ABSTRACT
The bacterial type IV secretion systems (T4SSs) are a functionally diverse superfamily of secretion systems found in many species of bacteria. Collectively, the T4SSs translocate DNA and monomeric and multimeric protein substrates to bacterial and eukaryotic cell types. T4SSs are composed of two large subfamilies, the conjugation machines and the effector translocators that transmit their cargoes through establishment of direct donor-target cell contacts, and a third small subfamily capable of importing or exporting substrates from or to the milieu. This review summarizes recent mechanistic and structural findings that are shedding new light on how T4SSs have evolved such functional diversity. Translocation signals are now known to be located C terminally or embedded internally in structural folds; these signals in combination with substrate-associated adaptor proteins mediate the docking of specific substrate repertoires to cognate VirD4-like receptors. For the Legionella pneumophila Dot/Icm system, recent work has elucidated the structural basis for adaptor-dependent substrate loading onto the VirD4-like DotL receptor. Advances in definition of T4SS machine structures now allow for detailed comparisons of nanomachines closely related to the Agrobacterium tumefaciens VirB/VirD4 T4SS with those more distantly related, e.g., the Dot/Icm and Helicobacter pylori Cag T4SSs. Finally, it is increasingly evident that T4SSs have evolved a variety of mechanisms dependent on elaboration of conjugative pili, membrane tubes, or surface adhesins to establish productive contacts with target cells. T4SSs thus have evolved extreme functional diversity through a plethora of adaptations impacting substrate selection, machine architecture, and target cell binding.
INTRODUCTION
The bacterial type IV secretion systems (T4SSs) are a large, versatile family of macromolecular translocation systems functioning in Gram-negative (G−) and Gram-positive (G+) bacteria (1). These systems mediate the transfer of DNA or monomeric or multimeric protein substrates to a large range of prokaryotic and eukaryotic cell types (Fig. 1A). Conjugation systems, the earliest described subfamily of T4SSs (2), transfer mobile genetic elements (MGEs) between bacteria. They pose an enormous medical problem because MGEs often harbor cargoes of antibiotic resistance genes and fitness traits that endow pathogens with antibiotic resistance and other growth advantages under selective pressures (3–5). Effector translocators, a more recently described T4SS subfamily (6, 7), are deployed by pathogenic bacteria to deliver effector proteins to eukaryotic cells during the course of infection (8–11). The conjugation and effector translocator systems, as well as newly discovered interbacterial killing systems, transmit their cargos through direct donor-target cell contact (12–14). A few other T4SSs designated uptake or release systems acquire DNA substrates from the milieu or release DNA or protein substrates into the milieu (Fig. 1A) (1, 6).
FIGURE 1.
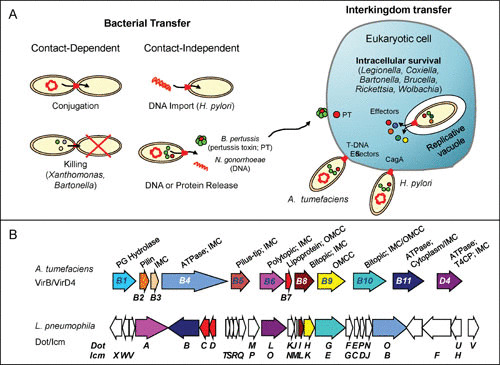
Functional and compositional diversity of the bacterial type IV secretion systems (T4SSs). (A) (Left) Contact-dependent conjugation systems and recently described killing systems deliver DNA or protein substrates directly to bacterial target cells. Contact-independent systems mediate DNA import, DNA export, or export of the multimeric pertussis toxin. (Right) Various pathogenic bacteria and symbionts have evolved T4SSs to deliver effector proteins or DNA-protein complexes into eukaryotic host cells to subvert host physiological processes. (B) Gene arrangements and architectures of the A. tumefaciens VirB/VirD4 and L. pneumophila Dot/Icm secretion systems, with color-coding of the genes encoding homologous subunits; unshaded genes are unique to the Dot/Icm system. The VirB/VirD4 subunit enzymatic functions and associations with inner membrane complex (IMC), outer membrane core complex (OMCC), or pilus are listed. PG Hydrolase, peptidoglycan hydrolase; T4CP, type IV coupling protein.
