Abstract
Background
Healthcare workers (HCWs) are significant vectors for transmission of multidrug- resistant organisms among patients in intensive care units (ICUs). We studied ICU patients on Contact Precautions, colonized with vancomycin-resistant Enterococcus (VRE) to assess whether bacterial burden is associated with transmission to HCWs’ gloves or gowns, a surrogate outcome for transmission to subsequent patients.
Methods:
From this prospective cohort study we analyzed 96 VRE colonized ICU patients and five HCWs per patient. We obtained samples from patients’ perianal, skin, and stool to assess bacterial burden and cultured HCWs’ gloves and gowns for VRE after patient care.
Results
Seventy-one of 479 (15%) HCW-patient interactions led to HCWs’ glove or gown contamination with VRE. HCW contamination was associated with VRE burden on the perianal swab (OR: 1.37 [95% CI 1.19, 1.57]); skin swabs (OR: 2.14 [95% CI: 1.51, 3.02)]; and in stool (OR: 1.95 [95% CI: 1.39, 2.72]). Compared to colonization with E. faecalis, colonization with E. faecium was associated with higher bacterial burden and higher odds of transmission to HCWs.
Discussion
We show that ICU patients with higher bacterial burden are more likely to transmit VRE to HCWs. These findings have implications for VRE de-colonization and other infection control interventions.
Keywords: vancomycin-resistant Enterococcus, bacterial burden, healthcare workers, Contact Precautions
Summary:
We found that the risk of gown and glove contamination increases as patient vancomycin-resistant Enterococcus burden increases. Further, we found that patients colonized with E. faecium were nine times as likely to transmit as those colonized with E. faecalis.
Background
Enterococcus makes up 14% of all hospital-associated infections (HAI) reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention [1]. Roughly 35.5% of all enterococcal HAIs are resistant to vancomycin (VRE) [1, 2]. VRE is responsible for bacteremia, surgical site infections, and urinary tract infections [2] and the cause of approximately 1,300 deaths annually [3]. The two predominant species of VRE are E. faecium and E. faecalis, which make up roughly 77% and 9% of isolates, respectively [3].
The majority of patient colonization remains unknown in the absence of active surveillance [4]. However, VRE has a demonstrated propensity for skin colonization, which can increase the risk of catheter-related bacteremia and cross-infection [5]. Patients’ skin colonization can also lead to transmission of VRE to the healthcare workers’ (HCWs’) hands and clothing while providing patient care. This is especially concerning as VRE has been shown to last up to 60 minutes on HCWs’ hands in the absence of hand hygiene [6]. This contamination of HCWs can lead to further transmission of VRE to patients’ other body sites (cross contamination), the patient’s environment, or to other patients in the HCWs’ care [7].
Whether the bacterial burden of VRE increases the risk of transmission is thus far unknown. While several studies have found that colonized patients can transmit MDROs, including VRE, to HCWs’ gloves and gowns, these studies suffered from small samples sizes [8–11] and associations between patient bacterial burden or colonizing species and HCW contamination were not estimated. Therefore, we conducted a study of ICU patients colonized with VRE to assess the relationship between bacterial burden and transmission to HCWs’ gloves or gowns. These results could have major implications for infection prevention practices, such as the use of Contact Precautions and de-contamination policies for patients with high levels of bacterial colonization.
Methods
Study design and participants
We conducted a prospective cohort study to estimate the contribution of bacterial burden on the transmission of VRE from patients to HCWs’ gloves and gowns, a surrogate outcome for possible transmission to other patients. One hundred patients were recruited from the medical and surgical ICUs within the University of Maryland Medical Center between January 1 and November 15, 2017. The medical ICU is a 29-bed unit that provides care to adult patients with acute or potentially life-threatening medical conditions, while the surgical ICU is a 24-bed unit designed to care for adult surgical patients. These ICUs screen for VRE on rectal swabs at admission and once weekly as part of the VRE infection prevention active surveillance program. The research staff were notified each day of patients with recent (within 72 hours) VRE-positive rectal surveillance cultures via email alerts associated with hospital microbiology reports. The first five HCWs for each patient were approached sequentially for participation in the study before engaging in care activities. The University of Maryland Baltimore Institutional Review Board approved this study.
