Significance
Heparan sulfate (HS) belongs to the glycosaminoglycan family of carbohydrates anchored to core proteins and regulate many cell-cell signaling events. The HS large structural variability, orchestrated by sequential enzymatic modifications of the polymer, greatly regulates HS binding activities; however, the molecular mechanisms associated with these modification reactions are poorly documented. To decipher the mode of substrate binding and catalytic mechanism of the enzyme Glce, which converts the HS component GlcA into IdoA and dictates the epimerization profile for polymer activity, we combined glycoengineering, glycochemistry, biochemistry, and structural biology approaches. Our data unveil selective conformational distortions of the HS precursor chain on binding and document how a mechanistic strategy shared with mandelate racemase and HS lyases evolved toward a specific activity.
Keywords: C5 epimerization, heparan sulfate, catalytic mechanism, X-ray crystallography, substrate distortion
Abstract
Heparan sulfate (HS) is a linear, complex polysaccharide that modulates the biological activities of proteins through binding sites made by a series of Golgi-localized enzymes. Of these, glucuronyl C5-epimerase (Glce) catalyzes C5-epimerization of the HS component, d-glucuronic acid (GlcA), into l-iduronic acid (IdoA), which provides internal flexibility to the polymer and forges protein-binding sites to ensure polymer function. Here we report crystal structures of human Glce in the unbound state and of an inactive mutant, as assessed by real-time NMR spectroscopy, bound with a (GlcA-GlcNS)n substrate or a (IdoA-GlcNS)n product. Deep infiltration of the oligosaccharides into the active site cleft imposes a sharp kink within the central GlcNS-GlcA/IdoA-GlcNS trisaccharide motif. An extensive network of specific interactions illustrates the absolute requirement of N-sulfate groups vicinal to the epimerization site for substrate binding. At the epimerization site, the GlcA/IdoA rings are highly constrained in two closely related boat conformations, highlighting ring-puckering signatures during catalysis. The structure-based mechanism involves the two invariant acid/base residues, Glu499 and Tyr578, poised on each side of the target uronic acid residue, thus allowing reversible abstraction and readdition of a proton at the C5 position through a neutral enol intermediate, reminiscent of mandelate racemase. These structures also shed light on a convergent mechanism of action between HS epimerases and lyases and provide molecular frameworks for the chemoenzymatic synthesis of heparin or HS analogs.
Heparan sulfate (HS) belongs to a family of linear anionic polysaccharides, the glycosaminoglycans, which are present at the cell surface and within the extracellular matrix (1). Through their ability to bind a wide array of proteins and modulate their local concentration, stability, structure, and/or activity, HS participates in most major cellular processes, including cell-cell signaling (2, 3). From a structural standpoint, the HS-mediated binding activities, which are closely linked to the extensive structural variability of the polymer chain, are generated during a multistep biosynthesis process occurring predominantly in the Golgi apparatus. This process comprises the polymerization of a disaccharide repeat consisting of alternate GlcA and GlcNAc, followed by a series of modification reactions that include (i) GlcNAc N-deacetylation and N-sulfation, yielding N-sulfated glucosamine (GlcNS); (ii) C5-epimerization of GlcA into IdoA; and (iii) variable sulfation at O2 of IdoA and GlcA units, O6 of GlcNAc and GlcNS units, and occasionally O3 of GlcNS units (4, 5). Remarkably, these modifications occur not uniformly along the polymer, but rather within relatively short segments of usually three to six disaccharides (known as S-domains) that display tremendous variations in epimerization degree and sulfation patterning (ref. 6 and references therein). It is commonly known that such structural variability may confer critical and biologically relevant docking sites for protein ligands (6–8), yet the mechanisms by which the sulfation and epimerization patterns are defined and regulated remain unclear.
To better understand how HS functional motifs are built, we investigated structures of Glce, the key enzyme involved in reversible epimerization of GlcA into IdoA, the ratio of which greatly contributes to generate functional protein-binding motifs. Indeed, unlike most monosaccharides, IdoA can adopt two distinct ring conformations, 1C4 or 2SO, conferring to the polymer local plasticity with enhanced binding properties that are commonly associated with the biological properties of the HS chains (9, 10). In addition, IdoA is 2-O sulfated more efficiently than GlcA (11), a modification that prevents reverse epimerization into GlcA and strongly facilitates interaction with most HS-binding proteins. Not surprisingly, Glce-deficient mice (Glce−/−) display severe developmental defects resulting in a lethal phenotype, emphasizing the essential interplay between Glce and IdoA residues for polymer function (12, 13). In Caenorhabditis elegans, various HS-modifying enzymes have very specific effects in nervous system development, and a deficiency in Glce severely affects axonal development (14). In humans, the missense polymorphism SNP rs3865014 (A > G, Val597Ile) in the glce gene is associated with breast cancer susceptibility in Siberian women (15–17), consistent with the tumor-suppressor function of Glce in breast and lung carcinogenesis.
Crystal structures of zebrafish Glce (zGlce), unliganded and bound to a heparin-derived hexasaccharide, were recently reported (18). However, heparin is an inhibitor, not a substrate, of Glce, and the high level of 2- and 6-O sulfation might have impaired correct positioning of the hexasaccharide in the catalytic cleft. To decipher the mode of substrate binding and the catalytic mechanism of the epimerization reaction, we chemoenzymatically generated a series of GlcA-GlcNS disaccharide oligomers of various degrees of polymerization (dp6, dp8, and dp10) as natural substrates, and chemically synthesized the (IdoA-GlcNS)4 reaction product (19, 20). To document the mode of substrate binding and key residues involved in the epimerization mechanism, we solved crystal structures of human Glce (hGlce) in the unliganded form and of an inactive mutant as complexes with the dp10 substrate and the product, respectively, and recorded real-time NMR-based kinetics for hGlce and three active site mutants. We show that the O,3B/B1,4 constrained conformations of the GlcA/IdoA rings at the epimerization site (i) favor in-line abstraction/reprotonation on the axially oriented C5 hydrogen by two suitably poised acid/base residues; (ii) limit motion of the hexuronic acid carboxylic group; and (iii) facilitate the reaction by stabilizing high-energy boat conformers closely located on a conformational itinerary. Our data also reveal that Glce stabilizes a neutral enol intermediate, reminiscent of mandelate racemase, and preserves the glycosidic linkage while sharing part of its catalytic mechanism with heparin-sulfate lyases.