The T4SSs are defined by the presence of a minimum set of conserved or “signature” subunits (8). The Agrobacterium tumefaciens VirB/VirD4 T4SS, whose Vir subunit nomenclature is widely adopted in this field when referring to the conserved subunits of T4SSs, is assembled from VirB1 through VirB11 and VirD4 (Fig. 1B) (15). In G− species, T4SSs are composed of these Vir-like subunits, although many systems have appropriated other subunits or domains from unknown ancestries presumably for specialized functions (16–18). In G+ species, six VirB/D4-like subunits (VirB1, VirB3, VirB4, VirB6, VirB8, and VirD4) are required to build “minimized” systems spanning the single cytoplasmic membrane and cell wall (8, 19). Vir subunits can be grouped as (i) two or three conserved ATPases (VirB4, VirB11, and VirD4) that coordinate the recruitment and processing of substrates, catalyze structural changes in the T4SS channel necessary for substrate passage, and in some cases regulate pilus biogenesis (12, 20–23), (ii) integral inner membrane (IM) subunits (VirB3, VirB6, and VirB8) that presumptively form an IM channel, (iii) a transglycosylase (VirB1) that contributes to (G− species) or is required for (G+ species) assembly of the channel across the murein layer (8, 24, 25), and (iv) outer membrane (OM)-associated subunits (VirB7, VirB9, and VirB10) that form a structural scaffold for the portion of the channel spanning the periplasm and OM of G− species (26, 27). Here, we summarize results of recent mechanistic and high-resolution microscopy studies that are providing exciting new insights into the biogenesis and structural arrangement of T4SSs and how they have evolved such extreme biological diversity.
SUBSTRATE RECOGNITION: SUBSTRATE SIGNALS AND ADAPTORS/CHAPERONES
T4SSs recruit specific repertoires of substrates through recognition of translocation signals (TSs) and accessory factors bound to substrates. For conjugative DNA transfer, the recruitment and delivery of DNA substrates through cognate T4SSs can be summarized briefly as follows. First, an accessory factor binds the origin-of-transfer (oriT) sequence carried by an MGE. Accessory factors generally fall into the ribbon-helix-helix family of DNA binding proteins, as exemplified by the TraM protein encoded by F plasmid and TrwA encoded by plasmid R388 (hereafter, the origin of the named T4SS or protein will appear in subscript, e.g., TraMF and TrwAR388) (28–31). The accessory factor, through a combination of DNA bending and direct protein-protein interactions, recruits a protein termed the relaxase to oriT to form the catalytically active relaxosome (32, 33). The relaxase cleaves the DNA strand destined for transfer DNA (T-strand), and as a consequence of nicking, the relaxase remains covalently bound to the 5′ end of the T-strand. The accessory factor and relaxase together promote docking of the relaxosome with a cognate VirD4 substrate receptor. In the F plasmid transfer system, the TraMF accessory factor binds a short motif at the C terminus of VirD4-like TraD, whereas the TraIF relaxase carries internal TSs that form only when TraI adopts its tertiary structure (34). Other relaxases may additionally or alternatively carry C-terminal TSs, which are typically composed of clusters of positively charged or hydrophobic residues (35–40). These TSs mediate relaxase interactions with the VirD4 receptors, although the structural bases for these interactions are not yet defined. Other specialized accessory factors are members of the ParA superfamily of partitioning proteins; these proteins also physically couple the relaxosome with the VirD4 receptors through establishment of multiple protein-protein contacts (41–43). Once the relaxosome docks with the VirD4 substrate receptor, by mechanisms that are not yet known, the relaxase is unfolded (23) and the accessory factor(s) is released, and the relaxase pilots the covalently bound T-strand through the T4SS to recipient cells. In recipient cells, the relaxase catalyzes recircularization of the T-strand through a reversal of the strand-breaking reaction, followed by second-strand synthesis and replication of the transferred element.
Among the effector translocators, some systems translocate only one or a few effector proteins, whereas others deliver several hundred into eukaryotic target cells, where they function in a myriad of ways to subvert host cell physiologies (Fig. 1A) (1, 6, 9). As with the conjugation systems, effector translocators recognize their substrate repertoires through a combination of internal and C-terminal TSs carried by the effectors and binding of adaptors or chaperones associated with the effectors. In addition to their role in physically coupling the effector with the VirD4 receptor, adaptors and chaperones block effector aggregation or prevent nonproductive protein interactions in the bacterium prior to effector translocation (44–49). Until recently, effector translocators were thought to function exclusively to deliver protein effectors to eukaryotic cells, where they disrupt host cell physiological processes that aid in infection. Recently, however, members of this subfamily were shown to translocate toxin components of toxin-antitoxin modules to kill other bacteria in the vicinity (Fig. 1A). Xanthomonas spp., for example, deploy a VirB/VirD4-like T4SS to deliver toxins whose bacteriolytic activities can degrade peptidoglycan in target cells lacking the corresponding antitoxin (13). In Bartonella spp., the VbhT toxin is similarly transmitted via a VirB-like T4SS to target bacteria (14). Interestingly, VbhT carries a C-terminal TS identical to that previously determined to be involved in T4SS trafficking of interkingdom effectors during the course of Bartonella infections. These findings establish an evolutionary link between toxins transmitted interbacterially for niche establishment and effectors delivered to eukaryotic cells for pathogenic ends (14).