Data collection
We cultured the patients’ perianal area, chest, antecubital fossa, and obtained a stool sample when available to measure patient VRE bacterial burden. The perianal area was sampled using aseptic technique with ESwab (Copan Diagnostics, Murrieta, CA). The swab was rubbed gently back and forth three times on the skin immediately around the anus, covering an area approximately four centimeters (cm) in diameter. The chest and antecubital fossa were chosen because these body sites are likely to be touched by HCWs and have been examined in previous studies [5, 8, 10]. These skin sites were sampled using a 10×10 cm2 template, rubbing the swab within the template with a twirling motion to ensure all sides of the swab came in contact with the skin. Stool samples were collected, when available, in Dynarex (Orangeburg, NY) sterile stool specimen containers.
Five interactions between HCWs and patients were observed shortly after (within a few hours) obtaining the patient swabs. Following patient care, but prior to doffing, the gloves and gown of each HCW were cultured for the presence of VRE. The BBL dual Culturette swab (BBL, BD, Sparks, MD) was rubbed gently with a twirling motion along the dorsum of each finger and the palm of both the right and left hand. HCWs’ gowns were sampled with a separate swab, using a twirling motion twice on each forearm and then in a “W” pattern along the beltline.
The HCW role and duration of time each spent in the room was observed by the study researchers. Patient characteristics including the presence or absence of an artificial airway (endotracheal or tracheostomy tube), Foley catheter, central venous catheter (central line), chest tube, surgical drain, rectal tube, nasogastric tube, diarrhea, and wound were also collected. International Classification of Diseases, 10th Revision, (ICD-10) codes, age, sex, and race, were abstracted from the electronic medical record of each patient. The ICD-10 codes were used to calculate the Elixhauser Index, a validated comorbidity score for hospital inpatients [12].
Laboratory procedures
HCWs’ gown and glove swabs were cultured for the presence of VRE. The swabs were placed into tryptic soy broth with 6.5% NaCl and incubated for 24 hours at 35± 2°C. After incubation, 50 μL from each broth tube were inoculated onto a Bile Esculin Azide Agar with 6 μg/ml vancomycin (BEAV; Remel, Lenexa, KS) plate for isolation. The BEAV plates were incubated aerobically at 35± 2°C for 48 hours. All enterococcal isolates were frozen in tryptic soy broth with 15% glycerol and stored at −80°C.
Patient swabs and stool samples were placed into collection tubes and the swabs from the skin and perianal samples were vortexed separately for one minute. One gram of stool was extracted from the stool container, added to 1mL of 0.9% saline in an Eppendorf tube, and vortexed until well mixed (at least one minute). One mL of each patient sample was serially diluted using Butterfield’s Buffer. BEAV was inoculated with 100 μL of each serial dilution and distributed evenly onto the agar plate using a cell spreader. Also, 100 μL of the original sample was inoculated into tryptic soy broth with 6.5% NaCl. The plates and broth tubes were incubated for 48 hours aerobically at 35± 2°C, after which the number of bacterial colonies were counted. If there was no growth on the inoculated plates, 100 μL from the previously inoculated tryptic soy broth with 6.5% NaCl tubes were inoculated onto a BEAV plate. If there was growth on the BEAV agar after 48 hours at 35± 2°C, a count of one colony forming unit (CFU) was given. Following incubation, colonies were subcultured and identified to the species level by the VITEK II (BioMérieux, Marcy-l’Étoile, France). Antimicrobial susceptibility testing was conducted using the Kirby-Bauer disk diffusion method according to the Clinical Laboratory Standards Institute guidelines.
