Significance
The production of unsaturated fatty acids (UFAs) is essential to all life. Escherichia coli produces UFAs during fatty acid biosynthesis, requiring the action of two dehydratases, FabA and FabZ. While both enzymes can prepare α,β-trans unsaturated fatty acid intermediates via dehydration, only FabA can further isomerize an intermediate to produce a β,γ-cis unsaturated species, the precursor to E. coli UFAs. Through the combined application of chemical biology tools, structural biology, and computer simulations, we have developed a more complete understanding of the divergent activity and substrate selectivities of FabA and FabZ. This work reveals the structure of E. coli FabZ and clarifies the molecular mechanisms responsible for the introduction of cis unsaturation in fatty acids, a long-standing mechanistic uncertainty.
Keywords: de novo unsaturated fatty acid biosynthesis, cell membrane homeostasis, chemical biology, structural biology, computational biology
Abstract
Fatty acid biosynthesis in α- and γ-proteobacteria requires two functionally distinct dehydratases, FabA and FabZ. Here, mechanistic cross-linking facilitates the structural characterization of a stable hexameric complex of six Escherichia coli FabZ dehydratase subunits with six AcpP acyl carrier proteins. The crystal structure sheds light on the divergent substrate selectivity of FabA and FabZ by revealing distinct architectures of the binding pocket. Molecular dynamics simulations demonstrate differential biasing of substrate orientations and conformations within the active sites of FabA and FabZ such that FabZ is preorganized to catalyze only dehydration, while FabA is primed for both dehydration and isomerization.
To maintain membrane phospholipid homeostasis and modulate membrane fluidity, all organisms must synthesize unsaturated fatty acids (UFAs) (1, 2). α- and γ-proteobacteria, including Escherichia coli, Yersinia pestis, and Pseudomonas aeruginosa produce UFAs during de novo fatty acid biosynthesis, requiring the coordinated action of two dehydratases (DHs), FabA and FabZ (3, 4). This system of UFA biosynthesis is unique to bacteria and is of interest as an antibiotic target. FabA and FabZ are part of the type II fatty acid synthase (FAS), in which discrete enzymes synthesize fatty acids in a stepwise, iterative manner. Throughout this process the growing fatty acid remains tethered to an acyl carrier protein (ACP), AcpP, for solubility and transport (Fig. 1A), and it has been demonstrated that protein-protein interactions between AcpP and partner enzymes are essential to fatty acid synthesis (5–9). Both FabZ and FabA catalyze the dehydration of β-hydroxyacyl-AcpP, but they have varying substrate selectivity, with C8–C12 substrates favored by FabA and a C6 substrate favored by FabZ (Fig. 1D) (4). FabA can also perform an additional allylic rearrangement that converts trans-dec-2-enoyl-AcpP, the immediate product of dehydration, into cis-dec-3-enoyl-AcpP, the precursor to E. coli UFAs. FabZ is incapable of this isomerization. While the role of FabA in production of unsaturated fatty acids has been the subject of intense mechanistic study for decades (10), differences to the more recently discovered FabZ (11) and the structure–function relationships of the two enzymes have not been established definitively. E. coli FabA and FabZ share 23% amino acid identity (41% similarity), but a crystal structure is available for only FabA (12). Crystal structures have been reported for both apo-FabA and apo-FabZ from P. aeruginosa (13, 14), but the similar active site pockets do not reveal the molecular basis for the FabA exclusive activity as an isomerase. Further, while the dehydration mechanism (Fig. 1B) of FabA is known (15–18), the mechanism of isomerization (Fig. 1C), the second step in the transformation, remains speculative (12, 17, 19). Recently, the crystal structure of a complex of phosphopantetheinylated AcpP (holo-AcpP) and FabZ from Helicobacter pylori was described (20), but the ACP lacked an acyl group, limiting conclusions regarding the distinct activities and substrate preferences of FabA and FabZ.
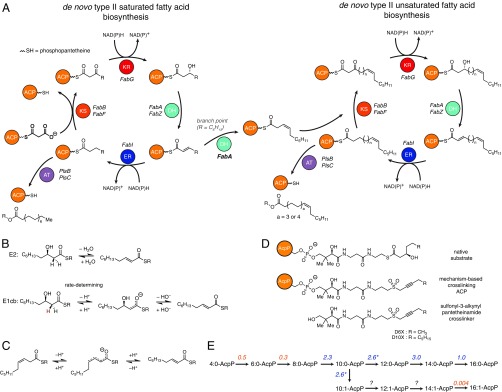
Fig. 1.
Fatty acid biosynthesis in E. coli. (A) Elongation phase of de novo fatty acid production in E. coli. Note that AT in this diagram refers to PlsB and PlsC, glycerol-3-phosphate acyl transferases responsible for offload fatty acids from AcpP. (B) Mechanisms for the dehydration of β-hydroxydecanoyl-AcpP. Evidence suggests that an E1cb (not E2) mechanism is operative. Although the reaction pathways for β-hydroxydecanoyl-AcpP are shown, these mechanisms are plausible for any β-hydroxyacyl-AcpP substrate. Note the hydrogen colored red is the pro-R hydrogen abstracted by FabA (4). (C) Proposed mechanism for the FabA-catalyzed isomerization of trans-2-decenoyl-AcpP to cis-3-decenoyl-AcpP. The R group represents the omitted portion of the acyl-AcpP thioester. (D) Substrates and probes used to characterized AcpP-DH interactions. (E) The experimentally determined substrate preferences of FabA and FabZ. The number shown above the arrow is the ratio of the specific activities of FabA and those of FabZ. Blue and orange colors indicate a substrate’s preference for FabA and FabZ, respectively. Note that the rate at which FabA processes β-hydroxydecanoyl-AcpP includes both rate of the formation of the dehydration and isomerization products. The nomenclature used in E is defined in SI Appendix, Fig. S7.
Visualizing complexes of acyl-AcpP with its enzymatic partners has posed a challenge because such complexes are short-lived. We previously overcame this problem by employing mechanism-based cross-linking probes to covalently trap FabA in functional association with acyl-AcpP to solve the crystal structure of the complex (AcpP=FabA) (21). Here, we applied a similar cross-linking strategy in combination with fusion-protein methods to trap E. coli FabZ with acyl-AcpP and report the crystal structure of the resulting complex (AcpP=FabZ). Having both AcpP=FabA and AcpP=FabZ structures in hand enabled us to perform molecular dynamics (MD) simulations of the acyl-AcpP•FabA and acyl-AcpP•FabZ complexes to evaluate the mechanisms of dehydration catalyzed by FabA and FabZ. The simulation results recapitulated the established substrate preferences of FabA and FabZ and additionally provided a complete structural rationale for the unique isomerase activity of FabA. Together these findings reveal the molecular basis for the differential activity of FabA and FabZ, accounting for the de novo synthesis of UFAs, a critical component of phospholipid metabolism in E. coli and other bacteria.
