Abstract
Background
Long noncoding RNA (lncRNA) is a key part of noncoding RNA class and increasing evidences have manifested that it plays a significant role in the physiology and pathology. The growth arrest-specific transcript 5 (GAS5) is a vital tumor suppressor in some types of cancers. However, the function of GAS5 in lung cancer remains largely no clear. The purpose of the current study was to identify the biological role of GAS5 in non-small cell lung cancer (NSCLC).
Material/Methods
To study the role of GAS5 in the NSCLC, the RT-PCR, Western Blot, Luciferase assay, and RNA immunoprecipitation assay was employed to determine the relationship of GAS5, miR-205, and PTEN. CCK8 assay, Cell migration and invasion assay was used for the role of GAS5 in lung cancer cell proliferation and metastasis.
Results
The results indicated that GAS5 was drastically downregulated in lung cancer cell lines. Further functional analysis showed that down-expression of GAS5 remarkably induced NSCLC growth, migration, and invasion. The luciferase reporter assays determined that miR-205 was a direct target of GAS5 in lung cancer. Moreover, the Phosphatase and tensin homologue (PTEN) was known as a direct target of miR-205 and miR-205/PTEN rescued the effects of GAS5 in NSCLC cells.
Conclusions
To sum up, our results illustrate that upregulation of GAS5 in NSCLC suppresses its growth, migration, and invasion via the miR-205/PTEN axis.
MeSH Keywords: Carcinoma, Non-Small-Cell Lung; MicroRNAs; PTEN Phosphohydrolase; RNA, Long Noncoding
Background
Lung cancer, also known as carcinoma of the lung, is one of the most common malignant tumors, which is the second most common cause of cancer and the second largest cause of cancer related mortality in the United States [1]. Non-small cell lung cancer (NSCLC) is the most familiar histological type of lung cancer, which accounts for 84% of lung cancers [2]. Because NSCLC patients, whose five-year survival rate was 18%, have been diagnosed already at the end stage. In the early stage of NSCLC, it is hard to detect and this cancer is difficult to treat. NSCLC may result from comprehensive action of multiple factors and the pathogenesis remains largely unknown.
Long noncoding RNA (lncRNA) is a class of noncoding RNA transcripts, which the length is about greater than 200 nts [3]. Recent studies have demonstrated that lncRNA was involved in transcription, post-transcriptional, and epigenetic mechanisms underlie that a variety of pathophysiological processes of the body is regulated by it [4–7]. Many lncRNAs were found to be involved in the context of NSCLC [8–12], including lncRNA growth arrest-specific 5 (GAS5). Recent results showed that the level of GAS5 was increased in tumor tissues of NSCLC patients as an oncogene; it suppressed tumorigenesis in NSCLC [13]. Therefore, it was interesting that whether GAS5 could be the important factor in NSCLC cells.
Here, we used qRT-PCR to identify GAS5 downregulated and miR-205 downregulated in NSCLC cells in the research. Our results indicate that GAS5 is as a ceRNA of miR-205, and that the GAS5 regulation of miR-205 plays a key role in controlling NSCLC progression by targeting and downregulating PTEN. Moreover, the GAS5-miR-205-PTEN axis may offer a new candidate network for NSCLC therapy.
Material and Methods
Cell culture and treatment
The normal cell line 16-HBE and the 6 lung cancer cell lines – A549, H460, 95D, H1299, SPC-A-1, and H522 – were purchased from ATCC. All cell lines were cultured in RPMI 1640 medium (Gibco, China) with 10% fetal bovine serum (FBS) and 1*PS, which is composed of 100 IU/ml penicillin and 100 U/ml streptomycin. The cells were cultured in an incubator with 5% CO2 at 37°C. RNA interference and cell transfection were performed 3 times each.
RNA interference and cell transfection
The plasmid vectors containing GAS5 sequence (pcDNA-GAS5) were constructed by Qiagen (USA). The specific RNA interference sequences for GAS5 (GAS5 siRNAs) were purchased from RiboBio (Qiagen, USA). NSCLC cells (2×105 cells/well) were placed in 6-well plates for 24 h. NSCLC cells were transfected by the plasmid vectors or GAS5 siRNAs according to the manufacturer’s instructions, using Lipofectamine 2000 transfection reagent (Invitrogen). The concentration of GAS5 siRNAs was 20 nM.