Statistical analysis
Frequencies with proportions and means with standard deviations (SD) were calculated to describe the demographics, clinical characteristics, and VRE species of the sample. Bacterial counts were logged transformed (log10 [VRE bacterial burden +1]) so that those without recovered VRE were included as 0, and counts were modeled in log10 CFU/mL for the perianal and stool samples, and log10 CFU/cm2 in the skin samples. The chest and antecubital fossa skin swabs were combined into one variable by taking the higher of the two measurements. The association between patient bacterial burden and VRE species with HCWs’ glove or gown contamination was estimated using logistic regression fit with generalized estimating equations with an exchangeable correlation matrix to account for within-patient correlation. These associations were expressed in odds ratios (OR) and 95% CI. The final model was selected by testing all covariates in the model and the model with the smallest QIC (quasi- Akaike’s Information Criterion) was chosen. Separate models were constructed for VRE burden recovered from each of the cultured patient sites. The following covariates were selected a priori as potential confounders: patient age, race, comorbidities, presence of invasive devices, diarrhea, type of HCW (physician/nurse practitioner, nurse, patient care technician, physical/occupational therapist, respiratory technician, or other) and duration of time HCW spent in the room. We estimated the mean bacterial burden and 95% confidence interval (CI) found in each of the samples by species. Pearson’s correlations were calculated to compare the bacterial burden between each sample.
All analyses were conducted using SAS version 9.4 (The SAS Institute, Cary, NC).
Results
Of the 100 patients enrolled, chest and antecubital fossa skin samples were available from 96 patients, perianal swabs from 94, and stool samples from 43. The demographics and clinical characteristics of the patients are presented in Table 1. The mean patient age was 61 years (SD: 13), 51/97 (53%) were white, 50/97 (52%) were men, 58/97 (60%) were from the medical ICU, and the median number of comorbidities was 6 (range 0 to 14) as measured by the Elixhauser Index. Most patients had at least one invasive device (93%), with a mean of 3 devices (SD: 1.6).
Table 1.
Demographics and characteristics of VRE colonized ICU patients enrolled between January 1, 2017 and November 15, 2017
| Characteristic | N=96 |
|---|---|
| Age in years, mean (SD) | 60.8 (13) |
| White race | 51 (53) |
| Male sex, n (%) | 50 (52) |
| ICU location, n (%) | |
| Medical ICU | 58 (60) |
| Surgical ICU | 38 (40) |
| Elixhauser Index, median (range) | 6 (0 – 14) |
| Diarrhea, n (%) | 29 (30) |
| Wound, n (%) | 56 (58) |
| Endotracheal tube, n (%) | 50 (52) |
| Central line, n (%) | 68 (71) |
| Foley catheter, n (%) | 55 (57) |
| Chest tube, n (%) | 7 (7) |
| Surgical drain, n (%) | 25 (26) |
| Rectal tube, n (%) | 32 (33) |
| Nasogastric tube, n (%) | 53 (55) |
| Number of devices, mean (SD) | 3 (1.6) |
Abbreviations: ICU, intensive care unit; n, number; SD, standard deviation; and VRE, vancomycin-resistant Enterococcus...