Results
Characterization of FabZ Interaction with AcpP.
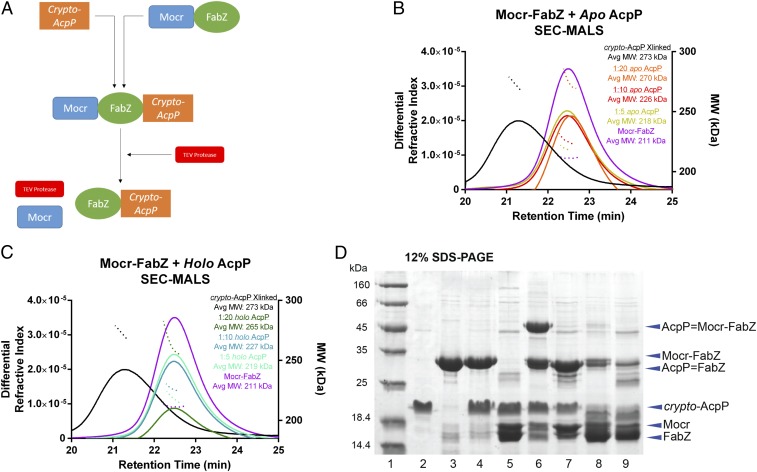
To compare catalytic selectivities and probe the structural basis of catalysis by natural FabA and FabZ partners, we sought to examine the E. coli FabZ in complex with AcpP. However, when highly purified for structure analysis, recombinant FabZ was not stable or monodisperse (SI Appendix, Table S1). Thus, we prepared FabZ with the Mocr protein fused to the N terminus (Fig. 2 and SI Appendix, Fig. S1) (22). The Mocr-FabZ fusion protein was purified to homogeneity in a stable and soluble form, and the Mocr fusion did not interfere with the expected hexameric association of FabZ (Fig. 2). After proteolytic cleavage of the Mocr fusion partner, FabZ remained soluble (as assessed by turbidity) only when the cleavage reaction included equimolar or greater concentrations of AcpP, implying that, like the H. pylori FabZ, the E. coli FabZ demonstrates inherent affinity for its carrier protein (SI Appendix, Table S1) (23). We then used size exclusion chromatography (SEC) with multiangle light scattering (MALS) to characterize the stoichiometry of complexes of Mocr-FabZ and holo-AcpP (with the phosphopantetheine cofactor linked at Ser36) or apo-AcpP (lacking phosphopantetheine). All combinations of FabZ and AcpP resulted in complex formation with oligomeric states ranging from (AcpP1-FabZ6) to (AcpP6-FabZ6) (Fig. 2). The oligomer state was dependent on AcpP concentration, with a 20-fold molar excess of either apo- or holo-AcpP fully saturating Mocr-FabZ. The complexes had similar elution volumes to Mocr-FabZ in absence of AcpP, indicating a dynamic mixture of transient oligomer states. This observation is consistent with a type II FAS system in which the AcpP must find and interact with each enzyme; should a tight, long-lasting complex form with any one enzyme, FAS throughput would drop significantly.
Fig. 2.
FabZ and AcpP cross-linking and biophysical analysis. (A) Schematic of FabZ and crypto-AcpP cross-linking reaction. FabZ was insoluble when released from the Mocr fusion partner, but solubility was retained by cross-linking to acidic AcpP before cleavage of the fusion partner. (B) Analysis of Mocr-FabZ and AcpP complexes by MALS coupled with SEC. SEC elution profiles (differential refractive index) are shown in solid lines and MALS average molecular weight plots as dotted lines. Calculated molecular weights are Mocr-FabZ, 201 kDa; apo-AcpP, 11 kDa; and Mocr-FabZ:(AcpP)6, 266 kDa. In both panels, standards of Mocr-FabZ (purple) and AcpP=Mocr-FabZ (black) are shown for comparison. Mocr-FabZ with apo-AcpP: 1:5 Mocr-FabZ:apo-AcpP (gold), 1:10 (red), 1:20 (orange). (C) Mocr-FabZ with holo-AcpP. 1:5 Mocr-FabZ:holo-AcpP (cyan), 1:10 (teal), 1:20 (dark green). For both apo- and holo-AcpP, a concentration-dependent increase in average MW is observed, with full saturation at a 20-fold excess of AcpP. (D) Analysis of cross-linking reactions of AcpP and Mocr-FabZ by denaturing polyacrylamide gel electrophoresis (12% SDS). Lane 1, MW standards; lane 2, apo-AcpP; lane 3, Mocr-FabZ; lane 4, apo-AcpP and Mocr-FabZ in cross-linking buffer; lane 5, apo-AcpP and Mocr-FabZ in cross-linking buffer with TEV protease; lane 6, crypto-DH6-AcpP and Mocr-FabZ cross-linking reaction; lane 7, crypto-DH6-AcpP and Mocr-FabZ cross-linking reaction with TEV protease; lane 8, crypto-DH10-AcpP and Mocr-FabZ cross-linking reaction; lane 9, crypto-DH10-AcpP and 20 Mocr-FabZ cross-linking reaction with TEV protease.
FabZ-AcpP Cross-Linking.
We then sought to generate a soluble AcpP=FabZ complex suitable for crystallography by cross-linking AcpP before proteolytic cleavage of the Mocr fusion partner. As previously reported, 3-alkynyl-sulfone-pantetheinamide probes can link AcpP and DHs (21, 24). Here, we chose a 6-carbon probe (DH6) to mimic the optimal FabZ substrate and appended it to AcpP (crypto-DH6-AcpP, SI Appendix, Fig. S2B) (4). Cross-linking trials with Mocr-FabZ and crypto-DH6-AcpP proceeded nearly to completion (Fig. 2D). In agreement with the previously reported substrate preference of FabZ (4), less than 10% cross-linked complex formed with a probe mimicking the least-favored substrate, crypto-DH10-AcpP (AcpP loaded with a 10-carbon probe, Fig. 2D and SI Appendix, Fig. S2A). SEC-MALS analysis of DH6-AcpP=Mocr-FabZ indicated a stable complex with shorter retention time than the noncovalent complexes with apo-AcpP or holo-AcpP and a molecular weight corresponding to (AcpP6=Mocr-FabZ6) (Fig. 2 B and C). AcpP=FabZ was stable upon proteolytic cleavage of the Mocr fusion partner and had an apparent molecular weight of 160 kDa by SEC (SI Appendix, Fig. S3), again corresponding to an AcpP6=FabZ6 complex.
