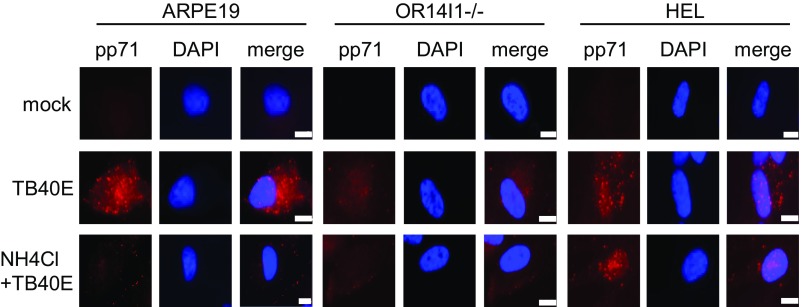
HCMV enters into epithelial cells by endocytosis followed by low pH-dependent fusion (12, 42). Uptake of fluorescently labeled transferrin was used to monitor endocytosis activity (43, 44). HCMV infection increases endocytosis, as measured by transferrin uptake in ARPE-19 epithelial cells by 2 hpi (SI Appendix, Fig. S9A). Moreover, HCMV uptake tracked with transferrin, as measured by the intracellular appearance and apparent colocalization of virion-derived pp71 with transferrin (SI Appendix, Fig. S9B), further confirming that HCMV enters epithelial cells through endocytosis. Both transferrin uptake and HCMV internalization were reduced in cells lacking OR14I1, suggesting that this receptor contributes to HCMV infection of epithelial cells by endocytosis. Many viruses that are internalized by endocytosis subsequently require the low-pH environment of endosomes to trigger viral membrane glycoproteins to promote fusion (45–47). Lysosomotropic agents such as ammonium chloride interfere with endosome acidification by buffering endosomal pH, which has been shown to inhibit infection by HCMV and other viruses that require low endosomal pH (12, 45, 48). When ARPE-19, HEL, and OR14I1−/− cells were pretreated with ammonium chloride, HCMV entry was reduced in ARPE-19 cells and to a lesser extent in OR14I1−/− ARPE-19 cells, but not affected in HEL cells (Fig. 7). These observations are consistent with a previous report showing that HCMV enters fibroblasts by pH-independent fusion at the plasma membrane (11) instead of endocytosis. Of note, the low levels of HCMV entry in OR14I1−/− cells are further reduced by ammonium chloride pretreatment (Fig. 7), thereby raising the possibility that a second endocytic pathway exists in ARPE-19 that can mediate viral entry.
Fig. 7.
OR14I1 mediates endocytosis for entry of HCMV. ARPE-19, OR14I1−/− ARPE-19, and HEL cells were pretreated with ammonium chloride (30 mM) for 2 h. Cells and virus were cooled on ice for 30 min and infected with cold TB40E (MOI 2.0) in the presence or absence of ammonium chloride for 1 h on ice. Cells were transferred to 37 °C. At 2 hpi, cells were washed and treated briefly with trypsin to remove surface-bound virus. The cells were then fixed, permeabilized, stained with anti-pp71, and imaged (63×). (Scale bar, 10 µm.)
Discussion
Here we used CRISPR/Cas9 screens to identify an olfactory receptor, OR14I1, required for HCMV infection of epithelial cells. This cell type is central to interhost HCMV transmission and pathogenesis. Further characterization revealed that OR14I1 functions as a host receptor for the viral pentameric complex and plays an essential role in epithelial cell tropism of HCMV. Our data also suggest that OR14I1 and PDGFR-α serve nonredundant roles as coreceptors for HCMV in epithelial cells. In the absence of the PC, HCMV no longer interacts with OR14I1 or efficiently infects epithelial cells. In contrast, HCMV infection of fibroblasts relies on the interaction of the viral trimeric complex with receptors (PDGFR-α) and coreceptors (integrins) for infection (SI Appendix, Fig. S10). A primary route of HCMV transmission involves infection of epithelial cells in the oral cavity (49), and epithelia are initial sites of HCMV replication and pathogenesis. Thus, the identification of an epithelial tropism receptor for the HCMV PC and the role of an adenylate cyclase and PKA signaling pathway in epithelial infection improve our understanding of HCMV pathogenesis and suggest previously unappreciated strategies for the prevention and treatment of HCMV infection. Importantly, the N terminus of OR14I1 is identified as a potential drug target to prevent HCMV infection.