Total RNA extraction and RT-PCR
For all cell lines, TRIzol reagent (Invitrogen, CA, USA) was used for extracting the total RNA using an RNA Plus kit from Tiangen (Beijing, China) according to the manufacturer’s protocols. We reversely transcribed 1 μg RNA into cDNA by superscript reverse transcriptase (Tiangen, Beijing, china) at 42°C for 1 h. After that, all the samples were assessed by FastFire qPCR PreMix (Tiangen, Beijing, China). Reaction condition was set at 37°C for 30 min and 5 s at 85°C and then maintained at 4°C. The melting curves were examined to measure the specificity of the products. U6 was used as internal control. The primers were: miR-205 forward, 5′-CTTGTCCTTCATTCCACCGGA-3′ and reverse, 5′-TGCCGCCTGAACTTCACTCC-3′. GAS5 forward, 5′-ACAGGCATTAGACAGAAAGC-3′ and reverse, 5′-TACCCAAGCAAGTCATCCA-3′. GAPDH: Forward, 5′-CAA AGG TGG ATC AGA TTC AAG-3′ and reverse, 5′-GGT GAG CAT TAT CAC CCA GAA-3′. The expression of target genes was calculated using the 2−ΔΔCt method.
Western blot analysis
The level of PTEN protein in all the cell lines was measured using Western blot assay. Total protein (25 μg/sample) was separated by SDS/PAGE, then transferred onto PVDF membranes (Millipore). Blocking liquid was used for blocking the membranes for 1 h at room temperature. Specific primary antibodies were used for incubating the PVDF membranes at 4°C overnight, and then the secondary antibodies were added, followed by incubation at room temperature for 2 h. Anti-PTEN antibodies (1: 1000), anti-GAPDH antibodies (1: 5000), and secondary antibodies (1: 10 000) were purchased from Abcam. The protein bands were visualized by ECL reagents (Thermo Biotech) and the relative density was quantitated via ImageJ software.
Cell migration and invasion assay
Both of the kits were purchased from Biyuntian. The migration and invasion assays of normal cell culture serum were done according to the manufacturer’s protocols. The cells were incubated in the Transwell chambers for 1 h, and then the cells in the chambers without and with Matrigel were fixed with 3.7% formalin and washed twice. Cells were transferred to 100% methanol, and washed twice again. Lastly, the cells were stained by Giemsa at room temperature for 15 min in the dark. Photographs were taken, and the number of cells in the plate was quantitated by ImageJ software.
CCK8 assay
CCK8 was used to estimate cell proliferation. We added 10 ul of CCK8 solution to every well and then incubated the plates for 2 h at 37°C. The cell viability was analyzed by absorbance determined at 450 nm.
Luciferase assay
The dual-luciferase assay was used to assess the relationship of PTEN, GAS5, and miR-205, according to the manufacturer’s instructions. The 3′-untranslated regions (UTR) of PTEN and GAS5 containing the wild and miR-205 binding sequences and its mutant were synthesized by Qiagen (USA).
RNA immunoprecipitation
RNA immunoprecipitation (RIP) was performed using a kit from Millipore. The complete RIP lysis buffer was used to lysis the cells transfected with GAS5 or miR-205 vector. The human anti-Ago2 antibody (1: 1000) or negative control were employed to conjugate with magnetic beads in RIP buffer, then RIP buffer was used to incubated with 100 ml of cell lysates. Proteinase K buffer was used for digesting the lysates. Afterwards, immunoprecipitated RNA was extracted and purified. Finally, we performed RT-PCR to analyze the levels of miR-205 and GAS5.
Statistical analysis
All data were analyzed by using the Statistical Product and Service Solutions (SPSS) 21.0 software. The independent 2-tailed t test was used to assess differences between NSCLC cells and controls. Statistical analyses were done for pairs of samples by use of the paired-sample t test. P<0.05 was regarded as significant.
Results
miR-205 was upregulated but GAS5 was downregulated in NSCLC
Six cell lines were used for the NSCLC. The level of miR-205 and GAS5 in those 6 NSCLC cell lines and in the normal lung cell line 16-HBE was examined using the RT-PCR assay. As shown in Figure 1, the level of GAS5 in the 6 NSCLC cell lines was significantly higher than the level of 16-HBE, especially in A549, H460, and H522 cell lines. In contrast, miR-205 expression in those NSCLC cell lines was remarkably downregulated compared to 16-HBE, especially in A549, H460, and H522 cell lines. Our results show that A549, H460, and H522 cell lines were much more sensitive to the expression changes of GAS5 and miR-205 than were the other cell lines.