Crystal Structure of AcpP6=FabZ6.
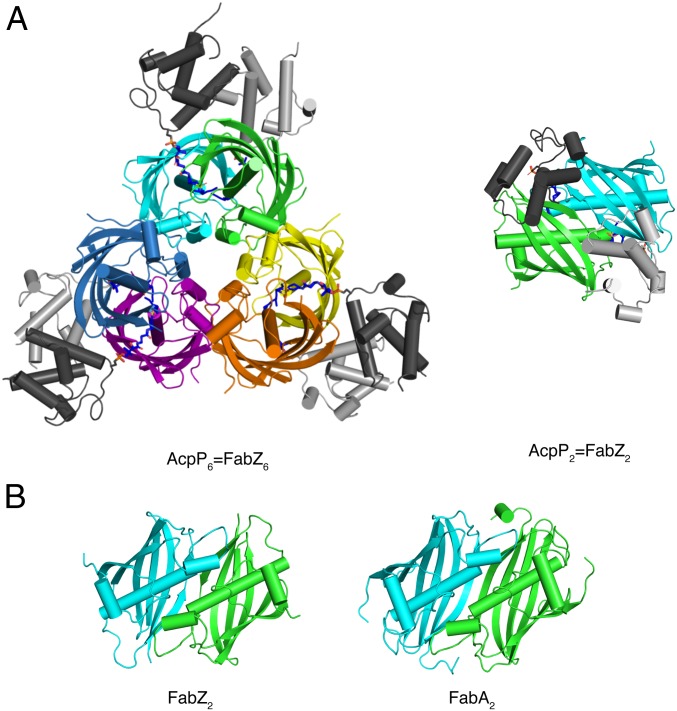
We obtained a 2.5-Å crystal structure of the AcpP6=FabZ6 complex (Fig. 3 and SI Appendix, Table S2). The FabZ hexamer is a trimer of FabA-like dimers, consistent with structures of other bacterial FabZ enzymes (13, 20, 25–27). In the hexamer, an AcpP is cross-linked to the catalytic His54 of each FabZ monomer and contacts both subunits of the FabZ dimer but makes no contacts with the other two “dimers” or with any other AcpP (Figs. 3 and 4). Continuous electron density is observed for the cross-linker and the catalytic His54 of FabZ (SI Appendix, Fig. S4).
Fig. 3.
Structure of AcpP=FabZ. (A) E. coli FabZ hexamer with six AcpPs. The FabZ hexamer is composed of three dimers (cyan and green, yellow and orange, purple and blue). (Left) The six AcpPs are at the periphery (dark and light gray), each cross-linked to a FabZ subunit. (Right) A “top-down” view of the cyan/green AcpP=FabZ dimer shows that the two bound AcpPs do not contact one another. The DH6 cross-linkers are rendered as sticks in atomic coloring (blue N, red O, yellow S, and dark or light gray C according to the tethered ACP). (B) Comparison of a FabZ dimer from the hexamer in A (Left) and E. coli FabA (Right), which is a dimer. FabZ and FabA have identical folds and dimer interfaces; FabZ is a trimer of FabA-like dimers, whereas FabA is a dimer. The FabZ cyan/green dimer in B is rotated 90° from the right view in A.
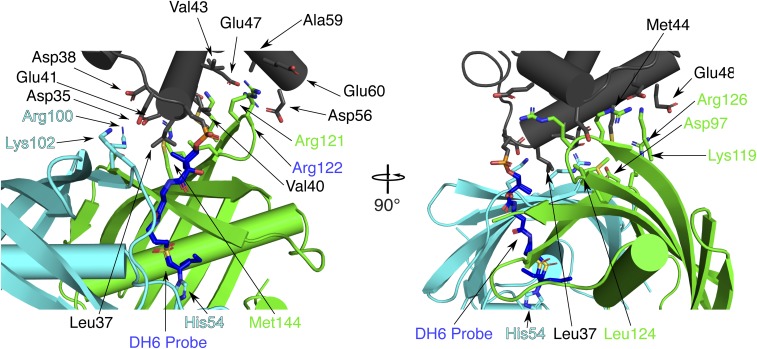
Fig. 4.
Interfacial contacts between AcpP and FabZ. The AcpP (gray) is cross-linked to the cyan FabZ subunit, but the protein-protein contacts with this subunit are limited to FabZ Arg100 and Lys102 (cyan sticks) and AcpP Asp35, Asp38, and Glu41 (gray sticks). AcpP has far more contacts with the partner subunit (green) of the FabZ dimer, where FabZ Lys119, Arg121, Arg122, Arg126, Leu124, Asp97, and Met144 (green sticks) contact AcpP Leu37, Val40, Glu41, Val43, Met44, Glu47, Glu48, Asp56, Ala59, and Glu60. The reciprocal situation pertains to the AcpP cross-linked to the green FabZ subunit.
The surface of AcpP helix α2 contacts the surface of the FabZ β-sheet via an interface comprised of several hydrophobic contacts surrounded by ionic interactions (Fig. 4 and SI Appendix, Table S3). As with FabA, acidic residues on AcpP contact basic residues on FabZ surrounding the active site entrance. The majority of protein-protein contacts occur between AcpP and the subunit of the FabZ dimer to which it is not cross-linked (AcpP subunit A to FabZ subunit B, and AcpP subunit B to FabZ subunit A). The cross-linked subunit also contributes to the interface, which buries 16% (745 Å2) of the AcpP surface. The interface is significantly larger than the AcpP=FabA interface (∼530 Å2) (21, 28), in which AcpP has no contacts with the FabA monomer to which it is cross-linked (SI Appendix, Table S3). The six AcpP-FabZ interfaces are nearly identical, although two of the six AcpPs are shifted slightly due to crystal contacts (SI Appendix, Fig. S5).
