Significance
Helicobacter pylori infects nearly half of the world’s population and is the primary cause of various gastric diseases. It has evolved various virulence factors to aid its host colonization and infection, including the vacuolating cytotoxin A (VacA) that is responsible for the pathogenesis of H. pylori-related diseases. Here, we resolve multiple structures of the water-soluble VacA oligomeric assemblies using cryoelectron microscopy (cryo-EM) at near-atomic resolution. These studies suggest a model of structural changes of functional VacA hexamer needed for the pore-formation process across the membrane and highlight the capability of cryo-EM to resolve multiple structure snapshots from a single specimen at near-atomic resolution.
Keywords: cryoelectron microscopy, Helicobacter pylori, vacuolating cytotoxin A, pore-forming toxin, oligomerization
Abstract
Human gastric pathogen Helicobacter pylori (H. pylori) is the primary risk factor for gastric cancer and is one of the most prevalent carcinogenic infectious agents. Vacuolating cytotoxin A (VacA) is a key virulence factor secreted by H. pylori and induces multiple cellular responses. Although structural and functional studies of VacA have been extensively performed, the high-resolution structure of a full-length VacA protomer and the molecular basis of its oligomerization are still unknown. Here, we use cryoelectron microscopy to resolve 10 structures of VacA assemblies, including monolayer (hexamer and heptamer) and bilayer (dodecamer, tridecamer, and tetradecamer) oligomers. The models of the 88-kDa full-length VacA protomer derived from the near-atomic resolution maps are highly conserved among different oligomers and show a continuous right-handed β-helix made up of two domains with extensive domain–domain interactions. The specific interactions between adjacent protomers in the same layer stabilizing the oligomers are well resolved. For double-layer oligomers, we found short- and/or long-range hydrophobic interactions between protomers across the two layers. Our structures and other previous observations lead to a mechanistic model wherein VacA hexamer would correspond to the prepore-forming state, and the N-terminal region of VacA responsible for the membrane insertion would undergo a large conformational change to bring the hydrophobic transmembrane region to the center of the oligomer for the membrane channel formation.
The bacterium Helicobacter pylori (H. pylori) infects nearly half of the world’s population and is the primary causative agent of gastric diseases such as gastritis, peptic ulcer, and gastric adenocarcinoma (1, 2). The pathogen has evolved a repertoire of virulence factors to aid its host colonization and infection. The vacuolating cytotoxin A (VacA) is one of the key virulence factors and is tightly associated with the pathogenesis of H. pylori-related diseases (3). VacA was first identified as a secreted toxin with the ability to induce intracellular vacuolation (4). In addition, VacA can induce multiple cellular effects, including membrane depolarization, autophagy, and inhibition of T cell proliferation (5–9).
The mature, secreted 88-kDa VacA protomer consists of two functional domains: an N-terminal p33 domain and a C-terminal p55 domain. The two domains are linked by a flexible loop which is sensitive to limited proteolysis in vitro (5). The p33 domain is critical for the pore-forming activity while the p55 domain is responsible for the receptor binding on host cells (10). To achieve efficient vacuolation, the entire p33 domain and the N-terminal 111 amino acid residues of the p55 domain are required (11). Once secreted, VacA assembles into water-soluble single-layer or double-layer flower-shaped oligomers, with each layer containing six or seven copies of protomers (12, 13). The current model of VacA-induced vacuolation proposes that VacA oligomers are first dissociated into monomers at low pH. This is then followed by the binding of VacA monomers to the host cell membrane, insertion into the lipid bilayer, oligomerization, and membrane channel formation (14). However, purified VacA without acid treatment also displays some degree of membrane binding (14, 15), suggesting that some VacA oligomers may exist as a “prepore” state for membrane binding before insertion. The assembly of VacA into oligomers is critical, as nonoligomerizing VacA mutants fail to induce vacuolation (16, 17). Previous biophysical studies have suggested that the VacA hexamer represents the functional state for channel formation (13, 18).
Since the discovery of VacA in the 1990s, it has been challenging to obtain an atomically detailed structure. Major milestones in understanding the VacA structure include the determination of the 2.4-Å crystal structure of the p55 domain, which reveals a predominant right-handed β-helix structure that is commonly seen in the autotransporter passenger domains of other bacterial toxins (19). The 19-Å cryoelectron microscopy (cryo-EM) structure of a VacA dodecamer further maps the p33 domain to the central core and the p55 domain to the peripheral arms of the flower-like structure (12). Recent cryo-EM structures of various VacA oligomers determined at ∼15-Å resolution also highlight the interactions of the p33 and the p55 domains for the assembly of VacA (13). Besides, a 4.5-Å crystallographic map of nonoligomerizing VacA mutant has been reported; however, no atomic model was built due to the insufficient resolution (16).
In this study, we resolve the cryo-EM structures of VacA in its water-soluble forms. The data reveal the near-atomic structural details of the VacA in its different oligomeric states. These structures suggest a molecular model for the membrane binding and insertion of VacA.
Results
Cryo-EM Structures of Multiple VacA Oligomeric Assembly States.
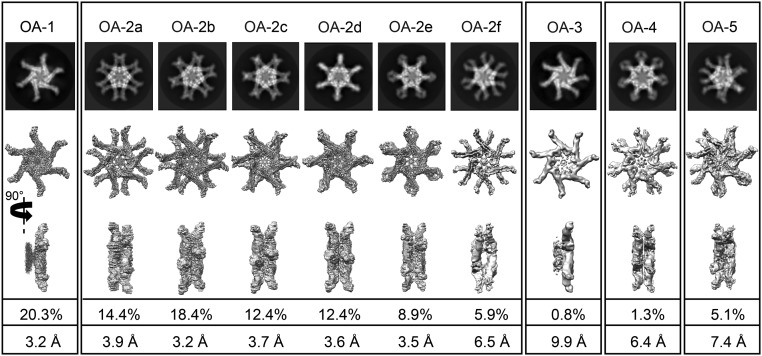
The s1i1m1 type of VacA was purified from the medium in which H. pylori strain 60190 was grown (SI Appendix, Fig. S1A) and subjected to cryo-EM analysis. A total of ∼540,000 particles out of ∼13,700 movie images were selected and the reference-free 2D class averages of these particles revealed that purified VacA adopts various oligomeric assembly (OA) states (Fig. 1 and SI Appendix, Fig. S1 B and C). Consistent with previous studies (12, 13), VacA hexamer and heptamer in one single chiral layer as well as dodecamer, tridecamer, and tetradecamer formed by two chiral layers stacking back to back were observed. Octameric and nonameric VacA oligomers (12, 20) were not found.