CRISPR screens of host genes have permitted the exploration of viral infection with generally greater versatility and fidelity in comparison with RNAi approaches. Interpretation is simplified because CRISPR approaches generate null cells whereas RNAi approaches can result in hypomorphic phenotypes. However, CRISPR-mediated gene ablation often limits readouts to initial virus–cell and proximal events. Both approaches are prone to false positive (and negative) results, so validation is a critical first step to studying individual hits. Given this caveat, we limited our analysis to validated hits and to pathway analysis with the assumption that it is less likely that multiple (or all of the) hits in a predicted pathway would be false positives. Experimentally, we find the relative strengths and weaknesses of the two approaches to be complementary, and exploited them in this study. We used a retinal pigment epithelial cell line (ARPE-19) and a primary cell line of human lung fibroblasts (HEL) for the screens, which are cell types targeted by HCMV in vivo. Using this approach, we identified both a known (PDGFR-α) and an unrecognized HCMV receptor (OR14I1).
OR14I1 is a GPCR and a member of the large olfactory receptor family. The recently recognized OR14 gene family (50) is part of the type II group of olfactory receptors (51). There are six OR14 genes and one pseudogene within the six subfamilies of human OR14, which are located on chromosome 1 (50). Traditionally, olfactory receptor families are thought of as chemosensors that are responsible for olfaction. As such, they are found in sensory organs that contain olfactory epithelium and neurons in the nasal cavity in mammals. In addition to this specialized role in the nose, olfactory receptors, including OR14I1, are widely distributed in organs and systems in mammals (52) with evidence for evolutionary constraint of these so-called ectopically expressed olfactory receptors, which is consistent with biologically significant function (53). Indeed, certain ectopically expressed olfactory receptors function in physiology, including roles in renal and blood pressure regulation (54, 55). However, no function has been ascribed to any member of the OR14 family.
That olfaction-related receptors can transmit infectious agents has been demonstrated previously (56–58). However, the specific epithelial cells responsible for HCMV transmission are not known. A report suggests that murine CMV (MCMV) exploits olfaction to enter new hosts (59). MCMV infects nasally rather than orally, both after experimental virus uptake and during natural transmission. HCMV can behave as a neurotropic virus with predilection for the retina and central nervous system (60). Murid herpesvirus 4 and herpes simplex virus 1 can use olfactory epithelium as an entry site (61, 62). Given the results presented here, it is tempting to speculate that HCMV could transmit via a similar mechanism.
Mammalian olfactory neurons possess a well-developed system of endocytic vesicles, endosomes, lysosomes, and endocytosis that function in the olfactory epithelium (63). As GPCRs, olfactory receptors engage ligands at the N terminus of the receptors and signal through AC and PKA pathways (64–66). Thus, HCMV interaction with the N terminus of OR14I1 and the use of downstream signaling to gain entry into cells are consistent with GPCR interactions with their native ligand. Unfortunately, like many olfactory receptors, OR14I1 is an orphan receptor. The absence of a known ligand limits our ability to perform comparative studies of the signaling processes and their relationship to entry.
Fibroblast infection involves interactions of the TC with PDGFR-α and gB-mediated fusion of the virion and plasma membranes (24). Infection of epithelial cells appears to be more complex with gB, and TC contributions plus the viral PC interactions with the host receptor. Viral entry in this context is thought to be via endocytosis/micropinocytosis (12). In addition to our identification of OR14I1 as a receptor for the PC, others have shown that CD147 functions as an entry mediator for PC+ HCMV into epithelial cells (29), although a direct interaction between CD147 and the PC was not observed. A recent PC interaction screen identified NRP2 as a receptor for the PC (30). The multipass membrane protein OR14I1 was apparently not detected. This may be due to an inherent bias of the biochemical screen that limited candidates to single-pass membrane proteins. Both CD147 and NRP2 appeared in our CRISPR screen. NRP2 was a lower-ranking hit, and neither was subjected to further analyses. The presence of at least three sets of virion glycoproteins and multiple host cell receptors demonstrates that virion–receptor interactions and infection of cells by HCMV are complex.
This report shows that the HCMV PC requires OR14I1 binding and activation of AC/PKA/AKT signaling to define epithelial tropism. These findings do not exclude roles for other coreceptors during HCMV infection, such as PDGFR-α/EGFR, integrins, and NRP2. HCMV infection of epithelial cells can be blocked by a synthetic peptide representing the N terminus of OR14I1 or inhibitors of intracellular signaling. Together, these findings answer questions regarding a mechanism for epithelial tropism, and offer antiviral strategies for the management of HCMV transmission and disease.