The AcpP-interacting surface of E. coli FabZ is strictly analogous to the AcpP-interacting surface of H. pylori FabZ (20). However, in contrast to our finding that the E. coli FabZ hexamer can interact productively with six AcpPs, a see-saw mechanism was proposed for the H. pylori FabZ in which only three AcpPs simultaneously engage the hexamer. This proposal was based on the crystal structure (FabZ6-AcpP3) where only one AcpP bound per dimer in the FabZ hexamer, together with analysis by SEC-MALS, small-angle X-ray scattering (SAXS), and microscale thermophoresis (MST) (23). However, the H. pylori FabZ6-AcpP3 stoichiometry in the crystal structure is more simply explained by crystal lattice contacts that block the three unoccupied AcpP sites. Previous surface plasmon resonance (SPR) characterization of the H. pylori FabZ-AcpP interaction indicated a single Kd of 120 nM (23), in contrast to the three weaker binding events deduced from MST data (20). In our SEC-MALS analysis of the E. coli FabZ-AcpP interaction, higher-order complexes existed with an excess of either apo- or holo-AcpP, and a 20-fold excess AcpP fully saturated the Mocr-FabZ hexamer (Fig. 2 B and C). The crystal lattice of the H. pylori AcpP3-FabZ6 is incompatible with an equimolar complex, so a higher-order complex may have been missed in the study. Alternatively, the different oligomer states observed for the E. coli AcpP6-FabZ6 complex (this work) and the H. pylori ACP3-FabZ6 complex (20) may reflect a real difference in the two FabZ proteins, perhaps arising from distinct strategies for dehydration of β-hydroxyacyl-AcpPs during fatty acid biosynthesis. Indeed, the dual FabA-FabZ DH system does not exist in ε-proteobacteria such as H. pylori, which possess only FabZ and use a desaturase to synthesize unsaturated fatty acids (29, 30).
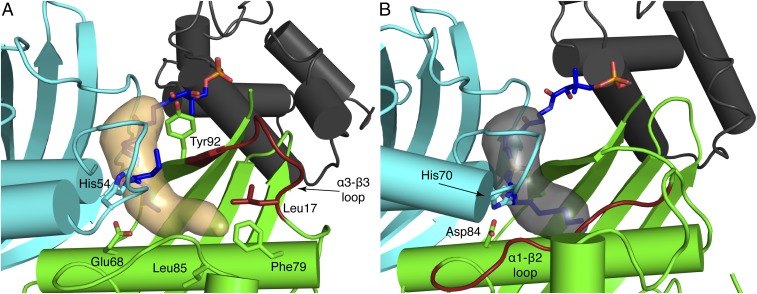
DHs possess a hydrophobic binding pocket that shields the fatty acyl substrate from solvent. In E. coli FabZ, the binding pocket touches the central “hotdog” helix (α3) and is bounded by amino acids in the α3-β3 loop (residues 85–90) and the α1-β1 loop. Phe79 in the hotdog helix (α3), Leu85 in the α3-β3 loop, and Leu17 on helix α1 form a hydrophobic surface at the innermost end of the pocket, where 12-carbon or longer substrates can bind (Fig. 5A). The binding pocket kinks at the C6 position and extends to a length which could easily accommodate longer substrates with a cis double bond installed by FabA. In contrast, the FabA substrate-binding pocket is straight and shorter, ideal to accommodate 10-carbon or shorter substrates (Fig. 5B). These differences in pocket length and shape between FabA and FabZ are due to the structures and lengths of their α1-β1 loop (longer in FabA) and α3-β3 loop (longer in FabZ) (Fig. 5 and SI Appendix, Fig. S6). The α1-β1 loop plays a much larger role in the substrate pocket of FabA, whereas the α3-β3 loop figures prominently in the FabZ pocket.
Fig. 5.
Comparison of E. coli FabZ and FabA substrate binding pockets. (A) FabZ (cyan and green) and AcpP (gray) with an ordered α3-β3 loop (red). The binding pocket (orange surface) is wide at the top with a narrow terminus and is long enough to accommodate C14 substrates. Key amino acids and the DH6 cross-linker are shown as sticks. (B) FabA (cyan and green) and AcpP (gray). The α1-β2 loop (red) defines the end of the binding pocket (gray surface), which is optimal for C10 substrates. Key amino acids and the DH10 cross-linker are shown as sticks.
The α3-β3 loop of FabZ connects the distal end of the pocket (Fig. 5A) and conserved Tyr92 at the substrate entrance. Tyr92 was proposed as a FabZ “gating residue” based on the reduced dehydration activity of an Ala substitution (26) and the “open” and “closed” conformations of the side chain in FabZ crystal structures (13, 20, 25, 27, 31). In the closed conformation, Tyr92 blocks the entrance of the substrate binding pocket (13, 25, 27, 31). In the AcpP=FabZ complex, the Tyr92 gate is open in all six FabZ monomers, and the acylated phosphopantetheine extends into the pocket. Interestingly, the α3-β3 loop is the least conserved region of FabZ and varies in length (7–10 amino acids, SI Appendix, Fig. S6). In FabZ structures with shorter α3-β3 loops, the loop is often poorly ordered or disordered. However, the shorter α3-β3 loop in the E. coli FabZ is well ordered with identical conformations in all six subunits, suggesting that FabZ interactions with AcpP or the acyl group facilitate loop ordering and formation of the substrate pocket. How this may occur is not apparent from the structure.
Comparative Simulations of FabZ and FabA Substrate Complexes.
We next used MD simulations to evaluate the mechanistic differences and substrate selectivities of the two enzymes. As AcpP=FabA and AcpP=FabZ represent the most accurate approximations of natural binding events at the moment of catalysis, these structures are ideally suited for MD simulations with modeled alternative substrates. Gaussian accelerated molecular dynamics (GaMD) (32–36) simulations of six acyl-AcpP2•FabA2 and acyl-AcpP2•FabZ2 were performed (SI Appendix, Fig. S7). The rmsds of the DH subunits and AcpPs indicate that AcpP motion accounts for most of the fluctuations of the acyl-AcpP•FabA complexes, whereas the DH subunits and AcpPs of acyl-AcpP•FabZ dimers have similar rmsds. AcpP undergoes not only internal motion but also translational and rotational motion with respect to the DH subunits, in agreement with the conformational flexibility observed in the AcpP=FabZ crystal structure (SI Appendix, Figs. S4 and S8–S13). However, subtle differences of FabA and FabZ subunits are evident from the root mean square fluctuations (rmsfs) sampled over the course these simulations. In all cases, the rmsfs of the residues of FabA are lower than those of FabZ (SI Appendix, Figs. S14 and S15). Further analysis more closely examined the two catalytic steps: β-hydroxy dehydration, as naturally catalyzed by both enzymes, and allylic rearrangement, as naturally catalyzed only by FabA. The dehydration of β-hydroxydecanoyl-AcpP to trans-2-decenoyl-AcpP can occur via two different mechanisms; a two-step E1cb mechanism or a one-step E2 concerted mechanism (Fig. 1B). The first step of the E1cb mechanism, deprotonation of the substrate Cα, occurs stereoselectively (15) and produces a resonance-stabilized enolate intermediate. This step is followed by β-elimination, in which the enolate intermediate collapses to the trans-2-decenoyl-AcpP product with elimination of the β-hydroxy group. In contrast, E2 dehydrations feature simultaneous α-deprotonation and β-elimination events. Additionally, E2 dehydrations require that the two groups being eliminated, here the Cα hydrogen and β-hydroxy group, must be antiperiplanar to each other, characterized by a C(O)-Cα-Cβ-Cγ dihedral angle of 180°. Previous mechanistic studies of FabA and the crystal structure of P. aeruginosa FabA in complex with a substrate mimic support the E1cb-like mechanism (14–18). Recent mechanistic studies of a FabA homolog in a type I polyketide synthase was interpreted in terms of a “single-base” mechanism, in which the active His abstracts a proton from the substrate Cα and also protonates the β-hydroxy leaving group. Nonetheless, the catalytic Asp/Glu plays an essential catalytic role, as it preorganizes the 3R-hydroxyacyl substrate for stereospecific dehydration via a hydrogen-bonding interaction and is essential for dehydration and isomerization activity (37).