Fig. 1.
Ten OAs of H. pylori VacA determined by cryo-EM. The 2D class average and corresponding 3D reconstruction of each conformation are shown. OA-1 is the hexamer. OA-2a–f are all conformers of the dodecamer, having different relative rotational orientations between the two layers. OA-3 is the heptamer. OA-4 is the tetradecamer. OA-5 is the tridecamer. The percentages and resolutions in the last two rows refer to the final dataset and final maps.
In line with the 2D class averaging analysis, results from the 3D reconstructions showed that populations of VacA contained single-layer complexes with six or seven protomers and bilayer complexes with 12, 13, or 14 protomers (Fig. 1). After 3D classification (SI Appendix, Fig. S2), 10 OAs were derived, including hexamer (OA-1), dodecamer (OA-2), heptamer (OA-3), tetradecamer (OA-4), and tridecamer (OA-5). Each of them displayed highly symmetric arrangements of the protomers although no symmetry was imposed in the 3D reconstruction (SI Appendix, Fig. S3A). The majority of the VacA oligomers in our images are dodecamers (72.4%), followed by hexamers (20.3%), tridecamers (5.1%), tetradecamers (1.3%), and heptamers (0.8%) (Fig. 1 and SI Appendix, Fig. S2). The cryo-EM maps of these oligomeric states were further refined individually. The dodecamers could be further subclassified into six conformers with different relative rotational orientations between the two layers, namely OA-2a–f (designated in the order of the relative angular difference) refined to 3.9-Å, 3.2-Å, 3.7-Å, 3.6-Å, 3.5-Å, and 6.5-Å resolutions, respectively (Fig. 1 and SI Appendix, Fig. S3B). In each of these maps, the resolutions are lower at the distal region of the p55 domain (SI Appendix, Fig. S3C), suggesting its flexibility. The remaining oligomers are hexamers, heptamers, tetradecamers, and tridecamers, which were refined to 3.2 Å, 9.9 Å, 6.4 Å, and 7.4 Å, respectively.
Conserved Structural Fold of VacA Protomer Among Different Oligomeric Assemblies.
Although the number of protomers varies among different VacA assembly structures, their protomers share the same structural fold. Noteworthy, there is a weak and discontinuous density region in the center of all VacA oligomers (SI Appendix, Fig. S3C), where no path of chain trace can be established. To illustrate the structural details of VacA, we describe the de novo model built for the hexamer (OA-1), which yields the highest resolution map among all of the oligomers and has the largest number of particles in our images. Densities for the secondary structure elements, most loops, and most of the side chains are clearly visible in reference to the sequence assignment (SI Appendix, Fig. S4A). We validated the resulting model (Fig. 2 A and B) using MolProbity (21) (SI Appendix, Table S1) and the agreement between map and model by cross-correlation coefficients (SI Appendix, Fig. S4B). Atom position accuracy and side-chain resolvability were assessed by atomic displacement parameters (22) (SI Appendix, Fig. S4B) and the side-chain Z scores (23) (SI Appendix, Fig. S4A), respectively.
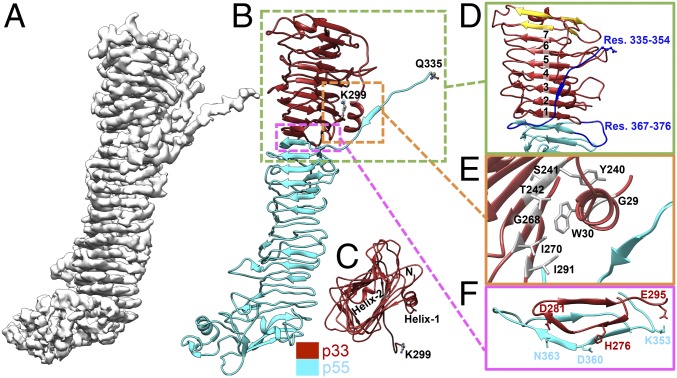
Fig. 2.
Near-atomic cryo-EM structure of VacA protomer in the hexamer (OA-1). (A) Cryo-EM map of the VacA protomer. (B) Model of the VacA protomer. The missing interdomain region is between K299 of p33 and Q335 of p55. K299 and Q335 are colored by element (carbon, gray; oxygen, red; nitrogen, blue) for clarity. (C) Top view of the p33 domain, with two annotated α-helices [helix30–37 (helix-1) and helix68–74 (helix-2)]. N refers to N terminus. (D) The Right-handed β-helix of p33, composed of seven complete turns (numbers 1–7) and four additional β-strands (gold). Two regions (blue) not built in the p55 crystal structure (19) are also shown. (E) The interactions of p33 N terminus with other regions. The indole ring of W30 is located in a pocket formed by T242, G268, I270, and I291. (F) The interface between p33 and p55.