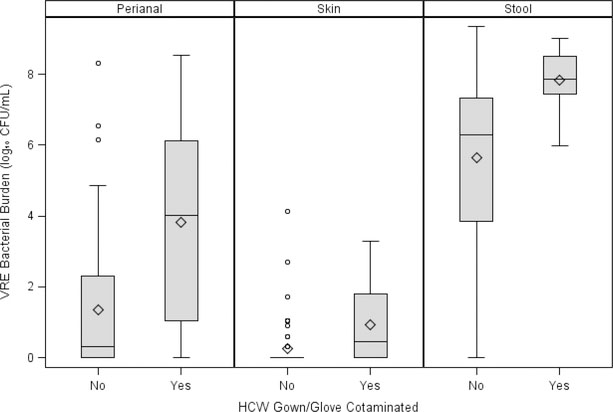
We observed 479 HCW-patient interactions, of which 71/479 (15%) led to HCWs’ glove or gown contamination with VRE. Figure 1 shows that patients who transmit VRE to HCWs have higher bacterial distributions in their perianal, skin, and stool samples. Table 2 presents the adjusted odds ratios and 95% CI for HCWs’ glove and gown contamination for each patient sample type, controlling for nasogastric tube, diarrhea, age, and amount of time the HCW spent in the room. In each patient sample there is an association between increasing bacterial burden and HCWs’ glove or gown contamination: adjusted OR (aOR) 1.37 (95% CI 1.19, 1.57) for the perianal, aOR: 1.95 (95% CI: 1.39, 2.72), for the stool, and aOR: 2.14 (95% CI: 1.51, 3.02) for the skin samples. This association between skin colonization and HCWs’ glove or gown contamination did not change after adjusting for the stool bacterial burden (data not shown).
Figure 1. Bacterial distributions of each sample by transmission to HCWs’ gloves or gowns.
Abbreviations: CFU, colony forming units; HCW, healthcare worker; mL, milliliter; and VRE, vancomycin-resistant Enterococcus.
Table 2.
Adjusted associations between VRE bacterial burden and HCWs’ glove or gown contamination by patient sample type
| Patient sample type | OR (95% CI) |
|---|---|
| Perianal (log10 CFU/mL) | 1.37 (1.19, 1.57)* |
| Skin (log10 CFU/cm2) | 2.14 (1.51, 3.02)* |
| Stool (log10 CFU/mL) | 1.95 (1.39, 2.72)† |
Abbreviations: CFU, colony forming units; CI, confidence interval; CM, centimeter; HCW, healthcare worker; mL, milliliter; OR, odds ratio; and VRE, vancomycin-resistant Enterococcus.
Adjusted for nasogastric tube, diarrhea, age, and time spent in the room by the healthcare worker
Adjusted for nasogastric tube, age, and time spent in the room by the healthcare worker
The frequency and mean bacterial burden by VRE species and patient sample type is presented in Table 3. VRE was identified in 60 of the 94 perianal samples collected, ranging from 1 to 1.30 × 106 CFU/mL. Patients who were colonized with E. faecium had an average of 2.4 log10 (95% CI: 0.89, 3.91) colony counts higher than those with E. faecalis. VRE was identified on 42 of the 43 collected stool samples, ranging from 0 to 2.25 × 109 CFU/mL. Those colonized with E. faecium in the stool had 1.6 log10 (95% CI: −0.39, 3.52) higher bacterial burden than those with E. faecalis. VRE was identified in 18 of the 96 samples collected from the patients’ chest, with bacterial burden ranging from 1 to 1.91 × 103 CFU/cm2. VRE was identified on 23 of the antecubital fossa swabs obtained from 95 patients, ranging from 1 to 1.33 × 104 CFU/cm2. Patients colonized with E. faecium on the skin had 0.97 log10 (95% CI: 0.04, 1.89) higher than those with E. faecalis. The amount of bacteria found in the stool was moderately correlated with the amount found in the perianal sample (r=0.56, p<0.001) and mildly correlated with the amount found on the skin (r=0.31, p<0.001).
Table 3.