Results and Discussion
Overall Structures of hGlce.
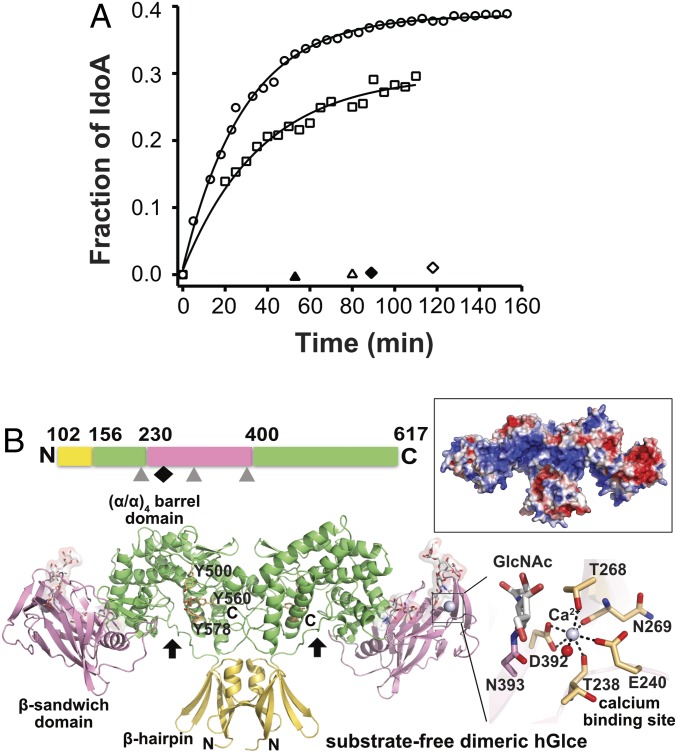
A recombinant, soluble form of hGlce (Val99-Asn617) was produced from a stabilized mammalian cell line and purified to homogeneity using complementary chromatography procedures (Materials and Methods). Real-time monitoring of the enzymatic activity of this recombinant hGlce by NMR spectroscopy using a 13C-labeled N-sulfated heparosan substrate revealed efficient conversion of GlcA to IdoA in the first 15 min to rapidly reach an equilibrium with a final IdoA content of ∼38% (Fig. 1A and SI Appendix, Table S1), a value consistent with the 1:3 IdoA/GlcA molar ratio reported for natural Glce from bovine liver or mouse mastocytoma (21, 22).
Fig. 1.
Enzymatic activity and overall structure of the substrate-free hGlce dimer. (A) 13C−1H correlation NMR assays of hGlce (40 nM; circles) and mutants Tyr500Phe (50 nM; squares), Tyr578Phe (40 nM; open triangles), and Glu499Gln (40 nM; open diamonds) incubated with 1 mg/mL of 13C-labeled N-sulfated heparosan (1 mg/mL). The GlcA-to-IdoA conversion, calculated by nonlinear fitting of the data to Equation S1 (SI Appendix, Supplementary Materials and Methods), reaches ∼38% for hGlce and ∼29% for the Tyr500Phe mutant (solid lines). No activity was detected for the Tyr578Phe and Glu499Gln mutants at 400 nM (filled triangle and diamond). (B, Top) Linear representation of the polypeptidic chain and three domains of soluble hGlce. The three N-glycosylation sites and the Ca2+ binding site are indicated by gray triangles and a black diamond, respectively. (B, Bottom) Overall view of the hGlce dimer with each subunit comprising a β-hairpin domain (yellow), a dominant (α/α)4-barrel domain (green), and a β-sandwich domain (pink) with a bound Ca2+ (violet sphere) and three N-glycans (white sticks, transparent surface). The three invariant Tyr residues possibly involved in catalysis are shown as orange sticks. The cleft is indicated by a black arrow. (Right) Close-up view of the pentagonal bipyramidal coordination geometry around the Ca2+ bound to the β-sandwich domain. (Top Right) Electrostatic potential distribution at the hGlce surface from −3 kT/e (red) to +3 kT/e (blue), showing the electropositive potential of the catalytic cleft.
The 2.5-Å resolution structure of hGlce in the apo form shows an overall dimeric assembly and well-defined electron densities for most of the protein regions, along with well-ordered glycan chains harboring up to six units at all three potential N-glycosylation sites (Asn225, Asn303, and Asn393) (Fig. 1B). Each subunit consists of three intertwined domains of a predominantly basic character, as observed for zGlce (18). The N-terminal 54-residue β-hairpin domain is composed of two upside-down/antiparallel β-hairpin motifs connected by a short helix. The 225-residue classical β-sandwich domain consists predominantly of a convex four-stranded β-sheet and a concave seven-stranded β-sheet. The dominant 285-residue C-terminal (α/α)4-barrel domain, which comprises the active site cleft, is composed of two noncontiguous regions and consists of four internal, mutually parallel α-helices interconnected by four external helices (Fig. 1B and SI Appendix, Fig. S1). The large β-sandwich domain is inserted between helices α1 and α2 from the α-barrel domain. The stable hGlce dimer forms by intersubunit swapping of the N-terminal β-hairpin and C-terminal α-helical barrel regions from each subunit. As a result, the subunit interface buries, to a 1.4-Å probe radius, a surface area of ∼3,100 Å2 on each subunit, a value consistent with the stable dimer of ∼130 kDa observed by gel filtration. The N-glycans are clustered at the distal ends of the dimer near the convex and concave faces of the β-sandwich domain. Opposite to the active site cleft, the β-sandwich and α-barrel domains are separated by a deep, narrow channel, 10 Å long and 3 Å wide, which could favor interdomain motions and provide functional plasticity to the overall assembly.
In each subunit, a Ca2+ is bound at the periphery of the β-sandwich domain, remote from the catalytic cleft but close to the Asn393-linked glycan, with a characteristic pentagonal bipyramidal geometry tightly coordinated by the invariant Glu240, Thr268, and Asp392 (bidentate ligation) side chains; the Thr238 and Asn269 main chain carbonyls; and a water molecule (Fig. 1B). Compared with zGlce, in which a water molecule occupies the same position as the bound Ca2+ in hGlce, large conformational adaptations of the β1-β2 loop region in the β-sandwich domain, with movements up to 9 Å, ideally position the hGlce Thr238 and Glu240 residues to satisfy the Ca2+ coordination geometry.