The VacA protomer model consists of amino acids 27–299 (p33 domain) and 335–811 (p55 domain) (Fig. 2B and Movie S1). To confirm the integrity of the secreted VacA sample used for the cryo-EM studies, SDS/PAGE and N-terminal sequencing were applied. Only one protein band migrating at ∼88 kDa was detected (SI Appendix, Fig. S1A), and residues 1AFF3 were sequenced (data not shown), suggesting that the protein sample did not undergo proteolysis. It is likely that the two missing regions, residues 1–26 in p33 domain and 300–334 between p33 and p55 domains, in our current map are highly flexible. Moreover, the missing residues 300–334 might contribute to the above-mentioned central density (SI Appendix, Fig. S5). As shown in Fig. 2 B–D, the core of p33 is a right-handed β-helix composed of seven complete turns, four additional β-strands, and two α-helices (helix30–37 and helix68–74, subscripts denote the residue numbers). The p33 β-helix extends seamlessly from the N terminus of p55, causing no obvious separation between the two domains. Interestingly, a structural homology search of p33 using DALI (24) revealed p55 as the top hit with a rmsd of 2.9 Å for 176 equivalently aligned Cα atoms. However, the p33 β-helix is formed by multiple ∼30-aa repeats, which is longer than each repeat (∼25 aa) of the p55 β-helix (19). While the C terminus end of the p33 β-helix is in contact with p55, the N terminus end is capped with an amphipathic α-helix helix68–74.
It has been proposed that the N-terminal 32 nonpolar amino acids of p33 are important for membrane channel formation through the formation of a membrane helix bundle (25). In our models, the N terminus of p33 can only be traced starting from residue L27. The visible N-terminal region of p33 consists of an α-helix helix30–37 followed by a long loop connected to the β-helix core. The hydrophobic face of helix30–37 is shielded from the solvent environment (Fig. 2E). Specifically, the indole ring of tryptophan W30 is located in a pocket formed by T242, G268, I270, and I291 via hydrophobic interactions. The orientation of helix30–37 is further stabilized by three sets of residues at a hydrogen bond distance (G29–Y240, W30–S241, W30–T242). It is worth mentioning that our cryo-EM model reveals the two regions, residues 335–354 and residues 367–376, that were not visible in the p55 crystal structure (19) (Fig. 2D). The former region, as discussed below, is critical for VacA oligomerization.
We further analyzed the interaction between p33 and p55 using PDBsum (26). The p33–p55 binding interface is about 1,300 Å2 and involves two salt bridges (H276–D360 and E295–K353) at the loops of the β-helix as well as three side-chain hydrogen bonds (H276–D360, D281–N363, and E295–K353) (Fig. 2F). Additionally, the main chains of the two domains (residues 275–293 and residues 352–380) are close enough to form multiple main-chain hydrogen bonds for the stabilization of the β-strands connected laterally. Hydrophobic interactions also appear to play a role in the p33–p55 interdomain association (e.g., I277, L280, L288, and I291 in p33; and F349, I359, I362, and L379 in p55). These structural findings explain previous biochemical studies that individually purified p33 and p55 polypeptides can interact with each other and function as the full-length VacA (27).
Diverse Types of VacA Oligomeric Assembly Structures.
Monolayer assemblies.
As mentioned above, in our study, water-soluble VacA exists in different quaternary structures. The broad spectrum of VacA oligomers depends on the formation of two basic “building blocks”—hexamer and heptamer. To understand the molecular basis of how VacA assembles into different oligomers, we first analyzed protomer–protomer contacts within the VacA hexamer (OA-1). As shown in Fig. 3 A–C and Movie S1, the molecular interface mainly lies on a segment (residues 338–350) that protrudes from the p55 domain of one protomer and interacts with the outermost β-strand (residues 47–75) of the p33 domain of the counterclockwise adjacent protomer. The interface area buries about 1,000 Å2 and involves two salt bridges (E338P:K47P+1 and D346P:K55P+1), three side-chain hydrogen bonds (T342P:R50P+1, Q343P:K75P+1, and D346P:K55P+1), and extensive main-chain hydrogen bonds (subscripts P and P+1 denote the protomer and its counterclockwise adjacent protomer, respectively) (Fig. 3C). We noted that the orientation of the protruding segment from p55 is likely governed by 293ProPro294 and 351GlyGly352 motifs (28) (Fig. 3D). It is likely that the nature of the flexible loop (residues 300–334) provides some plasticity for the inclusion of an extra protomer to form a heptamer instead of a hexamer. This notion is supported by previous studies that removal of residues 327–372 in VacA from H. pylori CCUG 17874 (corresponding residues 294–331 in VacA from our strain 60190) promotes the formation of VacA hexamer from 30% to 100% (29). Taken together, our findings are in line with previous mutagenesis studies, which reported that VacA mutants with deletion at residues 49–57 or residues 346–347 failed to oligomerize (16, 17, 30).
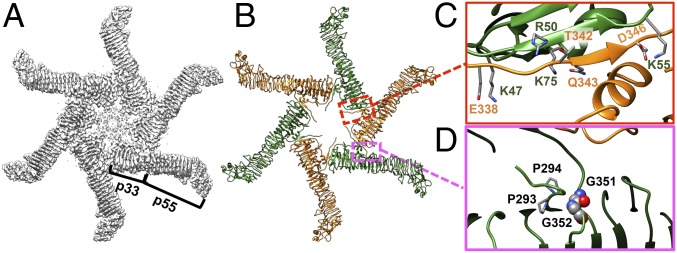
Fig. 3.
Interactions between VacA protomers in the hexamer (OA-1). (A) Cryo-EM map of the VacA hexamer. (B) Model of the VacA hexamer. (C) The protomer–protomer intermolecular interactions including multiple side-chain hydrogen bonds and salt bridges. (D) The 293ProPro294 and 351GlyGly352 motifs (28) probably control the orientation of the protruding segment from p55. The glycines are shown in sphere and colored by element for clarity.
Bilayer assemblies.
In addition to the hexamer and heptamer, eight different bilayer structures, including six dodecamers, one tridecamer, and one tetradecamer, were found in the same purified VacA sample (Fig. 1). It is interesting to note that different oligomeric assemblies are present in different proportions and resolved at different resolutions (SI Appendix, Table S1). Such variations may reflect their inherent stability and/or their different vulnerability during the cryospecimen freezing step. The prevalence of these higher-order oligomeric assemblies suggests favorable free energy comparable to the single-layer structures. Though the biological functions of these oligomers are not well established, it is conceivable that they may be the storage or inactive form of the secreted VacA in the culture media. Alternatively, they may merely be a consequence of the biochemical conditions.