Materials and Methods
Cell Lines.
ARPE-19 epithelial cells, human embryonic lung (HEL) fibroblasts, A549 epithelial cells, HEK293T cells, H1HeLa cells, MRC5 cells, and Sf9 insect cells were obtained from the ATCC. Detailed information on culture conditions is provided in SI Appendix, Materials and Methods.
Viruses.
HCMV AD169 (ATCC) was propagated in HEL fibroblasts by infecting at a multiplicity of infection (MOI) of 0.01 followed by culturing the cells until a cytopathic effect was well-developed. Virus was prepared by sonicating the cells, followed by centrifugal clarification at 2,000 × g. Titering was done using a standard plaque assay on HEL fibroblasts. HCMV TB40E-GFP virus was generated from a bacterial artificial chromosome (BAC), which was a kind gift of Eain Murphy, Cleveland Clinic, Cleveland, OH. This version of TB40E-GFP expresses GFP as a transgene under the control of an SV40 origin/promoter cassette. Infectious virus was recovered by electroporation of BAC DNA into HEL fibroblasts as described (67). TB40E-GFP was adapted to ARPE-19 epithelial cells through eight passages, infecting at an MOI of 0.1 for each passage. Viral stocks of TB40E-GFP were generated and titered as described for AD169. TR5 and TR5∆UL128–131 were described (68). TB40E∆UL128–131 virus was generated by BAC recombineering using the same primers and protocol used to create TR5∆UL128–131 and was kindly provided by Jay Nelson, Oregon Health & Science University, Beaverton, OR. BADwt (69) is derived from a BAC clone of HCMV AD169. BADrUL131 (17, 19) is a PC-expressing derivative of BADwt in which the UL131 ORF has been repaired. Both clones were kindly provided by Thomas Shenk, Princeton University, Princeton. Cell-free virions were purified by centrifugation (SW28 rotor; Beckman) at 23,000 rpm for 1 h through a sorbitol (Fisher Bioreagents) cushion, and then resuspended in serum-free medium. In each experiment, HEL or ARPE-19 cells were infected with HCMV at the noted MOI for 2 h at 37 °C, except for viral binding studies, which were done on ice.
Recombinant retroviruses and lentiviruses were produced in HEK293T cells, and titers were determined by standard colony formation assay on H1HeLa cells (70).
sgRNA Library Cells.
Streptococcus pyogenes Cas9 was subcloned from lentiCas9-Blast (Addgene; 52962) into the pHAGE-hygromycin lentiviral vector (Addgene). pHAGE-H-Cas9 lentivirus was used to transduce ARPE-19 cells and HEL fibroblasts (ARPE-19-Cas9, HEL-Cas9), which were selected with hygromycin B (200 μg/mL; Life Technologies). ARPE-19-Cas9 cells and HEL-Cas9 fibroblasts were transduced with the human GeCKO v.2 sgRNA library, part A or B (33) (Addgene; 1000000049; MOI 0.2), and selected with puromycin (2.5 μg/mL; Life Technologies).
CRISPR Screens.
ARPE-19-Cas9 cells expressing the GeCKO v.2 library were infected with TB40E-GFP (MOI 5.0). In a parallel screen, HEL-Cas9 fibroblasts expressing the GeCKO v.2 library were infected with AD169 (MOI 5.0). The surviving cells were expanded and genomic DNA was prepared. Screening details are described in SI Appendix, Materials and Methods.
sgRNA Sequencing and Analysis.
The host genome-integrated sgRNAs were amplified and subjected to NGS using an Ion Proton Sequencer (Life Technologies). After sequencing and analysis, selected candidate genes were chosen for further study if they had ≥20 reads per sgRNA across ≥3 independent sgRNAs. Detailed information is provided in SI Appendix, Materials and Methods.
ARPE-19−/− and HEL−/− Cells.
sgRNA sequences were cloned into the LentiGuide-Puro vector to generate knockout cells. For details, see SI Appendix, Materials and Methods.
ARPE-19 and HEL shRNA Cells.
shRNA sequences were cloned into the pLKO.1-blast lentiviral vector and used to generate shRNA cells. For details, see SI Appendix, Materials and Methods.
Immunoblotting.
Cells were processed for immunoblotting as described (71). Detailed information about antibodies is provided in SI Appendix, Materials and Methods.