Structures of the hGlce Inactive Mutant Tyr578Phe Bound with a (GlcA-GlcNS)5 Substrate and a (IdoA-GlcNS)4 Product.
Previous structure-guided mutagenesis of active site residues in zGlce identified the three invariant residues Tyr468, Tyr528, and Tyr546, corresponding to hGlce residues Tyr500, Tyr560, and Tyr578, as crucial for the enzyme activity (Fig. 1B) (18). Consistently, no catalytic activity was detected by real-time NMR spectroscopy for the hGlce Tyr578Phe mutant assayed at up to 400 nM, compared with hGlce at 40 nM (Fig. 1A). We used this inactive mutant to solve complementary structures of a complex with the chemoenzymatically prepared (GlcA-GlcNS)5 substrate and a complex with the chemically synthesized (IdoA-GlcNS)4 product (SI Appendix, Figs. S2 and S3).
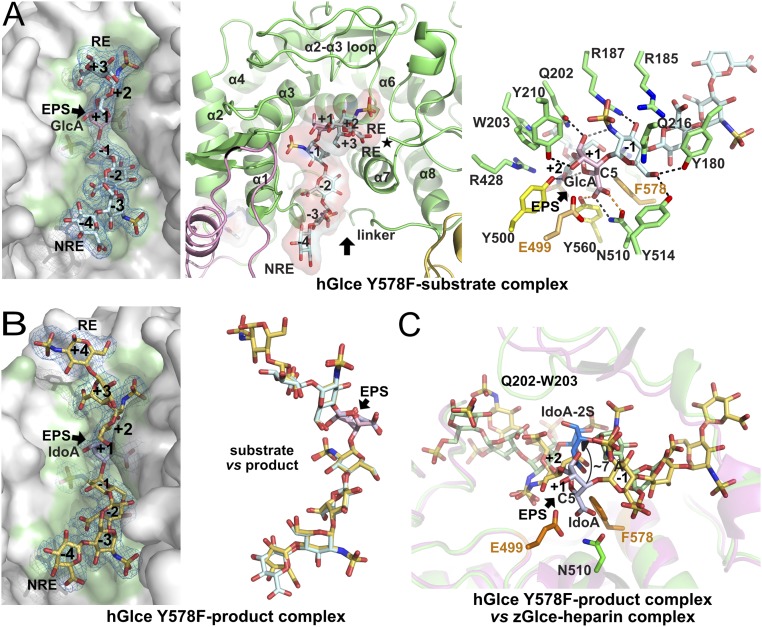
The 2.1-Å structure of the Tyr578Phe mutant bound with the dp10 substrate shows well-defined electron density maps for up to seven monosaccharide units (labeled −4 to +3 from the nonreducing end to the reducing end), indicative of a high occupancy for each of the two substrate molecules bound within the two active site clefts in the hGlce dimer (Fig. 2A). In each subunit, the negatively charged substrate is bound within a positively charged, elongated, and curved cleft that spans ∼25–30 Å in length (Figs. 1B and 2A). The cleft is formed at the N-terminal ends of the central helices from the α-barrel domain with strong participation of the sparsely structured Tyr158-Tyr210 linker that connects the β-hairpin to the α-barrel and helix α1. The composite nature of the catalytic cleft composed of residues from both subunits and intricate domains is illustrated by the absolute requirement of the C-terminal Asn617 residue at the end of helix α8 for substrate binding. In addition, an extended β-hairpin, which connects α2 and α3 from the α-barrel domain, projects over the active site cleft near the reducing end of the substrate (Fig. 2A). A similar overall conformation of the bound dp10 was observed in a 3.1-Å structure of hGlce also solved during this study, indicating that a lack of the hydroxyl group in the Phe578 mutant does not alter substrate binding.
Fig. 2.
Structures of hGlce bound to a substrate or a product and comparison with zGlce. (A, Left) Close-up view of the catalytic cleft of the Tyr578Phe mutant bound to heptasaccharide substrate with the unbiased electron density map contoured at 3.0 σ and the molecular surface of hGlce, highlighting the shape complementarity. (A, Center) Close-up view of the catalytic cleft showing the bound substrate and surrounding secondary elements. The asterisk denotes the C-terminus position. (A, Right) Close-up view of the substrate distortion in the GlcNS-1-GlcA+1-GlcNS+2 trisaccharide motif at the epimerization site with intra- (gray), inter- (black), and short H bonds (orange) interactions shown as dashed lines. (B) Close-up view of the bound (IdoA-GlcNS)4 product (light orange, violet sticks) (Left), same representation as in A, showing the conformational similarity with the substrate as shown in the overlay (Center). (C) Close-up view of the overlay of the hGlce-product complex, color-coded as in B, onto the zGlce-hexasaccharide heparin complex (pink cartoon and salmon heparin; PDB ID code 4PXQ), showing the large positional difference between the IdoA2S unit (blue) in zGlce and the corresponding IdoA unit (pink) at the hGlce epimerization site. The hGlce catalytic residues are shown as sticks.