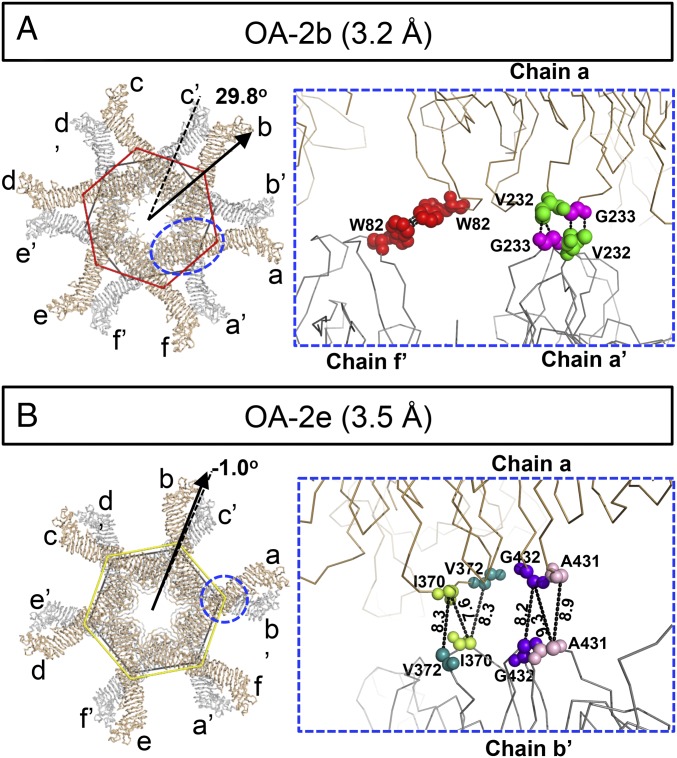
To characterize the structural details of these oligomeric assemblies, we used the 3.2-Å structure of the VacA dodecamer (OA-2b) as a representative of the bilayer model to study how VacA is organized into two layers (Fig. 4A and Movie S2). The protomers in one layer are designated as chains a, b, c, d, e, and f and those in the other layer as chains a′, b′, c′, d′, e′, and f′, each ordered counterclockwise, looking down on one face from the outside. (This annotation is used to enable the illustration of these complex structures in our figures and movies). Molecular interactions between the two VacA hexamers are mainly localized in the central regions of the p33 domains in these six conformers, with an interface area of ∼150 Å2 per VacA protomer, which indicates a weak or promiscuous interaction between the two layers. Using PISA and PDBsum structure bioinformatics software (26, 31) to further analyze our structure models, we identified the key residues likely to engage in interactions between the protomers across the two layers. As shown in Fig. 4A and Movie S3, two different patches of short-range hydrophobic interactions that may be important to stabilize the OA-2b dodecamer, which yielded the highest resolution structure, were found. The first hydrophobic patch lies on two W82 residues of chains a and f′ while the second patch is on V232 and G233 of chains a and a′. By the same token, chain n interacts with chains n′ and (n−1)′ to promote the back-to-back oligomerization. Our structure was determined at a resolution that water molecules and ions cannot be detected, but it is likely that they also play a role in stabilizing the two-layer structures. We also used the same structural bioinformatics web server to analyze all of the other VacA dodecamer assemblies except OA-2f due to its low-resolution map, as summarized in SI Appendix, Table S2. All of these assemblies are mainly stabilized through the long-range atom interactions or the long-range hydrophobic interactions (32) among multiple nonpolar residues (e.g., I370, V372, A431, and G432) between the same or adjacent protomers (e.g., Fig. 4B). These weak interactions would likely allow the switching between different conformations and the transition to other functional states (i.e., pore forming) upon binding to the membrane.
Fig. 4.
Hydrophobic interactions between two stacked VacA hexamers. (A and B) Models of two high-resolution dodecamers (OA-2b and OA-2e). In each figure, the Left shows the overall architecture of the lower (gray) and upper (sand) layers of the VacA dodecamers. The arrangements of individual layers of dodecamers are represented as polygons, whose sides are connected by the mass centers of individual domains. The rotation angles of the upper-layer hexamers (color coded from respective conformation) with respect to the lower-layer hexamers (black polygon) are shown. A, Right shows the two different patches of hydrophobic interactions that stabilize the OA-2b dodecamer. The key residues are shown in sphere. B, Right shows the residues that likely contribute to the stability of two stacked hexamers in OA-2e, with the closest distances (in angstroms) between Cα from different chains shown. The backbones of key residues are shown in sphere. W82 (red), V232 (green), G233 (magenta), I370 (lime green), V372 (dark cyan), A431 (light pink), G432 (purple).
Proposed Model for VacA Membrane Insertion.