Figure 1.
Level of miR-205 and GAS5 in NSCLC. (A, B) RT-PCR was performed to estimate the expression levels of miR-205 (A) and GAS5 (B) in cell lines. * P<0.05.
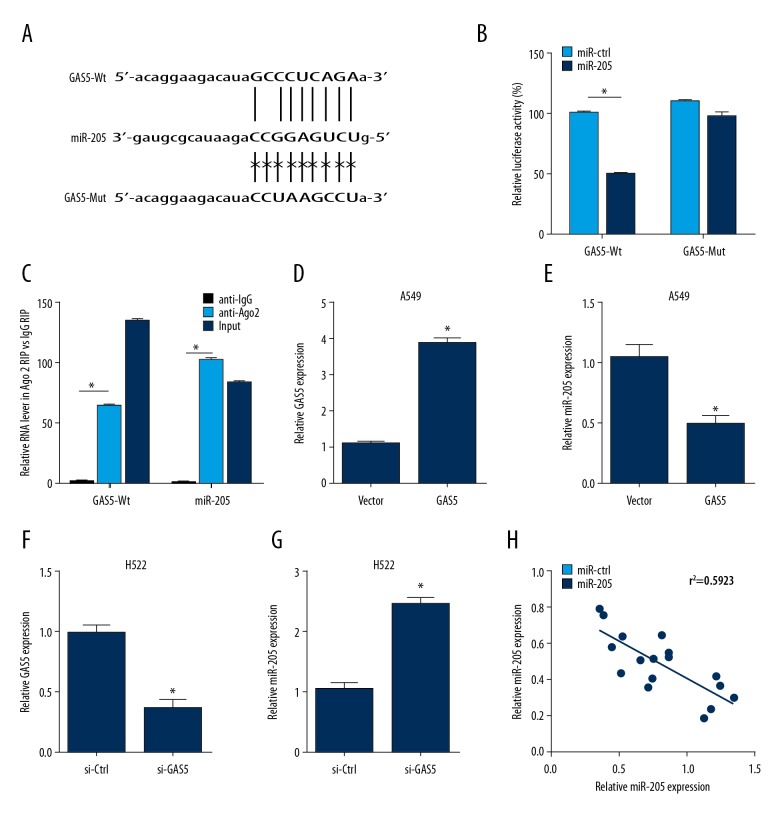
GAS5 is as a ceRNA of miR-205 in NSCLC cells
The has-pri-miR-205 is located at chromosome 1q32.2, and a putative Gas5-binding site is in its sequence (Figure 2A). For verification, GAS5-Wt, GAS5-Mut, miR-205, or miR-NC was transfected in A549 cells, and the luciferase reporter gene assays showed that GAS5 increased the fluorescence activity (Figure 2B). Moreover, RIP assay indicated that GAS5 and miR-205 upregulation led to increased level of Ago2 in GAS5 and miR-205 levels (Figure 2C). To further study the effect of GAS5 on miR-205, A549 cells were transfected with GAS5 and null vector, and H522 cells were transfected with si-ctr and si-GAS5 inhibitors. RT-PCR was performed to analyze the mRNA levels of GAS5 and miR-205. The results showed that GAS5 markedly decreased the mRNA expression of miR-205 in the A549 and H522 cells transfected with GAS5 (P<0.001) (Figure 2D–2G). Furthermore, our data demonstrated that GAS5 expression was negatively correlated with the level of miR-205 in the 6 NSCLC cell lines (Figure 2H).
Figure 2.
Relationship of GAS5 with miR-205 in NSCLC cells. (A) Wild-type miR-205 binding sites and the corresponding mutant in GAS5. (B) Luciferase activity of A549 cells transfected with GAS5-Wt, GAS5-Mut, miR-205, or miR-NC. (C) RIP assay was used in A549 cell, the co-precipitated RNAs of GAS5 and miR-205, which were determined by RT-PCR. (D–G) RT-PCR assay was used to measure the level of GAS5 and miR-205 in A549 cells (D, E) and H522 cells (F, G). (H) Negative correlation between GAS5 and miR-205 expression. * P<0.05.