The VirD4 Receptor
VirD4-like substrate receptors couple DNA or protein substrates to the T4SS; consequently, these subunits are also called type IV coupling proteins (T4CPs) (Fig. 2A and B) (50, 51). T4CPs are phylogenetically related to the SpoIIIE and FtsK ATPases functioning in DNA transport across membranes during sporulation and cell division, respectively (52, 53). These ATPases are typically configured as homohexamers with an N-terminal transmembrane domain and a nucleotide binding/hydrolysis domain (NBD), giving rise to an overall F1-ATPase architecture (53, 54). T4CPs also carry a sequence variable all-alpha domain (AAD) situated at the base of the NBD, and many carry a second, variable C-terminal domain (CTD) (51, 54). The AAD functions in substrate binding and specificity (55, 56), whereas the CTDs can also confer substrate specificity (28) or spatiotemporal control over substrate selection and translocation through the T4SS (57, 58). Two structural studies have shed important light on interactions between T4CP CTDs and cognate substrates. First, in the F plasmid transfer system, the accessory factor TraMF was shown to physically couple the F relaxosome to the TraD receptor through simultaneous binding of F’s oriT sequence and a 13-residue sequence at the end of TraD’s CTD (28, 59). Second, in the more complex Legionella pneumophila Dot/Icm system, which is capable of delivering over 300 effectors to eukaryotic target cells during infection, the VirD4-like DotL receptor has a long (∼200-residue) CTD that binds DotM, DotN, the IcmSW adaptor complex, and LvgA (Fig. 2B) (48, 49, 58, 60, 61). These bound factors collectively stabilize DotL and mediate recruitment of distinct subsets of effectors to the Dot/Icm T4SS. Modeling of the elongated CTD-adaptor subassembly onto DotL’s NBD hexameric sphere gives rise to a bipartitate bell-shape structure that presumably sits at the base of the translocation channel to recruit and feed substrates into the channel (Fig. 2B) (58). Interestingly, however, to date neither the DotL-adaptor complex nor other VirD4 hexamers have been visualized in association with cognate T4SS machines (see below) (18). These and other biochemical findings (62–64) have supported a model that T4CPs associate only transiently with cognate channels as a function of substrate binding.
FIGURE 2.
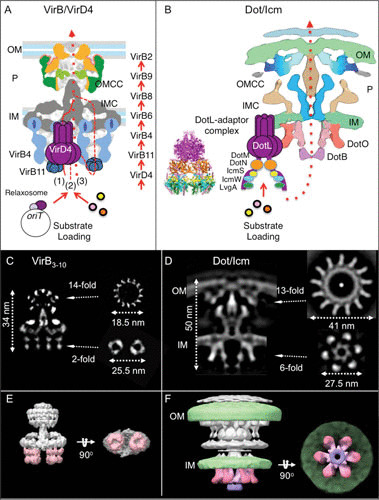
Architectures of the phylogenetically distant VirB/VirD4 and Dot/Icm T4SSs. (A) A schematic of the VirB3-10 structure elaborated by the TrwR388 T4SS and solved by single-particle nsEM. A hexamer of the VirD4 receptor is fitted between the two hexameric barrels of the VirB4 ATPase. The VirD4 receptor recruits MGEs, such as conjugative plasmids, through recognition of components of the relaxosome (relaxase and accessory factors) assembled at the origin-of-transfer (oriT) sequence. VirD4 recruits protein substrates (colored dots) through direct or adaptor-mediated contacts. Substrates engage with the VirD4 receptor and are then delivered sequentially through a translocation channel composed of the VirB proteins listed at the right, as deduced from the transfer DNA immunoprecipitation assay (78). The route of transfer across the IM is not known; substrates might be conveyed through the VirD4 hexamer (route 1, solid line), the VirB4 hexamer (route 2, small dashed line), or a channel composed of the VirB6 and VirB8 subunits (route 3, dotted line). Substrates then pass through the periplasm and across the OM via an OMCC channel. (B) A schematic of the L. pneumophila Dot/Icm T4SS solved by in situ cryo-ET (16). The centrally stacked hexamers of the VirB4-like DotO and DotB form the cytoplasmic entrance to a channel that spans the entire cell envelope. The bell-shaped DotL-adaptor complex is comprised of the hexameric nucleotide binding domain (purple) and C-terminal domain bound with DotN (brown), IcmS (yellow), IcmW (aqua), and LvgA (green); reprinted with permission by Kwak et al. (58). The DotL-adaptor receptor complex was not part of the visualized Dot/Icm T4SS (16) and is provisionally positioned adjacent to the Dot/Icm T4SS. Upon loading of substrates, the DotL-adaptor complex is postulated to present effectors to the DotB/DotO energy center for delivery through the central channel. (C to F) Comparison of the R388-encoded VirB3-10 substructure and the L. pneumophila Dot/Icm T4SS. (C) A central section through the longitudinal plane of the VirB3-10 single-particle reconstruction with cross sections of the OMCC and IMC at the positions indicated. (D) A central section through longitudinal plane of a global average structure of L. pneumophila Dot/Icm T4SS with cross sections at the positions indicated. (E) A three-dimensional (3D) surface rendering of the VirB3-10 substructure shown in side and bottom views. The side-by-side hexameric barrels of the VirB4 ATPase are colored pink. (F) A 3D surface rendering of the Dot/Icm T4SS shown in side and bottom views. The bacterial membranes are in green and the DotO and DotB hexameric ATPases comprising the entrance to the translocation channel are in shades of pink and purple, respectively.