At the epimerization site, the shape of the active site cleft imposes a sharply bent conformation that spans the central GlcNS-GlcA-GlcNS trisaccharide at the −1 to +2 subsites and constrains the GlcA unit at subsite +1 into a boat conformation in the vicinity of O,3B, as opposed to a predominant and more stable 4C1 chair conformation observed for the other GlcA rings. A dense network of interactions involving the GlcA endocyclic oxygen atom, the two transdiaxially oriented exocyclic O2-O3 hydroxyl groups, and the surrounding Gln202, Tyr210, Glu499, and Tyr500 side chains stabilize the distorted GlcA ring and bent trisaccharide conformation (Fig. 2A). The GlcA carboxyl group establishes short H bonds with the Asn510 Oδ and Tyr560 hydroxyl and faces the Asn510 side chain in a parallel arrangement. At subsite −1, the GlcNS moiety engages extensive polar interactions with a constellation of residues (Tyr180, Arg185, Arg187, Gln216, and Tyr514). Similarly, GlcNS at subsite +2 establishes numerous interactions with the Tyr500, Tyr560, mutated Phe578, and Arg575 side chains. As for subsite −1, the N-sulfate is anchored firmly to the Arg563 and Asn572 side chains and the Leu430 backbone carbonyl, highlighting the absolute requirement of N-sulfate groups vicinal to the epimerization site for substrate binding. At the reducing end, the carboxyl group of GlcA at subsite +3 interacts with the Gln202-Trp203 residue pair, and the O3 hydroxyl is tightly bound to the swapped Asn617 carboxyl terminus arising from the second subunit. The remaining three GlcA-GlcNS-GlcA units, which occupy the −2 to −4 subsites, respectively, at the nonreducing end, moderately interact with hGlce. At subsite-2, the GlcA ring stacks against the Tyr180 phenol ring and the O2 and carboxyl group interact with the Thr581 hydroxyl, Arg185 side chain, and Asn181 backbone amide. At subsite −3, only the N-sulfate group of GlcNS is bound to the backbone carbonyl of Gly179, leaving sufficient space to accommodate a GlcNAc unit, a feature consistent with an irreversible versus reversible mode of epimerization if a GlcNAc or GlcNS unit occupies site −3 (23). The distal GlcA residue at subsite −4 only establishes a weak van der Waals interaction with Val184. Overall, water-mediated contacts occur mainly near the substrate ends, highlighting the complementary shapes of the enzyme and the GlcNS-GlcA-GlcNS trisaccharide motif at the epimerization site.
The 2.45-Å structure of the Tyr578Phe mutant bound to the dp8 product shows well-defined electron density maps at the −4 to +4 subsites strikingly overlapping the corresponding units in the substrate complex, including the bent conformation of the central GlcNS-IdoA-GlcNS trisaccharide motif (Fig. 2B). Apart from small displacements of the ring atoms at both ends, the GlcA-to-IdoA epimerization process is not associated with significant conformational adaptation of the interacting residues. At the epimerization site, IdoA adopts a B1,4 boat conformation, close to the O,3B boat conformation of the corresponding GlcA in the hGlce-substrate complex (Figs. 2B and 3A). In contrast, the other IdoA rings adopt the standard and less strained 2SO skew-boat conformation. The short distance (∼3.0 Å) between the IdoA O2 hydroxyl and the adjacent GlcNS sulfamido group at subsite −1, along with the nearby Arg187 and Gln202 side chains, is incompatible with binding of a 2-O-sulfated IdoA-containing substrate, as reported previously (24). Similarly, the IdoA mode of binding at subsite −2 clearly prevents accommodation of a 2-O sulfate group, while it appears to be sterically tolerated at subsite +3. Consequently, the corresponding IdoA-2S in the zGlce-heparin structure is shifted by ∼7 Å away from the active site, associated with large movement of the loop tip harboring the Gln171-Trp172 residue pair (hGlce Gln202-Trp203) along with conformational adaptation of the nearby Arg156 guanidinium group (hGlce Arg187) (Fig. 2C) (18).
Fig. 3.
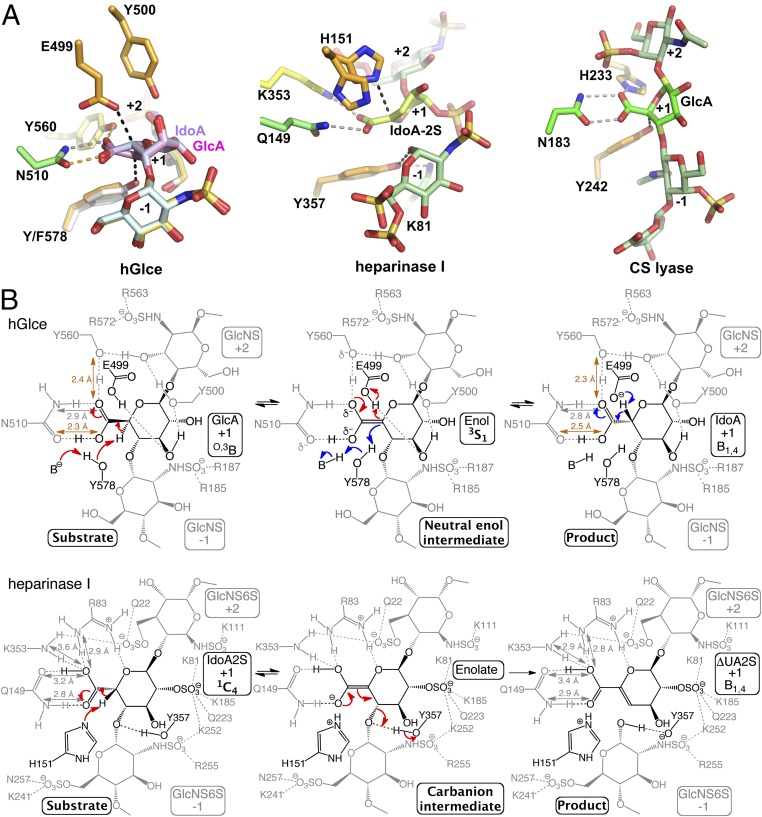
Proposed catalytic mechanism of Glce and comparison with lyases. (A) Structural comparison of the catalytic sites of hGlce (Left), heparinase I (Center; PDB ID code 3ILR) and chondroitin AC lyase (Right; PDB ID code 1RWG) viewed in similar orientation, showing the bound substrate/product and key active site residues. In all cases, the hGlce Asn510/Tyr560, heparinase I Gln149/Arg83/Lys353, and chondroitin AC lyase Asn183/His233 residues neutralize the uronic acid carboxyl group. (B) Comparison of the hGlce and heparinase I lyase catalytic mechanisms. (Top) hGlce catalyzes C5 epimerization of GlcA into IdoA (red arrows) by abstracting the GlcA C5 proton by the general base Tyr578 on one face of GlcA and donating a proton by the general acid Glu499 through a neutral enol intermediate on the opposite face. In the reverse reaction (blue arrows), the IdoA C5 proton is abstracted by deprotonated Glu499, while Tyr578 reprotonates the C5 atom on the opposite side of the uronic acid. (Bottom) Heparinase I catalyzes β-elimination of the glycosidic linkage between the −1 and +1 positions of the heparin substrate, with His151 and Tyr357 acting as a general base/acid through an anionic enolate intermediate (adapted with permission from ref. 33; permission conveyed through Copyright Clearance Center, Inc.). The H-bond network that neutralizes the carboxylate anion at the epimerization site and lowers the pKa value of the C5 hydrogen is evident for the two enzymes. The two low-barrier H bonds (orange) in hGlce are hallmarks of a neutral enol intermediate, while regular H bonds (grey) in heparinase I support an anionic enolate intermediate.