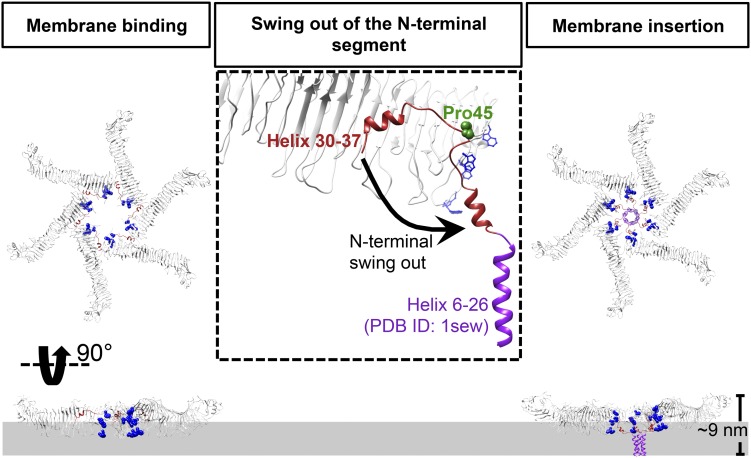
An important feature of VacA is that its N-terminal hydrophobic region inserts into the membrane for the formation of the anion channel (25). One model for VacA membrane assembly at low pH suggests the temporal order of membrane insertion, oligomerization, and pore formation (33). Besides, other experimental data demonstrated that VacA at neutral pH can also exhibit some weak binding to the cell membrane, suggesting an alternative pore-formation pathway at neutral pH with the oligomerization occurring before insertion and pore formation (14, 15). We also observed lower vacuolation activity (fewer vacuolated cells) of our purified VacA without acid treatment relative to that with acid treatment in HeLa cells (SI Appendix, Fig. S6). With the atomic details revealed by our cryo-EM structures, we propose a model to explain how VacA hexamer may directly participate in membrane binding and insertion for pore formation at neutral pH (Fig. 5). Our proposed model would provide a complementary mechanism for VacA-induced membrane channel formation.
Fig. 5.
Proposed model for membrane insertion of a VacA hexamer. The pore formation is illustrated as a three-step process, including membrane binding, swing out of the N-terminal segment, and membrane insertion. Interaction of water-soluble hexameric VacA with the membrane lipids induces helix30–37 to swing out from the protomer–protomer interface and then exposes the hydrophobic N terminus to form the helix bundle within the membrane (the helix 6–26 derived from PDB ID code 1sew is shown in purple). Tryptophans are shown in blue and proline in the hinge is shown in sphere with forest green. The side view shows only chains b–e.
A striking feature of the p33 N-terminal region is the burial of helix30–37 in the protomer–protomer interface (Fig. 2D). We noted the presence of a proline residue P45 at the loop connected to helix30–37, which may act as a hinge for the movement of helix30–37. Another feature of the p33 domain is the cluster of tryptophan residues (W49, W80, W82, W90, and W96) near the inner rim of the hexamer and helix30–37. This tryptophan-rich region may play an important role in the membrane-binding process as shown in other bacterial pore-forming toxins such as listeriolysin O, pneumolysin, and equinatoxin II (34–37). Membrane association may also be facilitated by simple electrostatic attraction, as one face of VacA hexamer is more positively charged (SI Appendix, Fig. S7) and is the membrane-facing surface that favors the contact with anionic phospholipids. Taken together, the water-soluble hexameric VacA may be a mimic of the membrane-bound state. To model the membrane insertion state, the N-terminal region (residues 27–44) would flip out and combine with the previously reported helix bundle (PDB ID code 1sew; residues 6–29) (19). We propose that interaction with the membrane lipids will induce helix30–37 to swing out from the protomer–protomer interface and subsequently lead to the exposure of the hydrophobic N-terminal region to form the helix bundle within the membrane. Notably, the majority of the VacA protomer remains unchanged; this is consistent with previous findings that the top view of the water-soluble form and membrane-inserted form of VacA are similar in shape (33, 38). Furthermore, the height of our proposed structure is about 9.0 nm, close to the height of the VacA oligomer associated with anionic lipid bilayers measured by atomic force microscopy (39). In this model, the double-layer oligomers would not be able to participate in the pore formation. It is conceivable that they may have other functional roles invcells.
Discussion
A number of α-pore-forming proteins have been solved to high resolution (40–43). The nature of VacA oligomerization has been a long-standing question for more than two decades. So far, only the p55 domain of a VacA monomer has been solved structurally to high resolution (19), but it did not provide any details about VacA oligomeric organization. Here, our cryo-EM structures provide near-atomic details of water-soluble VacA assemblies made from full-length VacA protomers. These structures reveal many previously unseen features. The protruding segment from the p55 domain of one protomer forms a β-strand extension from the outermost β-strand of the p33 domain of another protomer (Fig. 3C and Movie S1). In addition, we also identify a diverse spectrum of weak interactions in different assemblies (Fig. 4, SI Appendix, Table S2, and Movie S3). Such interactions form the basis for the VacA dodecameric assemblies.
It is believed that the N-terminal 32 uncharged residues containing multiple GXXXG motifs form a helical bundle for membrane insertion (25). Although residues 1–26 are not built in our cryo-EM models, the orientation of helix30–37 and the spatial disposition of the tyrosine-rich region in the p33 domain (Figs. 2E and 5) suggest a model for VacA membrane insertion under neutral pH condition. Notably, swinging out of helix30–37 would leave the VacA protomer largely unchanged (Fig. 5). This model is consistent with previous findings that the top view structure of the VacA water-soluble form, in general, resembles that of its membrane-inserted form (33, 38). Interestingly, the movement of an α-helix for membrane insertion has also been observed in other pore-forming toxins like FraC, ClyA, and XaxAB (40–43) (SI Appendix, Fig. S8). This suggests that α-pore-forming toxins may adopt a common mode of membrane insertion despite low sequence conservation among them. On the other hand, we also compared the Cα trace of our VacA protomer with that of the nonoligomerizing VacA mutant containing a partial p33 domain (16). We found that the two structures share a very similar β-helix fold, indicating no significant change in the overall structure between the monomer and the protomer of our structures. A previous assembly model proposed that membrane insertion requires dissociation of VacA oligomers into monomers at low pH condition (14, 15). It is therefore tempting to speculate that acid activation of VacA promotes both monomer formation and exposure of the N-terminal helix for membrane insertion. Clustering of VacA monomers to lipid rafts and the intrinsic nature of VacA oligomerization further lead to the formation of membrane pore. However, it is yet unclear why hexameric VacA is exploited as the membrane-bound state while oligomerization to heptamer in the water-soluble state is allowed. Determination of the membrane-inserted state of VacA structure will be essential for elucidating the assembly mechanism for VacA anion channel formation.