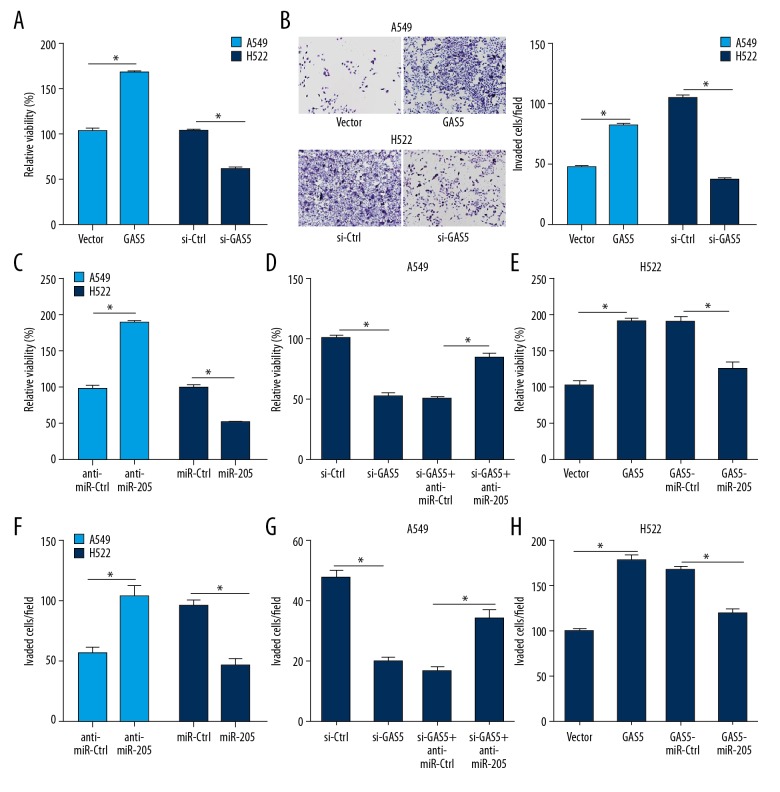
GAS5 regulated NSCLC proliferation and cell invasion, partially through miR-205
To elucidate the relationship between GAS5 and miR-205, assessed whether GAS5 inhibits cell proliferation via miR-205 in NSCLC cells. Our results indicated that GAS5 inhibited cell proliferation in the A549 and H522 cells (Figure 3A). As shown in Figure 3B, GAS5 suppressed cell migration and cell invasion in the A549 and H522 cells (Figure 3B). Furthermore, we found that miR-205 induced cell proliferation in the A549 and H522 cells (Figure 3C), and GAS5 suppressed cell proliferation via miR-205 in the A549 and H522 cells (Figure 3D, 3E). As shown in Figure 3F, miR-205 promoted cell migration and cell invasion in the A549 and H522. Similarly, GAS5 inhibited the cell migration and cell invasion via miR-205 in the A549 and H522 cells (Figure 3G, 3H).
Figure 3.
NSCLC cell viability and invasion were regulated by GAS5. (A) CCK-8 assay was used in A549 cells transfected with si-NC or si-GAS5 and H522 cells transfected with Vector or GAS5 for NSCLC proliferation. (B) Cell invasion assay was used in A549 cells. (C–E) CCK-8 assay was used to assess cell viability in A549 and H522 cells lines. (C) miR-ctrl or miR-205 was transfected in A549 cell lines, anti-miR-Ctrl, or anti-miR-205 in H522 cells; (D) Vector, GAS5, or together with miR-NC or miR-205 in A549 cell lines; and (E) si-NC, si-GAS5, or combined with anti-miR-NC or anti-miR-205 in H522 cells. (F–H) Cell invasion assay was used to assess invasiveness in A549 and H522 cells. (F) miR-Ctrl or miR-205 was transfected in A549 cell lines, anti-miR-Ctrl, or anti-miR-205 in H522 cells; (G) Vector, GAS5, or together with miR-NC or miR-205 in A549 cells; and (H) si-NC, si-GAS5, or combined with anti-miR-NC or anti-miR-205 in H522 cells. * P<0.05.