GaMD simulations of both β-hydroxydecanoyl-AcpP2•FabA2 and β-hydroxydecanoyl-AcpP2•FabZ2 support the proposed E1cb dehydration as well. Both simulations indicated a clear bias for a +60 °C(O)-Cα-Cβ-Cy dihedral angle, which positions the substrate proR Hα and β-hydroxy group ideally for E1cb elimination (SI Appendix, Fig. S16).
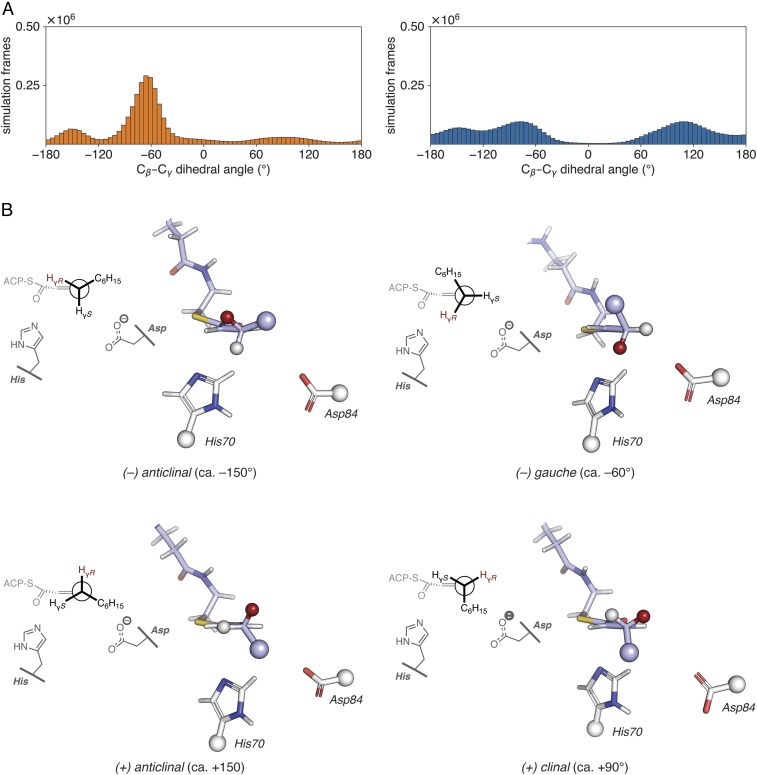
The second mechanistic capability of FabA to isomerize trans-2-decenoyl-AcpP to cis-3-decenoyl-AcpP is poorly characterized compared with dehydration. Analysis of FabA and FabZ crystal structures have provided no clear rationale for the isomerization (in)ability of each enzyme. Mechanistic studies of the production of cis-3-decenoyl-AcpP via allylic rearrangement by E. coli FabA have unequivocally demonstrated the intermediacy of trans-2-decenoyl-AcpP (10, 16, 17). NMR isotope studies further showed that FabA abstracts the proR Hγ of trans-2-decenoyl-AcpP to produce cis-3-decenoyl-AcpP (19). Beyond these well-established findings, few mechanistic details are known. The most plausible isomerization mechanism would occur in a two-step fashion, wherein deprotonation at Cγ precedes (re)protonation at Cα. Such a mechanism would involve intermediacy of a relatively stable dienolate anion (Fig. 1C). Consequently, we performed GaMD simulations of trans-2-decenoyl-AcpP2•FabA2 and trans-2-decenoyl-AcpP2•FabZ2 to understand how the product of dehydration is accommodated in each active site. Given that the proR Hγ is stereoselectively abstracted by FabA, analysis of the rotameric preferences about the substrate Cβ–Cγ bond within the active sites of the two dehydratases was expected to provide insights into the mechanism of action of FabA as an isomerase. In the simulations, the substrate predominantly assumed one of four possible Cβ–Cγ rotameric states (Fig. 6), the (−) gauche (−60°) conformation. Remarkably, of the rotameric states computationally sampled, only in the (−) gauche conformer is the proR Hγ suitably positioned within the active site to be abstracted by the catalytic His70 to initiate the allylic rearrangement. While both FabA and FabZ sampled the same four conformers, only FabA showed a distinct preference for the (−) gauche conformer compared with FabZ (Fig. 6A). Thus, FabA isomerization may be a consequence of preorganizing the allylic substrate for proR Hγ abstraction after dehydration. These findings are qualitatively consistent with a hypothesis of Naismith and coworkers that the active site is shaped to sterically occlude alternative conformations, preventing their formation (14) However, the protein is inherently dynamic so that nonproductive conformations are not entirely excluded. The simulations reveal a 20-fold preference for the productive conformation in FabA.
Fig. 6.
Analysis of MD simulations of trans–2-decenoyl-AcpP•DH complexes. (A) Distribution of dihedral angles for the Cɑ–Cβ–Cγ–Cδ torsion of the trans-2-decenoyl substrate sampled throughout the course of GaMD simulations of trans-2-decenoyl-AcpP•FabA (Left, in orange) and trans-dec-2-enoyl-AcpP•FabZ (Right, in blue). Simulation data written every 0.5 ps were binned to prepare histograms using a bin width of 5°. (B) Representative structures of the four Cβ–Cγ rotamers sampled computationally. These structures were identified from the larger simulated ensemble of the trans-2-decenoyl-AcpP•FabA complex as described in SI Appendix. Structures are oriented as Newman projections along the Cγ–Cβ bond of the trans-2-decenoyl substrate (shown in stick representation with carbon, oxygen, and nitrogen atoms in violet, red, and blue, respectively) within the active site of FabA. Catalytic histidine and aspartate are also shown in stick form with white carbon, red oxygen, and blue nitrogen. Large spheres indicate the portions of the substrate and catalytic residues omitted in the 3D images. The smaller spheres found at Cγ show the proR (red) and proS (white) γ-hydrogens; the proR Hγ is abstracted in the isomerization reaction. A 2D representation of each structure is shown on the Left of each panel.