Frequency, mean bacterial burden, and odds ratios for transmission to HCWs’ gloves and gowns by VRE species and patient sample type
| Patient sample type | VRE species* | n/Total (%) | Mean burden† (95% CI) | OR (95% CI) for transmission to HCWs‡ |
|---|---|---|---|---|
| Perianal | E. faecium | 49/59 (83%) | 3.92 (3.30, 4.55) | 1.98 (0.53, 7.41) |
| E. faecalis | 10/59 (17%) | 1.52 (0.16, 2.89) | Reference | |
| Stool | E. faecium | 36/40 (90%) | 7.09 (6.49, 7.69) | 1.61 (0.37, 6.89) |
| E. faecalis | 4/40 (10%) | 5.53 (3.72, 7.34) | Reference | |
| Skin | E. facecium | 26/31 (84%) | 1.68 (1.31, 2.06) | 9.32 (1.32, 66.03) |
| E. faecalis | 5/31 (16%) | 0.72 (−0.13, 1.57) | Reference |
Abbreviations: CFU, colony forming units; CI, confidence interval; CM, centimeter; HCW, healthcare worker; mL, milliliter; n, number; OR, odds ratio; and VRE, vancomycin-resistant Enterococcus.
Other species not listed were identified as E. avium and E. casseliflavus
Perianal and stool bacterial loads are expressed in Log10 CFU/mL. Skin bacterial load is expressed in Log10 CFU/cm2
Adjusted for bacterial burden found in that sample
VRE species was also associated with increased HCWs’ glove and gown contamination, controlling for bacterial burden (Table 3). Patients colonized with E. faecium on the skin were 9.32 (95% CI: 1.32, 66.03) times as likely to contaminate HCWs’ gowns or gloves as those colonized with E. faecalis. There was increased odds of contamination for those colonized with E. faecium compared to E. faecalis in the perianal and stool samples as well, but these associations did not reach statistical significance.
Discussion
This study is the first of its size to quantify VRE bacterial burden and examine its role in the transmission of VRE from colonized patients to HCWs’ gloves or gowns. As bacterial burden in all patient samples increases so does the likelihood of HCWs’ glove and gown contamination, a potential source of transmission to other patients in the ICU. Previous studies have not examined the association between VRE species and transmission potential. Our results indicate the main driver of this transmission is likely due to E. faecium, which is associated with higher colony counts than E. faecalis. Patients colonized with E. faecium on their skin were nine times as likely to transmit the bacteria to HCWs as those colonized with E. faecalis after adjusting for bacterial burden.
Though we were only able to recover VRE from a small proportion of skin samples (27% from the antecubital fossa and 19% from the chest), there was a 114% increase in HCWs’ glove and gown contamination for each Log10 increase in skin bacterial burden. This strong association remained even after adjustment for stool bacterial burden. These results indicate that skin may be an efficient means of VRE transfer. Duckro et al. found that the antecubital fossa was the most efficient body site for VRE transmission [10]. In that study, HCWs contaminated their gloves 100% of the time after touching the patient’s antecubital fossa, compared to 60% after contacts with the patient’s chest [10]. Our study also found higher bacterial burden on the patients’ arms than on their chests. The patient’s antecubital fossa is touched often by HCWs during clinical care (e.g. for blood draws and blood pressure measurement), resulting in increased bacterial burden and transmission efficiency. As has been suggested previously, this area may also be a habitable environment for VRE [5].
These results show skin contamination increases the odds of VRE transmission and highlight the need for ICUs to invest in decontamination protocols, such as bathing with chlorhexidine gluconate (CHG), for their VRE colonized patients. CHG bathing has been shown to be associated with reductions in the incidence of VRE acquisition and gram-positive bacteremias [7, 13, 14]. In 2007, Vernon et al. [7] performed a single center clinical trial comparing the effect of three types of bathing routines on VRE acquisition. Compared to soap and water, daily bathing of patients with 2% CHG-saturated clothes resulted in a 60% decrease in VRE acquisition, a 40% reduction of HCWs’ glove contamination, and 70% reduction in environmental contamination [7]. Further, the use of CHG bathing resulted in a decrease of inguinal bacterial burden by 2.5 log10 colony counts [7]. The multicenter, cluster-randomized trial conducted by Climo et al. [13] in 2013 similarly found that daily CHG bathing reduced overall MDRO acquisition by 23% and bloodstream infections by 28% compared to cleansing with non-antimicrobial washcloths. Bleasdale et al. [14], also found that daily CHG bathing compared to soap and water baths leads to a reduction in hospital-acquired bloodstream infections. However, this trial and one by Noto et al. [15] did not see a reduction in other HAIs such as urinary tract infection, ventilator-associated pneumonia, or Clostridium difficile. There was no reduction in incidence of bloodstream infections either, though duration of the CHG intervention was only 10 weeks compared to the 24+ weeks in other trials [15]. The longer duration of these other trials may have reduced colonization pressure in the ICU by reducing the amount of bacteria in the environment over time. Patients in MICU and SICU in the present study are bathed with CHG once every 24 hours per hospital policy. The results of the above trials imply that the low levels of patient skin contamination in our study may be due to frequent CHG bathing..