Our data also show that the two-hexamer bilayer VacA structures have at least six possible relationships between the hexamer layers (Movie S2), though arguably, the more populated and higher resolution OA-2b dodecamer state may be marginally more stable. These structures must be energetically favorable to make them amenable to be studied from subnanometer to near-atomic resolutions. In our analysis, the two equivalent rings are assembled with limited hydrophobic interactions (Movie S3). The promiscuous switching of the hydrophobic interactions among the nonpolar residues from the same or adjacent subunit generated different oligomeric forms. In fact, the arms of the tridecamer (hexamer on heptamer) structure (OA-5, Fig. 1) looks similar to the arms of the different dodecameric conformations (OA-2a–f, Fig. 1). The biological functions of these various assemblies remain uncertain, though they may be speculated as inactive states before transitioning to the prepore-forming state (e.g., hexamer). Nevertheless, the discovery and analysis of such heterogeneity add another example to several recent reports which reveal multiple conformers of the same macromolecular complex by cryo-EM (44, 45). It is a recurring observation that macromolecular assemblies are structurally dynamic and that cryo-EM is the right tool to obtain structure snapshots of their structures which can generate further studies of dynamics and functions.
Materials and Methods
H. pylori Culture.
H. pylori strain 60190 (ATCC no. 49503) was used to produce VacA in this study. The H. pylori was cultured as previously described (46, 47). Briefly, H. pylori was cultured at 37 °C on Columbia agar base (Thermo Fisher Scientific) supplemented with 5% horse blood. CampyGen (Thermo Fisher Scientific) was used to generate the microaerophilic environment necessary for H. pylori growth. To produce VacA, liquid culture was used instead. The liquid culture was composed with Brucella broth (Becton Dickinson) supplemented with cholesterol (Thermo Fisher Scientific).
Preparation of VacA.
VacA, secreted by H. pylori strain 60190, was directly purified from the medium (16). Briefly, VacA was produced in the liquid culture medium for about 48 h. The medium was then spun down to remove the cell debris. The medium supernatant was concentrated by about 10-fold using concentrator with molecular weight cutoff of 100 kDa. The concentrated medium was then precipitated with the same volume of saturated ammonia sulfate. The sample was further spun down at 20,000 rpm for 1 h using JA20 rotor (Beckman Coulter). The pellet was resuspended using PBS and subjected to gel filtration using HiLoad Superdex, 200 pg (GE Healthcare). The fractions were examined on SDS/PAGE. Peak fractions were concentrated to about 0.5 mg/mL for further experiments.
Cryo-EM Data Acquisition.
The VacA sample was diluted at a final concentration of around 0.2 mg/mL. Three microliters of the samples were applied onto glow-discharged 200-mesh R2/1 Quantifoil grids coated by homemade thin continuous carbon. The grids were blotted for 2.5 s and rapidly cryocooled in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) at 4 °C and 100% humidity. The samples were screened using a Talos Arctica cryoelectron microscope (Thermo Fisher Scientific) operated at 200 kV and then imaged in a Titan Krios cryoelectron microscope (Thermo Fisher Scientific) with GIF energy filter (Gatan) at a magnification of 130,000× (corresponding to a calibrated sampling of 1.06 Å per pixel). Micrographs were recorded by EPU software (Thermo Fisher Scientific) with a Gatan K2 Summit direct electron detector, where each image is composed of 30 individual frames with an exposure time of 6 s and a dose rate of seven electrons per second per Å2. A total of 13,708 movie stacks were collected with a defocus range of 1.3–2.5 μm.
Single-Particle Image Processing and 3D Reconstruction.
All micrographs were motion corrected using MotionCor2 (48) and the contrast transfer function (CTF) was corrected using CTFFIND4 (49). All particles were autopicked using NeuralNet option in EMAN2 (50), and further checked manually. A total of 540,214 particle images were derived from the selected 13,587 micrographs. Then, particle coordinates were imported to Relion (51), where three rounds of 2D classification were performed to remove poor 2D class averages. Meanwhile, ∼2,000 particle images were selected to generate the 2D averages in EMAN2 and further used to build the initial model. A total of 299,174 particles were used for 3D classification in Relion (version 2.1) without symmetry. Multiple oligomeric assemblies came out from the 3D classification, and 10 selected oligomeric assemblies with their particles were transferred to cryoSPARC V2 (52) for further heterogeneous particle image refinement. Ten final maps were achieved after the homogeneous refinement in cryoSPARC V2 (see more information in SI Appendix, Fig. S2 and Table S1).
Docking and de Novo Model Building.
Model building was first conducted based on the 3.2-Å map of the OA-1. The asymmetric unit (ASU) of VacA contains p33 and p55 domains. Two ASUs were computationally extracted from our map (53). The partial p55 crystal structure (2qv3), containing residues 355–366 and 377–811, was used for docking into the computationally extracted map. Based on the cryo-EM map, the de novo model building of p33 (residues 27–299) and the p55 (residues 335–354 and 367–376) that were not previously resolved was performed using Coot (51, 54). Amino acid sequence registration was assigned initially to the cryo-EM density of bulky residues (Trp, Lys, Arg, Phe, and Tyr) and then to the rest of the residues. An atomic model composed of p33 and partially unresolved p55 (residues 27–299, 335–354, and 367–376) was obtained. The resulting model was refined using phenix.real_space_refine (55) application with secondary structure and geometry restraints. Coot was used to manually optimize the model. Finally, 750 of 821 residues of the full-length VacA protomer were modeled. The atomic model of ASU was then fitted into the cryodensity of the other protomers in VacA oligomers with Chimera (56), followed by the optimization using phenix.real_space_refine. The final models were evaluated by MolProbity (21) and side-chain Z scores (23). Statistics of the map reconstruction and model optimization are shown in SI Appendix, Table S1. PISA and PDBsum structure bioinformatics software (26, 31) were used to analyze our structure models to identify the key residues involved in interactions between the protomers. All figures were prepared using PyMol (57) or Chimera (56).