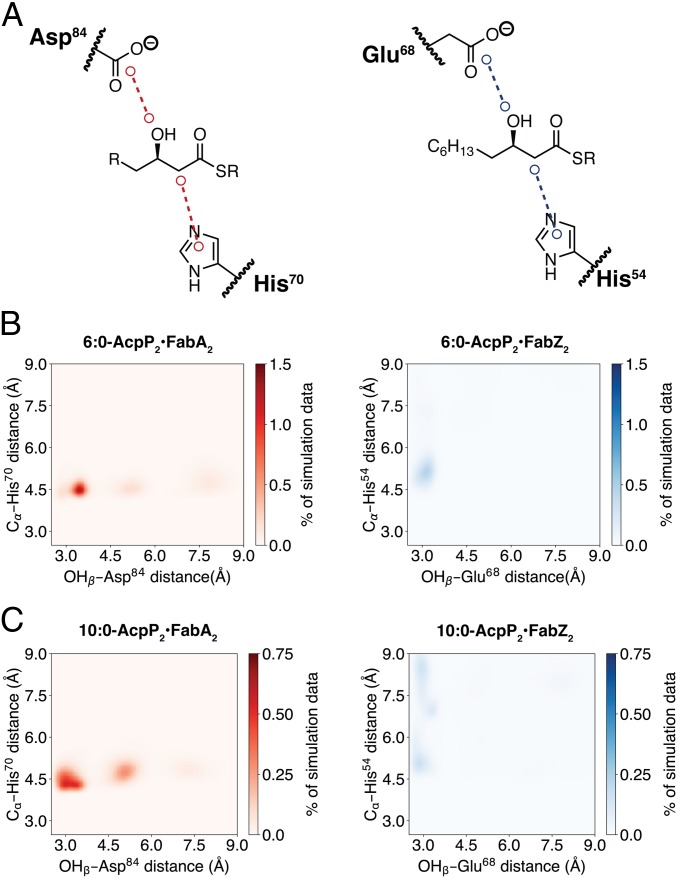
To evaluate the origins of substrate chain length preference, we performed simulations of β-hydroxyhexanoyl-AcpP2•FabA2/Z2 and β-hydroxydecanoyl-AcpP2•FabA2/Z2, the preferred substrates of FabZ (C6) and FabA (C10), respectively. Analysis of the simulation data demonstrates active site preorganization, as visualized by the distances between the catalytic His and substrate α-carbon as well as the distance between the catalytic Asp/Glu and the substrate β-hydroxy (Fig. 7). Throughout the simulation, FabZ maintained a hydrogen bond (d < 3.0 Å) between the carboxylate of Glu68 and the β-hydroxy substituent of β-hydroxyhexanoyl-AcpP, in agreement with the proposed Glu68 function as a general acid. In constrast, the FabA catalytic Asp84 formed a longer hydrogen bond (3.0 Å < d < 3.5 Å) with the β-hydroxy substituent in the simulations of β-hydroxyhexanoyl-AcpP2•FabA2. While the Asp/Glu of FabA and FabZ could form a hydrogen bond with the β-hydroxydecanoyl-AcpP, the C10-preferred FabA substrate, the FabA catalytic His70 maintained contact with the substrate α-carbon throughout the simulation, whereas the FabZ His54 often disengaged (d > 6.0 Å) the substrate (Fig. 7).
Fig. 7.
Distances between catalytic residues and key substrate atoms in MD simulations of 6:0-AcpP•DH and 10:0-AcpP•DH complexes. (A) Illustrations of distances analyzed. A 2D histogram of simulation data of the complexes of (B) β-hydroxyhexanoyl-AcpP•DHs (preferred FabZ substrate) and (C) β-hydroxydecanoyl-AcpP•DHs (preferred FabA substrate). Simulations with FabA are on the Left and with FabZ on the Right. Simulation data are binned along two coordinates (dimensions): the distance between the substrate β-hydroxy oxygen and the side chain carboxylate of Asp84 (FabA) or Glu68 (FabZ), and the distance between the imidazole of catalytic His70 (FabA) or His54 (FabZ) and the substrate α-carbon. Data were sorted into 0.05 Å × 0.05 Å bins. Color bars indicate the population of each bin as a percentage of the total simulation data collected.
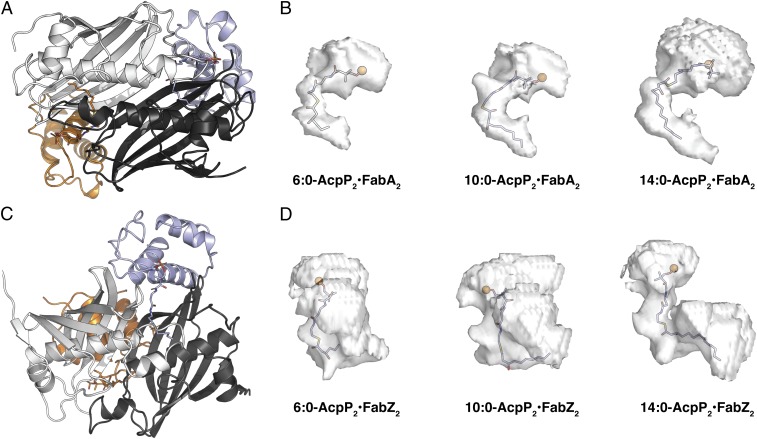
We further analyzed the shapes of the substrate binding pockets, as sampled during MD simulations (Fig. 8). All of the C10 substrates, decanoyl-AcpP, trans-2-decenoyl-AcpP, and cis-3-decenoyl-AcpP, were accommodated within the FabA binding pocket with minimal reorganization of the pocket and were of ideal length to occupy its entirety. When hexanoyl-AcpP was bound, the pocket was similar in size and shape to that sampled during simulations of the decanoyl-AcpP•FabA complexes; but the shorter 6-carbon alkyl chain left a void in the binding cavity that may be responsible for its lower activity with FabA (Fig. 8 and SI Appendix, Figs. S20, S21, and S24). In contrast, the 14-carbon substrates tetradecanoyl-AcpP and cis-7-tetradecenoyl-AcpP were too large for the FabA active site. These substrates not only forced the cavity to expand, but also increased the space between AcpP and FabA (Fig. 8B and SI Appendix, Figs. S20 and S21; large upper isosurfaces of tetradecanoyl-AcpP•FabA and cis-7-tetradecenoyl-AcpP•FabA), presumably reducing their affinity. Analysis of simulation data for acyl-AcpP•FabZ complexes demonstrated that the substrate binding pocket of FabZ was generally larger in volume and more dynamic than the FabA pocket (Fig. 8 C and D). The relationship between simulated pocket volumes and substrate specificities is less obvious for FabZ than for FabA, as the preferred C6 substrate and intermediate C10 substrate (Fig. 8) occupied similar pockets during simulations. In general, the pockets of FabZ sampled through MD simulations are larger (SI Appendix, Fig. S24), easily accommodating shorter chain substrates. Interestingly, the simulations recapitulated the proposed FabZ reorganization of the substrate pocket (i.e., the α3-β3 loop) when bound to C14 substrates (Fig. 8D and SI Appendix, Figs. S22–S24). This reorganization results in a longer, narrower U-shaped cavity volume compared with the larger globular cavities observed in simulations of β-hydroxyhexanoyl-AcpP•FabA and β-hydroxydecanoyl-AcpP•FabA (SI Appendix, Figs. S22 and S23). The simulations also illustrated a better fit of the C14 substrates with FabZ, as the upper isosurfaces were smaller than those observed for FabA and the C14 substrate penetrated into the depths of the FabZ pocket (Fig. 8D and SI Appendix, Figs. S22 and S23).