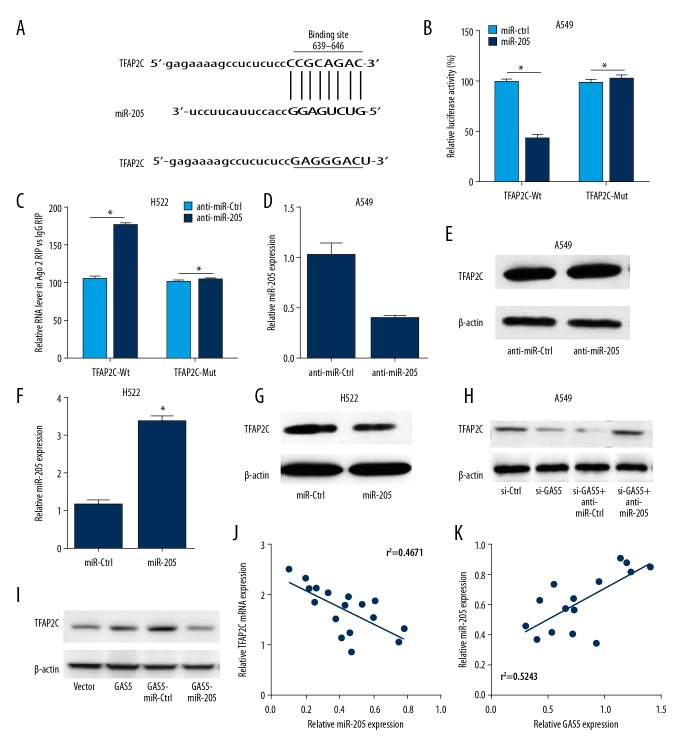
GAS5 upregulated the PTEN level via sponging miR-205 in NSCLC cells
Figure 4A shows the putative miR-205 targets of 3′UTR of the PTEN mRNA as shown by luciferase reporter plasmid, with a PTEN 3′-UTR containing the putative miR-205 targeting sites. The luciferase activity of the pMir-PTEN-3′-UTR-WT plasmid was significantly reduced by about 45% (P<0.01; Figure 4B, 4C), but overexpression of miR-205 did not regulate the luciferase activity in NSCLC cells transfected with the pMirPTEN-3′-UTR-Mut plasmid, which contains the miR-205 mutated targeting site (Figure 4B, 4C). In addition, we found that the level of PTEN protein was significantly suppressed and increased in NSCLC cells transfected with miR-205 and anti-miR-205, respectively (Figure 4D–4G). Moreover, GAS regulates the miR-205 expression in lung cancer cells (Figure 4H, 4I). Figure 4J and 4K show the PTEN mRNA expression had a significant negative relationship with the level of miR-205 in various lung cancer cell lines. Our results show that miR-205 inhibits PTEN expression by specifically interacting with PTEN mRNA and triggering its translational regulation.
Figure 4.
PTEN expression was upregulated by GAS5 via sponging miR-205 in NSCLC cells. (A) The putative wild-type miR-205 binding sites and the mutated miR-205 binding sites in the 3′UTR of PTEN mRNA. (B, C) Luciferase activity was detected by use of luciferase reporter assay in A549 and H522 cells. (B) The A549 cell lines were co-transfected with PTEN-3′UTR-Wt or PTEN-3′UTR-Mut and miR-NC or miR-205. (C) H522 cells with PTEN-3′UTR-Wt or PTEN-3′UTR-Mut and anti-miR-NC or anti-miR-205. (D, F) RT-PCR was employed to quantify the expression of miR-205 in A549 and H522 cells transfected with anti-miR-205 or anti-miR-NC (D) and miR-205 or miR-NC. (E, G–I) Western blot assay was conducted to detect the protein level of PTEN in A549 cells and H522 cells, (E) anti-miR-205 or anti-miR-NC in A549 cells, (G) miR-205 or miR-NC in H522 cells, (H) si-NC, si-GAS5, or co-transfected with anti-miR-NC or anti-miR-205 in A549 cells, and (I) Vector, GAS5, or co-transfected with miR-NC or miR-205 in H522 cells. (J, K) Correlation between miR-205 and PTEN mRNA expression in 6 NSCLC cell lines (J) is negative or (K) positive. *P<0.05.