Structural Advances: Subunits and Subassemblies
Structures now exist for intact or soluble domains of the conserved ATPases (VirD4, VirB4, and VirB11) and several channel/pilus subunits (VirB5, VirB8, VirB9, and VirB10), obtained by X-ray crystallography, nuclear magnetic resonance, or negative-stain electron microscopy (nsEM) (54, 65–72). Larger subassemblies termed outer membrane core complexes (OMCCs) have also been visualized by nsEM from systems phylogenetically related to the VirB/VirD4 T4SS (27, 73) as well as more distantly related systems, e.g., the L. pneumophila Dot/Icm and H. pylori Cag T4SSs (74, 75). The OMCCs of the VirB/VirD4-like systems are ∼1.1-MDa complexes composed of 14 copies of the VirB7-, VirB9-, and VirB10-like subunits. These complexes are arranged as large barrels of ∼185 Å in width and height (27). The outer layer of the OMCC from the pKM101-encoded T4SS (TrapKM101) was solved by X-ray crystallography, revealing a network of intra- and intersubunit contacts and a distal cap composed of 14 copies of a helix-loop-helix domain of VirB10 termed the antenna projection (26). The cap is postulated to span the OM, and its central pore of ∼32 Å is postulated to comprise the OM channel through which substrates pass to the cell exterior. Interestingly, chimeric T4SSs composed of the IM-spanning portion of the TrapKM101 T4SS joined to heterologous OMCCs from other VirB/VirD4-like T4SSs are capable of translocating DNA substrates between bacteria, confirming that the observed structural conservation of OMCCs from these systems extends to the level of function (73). Very recently, a structure of the entire OMCC of a Xanthomonas citri T4SS was solved at 3.3 Å by cryo-electron microscopy (cryo-EM); it shows in unprecedented atomic detail an extensive VirB7-VirB9-VirB10 interaction network and also identifies flexible linkers and weak contacts that are postulated to account for intrinsic flexibility of the OMCC necessary for signal-activated channel gating (20, 76, 77).
The VirB3-10 structure
A much larger substructure elaborated by the TrwR388 T4SS was solved by nsEM at a resolution of 20 Å (78). This structure is composed of homologs of the VirB3-VirB10 subunits and was designated the VirB3-10 or T4SS3-10 complex (Fig. 2C and E). The ∼3.5-MDa structure consists of the OMCC joined by a thin stalk to an even larger IM complex, designated the IMC, of 25.5 nm in diameter and 10.5 nm in thickness. The entire structure, with a length of ∼34 nm, spans the cell envelope such that the OMCC’s cap forms the OM pore and the upper portion of the IMC spans the IM. The IMC platform is composed of 12 copies each of VirB3, VirB5, and VirB8 and 24 copies of VirB6. This platform connects to two barrel-like structures of 10.5 nm in width and 13.5 nm in height that correspond to two side-by-side hexamers of VirB4 extending into the cytoplasm (Fig. 2C and E). The VirB3-10 structure lacks the conjugative pilus elaborated by conjugation machines in G− species (see below), as well as the VirB2, VirB11, and VirD4 homologs required for substrate transfer. Interestingly, however, in a recent update a VirB3-10 structure was solved that additionally has one or two dimers of VirD4 situated between the VirB4 hexameric barrels (79). These dimers might correspond to an assembly intermediate of the T4CP that engages with the channel in the absence of bound substrate. It is also intriguing to consider that an early X-ray structure of a soluble domain of the TrwBR388 coupling protein (54), which has guided our thinking for many years regarding the hexameric structure of T4CPs, might not reflect the oligomeric and active states of the VirD4 ATPases assembled in vivo (79).
The overall VirB3-10/VirD4 dimer structure lacks a detectable channel. However, results of a chromatin immunoprecipitation-based cross-linking assay termed transfer DNA immunoprecipitation using the model A. tumefaciens VirB/VirD4 system allowed for provisional assignments of channel composition. As DNA substrates are translocated through the VirB/VirD4 channel, they can be formaldehyde cross-linked sequentially with the VirD4 and VirB11 ATPases, then with the VirB6 and VirB8 IMC subunits, and finally with the VirB9 and VirB2 pilin subunits in the periplasm (Fig. 2A) (20, 80–83). In subsequent studies, evidence also was presented for DNA substrate close contacts with VirB4-like subunits (71, 84). Thus, seven VirB/VirD4 subunits depicted in Fig. 2A are envisioned to comprise the translocation channel, while the remaining VirB components contribute indirectly to channel assembly or function.