Fig. 8.
Cavities of the acyl-AcpP•FabA complexes sampled computationally. (A) Cartoon representation of the AcpP=FabA dimer oriented in a suitable manner (Bottom image) for the visualization of the acyl substrate chain within the FabA active site. (B) Isosurfaces of the acyl-AcpP•FabA complexes sampled via MD simulation. Ninety percent simulation data of each of these complexes possess an active site cavity that assumes a volume residing within the rendered isosurface. The phosphantethiene prosthetic group and acyl substrate are shown as sticks with the phosphantiethiene P as an orange sphere. Note that the computed cavities include unoccupied space (Upper regions of isosurfaces) between the carrier protein and the dehydratase (i.e., AcpP•DH interface). (C) Cartoon representation of the AcpP=FabZ dimer oriented in a suitable manner (Bottom image) for the visualization of the acyl substrate chain within the FabZ active site. (D) Isosurfaces of the acyl-AcpP•FabZ complexes sampled via MD simulation, rendered as in B.
Conclusions
In this study, we present a 2.5-Å crystal structure of the E. coli AcpP=FabZ complex, the structure of the E. coli FabZ, with six AcpPs in functional association with the FabZ hexamer. The E. coli AcpP=FabZ crystal structure used the power of covalent inhibitor-based mechanistic cross-linking probes to trap the native protein-protein interaction, clearly demonstrating that all six FabZ monomers can functionally and simultaneously interact with AcpP. The structure reveals a broad FabZ substrate-binding pocket that contrasts the narrow and straight binding cavity of FabA. The α3-β3 loop of FabZ that lines the distal end of the substrate pocket is well ordered, forming a hydrophobic cleft that may allow the dehydration of long-chain acyl substrates, especially those with cis-unsaturation. Analysis of molecular dynamics simulations supports these qualitative characterizations of the FabA and FabZ substrate binding pockets and provides a complimentary time-resolved understanding of the binding pocket as it responds to the internal motion of bound substrates. GaMD further revealed how both enzyme pockets stabilize substrate rotamers to allow dehydration via an E1cb mechanism, demonstrating a possible mode by which these dehydratases function. Simulations additionally showed that FabA, but not FabZ, selectively sampled the (−) gauche conformer of trans-2-decenoyl-AcpP for allylic rearrangement, in which the proR γ-hydrogen is suitably positioned for abstraction to form cis-3-decenoyl-AcpP. This illustrates the unique role of substrate preorganization in FabA catalysis to preferentially stabilize the only conformer that allows allylic rearrangement and ultimately the formation of UFAs. These structural and computational findings shed light on the divergent substrate preferences of E. coli FabA and FabZ, provide clarity for how the structure of each enzyme dictates mechanistic specificity, and illustrate the immense utility of the synthesis of chemical, structural, and computational biology in resolving the mechanisms by which biomolecules function.
Supplementary Material
Acknowledgments
We thank Drs. Anthony Mrse and Yongxuan Su for assistance with NMR and MS data acquisition, respectively; and the San Diego Supercomputer Center (SDSC) for access to computing resources (Triton Shared Computing Cluster). This research was supported by NIH Grants GM31749, GM095970, and DK042303; the SDSC; and the National Biomedical Computation Resource. The GM/CA at APS beamlines are funded in whole or in part with federal funds from the National Institute of General Medical Sciences (GM) (AGM-12006) and the National Cancer Institute (CA) (ACB-12002). This research used resources of the Advanced Photon Source (APS), a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6N3P).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818686116/-/DCSupplemental.