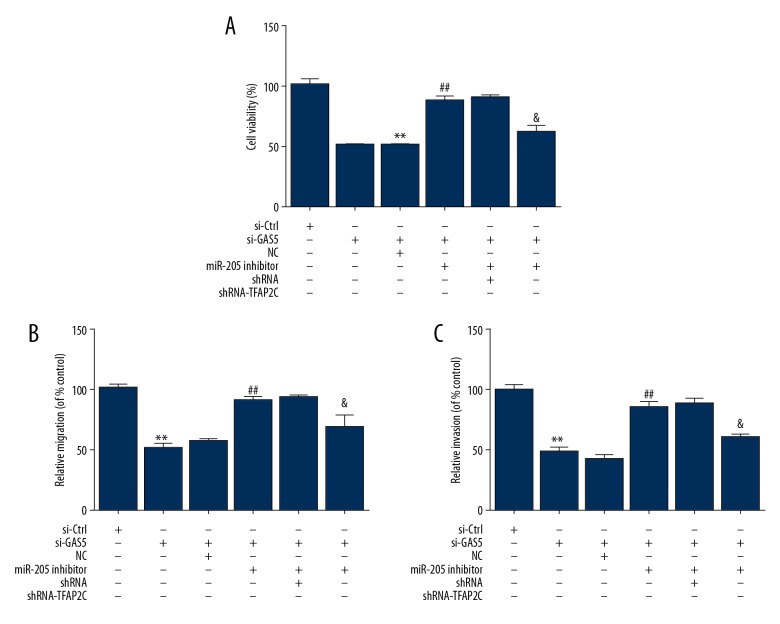
Upregulation of GAS5 inhibited NSCLC cell proliferation, migration, and invasion by miR-205/PTEN axis
To further study whether the GAS5-miR-205-PTEN network is involved in regulating NSCLC cell function, we performed CCK8 assay, Transwell migration assay, and Transwell invasion assay, showing that GAS5 transfection significantly reduced the cell viability, cell migration, and cell invasion of A549 cells, whereas overexpression of miR-205 partially countered these repressive effects (Figure 5A–5C). We also determined that overexpression of PTEN significantly decreased the cell viability, cell migration, and cell invasion of NSCLC cells (Figures 5A–5C).
Figure 5.
NSCLC cell proliferation, migration, and invasion were suppressed via upregulation of GAS5 via miR-205/PT axis EN. (A) CCK-8 assay was used to assess cell viability in A549 cells transfected with GAS5, miR-205, and PTEN. (B, C) A549 cells were transfected with GAS5, miR-205, and PTEN for 72 h, and cell migration and cell invasion were analyzed. ** P<0.01.
Discussion
Based on the abnormal level and multiple factors of lncRNAs in lung diseases and the roles of lncRNAs in human cancers, lncRNAs were confirmed to be a new therapeutic target for NSCLC. The specific one is lncRNA GAS5, which acts as a tumor suppressor that inhibits growth and suppresses the tumor cell migration and cell invasion in multiple cancer cell types [14–17]. In the present study, our results showed a novel role of GAS5 in human NSCLC cells. We demonstrated that GAS5 is decreased and miR-205 expression is increased in these 6 NSCLC cell lines. Overexpression of GAS5 can attenuate NSCLC cells induced via downregulation of the miR-205 level. GAS5 is a ceRNA of miR-205 that regulates the PTEN signaling pathway.
The expression and biological factors of GAS5 were also examined in NSCLC cells. Its endogenous level was downregulated in cancerous tissues and is associated with worse clinicopathological factors. Gain-of-function and loss-of-function experiments both revealed that GAS5 was vital for growth arrest and apoptosis of NSCLC cell lines [18]. In the present study, our data show that miR-205 plays a significant role in NSCLC progression, and also is a downstream target of GAS5. Moreover, we found that the level of miR-205 decreased in GAS5 overexpressed cells compared to controls. Thus, we chose miR-205 as the target for further study.
To study whether miR-205 can influence PTEN expression in lung cancer cells, we found that PTEN is a possible target gene of miR-205. The genes of PTEN are always deleted, mutated, and suppressed in many kinds of cancers [19]. Regulation of PTEN levels in animal models of cancer has been broadly analyzed, and it is proven to be haplo-insufficient [20] for preventing tumor initiation and progression [21,22]. Remarkably, the protein of PTEN has been discovered to be lost or low in many kinds of cancers [23]. Recent research found that the level of miR-205 affects ovarian cancer progression via suppressing PTEN by the direct targeting of the 3′-UTR of PTEN [24]. Our results strongly suggest that miR-205 suppresses PTEN in lung cancer cells by targeting the mRNA 3′-UTR to inhibits its translation. Additionally, overexpression of PTEN significantly prevents migration and invasion of lung cancer cells in vitro when transfected with miR-205.