Our results indicate that the association with perianal bacterial burden and transmission to HCWs is not as strong as the association seen in the stool or skin samples. The odds of HCWs’ glove or gown contamination increases by 37% for each Log10 increase in VRE isolated from the perianal swab. Even though a greater number of colonies were isolated from the perianal sample than the skin, this area is likely not often accessed by HCWs for routine procedures in the ICU. We found a nearly a two-fold increase in the odds of HCWs’ contamination for each Log10 increase in stool bacterial burden. Large concentrations of VRE in the stool have been previously found to correlate with skin and environmental contamination [16].
Potential limitations of this study include the fact that we sampled only a portion of the gloves and gowns instead of using a using alternative methods to culture the entire surface area. The transmission rate of VRE may be much higher than we were able to detect. However, we did culture from areas most likely to come in contact with the patient (glove fingers and gown arms), which are most likely involved in transmission. In addition, we did not quantify the bacterial burden recovered from the HCWs’ gloves and gowns. As such, we do not know how much VRE was transferred to HCWs or what amount of VRE is needed for transmit to future patients. Though, had these HCWs not been wearing personal protective equipment (i.e. gowns and gloves), they would have had VRE on their hands and clothing which could lead to a subsequent transmission to other patients. This study was conducted at a single site and in two ICUs. Transmission of VRE between patients and HCWs may vary in other acute care settings due to differences in patient care practices. However, these findings will likely be generalizable to other large-sized, academic hospitals and ICUs with similar patient case-mix. Furthermore, the findings of this study may not be generalizable to other organisms as transmission mechanisms may differ between pathogens.
This study is the first of its size to study the role of VRE bacterial load in transmission to HCWs. We only sampled from ICUs in our hospital that conducted active surveillance to minimize selection bias. Clinical cultures are often ordered when a patient shows clinical signs and symptoms of an infection. Bias may be introduced if patients identified from clinical cultures are included as these patients may have different risk factors for transmission than patients identified by surveillance cultures. The former patients may be sicker, have greater number of comorbidities and devices, and may be higher transmitters than patients identified through surveillance cultures.
This study demonstrates the role bacterial burden plays in transmission. In the absence of active surveillance, the majority of VRE colonized patients are unknown to HCWs. Those with higher bacterial burden are more likely to transmit VRE to HCWs in the absence of personal protective equipment. The role of E. faecium in transmission should be explored further. These results also have major implications for infection prevention practices that aim to lower VRE levels and decrease transmission. Examples include increased CHG bathing for VRE colonized patients in the ICU or for future work on alteration of the human microbiome that could lower levels of VRE colonization.
Acknowledgments.
We thank the study participants, without whom this research would not be possible; Lisa Pineles for her help with the study logistics; and Shirley Goodman and Alyssa Sbarro for their help with data collection; and Nicole Karikari, Licheng Zhao, and Gwen Paszkiewicz for specimens processing.
Financial support. This work was supported by the National Institute of Allergy and Infectious Disease at the National Institutes of Health [grant number 5R01AI121146] and the Centers for Disease Control and Prevention [grant number 1U54CK000450], US Department of Health and Human Services.