The L. pneumophila Dot/Icm Structure
The L. pneumophila Dot/Icm T4SS is assembled from VirB-like subunits as well as approximately 20 additional subunits (Fig. 1B) (85, 86). Not surprisingly, therefore, the Dot/Icm structure recently visualized by in situ cryo-electron tomography (cryo-ET) is much larger than the VirB3-10 substructure (Fig. 2D and F) (18, 87). The OMCC is 42 nm wide and 31 nm high and presents as a wheel-like structure with 13-fold symmetry, as opposed to the 14-fold symmetries of the VirB/VirD4 OMCCs (18). The entire wheel is embedded in the inner leaflet of the OM, and a central pore of 6 nm projects across the outer leaflet of the OM. The wheel extends into the periplasm, where it is connected to a cylinder that extends to the IM, establishing contact with the IMC. Most strikingly, refinement of the IMC showed that it adopts a 6-fold symmetry and forms two concentric rings of 16 nm and 27.5 nm at the cytoplasmic entrance to the translocation channel (Fig. 2D) (18). In side view, 6 inverted V structures extend into the cytoplasm, such that the inner arms of the V’s form the inner ring and the outer arms form the outer ring. These V structures are composed of VirB4-like DotO, and thus, the cytoplasmic complex consists of a hexamer of 6 DotO dimers (Fig. 2F). Furthermore, VirB11-like DotB was shown to dynamically associate at the base of the DotO inner ring by a mechanism dependent on ATP hydrolysis. The cytoplasmic complex is therefore composed of a central DotO hexamer onto which the DotB hexamer binds, presumably when the machine is activated for substrate transfer. This symmetric IMC architecture differs strikingly from the asymmetric IMC of the VirB3-10 structure marked by side-by-side VirB4 barrels (Fig. 2E and F). Gratifyingly, the Dot/Icm structure identifies for the first time a continuous T4SS channel extending from the cytoplasmic entrance (marked by the DotB lumen) to the cell surface (marked by the OMCC pore) (Fig. 2B, D, and F) (18). It is also interesting to note that the Dot/Icm T4SS assembles at the cell poles and that polar delivery of effectors into the eukaryotic host cell evidently is required for successful L. pneumophila infection (88).
T4SS-Associated Mechanisms for Target Cell Attachment
Conjugation systems of G− species elaborate conjugative pili that extend for as long as 20 μm from the donor cell (12). Flexible pili elaborated by F plasmids extend or retract to draw potential recipients into direct contact (89), whereas more brittle pili produced by other conjugative plasmids are either sloughed or broken from the cell, where they accumulate and induce cellular aggregation (12, 90). To date, only one effector translocator system, the H. pylori Cag T4SS, also has been shown to elaborate pili in the presence of host epithelial cells (91–93). Interestingly, this Cag T4SS additionally elaborates large sheathed structures, or “membrane tubes” (94), that were recently visualized by in situ cryo-ET. Features of these tube structures led the authors to suggest they might arise by the extension of a pilus from an IM platform. As the pilus protrudes across the OM, the distorted membrane surrounds the pilus, forming a sheath or tube that projects from the cell surface (95).
Surprisingly, the role of the pilus in substrate transfer is still not firmly established. On the one hand, there is some evidence in the F plasmid transfer system for DNA transfer between distant cells attached together by the F pilus, suggesting that the F pilus can function as a translocation channel (96). The structure of the F pilus was recently solved by cryo-EM, and strikingly, the lumen is lined with IM phospholipid (PL). This discovery has important implications regarding the mechanism of F pilus assembly and retraction, but the presence of PL also imparts an overall weak negative charge to the inner lumen of possible importance for conveyance of the DNA substrate through the pilus (97). On the other hand, several observations argue against a role for the pilus as a conduit for substrates. First, in the A. tumefaciens VirB/VirD4 T4SS and related T4SSs, “uncoupling” mutations have been isolated that selectively block pilus production without impeding substrate transfer, strongly indicating that extended pili are not required for DNA transfer (73, 83, 98). Second, conjugation systems functioning in G+ species do not elaborate pili yet can transfer DNA between cells at very high frequencies (8, 99, 100). Third, recently it was shown that the E. coli pKM101 conjugation system employs cell surface adhesins as an alternative to conjugative pili to mediate formation of donor-target cell contacts (101). This finding is of special interest given the paucity of evidence for pilus production by effector translocators other than the H. pylori Cag T4SS. It is enticing to propose that most members of the effector translocator subfamily have dispensed with energetically costly pilus production in favor of appropriating chromosomally encoded surface adhesins to establish productive bacterial-eukaryotic cell membrane contacts.
CONCLUDING REMARKS
Mechanistic and structural studies are rapidly shaping a deeper understanding of how the T4SS superfamily evolved such functional diversity. Most notably, recent advances have been made in structural definition of T4SSs that are phylogenetically distantly related to the “canonical” VirB/VirD4-like T4SSs, e.g., L. pneumophila Dot/Icm. Despite these advances, many fundamental questions remain: (i) How do substrates dock with the T4SS, and how are they processed for transfer? (ii) What is the route of translocation, and what are the signaling requirements for channel activation? (iii) What mechanisms mediate productive donor-target cell contacts, and what is the architecture of the mating junction? As basic studies continue to investigate these questions, we also note with excitement the emergence of translational studies aimed at designing T4SS machine inhibitors or repurposing T4SSs as therapeutic delivery systems (102–104). The integration of basic and translational approaches ensures a bright and vibrant future for the fascinating T4SS nanomachines as well as the scientists devoted to their study.