In this study, our data showed the level of GAS5, miR-205, and PTEN level in 6 NSCLC cell lines and normal lung cell line. Statistically significant differences in GAS5 and PTEN levels between high and low miR-205 classes were found. The roles of GAS5 and miR-205 may be cell background-dependent in regulating human NSCLC cells. These results also indicate that the GAS5-miR-205-PTEN axis in the regulation of human NSCLC cells may be affected by cell background.
Conclusions
Our results show that overexpression of long noncoding RNA GAS5 downregulates miR-205 to suppress lung cancer progression-related phenotypes by downregulating PTEN and that GAS5 relays the induced effects of miR-205 via PTEN in NSCLC cells. We demonstrated a GAS5-controlled regulatory network in lung cancer, and found that the GAS5-miR-205-PTEN axis may provide a target pathway for lung cancer therapy.
Footnotes
Source of support: National Natural Science Foundation of China (NSFC, No. 81672650)
Conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Foss KM, Sima C, Ugolini D, Weiss GJ, et al. miR-1254 and miR-574-5p: Serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6:482–88. doi: 10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 3.Spizzo R, Almeida MI, Colombatti A, et al. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene. 2012;31:4577–87. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li G, Zhang H, Wan X, et al. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014 doi: 10.1155/2014/780521. 780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol. 2013;425:3723–30. doi: 10.1016/j.jmb.2012.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta. 2013;1832:1613–23. doi: 10.1016/j.bbadis.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Shao Y, Ye M, Jiang X, et al. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120(21):3320–28. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 9.Isin M, Ozgur E, Cetin G, et al. Investigation of circulating lncRNAs in B cell neoplasms. Clin Chim Acta. 2014;431:255–59. doi: 10.1016/j.cca.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 10.Ren S, Wang F, Shen J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49:2949–59. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 11.Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. Biomed Res Int. 2013;2013 doi: 10.1155/2013/136106. 136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber DG, Johnen G, Casjens S, et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6:518. doi: 10.1186/1756-0500-6-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi X, Sun M, Liu H, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer’. Mol Carcinog. 2015;54(Suppl 1):E1–2. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 14.Hajjari M, Salavaty A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12(1):1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y, Zhang L, Wang Y, et al. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014;35(10):9531–38. doi: 10.1007/s13277-014-2523-7. [DOI] [PubMed] [Google Scholar]
- 16.Yan L, Zhou J, Gao Y, et al. Regulation of tumor cell migration and invasion by the H19/let-7 axis is antagonized by metformin-induced DNA methylation. Oncogene. 2014;34(23):3076–84. doi: 10.1038/onc.2014.236. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Wang Q, Qiang Q, et al. Sp1-mediated transcriptional regulation of MALAT1 plays a critical role in tumor. J Cancer Res Clin Oncol. 2015;141(11):1909–20. doi: 10.1007/s00432-015-1951-0. [DOI] [PubMed] [Google Scholar]
- 18.Shi X, Sun M, Liu H, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog. 2015;54(Suppl 1):E1–12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 19.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–69. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwabi-Addo B, Giri D, Schmidt K, et al. Haploinsufficiency of the PTEN tumor suppressor gene promotes prostate cancer progression. Proc Natl Acad Sci USA. 2001;98:11563–68. doi: 10.1073/pnas.201167798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trotman LC, Niki M, Dotan ZA, et al. PTEN dose dictates cancer progression in the prostate. PLoS Biol. 2003;1(3):E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leslie NR, Foti M. Non-genomic loss of PTEN function in cancer: Nin my genes. Trends Pharmacol Sci. 2011;32:131–40. doi: 10.1016/j.tips.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Chu P, Liang A, Jiang A, Zong L. miR-205 regulates the proliferation and invasion of ovarian cancer cells via suppressing PTEN/SMAD4 expression. Oncol Lett. 2018;15(5):7571–78. doi: 10.3892/ol.2018.8313. [DOI] [PMC free article] [PubMed] [Google Scholar]