Catalytic Mechanism and Mutational Analysis.
Several C5-epimerases have been cloned and biochemically characterized (18, 25–28), and a reaction mechanism occurring via reversible abstraction and readdition of a proton at the GlcA/IdoA C5 position/atom, through an α-carbanion or enolate intermediate, has been proposed (29). Identification of the catalytic residues of Glce and of unique structural features in the substrate/product binding mode provides insight into the C5-epimerization mechanism marked by low-barrier H bonds (30). Moreover, stabilization of a neutral enol intermediate coupled with selective distortion of carbohydrate ring geometry along a particular catalytic puckering itinerary provide driving forces for the epimerization process. This study also reveals that the hGlce Tyr168 and Tyr222 residues, both of which are reportedly involved in catalysis (31), do not participate in substrate binding, but substitutions at these positions might affect protein folding.
The structures indicate that the Tyr578 phenolic oxygen is poised (OH-C5 distance, ∼2.7 Å) to function as a general base for abstracting the C5 proton of the GlcA unit (Fig. 3A), consistent with inactivity of the Tyr578Phe mutant (Fig. 1A). The epimerization mechanism requires neutralization of the negative charge on the GlcA carboxylate anion to reduce the pKa value of the C5 hydrogen (α-proton). In this context, hGlce uses a strategy similar to that of the chondroitin AC lyase (32) and heparin lyase I (33), with the Asn510 side chain and Tyr560 hydroxyl nearly coplanar to the GlcA carboxylic group (Fig. 3A). Such a topology requires a protonated acid group from which a neutral enol intermediate can efficiently form (Fig. 3B). Thus, the Tyr560 hydroxyl, which is not ideally positioned to act as a general base, is poised to provide a proton relay path to the solvent, consistent with inactivity of the corresponding Tyr528Phe mutant in zGlce (18). This mechanistic pathway resembles the Mg2+-dependent α-proton transfer reaction of carboxylic acid substrate catalyzed by mandelate racemase (34) and is kinetically more competent than the previously proposed reaction pathway involving formation of a α-carbanion or enolate intermediate (18, 21). On the opposite side of the GlcA/IdoA pyranose ring, the glutamate of the Glu499-Tyr500 residue pair, with an Oε-C5 distance of ∼2.8 Å compared with the OH-C5 distance of ∼3.8 Å in the product-bound structure, is the best candidate to serve as a general acid (Fig. 3A). The nearby Tyr500, Arg428, and possibly Tyr210 side chains could provide a proton relay with the solvent. This active site topology is consistent with the inactivity of the Glu499Gln mutant relative to hGlce, compared with a decreased activity of only ∼29% for the Tyr500Phe mutant (Fig. 1A and SI Appendix, Table S1). Most of the hGlce residues interacting with the substrate/product oligosaccharide, as well as the two catalytic glutamate and tyrosine residues, are conserved in eukaryotic d-glucuronyl C5-epimerase family and in the bacterial d-glucuronyl C5-epimerase (RED65) and the heparosan-glucuronate 5-epimerase (HG-5epi) (SI Appendix, Fig. S4) (35, 36), arguing for a similar C5 proton abstraction/readdition mechanism.
Structural comparison of the substrate- and product-bound mutant reveals a nearly identical conformation of the two GlcA/IdoA rings at the epimerization site, besides the two C5 atoms, distant by 0.8 Å. Thus, the active site of Glce has evolved to enable selective motion of the C5-epimerized center through the improper C5-O5-C6-C4 dihedral angle (Fig. 3A) while maintaining tighter conformation of the other GlcA/IdoA ring atoms and the two flanking GlcNS units. Most notably, ring distortion appears to act as an additional driving force for Glce activity. Indeed, the Cremer-Pople puckering parameters (37) of the substrate and product rings reveal unusual conformations in the vicinity of O,3B and B1,4 for the respective GlcA and IdoA units at the catalytic site, while the other GlcA/IdoA units fall within the most stable and populated 4C1/2SO conformations. The O,3B- and B1,4-constrained conformations of GlcA/IdoA not only destabilize the ground states, but also preactivate the substrate/product rings at subsite +1 by bringing them into closely related conformations while maintaining the collinearity and axial conformation of the C5-H5 bond. Indeed, the O,3B and B1,4 ring conformations lie on a conformational itinerary interconverting 4C1 GlcA to 2SO IdoA through the less-constrained skew boat 3S1 conformation suitable for a neutral enol intermediate (Fig. 3B and SI Appendix, Fig. S5A). Taken together, these data support previous kinetic isotope effects showing that readdition of a proton to the intermediate is the rate-limiting step of the epimerization reaction (21). Indeed, the higher-energy O,3B/B1,4 conformations of the GlcA/IdoA rings should permit a decrease in the activation energy needed for carboxylic acid tautomerization into a neutral enol intermediate that could adopt a more relaxed 3S1 conformation stabilized by low-barrier H bonds (SI Appendix, Fig. S5B). In fact, several oligosaccharide-degrading enzymes (e.g., lyases such as chondroitin AC lyase) are known to force the substrate to bind with the key uronic acid in an energetically less favorable nonchair conformation to improve catalytic efficiency.