Imaging.
Phase, GFP, transferrin, HCMV pp71, and early protein images were captured on a Zeiss microscope (AxioObserver Z1) at 4×, 10×, or 63× magnification. For quantitation of fixed cells, cells were fixed and DNA was stained. The fixed cells were imaged and analyzed with MetaXpress imaging software (Molecular Devices) to determine the total cells per well and the percentage of infected cells (GFP-positive) in each well. Vignetting in images is due to edge effects resulting from capturing entire wells. Detailed information is provided in SI Appendix, Materials and Methods.
Plasmids.
PQCXIN-OR14I1, PQCXIN-Flag-OR14I1, and PQCXIN-OR14I1 rescue were constructed using the PQCXIN retroviral vector (Clontech). Cloning details are included in SI Appendix, Materials and Methods.
Transient Transfections and Confocal Imaging of CD71 and Flag-OR14I1.
Flag-OR14I1 plasmid was transiently transfected into ARPE-19 cells. Thirty-six hours after transfection, cells were fixed, permeabilized, and costained with anti-Flag (F1804; Sigma; 1:100) and anti-CD71 (555534; BD Pharmingen; 1:100) antibodies. Samples were imaged using a Nikon A1 inverted confocal microscope. Detailed information is provided in SI Appendix, Materials and Methods.
Cell-Surface Protein Detection.
The Pierce Cell Surface Protein Isolation Kit (Thermo Scientific) was used for isolation and collection of surface proteins, generally following the manufacturer’s protocol. For further details, see SI Appendix, Materials and Methods. Immunoblotting was performed with antibodies specific for OR14I1 (ARP71293-P050; Aviva Systems Biology) and calnexin (sc-23954; Santa Cruz Biotechnology).
Flow Cytometry.
ARPE-19 cells were infected with GFP-expressing virus. Flow cytometry was performed on a MACSQuant analyzer (Miltenyi Biotec) to quantify GFP-positive cells. For details, see SI Appendix, Materials and Methods.
Virus Binding Assay.
ARPE-19 cells were chilled on ice for 20 min and then incubated with chilled TB40E-GFP at the indicated MOI on ice for 1 h. The cells were then washed 10 times with cold PBS. Cellular DNA and cell-associated viral DNA were isolated using a DNeasy Blood & Tissue Kit (Qiagen). Viral DNA (UL83) and host DNA (β-ACTIN) were quantified by real-time quantitative PCR as described (72).
Generation of ARPE OR14I1 Rescue and Overexpression Cells.
PQCXIN-OR14I1 rescue plasmid was packaged into retroviruses which were used to transduce ARPE-19 OR14I1−/− cells (for rescue), or PQCXIN-OR14I1 was packaged into retroviruses which were used to transduce ARPE-19 cells (for overexpression) as previously described (72). OR14I1 expression was confirmed by immunoblotting.
Expression of Human OR14I1 in Sf9 Cells.
Human OR14I1 cDNA was Flag-tagged at the N terminus and subcloned into the pFasBac/CT-Topo baculovirus vector to generate Sf9 cells that express human OR14I1. Detailed information is provided in SI Appendix, Materials and Methods.
Membrane Vesicle Preparation.
Sf9 cells were harvested at 4 dpi with the recombinant OR14I1-expressing baculovirus. Pellets were washed twice with PBS and resuspended in hypotonic lysis buffer. Details of the preparation and processing of membrane vesicles are available in SI Appendix, Materials and Methods.
Neutralization Assay.
Purified TB40E-GFP virus (30-µL volume, 1 × 105 pfu) was incubated with either anti-pUL128 rabbit polyclonal antibody (1 µg/30 μL), anti-pUL130 3E3 murine monoclonal antibody (2 µg/30 μL), or isotype-matched control Ig antibodies (Abcam) for 1 h at 700 rpm in a ThermoMixer C (Eppendorf) at room temperature. Both the anti-UL128 and anti-UL130 antibodies bind to the PC and neutralize HCMV (19) (antibodies were provided by Thomas Shenk). After incubation with the noted antibodies, the virus preparation was used for membrane flotation assays.
Membrane Flotation Assay.
Purified TB40E-GFP virus, or virus incubated with the antibodies noted above (Neutralization Assay), was mixed with either Sf9-control or Sf9-Flag-OR14I1 membrane vesicles. After incubation, the membrane vesicle/virus sample was then mixed with sucrose solution. The samples were then ultracentrifuged. Fractions were collected from top to bottom and analyzed by immunoblotting. Details of this assay are provided in SI Appendix, Materials and Methods.