References
- 1.Cronan JE, Jr, Gelmann EP. Physical properties of membrane lipids: Biological relevance and regulation. Bacteriol Rev. 1975;39:232–256. doi: 10.1128/br.39.3.232-256.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 3.Cronan JE, Jr, Rock CO. Biosynthesis of membrane lipids. Ecosal Plus. 2008;3 doi: 10.1128/ecosalplus.3.6.4. [DOI] [PubMed] [Google Scholar]
- 4.Heath RJ, Rock CO. Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J Biol Chem. 1996;271:27795–27801. doi: 10.1074/jbc.271.44.27795. [DOI] [PubMed] [Google Scholar]
- 5.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 6.Crosby J, Crump MP. The structural role of the carrier protein–Active controller or passive carrier. Nat Prod Rep. 2012;29:1111–1137. doi: 10.1039/c2np20062g. [DOI] [PubMed] [Google Scholar]
- 7.Beld J, Lee DJ, Burkart MD. Fatty acid biosynthesis revisited: Structure elucidation and metabolic engineering. Mol Biosyst. 2015;11:38–59. doi: 10.1039/c4mb00443d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finzel K, Lee DJ, Burkart MD. Using modern tools to probe the structure-function relationship of fatty acid synthases. ChemBioChem. 2015;16:528–547. doi: 10.1002/cbic.201402578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roujeinikova A, et al. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J Mol Biol. 2007;365:135–145. doi: 10.1016/j.jmb.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 10.Kass LR, Bloch K. On the enzymatic synthesis of unsaturated fatty acids in Escherichia coli. Proc Natl Acad Sci USA. 1967;58:1168–1173. doi: 10.1073/pnas.58.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohan S, Kelly TM, Eveland SS, Raetz CR, Anderson MS. An Escherichia coli gene (FabZ) encoding (3R)-hydroxymyristoyl acyl carrier protein dehydrase. Relation to fabA and suppression of mutations in lipid A biosynthesis. J Biol Chem. 1994;269:32896–32903. [PubMed] [Google Scholar]
- 12.Leesong M, Henderson BS, Gillig JR, Schwab JM, Smith JL. Structure of a dehydratase-isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: Two catalytic activities in one active site. Structure. 1996;4:253–264. doi: 10.1016/s0969-2126(96)00030-5. [DOI] [PubMed] [Google Scholar]
- 13.Kimber MS, et al. The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa. J Biol Chem. 2004;279:52593–52602. doi: 10.1074/jbc.M408105200. [DOI] [PubMed] [Google Scholar]
- 14.Moynié L, et al. Structural insights into the mechanism and inhibition of the β-hydroxydecanoyl-acyl carrier protein dehydratase from Pseudomonas aeruginosa. J Mol Biol. 2013;425:365–377. doi: 10.1016/j.jmb.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Schwab JM, Habib A, Klassen JB. A thorough study of the stereochemical consequences of the hydration/dehydration reaction catalyzed by β-hydroxydecanoyl thioester dehydrase. J Am Chem Soc. 1986;108:5304–5308. [Google Scholar]
- 16.Brock DJ, Kass LR, Bloch K. β-hydroxydecanoyl thioester dehydrase. II. Mode of action. J Biol Chem. 1967;242:4432–4440. [PubMed] [Google Scholar]
- 17.Rando RR, Bloch K. Mechanism of action of β-hydroxydecanoyl thioester dehydrase. J Biol Chem. 1968;243:5627–5634. [PubMed] [Google Scholar]
- 18.Schwab JM, Klassen JB, Habib A. Stereochemical course of the hydration reaction catalysed by β-hydroxydecanoylthioester dehydrase. J Chem Soc Chem Comm. 1986;0:357–358. [Google Scholar]
- 19.Schwab JM, Klassen JB. Steric course of the allylic rearrangement catalyzed by β-hydroxydecanoylthioester dehydrase. Mechanistic implications. J Am Chem Soc. 1984;106:7217–7227. [Google Scholar]
- 20.Zhang L, et al. Crystal structure of FabZ-ACP complex reveals a dynamic seesaw-like catalytic mechanism of dehydratase in fatty acid biosynthesis. Cell Res. 2016;26:1330–1344. doi: 10.1038/cr.2016.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen C, et al. Trapping the dynamic acyl carrier protein in fatty acid biosynthesis. Nature. 2014;505:427–431. doi: 10.1038/nature12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DelProposto J, Majmudar CY, Smith JL, Brown WC. Mocr: A novel fusion tag for enhancing solubility that is compatible with structural biology applications. Protein Expr Purif. 2009;63:40–49. doi: 10.1016/j.pep.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, et al. Helicobacter pylori acyl carrier protein: Expression, purification, and its interaction with β-hydroxyacyl-ACP dehydratase. Protein Expr Purif. 2007;52:74–81. doi: 10.1016/j.pep.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa F, Haushalter RW, Lee DJ, Finzel K, Burkart MD. Sulfonyl 3-alkynyl pantetheinamides as mechanism-based cross-linkers of acyl carrier protein dehydratase. J Am Chem Soc. 2013;135:8846–8849. doi: 10.1021/ja4042059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGillick BE, Kumaran D, Vieni C, Swaminathan S. β-Hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Francisella tularensis and Yersinia pestis: Structure determination, enzymatic characterization, and cross-inhibition studies. Biochemistry. 2016;55:1091–1099. doi: 10.1021/acs.biochem.5b00832. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, et al. Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) J Biol Chem. 2008;283:5370–5379. doi: 10.1074/jbc.M705566200. [DOI] [PubMed] [Google Scholar]
- 27.Kostrewa D, Winkler FK, Folkers G, Scapozza L, Perozzo R. The crystal structure of PfFabZ, the unique β-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum. Protein Sci. 2005;14:1570–1580. doi: 10.1110/ps.051373005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finzel K, et al. Probing the substrate specificity and protein-protein interactions of the E. coli fatty acid dehydratase, FabA. Chem Biol. 2015;22:1453–1460. doi: 10.1016/j.chembiol.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Cronan JE. Functional replacement of the FabA and FabB proteins of Escherichia coli fatty acid synthesis by Enterococcus faecalis FabZ and FabF homologues. J Biol Chem. 2004;279:34489–34495. doi: 10.1074/jbc.M403874200. [DOI] [PubMed] [Google Scholar]
- 30.Bi H, Zhu L, Jia J, Zeng L, Cronan JE. Unsaturated fatty acid synthesis in the gastric pathogen Helicobacter pylori proceeds via a backtracking mechanism. Cell Chem Biol. 2016;23:1480–1489. doi: 10.1016/j.chembiol.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirkpatrick AS, Yokoyama T, Choi K-J, Yeo H-J. Campylobacter jejuni fatty acid synthase II: Structural and functional analysis of β-hydroxyacyl-ACP dehydratase (FabZ) Biochem Biophys Res Commun. 2009;380:407–412. doi: 10.1016/j.bbrc.2009.01.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamelberg D, Mongan J, McCammon JA. Accelerated molecular dynamics: A promising and efficient simulation method for biomolecules. J Chem Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, McCammon JA. Accelerated molecular dynamics: Theory, implementation and applications. AIP Conf Proc. 2012;1456:165–172. [Google Scholar]
- 34.Pierce LCT, Salomon-Ferrer R, Augusto F de Oliveira C, McCammon JA, Walker RC. Routine access to millisecond time scale events with accelerated molecular dynamics. J Chem Theory Comput. 2012;8:2997–3002. doi: 10.1021/ct300284c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miao Y, Feher VA, McCammon JA. Gaussian accelerated molecular dynamics: Unconstrained enhanced sampling and free energy calculation. J Chem Theory Comput. 2015;11:3584–3595. doi: 10.1021/acs.jctc.5b00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miao Y, et al. Improved reweighting of accelerated molecular dynamics simulations for free energy calculation. J Chem Theory Comput. 2014;10:2677–2689. doi: 10.1021/ct500090q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie X, Cane DE. pH-rate profiles establish that polyketide synthase dehydratase domains utilize a single-base mechanism. Org Biomol Chem. 2018;16:9165–9170. doi: 10.1039/c8ob02637h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.