The unified mechanism of action proposed for the HS lyases and epimerases (38), which involves a similar C5 proton abstraction, raises an intriguing question as to how Glce prevents glycosidic bond cleavage through a β-elimination reaction (31, 39). In the lytic reaction, the proton must be donated to the leaving group, while for the epimerase, it should occur on the C5 of the intermediate, emphasizing the need for accurate positioning of the general acid for proton donation (Fig. 3B). In Glce, the optimal position of the two catalytic residues on both sides of the GlcA/IdoA C5 atom and the lack of a general acid residue proximal to the otherwise “scissile” glycosidic bond support the concept of aborted β-elimination for uronate epimerases by preventing protonation of the glycosidic bond oxygen (Fig. 3B). Moreover, the dense intramolecular and intermolecular interaction network stabilizes the conformation of the target uronic acid along the O,3B-3S1-B1,4 conformational itinerary (Fig. 2A). Thus, the tetrahedral geometry of the C4 carbon is preserved, which prevents this carbon from converting into the flat form that would be required for the anionic E1cB elimination mechanism of lyases. However, Glce shows no significant sequence homology with Mg2+-dependent dermatan sulfate epimerase and Ca2+-dependent alginate epimerase, the other two types of epimerases acting on HexA residues in polysaccharides, suggesting that these epimerases have acquired a similar function by convergent evolution (40–42).
Molecular Bases of the N- and O-Sulfation Pattern Specificity of Glce.
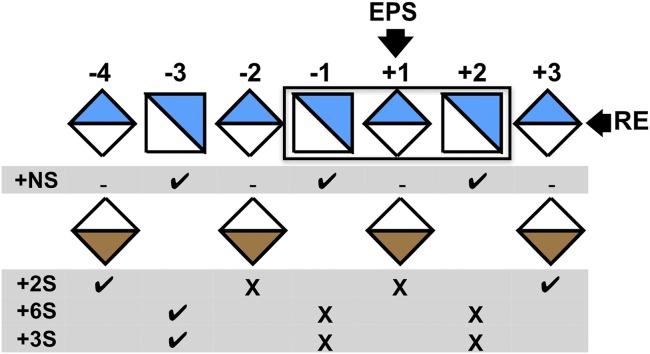
The structure of the hGlce-substrate complex is consistent with an octasaccharide being a substrate of minimum size for the epimerization reaction, since no activity was detected for a tetrasaccharide or hexasaccharide (24, 28). Moreover, the topology and distribution of polar and aromatic residues in the active site cleft impose unique structural constraints on the substrate/product conformations at the epimerization site, emphasizing the stringent substrate specificity of Glce relative to GlcN N-/6-O-sulfation and GlcA 2-O-sulfation on either side of the GlcA/IdoA units at subsite +1 (Fig. 4). Except for the GlcNS units located at the distal ends of the substrate/product, the N-sulfate groups of the two GlcNS units adjacent to the epimerization site are deeply anchored in the binding cleft, consistent with the absolute requirement for an N-sulfate group at subsite −1 for the epimerization reaction to occur (23, 43, 44). In contrast, O-sulfation at O2 of IdoA and or O6 or O3 of GlcN units at subsite −2 to +2 lead to HS ligand binding in a nonproductive, extended conformation only partially overlapping with substrate binding, as has been reported for the structure of the crystalline zGlce-heparin hexasaccharide complex (18), illustrating the molecular basis for substrate specificity and heparin inhibition by Glce (Fig. 2C).
Fig. 4.
Structure-based recognition pattern of N- and O-sulfated groups by hGlce. The scheme was drawn according to the Glce substrate specificity (24). The arrow points to the GlcA/IdoA unit at the epimerization site.
In summary, our structural analysis of apo hGlce and its substrate/product-bound inactive mutant has unveiled the catalytic mechanism of the HS C5 epimerases shared by other members of the family and clarified the structural bases by which Glce regulates HS structure and function. Although Glce appears to use many of the strategies used by polysaccharide lyases and mandelate racemases, including a distorted GlcA/IdoA ring geometry, we note subtle differences that likely evolved in response to unique chemical structures of the HS polymer. The structural variability observed for the HS precursor chain highlights the importance of conformational plasticity for HS structure-function activities. In addition, these structures provide a detailed atomic framework for future development of Glce inhibitors or chemoenzymatically synthesized heparin/HS analogs. Given the convergent mechanistic strategy between Glce and lyases, converting lyases into novel enzymes with epimerase activities could be considered, thereby extending current possibilities to alter uronate residues at the polymer level.
Materials and Methods
A soluble form of hGlce (Val99-Asn617; National Center for Biotechnology Information Gene ID 26035) and its Tyr500Phe and Tyr578Phe mutants were stably expressed in HEK-EBNA cells (from the National Research Center Canada, Biotechnology Research Institute), while the Glu499Gln mutant was transiently expressed from Expi293F cells. All proteins were purified to homogeneity using conventional procedures. The (GlcA-GlcNS)3–5 oligosaccharides were prepared from a mixture of dp6, dp8, and dp10 heparosan (GlcA-GlcAc) as described previously (45) and N-sulfated as described previously (28). The (IdoA-GlcNS)4 octasaccharide was prepared by chemical synthesis as described previously (20). Enzymatic activities of hGlce and its three mutants were recorded by real-time NMR spectroscopy using a 13C-labeled N-sulfated heparosan substrate as described previously (28). hGlce in the apo form and the Tyr578Phe mutant bound with the dp10 substrate or the dp8 product were crystallized by vapor diffusion. The structures were solved by molecular replacement or rigid-body motions and refined to 2.5 Å, 2.1 Å, and 2.45 Å, respectively.
Detailed descriptions of the procedures for protein expression and purification, preparation of oligosaccharides, recording of enzyme activity, and crystallization and X-ray crystallography are provided in SI Appendix, Materials and Methods. Statistics are summarized in SI Appendix, Table S2. The atomic structures and structure factors have been deposited in the Protein Data Bank (PDB ID codes 6HZZ, 6I01, and 6I02) (46–48).