In Vitro Blocking Assay.
TB40E-GFP virus was incubated with Sf9-control– or Sf9-Flag-OR14I1–containing membrane vesicles (40 μg/mL) for 2 h at 37 °C in a ThermoMixer. ARPE-19 cells were chilled and then incubated with the resultant chilled virus on ice for 1 h (MOI 3.0).
Three-Dimensional Structure Prediction and Peptide Synthesis.
A structure for OR14I1 was inferred using SWISS-MODEL (https://swissmodel.expasy.org) with 4yay.1.A (soluble cytochrome b562, type 1 angiotensin II receptor) as a G protein-coupled receptor prototype and visualized using Swiss-PdbViewer 4.1.0 (https://spdbv.vital-it.ch/download_prerelease.html). Regions of OR14I1 predicted to be on the cell surface were identified and corresponding peptides were synthesized (peptide sequences are listed in Fig. 4A). Peptides were synthesized by Bachem.
Peptide Blocking Assay.
The noted synthetic peptides (100 μg/mL each or as described) were mixed with TB40E-GFP virus (1.2 × 106 pfu) and incubated at 37 °C for 2 h with rocking. The resulting samples were then subjected to the cells for binding (Virus Binding Assay) or infection assay. For infection assay, the mix of peptide and TB40E was then added to the cells for 2 h. After incubation, the virus and peptide were removed and replaced with culture medium without peptide.
AC/PKA Signaling Pathway Modulation.
ARPE-19 cells expressing the indicated shRNAs were treated with H-89, SQ22536, or forskolin before and during infection with TB40E-GFP. Detailed information is available in SI Appendix, Materials and Methods.
Viral Entry Assays.
Cells were pretreated with PKA inhibitor H-89 (20 μM), AC antagonist SQ22536 (150 μM), AC agonist forskolin (20 μM), or DMSO solvent and then incubated with cold TB40E virus with the noted small molecules. Cells were then transferred to 37 °C, fixed, stained with anti-pp71 (red), and imaged (63×). Detailed information is provided in SI Appendix, Materials and Methods.
PKA/AKT Signaling Detection.
Cells were starved and infected with TB40E virus. The levels of p-AKT, total AKT, p-PKA, and actin were detected by immunoblotting. Detailed information is available in SI Appendix, Materials and Methods.
Endocytosis Assay.
A published protocol was employed that uses labeled transferrin to monitor uptake (43, 44). In addition, the lysosomotropic agent ammonium chloride, which inhibits endocytosis (12, 45, 48), was examined for its effect on viral entry. Detailed information is available in SI Appendix, Materials and Methods.
Statistical Analysis.
Statistical analyses were performed using unpaired t tests. Values are expressed as mean ± SD of three independent experiments. A P value of <0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank John Holik, Anne Mirza, and Olesea Cojohari for helpful discussions and reviewing the manuscript. This work was supported by NIH Grants AI109001 (to T.F.K.), AI056077 (to A.D.Y.), and AI127335 (to A.D.Y.). A.L.B. is grateful to the Bill and Melinda Gates Foundation and Gilead Pharmaceuticals for their support.
Footnotes
Conflict of interest statement: X.E., A.L.B., and T.F.K. are listed as coinventors in a patent application based in part on data from this study.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814850116/-/DCSupplemental.