Vacuolation Assay.
Vacuolation assay was performed as described in ref. 16. Briefly, HeLa cells were cultured in DMEM supplemented with 5% FBS at 37 °C in a 5% CO2 incubator. A total of ∼1 × 104 cells were seeded on a 24-well plate and cultured for 24 h. VacA was first acid activated using HCl at room temperature for 10 min. Then 5 μg VacA with or without acid treatment was added into DMEM supplemented with 10 mM ammonia chloride. Cells were cultured for another 3 h. The cells were then observed under an Olympus IX71 microscope using the 10× objective lens. Images were recorded using a CCD camera (ThorLabs).
Supplementary Material
Acknowledgments
This research is supported by NIH Grants P41GM103832, R01GM079429, and S10OD021600 (to W.C.), and GM123159 (to H.v.d.B.); Hong Kong Research Grants Council Grants CUHK460113 and N_CUHK454/13 (to S.W.N.A.); National Science Foundation Grant OIA-1738547 (to T.-C.M.); and National Natural Science Foundation of China-Research Grants Council (NSFC-RGC) Grant 31361163001 (to Q.Z.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Cryo-EM maps of 10 VacA oligomeric assemblies and six associated atomic models have been deposited in the Electron Microscopy Data Bank with EMD accession codes 0542, 0543, 0544, 0545, 0546, 0547, 0548, 0549, 0550, and 0551, and the Protein Data Bank under PDB ID codes 6NYF, 6NYG, 6NYJ, 6NYL, 6NYM, and 6NYN.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821959116/-/DCSupplemental.
References
- 1.Diaconu S, Predescu A, Moldoveanu A, Pop CS, Fierbinţeanu-Braticevici C. Helicobacter pylori infection: Old and new. J Med Life. 2017;10:112–117. [PMC free article] [PubMed] [Google Scholar]
- 2.Polk DB, Peek RM., Jr Helicobacter pylori: Gastric cancer and beyond. Nat Rev Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roesler BM, Rabelo-Gonçalves EMA, Zeitune JMR. Virulence factors of Helicobacter pylori: A review. Clin Med Insights Gastroenterol. 2014;7:9–17. doi: 10.4137/CGast.S13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leunk RD, Johnson PT, David BC, Kraft WG, Morgan DR. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol. 1988;26:93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 5.Torres VJ, Ivie SE, McClain MS, Cover TL. Functional properties of the p33 and p55 domains of the Helicobacter pylori vacuolating cytotoxin. J Biol Chem. 2005;280:21107–21114. doi: 10.1074/jbc.M501042200. [DOI] [PubMed] [Google Scholar]
- 6.Cover TL, Puryear W, Perez-Perez GI, Blaser MJ. Effect of urease on HeLa cell vacuolation induced by Helicobacter pylori cytotoxin. Infect Immun. 1991;59:1264–1270. doi: 10.1128/iai.59.4.1264-1270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain P, Luo Z-Q, Blanke SR. Helicobacter pylori vacuolating cytotoxin A (VacA) engages the mitochondrial fission machinery to induce host cell death. Proc Natl Acad Sci USA. 2011;108:16032–16037. doi: 10.1073/pnas.1105175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu P, et al. Helicobacter pylori VacA induces autophagic cell death in gastric epithelial cells via the endoplasmic reticulum stress pathway. Cell Death Dis. 2017;8:3207. doi: 10.1038/s41419-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winter J, Letley D, Rhead J, Atherton J, Robinson K. Helicobacter pylori membrane vesicles stimulate innate pro- and anti-inflammatory responses and induce apoptosis in Jurkat T cells. Infect Immun. 2014;82:1372–1381. doi: 10.1128/IAI.01443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat Rev Microbiol. 2005;3:320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- 11.Ye D, Willhite DC, Blanke SR. Identification of the minimal intracellular vacuolating domain of the Helicobacter pylori vacuolating toxin. J Biol Chem. 1999;274:9277–9282. doi: 10.1074/jbc.274.14.9277. [DOI] [PubMed] [Google Scholar]
- 12.El-Bez C, Adrian M, Dubochet J, Cover TL. High resolution structural analysis of Helicobacter pylori VacA toxin oligomers by cryo-negative staining electron microscopy. J Struct Biol. 2005;151:215–228. doi: 10.1016/j.jsb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Chambers MG, et al. Structural analysis of the oligomeric states of Helicobacter pylori VacA toxin. J Mol Biol. 2013;425:524–535. doi: 10.1016/j.jmb.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raghunathan K, et al. Determinants of raft partitioning of the Helicobacter pylori pore-forming toxin VacA. Infect Immun. 2018;86:e00872-17. doi: 10.1128/IAI.00872-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClain MS, Schraw W, Ricci V, Boquet P, Cover TL. Acid activation of Helicobacter pylori vacuolating cytotoxin (VacA) results in toxin internalization by eukaryotic cells. Mol Microbiol. 2000;37:433–442. doi: 10.1046/j.1365-2958.2000.02013.x. [DOI] [PubMed] [Google Scholar]
- 16.González-Rivera C, et al. A nonoligomerizing mutant form of Helicobacter pylori VacA allows structural analysis of the p33 domain. Infect Immun. 2016;84:2662–2670. doi: 10.1128/IAI.00254-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivie SE, et al. Helicobacter pylori VacA subdomain required for intracellular toxin activity and assembly of functional oligomeric complexes. Infect Immun. 2008;76:2843–2851. doi: 10.1128/IAI.01664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwamoto H, Czajkowsky DM, Cover TL, Szabo G, Shao Z. VacA from Helicobacter pylori: A hexameric chloride channel. FEBS Lett. 1999;450:101–104. doi: 10.1016/s0014-5793(99)00474-3. [DOI] [PubMed] [Google Scholar]
- 19.Gangwer KA, et al. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc Natl Acad Sci USA. 2007;104:16293–16298. doi: 10.1073/pnas.0707447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adrian M, Cover TL, Dubochet J, Heuser JE. Multiple oligomeric states of the Helicobacter pylori vacuolating toxin demonstrated by cryo-electron microscopy. J Mol Biol. 2002;318:121–133. doi: 10.1016/S0022-2836(02)00047-5. [DOI] [PubMed] [Google Scholar]