Supplementary Material
Acknowledgments
We thank R. Lebrun (Proteomics Facility, Mediterranean Institute of Microbiology) for protein analyses, G. Sulzenbacher (Architecture et Fonction des Macromolécules Biologiques) for assistance with data collection, B. Henrissat and P. Marchot (Architecture et Fonction des Macromolécules Biologiques) for fruitful discussions, S. Fushinobu for sharing his web tool for calculating Cremer-Pople puckering parameters, and the European Synchrotron Radiation Facility and SOLEIL synchrotrons for beam time allocation and their beam line staff for assistance with data collection. This work was supported in part by the CNRS in the framework of the Groupe de Recherche sur les Glycosaminoglycanes (GDR 3739), Aix-Marseille University, the French Infrastructure for Integrated Structural Biology (ANR-10-INBS-05), the LabEx Grenoble Alliance for Integrated Structural & Cell Biology (ANR-10-LABX-49-01), and the LabEx Research on Medication and Innovative Therapeutics (ANR-10-LABX-33) under the Idex Paris-Saclay (ANR-11-IDEX-0003-01).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. U.L. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6HZZ, 6I01, and 6I02).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818333116/-/DCSupplemental.
References
- 1.Lindahl U, Couchman J, Kimata K, Esko JD. In: Proteoglycans and Sulfated Glycosaminoglycans. 3rd Ed. Varki A, et al., editors. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 2015. pp. 207–222. [Google Scholar]
- 2.Poulain FE, Yost HJ. Heparan sulfate proteoglycans: A sugar code for vertebrate development? Development. 2015;142:3456–3467. doi: 10.1242/dev.098178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3:a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esko JD, Selleck SB. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 5.Kreuger J, Kjellén L. Heparan sulfate biosynthesis: Regulation and variability. J Histochem Cytochem. 2012;60:898–907. doi: 10.1369/0022155412464972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu D, Esko JD. Demystifying heparan sulfate–protein interactions. Annu Rev Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imberty A, Lortat-Jacob H, Pérez S. Structural view of glycosaminoglycan–protein interactions. Carbohydr Res. 2007;342:430–439. doi: 10.1016/j.carres.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Kjellén L, Lindahl U. Specificity of glycosaminoglycan–protein interactions. Curr Opin Struct Biol. 2018;50:101–108. doi: 10.1016/j.sbi.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Casu B, Lindahl U. Structure and biological interactions of heparin and heparan sulfate. Adv Carbohydr Chem Biochem. 2001;57:159–206. doi: 10.1016/s0065-2318(01)57017-1. [DOI] [PubMed] [Google Scholar]
- 10.Capila I, Linhardt RJ. Heparin–protein interactions. Angew Chem Int Ed Engl. 2002;41:391–412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 11.Rong J, Habuchi H, Kimata K, Lindahl U, Kusche-Gullberg M. Substrate specificity of the heparan sulfate hexuronic acid 2-O-sulfotransferase. Biochemistry. 2001;40:5548–5555. doi: 10.1021/bi002926p. [DOI] [PubMed] [Google Scholar]
- 12.Li J-P, et al. Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking l-iduronic acid and in neonatal lethality. J Biol Chem. 2003;278:28363–28366. doi: 10.1074/jbc.C300219200. [DOI] [PubMed] [Google Scholar]
- 13.Feyerabend TB, Li J-P, Lindahl U, Rodewald H-R. Heparan sulfate C5-epimerase is essential for heparin biosynthesis in mast cells. Nat Chem Biol. 2006;2:195–196. doi: 10.1038/nchembio777. [DOI] [PubMed] [Google Scholar]
- 14.Bülow HE, Hobert O. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron. 2004;41:723–736. doi: 10.1016/s0896-6273(04)00084-4. [DOI] [PubMed] [Google Scholar]
- 15.Grigorieva E, et al. Decreased expression of human d-glucuronyl C5-epimerase in breast cancer. Int J Cancer. 2008;122:1172–1176. doi: 10.1002/ijc.23203. [DOI] [PubMed] [Google Scholar]
- 16.Prudnikova TY, et al. Antiproliferative effect of d-glucuronyl C5-epimerase in human breast cancer cells. Cancer Cell Int. 2010;10:27. doi: 10.1186/1475-2867-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belyavskaya VA, et al. GLCE rs3865014 (Val597Ile) polymorphism is associated with breast cancer susceptibility and triple-negative breast cancer in Siberian population. Gene. 2017;628:224–229. doi: 10.1016/j.gene.2017.07.054. [DOI] [PubMed] [Google Scholar]
- 18.Qin Y, et al. Structural and functional study of d-glucuronyl C5-epimerase. J Biol Chem. 2015;290:4620–4630. doi: 10.1074/jbc.M114.602201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dilhas A, Lucas R, Loureiro-Morais L, Hersant Y, Bonnaffé D. Mixture synthesis and “charge tagging”-based demixing: An efficient strategy for the preparation of heparan sulfate libraries. J Comb Chem. 2008;10:166–169. doi: 10.1021/cc8000019. [DOI] [PubMed] [Google Scholar]
- 20.Lubineau A, Lortat-Jacob H, Gavard O, Sarrazin S, Bonnaffé D. Synthesis of tailor-made glycoconjugate mimetics of heparan sulfate that bind IFN-gamma in the nanomolar range. Chemistry. 2004;10:4265–4282. doi: 10.1002/chem.200306063. [DOI] [PubMed] [Google Scholar]
- 21.Hagner-Mcwhirter A, Lindahl U, Li Jp. Biosynthesis of heparin/heparan sulphate: Mechanism of epimerization of glucuronyl C-5. Biochem J. 2000;347:69–75. [PMC free article] [PubMed] [Google Scholar]
- 22.Malmström A, et al. Biosynthesis of heparin: Partial purification of the uronosyl C-5 epimerase. J Biol Chem. 1980;255:3878–3883. [PubMed] [Google Scholar]