References
- 1.Britt W. Manifestations of human cytomegalovirus infection: Proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol. 2008;325:417–470. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- 2.Dollard SC, Grosse SD, Ross DS. New estimates of the prevalence of neurological and sensory sequelae and mortality associated with congenital cytomegalovirus infection. Rev Med Virol. 2007;17:355–363. doi: 10.1002/rmv.544. [DOI] [PubMed] [Google Scholar]
- 3.Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol. 2007;17:253–276. doi: 10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- 4.Sinzger C, Digel M, Jahn G. Cytomegalovirus cell tropism. Curr Top Microbiol Immunol. 2008;325:63–83. doi: 10.1007/978-3-540-77349-8_4. [DOI] [PubMed] [Google Scholar]
- 5.Chan G, Nogalski MT, Yurochko AD. Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proc Natl Acad Sci USA. 2009;106:22369–22374. doi: 10.1073/pnas.0908787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q, Fischer E, Cohen JI. Cell surface THY-1 contributes to human cytomegalovirus entry via a macropinocytosis-like process. J Virol. 2016;90:9766–9781. doi: 10.1128/JVI.01092-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Huang DY, Huong SM, Huang ES. Integrin alphavbeta3 is a coreceptor for human cytomegalovirus. Nat Med. 2005;11:515–521. doi: 10.1038/nm1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campadelli-Fiume G, Collins-McMillen D, Gianni T, Yurochko AD. Integrins as herpesvirus receptors and mediators of the host signalosome. Annu Rev Virol. 2016;3:215–236. doi: 10.1146/annurev-virology-110615-035618. [DOI] [PubMed] [Google Scholar]
- 9.Soroceanu L, Akhavan A, Cobbs CS. Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature. 2008;455:391–395. doi: 10.1038/nature07209. [DOI] [PubMed] [Google Scholar]
- 10.Wille PT, Wisner TW, Ryckman B, Johnson DC. Human cytomegalovirus (HCMV) glycoprotein gB promotes virus entry in trans acting as the viral fusion protein rather than as a receptor-binding protein. MBio. 2013;4:e00332-13. doi: 10.1128/mBio.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Compton T, Nepomuceno RR, Nowlin DM. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology. 1992;191:387–395. doi: 10.1016/0042-6822(92)90200-9. [DOI] [PubMed] [Google Scholar]
- 12.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol. 2006;80:710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plachter B, Sinzger C, Jahn G. Cell types involved in replication and distribution of human cytomegalovirus. Adv Virus Res. 1996;46:195–261. doi: 10.1016/s0065-3527(08)60073-1. [DOI] [PubMed] [Google Scholar]
- 14.Wille PT, Knoche AJ, Nelson JA, Jarvis MA, Johnson DC. A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells. J Virol. 2010;84:2585–2596. doi: 10.1128/JVI.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerna G, et al. Dendritic-cell infection by human cytomegalovirus is restricted to strains carrying functional UL131-128 genes and mediates efficient viral antigen presentation to CD8+ T cells. J Gen Virol. 2005;86:275–284. doi: 10.1099/vir.0.80474-0. [DOI] [PubMed] [Google Scholar]
- 16.Hahn G, et al. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol. 2004;78:10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Shenk T. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol. 2005;79:10330–10338. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vanarsdall AL, Johnson DC. Human cytomegalovirus entry into cells. Curr Opin Virol. 2012;2:37–42. doi: 10.1016/j.coviro.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci USA. 2005;102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Huong SM, Chiu ML, Raab-Traub N, Huang ES. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature. 2003;424:456–461. doi: 10.1038/nature01818. [DOI] [PubMed] [Google Scholar]
- 21.Isaacson MK, Feire AL, Compton T. Epidermal growth factor receptor is not required for human cytomegalovirus entry or signaling. J Virol. 2007;81:6241–6247. doi: 10.1128/JVI.00169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JH, Collins-McMillen D, Buehler JC, Goodrum FD, Yurochko AD. Human cytomegalovirus requires epidermal growth factor receptor signaling to enter and initiate the early steps in the establishment of latency in CD34+ human progenitor cells. J Virol. 2017;91:e01206-16. doi: 10.1128/JVI.01206-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanarsdall AL, Wisner TW, Lei H, Kazlauskas A, Johnson DC. PDGF receptor-α does not promote HCMV entry into epithelial and endothelial cells but increased quantities stimulate entry by an abnormal pathway. PLoS Pathog. 2012;8:e1002905. doi: 10.1371/journal.ppat.1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, et al. Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry. PLoS Pathog. 