ACKNOWLEDGMENTS
We thank members of the Christie and Hu laboratories for helpful discussions.
Work in the Christie laboratory is supported by NIH grants R01GM48476 and 1R21AI137918. Work in the Hu laboratory is supported by grant AU-1953-20180324 from the Welch Foundation. Work in the Christie and Hu laboratories is supported by R21AI142378 and in part by NIH grant DK056338, which supports the Texas Medical Center Digestive Diseases Center.
REFERENCES
- 1.Grohmann E, Christie PJ, Waksman G, Backert S. 2018. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol 107:455–471. 10.1111/mmi.13896. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lederberg J, Tatum EL. 1946. Novel genotypes in mixed cultures of biochemical mutants of bacteria. Cold Spring Harb Symp Quant Biol 11:113–114. 10.1101/SQB.1946.011.01.014. [DOI] [Google Scholar]
- 3.Bellanger X, Payot S, Leblond-Bourget N, Guédon G. 2014. Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol Rev 38:720–760. 10.1111/1574-6976.12058. [PubMed] [DOI] [PubMed] [Google Scholar]
- 4.von Wintersdorff CJ, Penders J, van Niekerk JM, Mills ND, Majumder S, van Alphen LB, Savelkoul PH, Wolffs PF. 2016. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol 7:173. 10.3389/fmicb.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koraimann G. 2018. Spread and persistence of virulence and antibiotic resistance genes: a ride on the F plasmid conjugation module. EcoSal Plus 8:ESP-0003-2018. 10.1128/ecosalplus.ESP-0003-2018. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat Rev Microbiol 1:137–149. 10.1038/nrmicro753. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winans SC, Burns DL, Christie PJ. 1996. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol 4:64–68. 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatty M, Laverde Gomez JA, Christie PJ. 2013. The expanding bacterial type IV secretion lexicon. Res Microbiol 164:620–639. 10.1016/j.resmic.2013.03.012. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez-Rivera C, Bhatty M, Christie PJ. 2016. Mechanism and function of type IV secretion during infection of the human host. Microbiol Spectr 4:VMBF-0024-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu J, Luo ZQ. 2017. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 15:591–605. 10.1038/nrmicro.2017.67. [PubMed] [DOI] [PubMed] [Google Scholar]
- 11.Dehio C, Tsolis RM. 2017. Type IV effector secretion and subversion of host functions by Bartonella and Brucella species. Curr Top Microbiol Immunol 413:269–295. 10.1007/978-3-319-75241-9_11. [DOI] [PubMed] [Google Scholar]
- 12.Arutyunov D, Frost LS. 2013. F conjugation: back to the beginning. Plasmid 70:18–32. 10.1016/j.plasmid.2013.03.010. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Souza DP, Oka GU, Alvarez-Martinez CE, Bisson-Filho AW, Dunger G, Hobeika L, Cavalcante NS, Alegria MC, Barbosa LR, Salinas RK, Guzzo CR, Farah CS. 2015. Bacterial killing via a type IV secretion system. Nat Commun 6:6453. 10.1038/ncomms7453. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Harms A, Liesch M, Körner J, Québatte M, Engel P, Dehio C. 2017. A bacterial toxin-antitoxin module is the origin of inter-bacterial and inter-kingdom effectors of Bartonella. PLoS Genet 13:e1007077. 10.1371/journal.pgen.1007077. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger BR, Christie PJ. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol 176:3646–3660. 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie PJ. 2016. The mosaic type IV secretion systems. EcoSal Plus 7:ESP-0020-2015. 10.1128/ecosalplus.ESP-0020-2015. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christie PJ, Gomez Valero L, Buchrieser C. 2017. Biological diversity and evolution of type IV secretion systems. Curr Top Microbiol Immunol 413:1–30. 10.1007/978-3-319-75241-9_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chetrit D, Hu B, Christie PJ, Roy CR, Liu J. 2018. A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat Microbiol 3:678–686. 10.1038/s41564-018-0165-z. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grohmann E, Keller W, Muth G. 2017. Mechanisms of conjugative transfer and type IV secretion-mediated effector transport in Gram-positive bacteria. Curr Top Microbiol Immunol 413:115–141. 10.1007/978-3-319-75241-9_5. [DOI] [PubMed] [Google Scholar]
- 20.Cascales E, Atmakuri K, Sarkar MK, Christie PJ. 2013. DNA substrate-induced activation of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol 195:2691–2704. 10.1128/JB.00114-13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ripoll-Rozada J, Zunzunegui S, de la Cruz F, Arechaga I, Cabezón E. 2013. Functional interactions of VirB11 traffic ATPases with VirB4 and VirD4 molecular motors in type IV secretion systems. J Bacteriol 195:4195–4201. 10.1128/JB.00437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandran Darbari V, Waksman G. 2015. Structural biology of bacterial type IV secretion systems. Annu Rev Biochem 84:603–629. 10.1146/annurev-biochem-062911-102821. [DOI] [PubMed] [Google Scholar]