- 23.Sheng J, Xu Y, Dulaney SB, Huang X, Liu J. Uncovering biphasic catalytic mode of C5-epimerase in heparan sulfate biosynthesis. J Biol Chem. 2012;287:20996–21002. doi: 10.1074/jbc.M112.359885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobsson I, et al. Biosynthesis of heparin: Substrate specificity of heparosan N-sulfate d-glucuronosyl 5-epimerase. J Biol Chem. 1984;259:1056–1063. [PubMed] [Google Scholar]
- 25.Li J, et al. Biosynthesis of heparin/heparan sulfate: cDNA cloning and expression of d-glucuronyl C5-epimerase from bovine lung. J Biol Chem. 1997;272:28158–28163. doi: 10.1074/jbc.272.44.28158. [DOI] [PubMed] [Google Scholar]
- 26.Li JP, Gong F, El Darwish K, Jalkanen M, Lindahl U. Characterization of the d-glucuronyl C5-epimerase involved in the biosynthesis of heparin and heparan sulfate. J Biol Chem. 2001;276:20069–20077. doi: 10.1074/jbc.M011783200. [DOI] [PubMed] [Google Scholar]
- 27.Crawford BE, Olson SK, Esko JD, Pinhal MA. Cloning, Golgi localization, and enzyme activity of the full-length heparin/heparan sulfate-glucuronic acid C5-epimerase. J Biol Chem. 2001;276:21538–21543. doi: 10.1074/jbc.M100880200. [DOI] [PubMed] [Google Scholar]
- 28.Préchoux A, Halimi C, Simorre J-P, Lortat-Jacob H, Laguri C. C5-epimerase and 2-O-sulfotransferase associate in vitro to generate contiguous epimerized and 2-O-sulfated heparan sulfate domains. ACS Chem Biol. 2015;10:1064–1071. doi: 10.1021/cb501037a. [DOI] [PubMed] [Google Scholar]
- 29.Prihar HS, et al. Biosynthesis of heparin. Hydrogen exchange at carbon 5 of the glucuronosyl residues. Biochemistry. 1980;19:495–500. doi: 10.1021/bi00544a016. [DOI] [PubMed] [Google Scholar]
- 30.Cleland WW. Low-barrier hydrogen bonds and low fractionation factor bases in enzymatic reactions. Biochemistry. 1992;31:317–319. doi: 10.1021/bi00117a001. [DOI] [PubMed] [Google Scholar]
- 31.Li K, Bethea HN, Liu J. Using engineered 2-O-sulfotransferase to determine the activity of heparan sulfate C5-epimerase and its mutants. J Biol Chem. 2010;285:11106–11113. doi: 10.1074/jbc.M109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lunin VV, et al. High-resolution crystal structure of Arthrobacter aurescens chondroitin AC lyase: An enzyme-substrate complex defines the catalytic mechanism. J Mol Biol. 2004;337:367–386. doi: 10.1016/j.jmb.2003.12.071. [DOI] [PubMed] [Google Scholar]
- 33.Han Y-HH, et al. Structural snapshots of heparin depolymerization by heparin lyase I. J Biol Chem. 2009;284:34019–34027. doi: 10.1074/jbc.M109.025338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bearne SL, St Maurice M. A paradigm for CH bond cleavage: Structural and functional aspects of transition state stabilization by mandelate racemase. Adv Protein Chem Struct Biol. 2017;109:113–160. doi: 10.1016/bs.apcsb.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Raedts J, Lundgren M, Kengen SWM, Li J-P, van der Oost J. A novel bacterial enzyme with d-glucuronyl C5-epimerase activity. J Biol Chem. 2013;288:24332–24339. doi: 10.1074/jbc.M113.476440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochizuki H, Yamagishi K, Suzuki K, Kim YS, Kimata K. Heparosan-glucuronate 5-epimerase: Molecular cloning and characterization of a novel enzyme. Glycobiology. 2015;25:735–744. doi: 10.1093/glycob/cwv013. [DOI] [PubMed] [Google Scholar]
- 37.Cremer D, Pople JA. General definition of ring puckering coordinates. J Am Chem Soc. 1975;97:1354–1358. [Google Scholar]
- 38.Gacesa P. Alginate-modifying enzymes: A proposed unified mechanism of action for the lyases and epimerases. FEBS Lett. 1987;212:199–202. [Google Scholar]
- 39.Shaya D, et al. Crystal structure of heparinase II from Pedobacter heparinus and its complex with a disaccharide product. J Biol Chem. 2006;281:15525–15535. doi: 10.1074/jbc.M512055200. [DOI] [PubMed] [Google Scholar]
- 40.Valla S, Li J, Ertesvåg H, Barbeyron T, Lindahl U. Hexuronyl C5-epimerases in alginate and glycosaminoglycan biosynthesis. Biochimie. 2001;83:819–830. doi: 10.1016/s0300-9084(01)01313-x. [DOI] [PubMed] [Google Scholar]
- 41.Maccarana M, et al. Biosynthesis of dermatan sulfate: Chondroitin-glucuronate C5-epimerase is identical to SART2. J Biol Chem. 2006;281:11560–11568. doi: 10.1074/jbc.M513373200. [DOI] [PubMed] [Google Scholar]
- 42.Thelin MA, et al. Biological functions of iduronic acid in chondroitin/dermatan sulfate. FEBS J. 2013;280:2431–2446. doi: 10.1111/febs.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagner-McWhirter A, Li J-P, Oscarson S, Lindahl U. Irreversible glucuronyl C5-epimerization in the biosynthesis of heparan sulfate. J Biol Chem. 2004;279:14631–14638. doi: 10.1074/jbc.M313760200. [DOI] [PubMed] [Google Scholar]
- 44.Hagner-McWhirter A, et al. Biosynthesis of heparin/heparan sulfate: Kinetic studies of the glucuronyl C5-epimerase with N-sulfated derivatives of the Escherichia coli K5 capsular polysaccharide as substrates. Glycobiology. 2000;10:159–171. doi: 10.1093/glycob/10.2.159. [DOI] [PubMed] [Google Scholar]
- 45.Barreteau H, Richard E, Drouillard S, Samain E, Priem B. Production of intracellular heparosan and derived oligosaccharides by lyase expression in metabolically engineered E. coli K-12. Carbohydr Res. 2012;360:19–24. doi: 10.1016/j.carres.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 46.Debarnot C, et al. 2018 Structure of human d-glucuronyl C5 epimerase. Protein Data Bank. Available at https://www.rcsb.org/structure/6HZZ. Deposited October 25, 2018.
- 47.Debarnot C, et al. 2018 Structure of human d-glucuronyl C5 epimerase in complex with substrate. Protein Data Bank. Available at https://www.rcsb.org/structure/6I01. Deposited October 25, 2018.
- 48.Debarnot C, et al. 2018 Structure of human d-glucuronyl C5 epimerase in complex with product. Protein Data Bank. Available at https://www.rcsb.org/structure/6I02. Deposited October 25, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.