2017;13:e1006281. doi: 10.1371/journal.ppat.1006281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu K, Oberstein A, Wang W, Shenk T. Role of PDGF receptor-α during human cytomegalovirus entry into fibroblasts. Proc Natl Acad Sci USA. 2018;115:E9889–E9898. doi: 10.1073/pnas.1806305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maidji E, Genbacev O, Chang HT, Pereira L. Developmental regulation of human cytomegalovirus receptors in cytotrophoblasts correlates with distinct replication sites in the placenta. J Virol. 2007;81:4701–4712. doi: 10.1128/JVI.02748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Compton T. Receptors and immune sensors: The complex entry path of human cytomegalovirus. Trends Cell Biol. 2004;14:5–8. doi: 10.1016/j.tcb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, Wilkie AR, Weller M, Liu X, Cohen JI. THY-1 cell surface antigen (CD90) has an important role in the initial stage of human cytomegalovirus infection. PLoS Pathog. 2015;11:e1004999. doi: 10.1371/journal.ppat.1004999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanarsdall AL, et al. CD147 promotes entry of pentamer-expressing human cytomegalovirus into epithelial and endothelial cells. MBio. 2018;9:e00781-18. doi: 10.1128/mBio.00781-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez-Martin N, et al. An unbiased screen for human cytomegalovirus identifies neuropilin-2 as a central viral receptor. Cell. 2018;174:1158–1171.e19. doi: 10.1016/j.cell.2018.06.028. [DOI] [PubMed] [Google Scholar]
- 31.Tao L, et al. Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature. 2016;538:350–355. doi: 10.1038/nature19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perreira JM, Meraner P, Brass AL. Functional genomic strategies for elucidating human-virus interactions: Will CRISPR knockout RNAi and haploid cells? Adv Virus Res. 2016;94:1–51. doi: 10.1016/bs.aivir.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savidis G, et al. Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Rep. 2016;16:232–246. doi: 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 35.Lowe G, Nakamura T, Gold GH. Adenylate cyclase mediates olfactory transduction for a wide variety of odorants. Proc Natl Acad Sci USA. 1989;86:5641–5645. doi: 10.1073/pnas.86.14.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong ST, et al. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27:487–497. doi: 10.1016/s0896-6273(00)00060-x. [DOI] [PubMed] [Google Scholar]
- 37.Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- 38.Miazzi F, Hansson BS, Wicher D. Odor-induced cAMP production in Drosophila melanogaster olfactory sensory neurons. J Exp Biol. 2016;219:1798–1803. doi: 10.1242/jeb.137901. [DOI] [PubMed] [Google Scholar]
- 39.Echeverri CJ, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 40.Tsai B, et al. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J. 2003;22:4346–4355. doi: 10.1093/emboj/cdg439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker JD, Maier CL, Pober JS. Cytomegalovirus-infected human endothelial cells can stimulate allogeneic CD4+ memory T cells by releasing antigenic exosomes. J Immunol. 2009;182:1548–1559. doi: 10.4049/jimmunol.182.3.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bodaghi B, et al. Entry of human cytomegalovirus into retinal pigment epithelial and endothelial cells by endocytosis. Invest Ophthalmol Vis Sci. 1999;40:2598–2607. [PubMed] [Google Scholar]
- 43.Tacheva-Grigorova SK, Santos AJ, Boucrot E, Kirchhausen T. Clathrin-mediated endocytosis persists during unperturbed mitosis. Cell Rep. 2013;4:659–668. doi: 10.1016/j.celrep.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perreira JM, et al. RNASEK is a V-ATPase-associated factor required for endocytosis and the replication of rhinovirus, influenza A virus, and dengue virus. Cell Rep. 2015;12:850–863. doi: 10.1016/j.celrep.2015.06.076. [DOI] [PubMed] [Google Scholar]
- 45.Brindley MA, Maury W. Endocytosis and a low-pH step are required for productive entry of equine infectious anemia virus. J Virol. 2005;79:14482–14488. doi: 10.1128/JVI.79.23.14482-14488.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lakadamyali M, Rust MJ, Zhuang X. Endocytosis of influenza viruses. Microbes Infect. 2004;6:929–936. doi: 10.1016/j.micinf.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicola AV, McEvoy AM, Straus SE. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J Virol. 2003;77:5324–5332. doi: 10.1128/JVI.77.9.5324-5332.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dijkstra J, Van Galen M, Scherphof GL. Effects of ammonium chloride and chloroquine on endocytic uptake of liposomes by Kupffer cells in vitro. Biochim Biophys Acta. 1984;804:58–67. doi: 10.1016/0167-4889(84)90099-5. [DOI] [PubMed] [Google Scholar]