- 23.Trokter M, Waksman G. 2018. Translocation through the conjugative type 4 secretion system requires unfolding of its protein substrate. J Bacteriol 200:e00615-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arends K, Celik EK, Probst I, Goessweiner-Mohr N, Fercher C, Grumet L, Soellue C, Abajy MY, Sakinc T, Broszat M, Schiwon K, Koraimann G, Keller W, Grohmann E. 2013. TraG encoded by the pIP501 type IV secretion system is a two-domain peptidoglycan-degrading enzyme essential for conjugative transfer. J Bacteriol 195:4436–4444. 10.1128/JB.02263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laverde Gomez JA, Bhatty M, Christie PJ. 2014. PrgK, a multidomain peptidoglycan hydrolase, is essential for conjugative transfer of the pheromone-responsive plasmid pCF10. J Bacteriol 196:527–539. 10.1128/JB.00950-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015. 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fronzes R, Schäfer E, Wang L, Saibil HR, Orlova EV, Waksman G. 2009. Structure of a type IV secretion system core complex. Science 323:266–268. 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu J, Wong JJ, Edwards RA, Manchak J, Frost LS, Glover JN. 2008. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol Microbiol 70:89–99. 10.1111/j.1365-2958.2008.06391.x. [DOI] [PubMed] [Google Scholar]
- 29.Moncalián G, Grandoso G, Llosa M, de la Cruz F. 1997. oriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J Mol Biol 270:188–200. 10.1006/jmbi.1997.1082. [DOI] [PubMed] [Google Scholar]
- 30.Moncalián G, de la Cruz F. 2004. DNA binding properties of protein TrwA, a possible structural variant of the Arc repressor superfamily. Biochim Biophys Acta 1701:15–23. 10.1016/j.bbapap.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Tato I, Matilla I, Arechaga I, Zunzunegui S, de la Cruz F, Cabezon E. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J Biol Chem 282:25569–25576. 10.1074/jbc.M703464200. [DOI] [PubMed] [Google Scholar]
- 32.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 34:18–40. 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 33.Zechner EL, Lang S, Schildbach JF. 2012. Assembly and mechanisms of bacterial type IV secretion machines. Philos Trans R Soc Lond B Biol Sci 367:1073–1087. 10.1098/rstb.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redzej A, Ilangovan A, Lang S, Gruber CJ, Topf M, Zangger K, Zechner EL, Waksman G. 2013. Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems. Mol Microbiol 89:324–333. 10.1111/mmi.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vergunst AC, van Lier MC, den Dulk-Ras A, Stüve TA, Ouwehand A, Hooykaas PJ. 2005. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A 102:832–837. 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Kregten M, Lindhout BI, Hooykaas PJ, van der Zaal BJ. 2009. Agrobacterium-mediated T-DNA transfer and integration by minimal VirD2 consisting of the relaxase domain and a type IV secretion system translocation signal. Mol Plant Microbe Interact 22:1356–1365. 10.1094/MPMI-22-11-1356. [DOI] [PubMed] [Google Scholar]
- 37.Alperi A, Larrea D, Fernández-González E, Dehio C, Zechner EL, Llosa M. 2013. A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system. J Bacteriol 195:4999–5006. 10.1128/JB.00367-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lang S, Kirchberger PC, Gruber CJ, Redzej A, Raffl S, Zellnig G, Zangger K, Zechner EL. 2011. An activation domain of plasmid R1 TraI protein delineates stages of gene transfer initiation. Mol Microbiol 82:1071–1085. 10.1111/j.1365-2958.2011.07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang S, Zechner EL. 2012. General requirements for protein secretion by the F-like conjugation system R1. Plasmid 67:128–138. 10.1016/j.plasmid.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer R. 2015. Mapping type IV secretion signals on the primase encoded by the broad-host-range plasmid R1162 (RSF1010). J Bacteriol 197:3245–3254. 10.1128/JB.00443-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. 2007. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J 26:2540–2551. 10.1038/sj.emboj.7601696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamilton HL, Domínguez NM, Schwartz KJ, Hackett KT, Dillard JP. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol 55:1704–1721. 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- 43.Gruber CJ, Lang S, Rajendra VK, Nuk M, Raffl S, Schildbach JF, Zechner EL. 2016. Conjugative DNA transfer is enhanced by plasmid R1 partitioning proteins. Front Mol Biosci 3:32. 10.3389/fmolb.2016.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuïnk TJ, Hooykaas PJ. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979–982. 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Z, Sagulenko E, Ding Z, Christie PJ. 2001. Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J Bacteriol 183:3855–3865. 10.1128/JB.183.13.3855-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atmakuri K, Ding Z, Christie PJ. 2003. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol Microbiol 49:1699–1713. 