Footnotes
Potential conflicts of interest. All authors, no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Sievert DM, Ricks P, Edwards JR, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2009–2010. Infect Control Hosp Epidemiol, 2013; 34: 1–14. [DOI] [PubMed] [Google Scholar]
- 2.Weiner LM, Webb AK, Limbago B, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2011–2014. Infection Control & Hospital Epidemiology, 2016; 37: 1288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC). Antibiotic resistance threats in the united states. 2013.
- 4.Shadel BN, Puzniak LA, Gillespie KN, Lawrence SJ, Kollef M, Mundy LM. Surveillance for vancomycin-resistant enterococci: Type, rates, costs, and implications. Infect Control Hosp Epidemiol, 2006; 27: 1068–75. [DOI] [PubMed] [Google Scholar]
- 5.Beezhold DW, Slaughter S, Hayden MK, et al. Skin colonization with vancomycin-resistant enterococci among hospitalized patients with bacteremia. Clin Infect Dis, 1997; 24: 704–6. [DOI] [PubMed] [Google Scholar]
- 6.Noskin GA, Stosor V, Cooper I, Peterson LR. Recovery of vancomycin-resistant enterococci on fingertips and environmental surfaces. Infect Control Hosp Epidemiol, 1995; 16: 577–81. [DOI] [PubMed] [Google Scholar]
- 7.Vernon MO, Hayden MK, Trick WE, et al. Chlorhexidine gluconate to cleanse patients in a medical intensive care unit: The effectiveness of source control to reduce the bioburden of vancomycin-resistant enterococci. Arch Intern Med, 2006; 166: 306–12. [DOI] [PubMed] [Google Scholar]
- 8.Hayden MK, Blom DW, Lyle EA, Moore CG, Weinstein RA. Risk of hand or glove contamination after contact with patients colonized with vancomycin-resistant enterococcus or the colonized patients’ environment. Infect Control Hosp Epidemiol, 2008; 29: 149–54. [DOI] [PubMed] [Google Scholar]
- 9.Morgan DJ, Rogawski E, Thom KA, et al. Transfer of multidrug-resistant bacteria to healthcare workers’ gloves and gowns after patient contact increases with environmental contamination. Crit Care Med, 2012; 40: 1045–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duckro AN, Blom DW, Lyle EA, Weinstein RA, Hayden MK. Transfer of vancomycin-resistant enterococci via health care worker hands. Arch Intern Med, 2005; 165: 302–7. [DOI] [PubMed] [Google Scholar]
- 11.Snyder GM, Thom KA, Furuno JP, et al. Detection of methicillin-resistant staphylococcus aureus and vancomycin-resistant enterococci on the gowns and gloves of healthcare workers. Infect Control Hosp Epidemiol, 2008; 29: 583–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care, 2005; 43: 1130–9. [DOI] [PubMed] [Google Scholar]
- 13.Climo MW, Yokoe DS, Warren DK, et al. Effect of daily chlorhexidine bathing on hospital-acquired infection. N Engl J Med, 2013; 368: 533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bleasdale SC, Trick WE, Gonzalez IM, Lyles RD, Hayden MK, Weinstein RA. Effectiveness of chlorhexidine bathing to reduce catheter-associated bloodstream infections in medical intensive care unit patients. Arch Intern Med, 2007; 167: 2073–9. [DOI] [PubMed] [Google Scholar]
- 15.Noto MJ, Domenico HJ, Byrne DW, et al. Chlorhexidine bathing and health care-associated infections: A randomized clinical trial. JAMA, 2015; 313: 369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sethi AK, Al-Nassir WN, Nerandzic MM, Donskey CJ. Skin and environmental contamination with vancomycin-resistant enterococci in patients receiving oral metronidazole or oral vancomycin treatment for clostridium difficile-associated disease. Infect Control Hosp Epidemiol, 2009; 30: 13–7. [DOI] [PubMed] [Google Scholar]