- 49.Mayer BT, et al. Transient oral human cytomegalovirus infections indicate inefficient viral spread from very few initially infected cells. J Virol. 2017;91:e00380-17. doi: 10.1128/JVI.00380-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olender T, Lancet D, Nebert DW. Update on the olfactory receptor (OR) gene superfamily. Hum Genomics. 2008;3:87–97. doi: 10.1186/1479-7364-3-1-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan I, et al. Olfactory receptor subgenomes linked with broad ecological adaptations in Sauropsida. Mol Biol Evol. 2015;32:2832–2843. doi: 10.1093/molbev/msv155. [DOI] [PubMed] [Google Scholar]
- 52.Ferrer I, et al. Olfactory receptors in non-chemosensory organs: The nervous system in health and disease. Front Aging Neurosci. 2016;8:163. doi: 10.3389/fnagi.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De la Cruz O, et al. A signature of evolutionary constraint on a subset of ectopically expressed olfactory receptor genes. Mol Biol Evol. 2009;26:491–494. doi: 10.1093/molbev/msn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pluznick JL, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA. 2013;110:4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shepard BD, et al. A renal olfactory receptor aids in kidney glucose handling. Sci Rep. 2016;6:35215. doi: 10.1038/srep35215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menendez CM, Carr DJJ. Herpes simplex virus-1 infects the olfactory bulb shortly following ocular infection and exhibits a long-term inflammatory profile in the form of effector and HSV-1-specific T cells. J Neuroinflammation. 2017;14:124. doi: 10.1186/s12974-017-0903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mori I, Nishiyama Y, Yokochi T, Kimura Y. Olfactory transmission of neurotropic viruses. J Neurovirol. 2005;11:129–137. doi: 10.1080/13550280590922793. [DOI] [PubMed] [Google Scholar]
- 58.Reiss CS, Plakhov IV, Komatsu T. Viral replication in olfactory receptor neurons and entry into the olfactory bulb and brain. Ann N Y Acad Sci. 1998;855:751–761. doi: 10.1111/j.1749-6632.1998.tb10655.x. [DOI] [PubMed] [Google Scholar]
- 59.Farrell HE, et al. Murine cytomegalovirus exploits olfaction to enter new hosts. MBio. 2016;7:e00251-16. doi: 10.1128/mBio.00251-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agrawal R. Cytomegalovirus and eye. Eye (Lond) 2012;26:1152. doi: 10.1038/eye.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan CS, Stevenson PG. B cell response to herpesvirus infection of the olfactory neuroepithelium. J Virol. 2014;88:14030–14039. doi: 10.1128/JVI.02345-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gillet L, May JS, Stevenson PG. In vivo importance of heparan sulfate-binding glycoproteins for murid herpesvirus-4 infection. J Gen Virol. 2009;90:602–613. doi: 10.1099/vir.0.005785-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bannister LH, Dodson HC. Endocytic pathways in the olfactory and vomeronasal epithelia of the mouse: Ultrastructure and uptake of tracers. Microsc Res Tech. 1992;23:128–141. doi: 10.1002/jemt.1070230204. [DOI] [PubMed] [Google Scholar]
- 64.Bologna Z, Teoh JP, Bayoumi AS, Tang Y, Kim IM. Biased G protein-coupled receptor signaling: New player in modulating physiology and pathology. Biomol Ther (Seoul) 2017;25:12–25. doi: 10.4062/biomolther.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeMaria S, Ngai J. The cell biology of smell. J Cell Biol. 2010;191:443–452. doi: 10.1083/jcb.201008163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang X, Eggert US. Non-traditional roles of G protein-coupled receptors in basic cell biology. Mol Biosyst. 2013;9:586–595. doi: 10.1039/c2mb25429h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paredes AM, Yu D. Human cytomegalovirus: Bacterial artificial chromosome (BAC) cloning and genetic manipulation. Curr Protoc Microbiol. 2012;Chap 14:Unit 14E.4. doi: 10.1002/9780471729259.mc14e04s24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim JH, Collins-McMillen D, Caposio P, Yurochko AD. Viral binding-induced signaling drives a unique and extended intracellular trafficking pattern during infection of primary monocytes. Proc Natl Acad Sci USA. 2016;113:8819–8824. doi: 10.1073/pnas.1604317113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu D, Smith GA, Enquist LW, Shenk T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J Virol. 2002;76:2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kwon YJ, Hung G, Anderson WF, Peng CA, Yu H. Determination of infectious retrovirus concentration from colony-forming assay with quantitative analysis. J Virol. 2003;77:5712–5720. doi: 10.1128/JVI.77.10.5712-5720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.E X, et al. An E2F1-mediated DNA damage response contributes to the replication of human cytomegalovirus. PLoS Pathog. 2011;7:e1001342. doi: 10.1371/journal.ppat.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.E X, et al. A novel DDB2-ATM feedback loop regulates human cytomegalovirus replication. J Virol. 2014;88:2279–2290. doi: 10.1128/JVI.03423-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.