- 21.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hryc CF, et al. Accurate model annotation of a near-atomic resolution cryo-EM map. Proc Natl Acad Sci USA. 2017;114:3103–3108. doi: 10.1073/pnas.1621152114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pintilie G, Chiu W. Assessment of structural features in Cryo-EM density maps using SSE and side chain Z-scores. J Struct Biol. 2018;204:564–571. doi: 10.1016/j.jsb.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44:W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim S, Chamberlain AK, Bowie JU. Membrane channel structure of Helicobacter pylori vacuolating toxin: Role of multiple GXXXG motifs in cylindrical channels. Proc Natl Acad Sci USA. 2004;101:5988–5991. doi: 10.1073/pnas.0308694101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laskowski RA, Jabłońska J, Pravda L, Vařeková RS, Thornton JM. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018;27:129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.González-Rivera C, et al. Reconstitution of Helicobacter pylori VacA toxin from purified components. Biochemistry. 2010;49:5743–5752. doi: 10.1021/bi100618g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob J, Duclohier H, Cafiso DS. The role of proline and glycine in determining the backbone flexibility of a channel-forming peptide. Biophys J. 1999;76:1367–1376. doi: 10.1016/S0006-3495(99)77298-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burroni D, et al. Deletion of the major proteolytic site of the Helicobacter pylori cytotoxin does not influence toxin activity but favors assembly of the toxin into hexameric structures. Infect Immun. 1998;66:5547–5550. doi: 10.1128/iai.66.11.5547-5550.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Genisset C, et al. A Helicobacter pylori vacuolating toxin mutant that fails to oligomerize has a dominant negative phenotype. Infect Immun. 2006;74:1786–1794. doi: 10.1128/IAI.74.3.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Israelachvili J, Pashley R. The hydrophobic interaction is long range, decaying exponentially with distance. Nature. 1982;300:341–342. doi: 10.1038/300341a0. [DOI] [PubMed] [Google Scholar]
- 33.Pyburn TM, et al. Structural organization of membrane-inserted hexamers formed by Helicobacter pylori VacA toxin. Mol Microbiol. 2016;102:22–36. doi: 10.1111/mmi.13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Separovic F, Kozorog M, Sani M-A, Anderluh G. Role of the tryptophan-rich motif of listeriolysin O in membrane binding. Biophys J. 2017;112(Suppl 1):524a. [Google Scholar]
- 35.Tilley SJ, Orlova EV, Gilbert RJC, Andrew PW, Saibil HR. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell. 2005;121:247–256. doi: 10.1016/j.cell.2005.02.033. [DOI] [PubMed] [Google Scholar]
- 36.Weis S, Palmer M. Streptolysin O: The C-terminal, tryptophan-rich domain carries functional sites for both membrane binding and self-interaction but not for stable oligomerization. Biochim Biophys Acta. 2001;1510:292–299. doi: 10.1016/s0005-2736(00)00360-6. [DOI] [PubMed] [Google Scholar]
- 37.Hong Q, et al. Two-step membrane binding by equinatoxin II, a pore-forming toxin from the sea anemone, involves an exposed aromatic cluster and a flexible helix. J Biol Chem. 2002;277:41916–41924. doi: 10.1074/jbc.M204625200. [DOI] [PubMed] [Google Scholar]
- 38.Schibli DJ, Montelaro RC, Vogel HJ. The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry. 2001;40:9570–9578. doi: 10.1021/bi010640u. [DOI] [PubMed] [Google Scholar]
- 39.Czajkowsky DM, Iwamoto H, Cover TL, Shao Z. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proc Natl Acad Sci USA. 1999;96:2001–2006. doi: 10.1073/pnas.96.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka K, Caaveiro JMM, Morante K, González-Mañas JM, Tsumoto K. Structural basis for self-assembly of a cytolytic pore lined by protein and lipid. Nat Commun. 2015;6:6337. doi: 10.1038/ncomms7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzokov SB, et al. Structure of the hemolysin E (HlyE, ClyA, and SheA) channel in its membrane-bound form. J Biol Chem. 2006;281:23042–23049. doi: 10.1074/jbc.M602421200. [DOI] [PubMed] [Google Scholar]
- 42.Mueller M, Grauschopf U, Maier T, Glockshuber R, Ban N. The structure of a cytolytic α-helical toxin pore reveals its assembly mechanism. Nature. 2009;459:726–730. doi: 10.1038/nature08026. [DOI] [PubMed] [Google Scholar]
- 43.Schubert E, Vetter IR, Prumbaum D, Penczek PA, Raunser S. Membrane insertion of α-xenorhabdolysin in near-atomic detail. eLife. 2018;7:e38017. doi: 10.7554/eLife.38017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong Y, et al. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature. 2018;565:49–55. doi: 10.1038/s41586-018-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de la Peña AH, Goodall EA, Gates SN, Lander GC, Martin A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis–driven translocation. Science. 2018;362:eaav0725. doi: 10.1126/science.aav0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, et al. A putative spermidine synthase interacts with flagellar switch protein FliM and regulates motility in Helicobacter pylori. Mol Microbiol. 2017;106:690–703. doi: 10.1111/mmi.13829. [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, Au SWN. Helicobacter pylori does not use spermidine synthase to produce spermidine. Biochem Biophys Res Commun. 2017;490:861–867. doi: 10.1016/j.bbrc.2017.06.132. [DOI] [PubMed] [Google Scholar]
- 48.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 53.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J Struct Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2012;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 57.Rigsby RE, Parker AB. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ. 2016;44:433–437. doi: 10.1002/bmb.20966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.