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dym O, Albeck S, Unger T, Jacobovitch J, Branzburg A, Michael Y, Frenkiel-Krispin D, Wolf SG, Elbaum M. 2008. Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners. Proc Natl Acad Sci U S A 105:11170–11175. 10.1073/pnas.0801525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vincent CD, Friedman JR, Jeong KC, Sutherland MC, Vogel JP. 2012. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol 85:378–391. 10.1111/j.1365-2958.2012.08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutherland MC, Nguyen TL, Tseng V, Vogel JP. 2012. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog 8:e1002910. 10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabezón E, Sastre JI, de la Cruz F. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet 254:400–406. 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 51.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808. 10.1128/MMBR.00023-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burton B, Dubnau D. 2010. Membrane-associated DNA transport machines. Cold Spring Harb Perspect Biol 2:a000406. 10.1101/cshperspect.a000406. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomis-Rüth FX, Solà M, de la Cruz F, Coll M. 2004. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des 10:1551–1565. 10.2174/1381612043384817. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.Gomis-Rüth FX, Moncalián G, Pérez-Luque R, González A, Cabezón E, de la Cruz F, Coll M. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637–641. 10.1038/35054586. [PubMed] [DOI] [PubMed] [Google Scholar]
- 55.de Paz HD, Larrea D, Zunzunegui S, Dehio C, de la Cruz F, Llosa M. 2010. Functional dissection of the conjugative coupling protein TrwB. J Bacteriol 192:2655–2669. 10.1128/JB.01692-09. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitaker N, Chen Y, Jakubowski SJ, Sarkar MK, Li F, Christie PJ. 2015. The all-alpha domains of coupling proteins from the Agrobacterium tumefaciens VirB/VirD4 and Enterococcus faecalis pCF10-encoded type IV secretion systems confer specificity to binding of cognate DNA substrates. J Bacteriol 197:2335–2349. 10.1128/JB.00189-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whitaker N, Berry TM, Rosenthal N, Gordon JE, Gonzalez-Rivera C, Sheehan KB, Truchan HK, VieBrock L, Newton IL, Carlyon JA, Christie PJ. 2016. Chimeric coupling proteins mediate transfer of heterologous type IV effectors through the Escherichia coli pKM101-encoded conjugation machine. J Bacteriol 198:2701–2718. 10.1128/JB.00378-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwak MJ, Kim JD, Kim H, Kim C, Bowman JW, Kim S, Joo K, Lee J, Jin KS, Kim YG, Lee NK, Jung JU, Oh BH. 2017. Architecture of the type IV coupling protein complex of Legionella pneumophila. Nat Microbiol 2:17114. 10.1038/nmicrobiol.2017.114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong JJ, Lu J, Glover JN. 2012. Relaxosome function and conjugation regulation in F-like plasmids—a structural biology perspective. Mol Microbiol 85:602–617. 10.1111/j.1365-2958.2012.08131.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Meir A, Chetrit D, Liu L, Roy CR, Waksman G. 2018. Legionella DotM structure reveals a role in effector recruiting to the type 4B secretion system. Nat Commun 9:507. 10.1038/s41467-017-02578-x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu J, Xu D, Wan M, Yin L, Wang X, Wu L, Liu Y, Liu X, Zhou Y, Zhu Y. 2017. Structural insights into the roles of the IcmS-IcmW complex in the type IVb secretion system of Legionella pneumophila. Proc Natl Acad Sci U S A 114:13543–13548. 10.1073/pnas.1706883115. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schröder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol 184:2767–2779. 10.1128/JB.184.10.2767-2779.2002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schröder G, Lanka E. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J Bacteriol 185:4371–4381. 10.1128/JB.185.15.4371-4381.2003. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Larrea D, de Paz HD, Arechaga I, de la Cruz F, Llosa M. 2013. Structural independence of conjugative coupling protein TrwB from its type IV secretion machinery. Plasmid 70:146–153. 10.1016/j.plasmid.2013.03.006. [PubMed] [DOI] [PubMed] [Google Scholar]
- 65.Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G. 2000. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol Cell 6:1461–1472. 10.1016/S1097-2765(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 66.Yeo H-J, Yuan Q, Beck MR, Baron C, Waksman G. 2003. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc Natl Acad Sci U S A 100:15947–15952. 10.1073/pnas.2535211100. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Terradot L, Bayliss R, Oomen C, Leonard GA, Baron C, Waksman G. 2005. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci U S A 102:4596–4601. 10.1073/pnas.0408927102. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hare S, Bayliss R, Baron C, Waksman G. 2006. A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J Mol Biol 360:56–66. 10.1016/j.jmb.2006.04.060. [PubMed] [DOI] [PubMed] [Google Scholar]
- 69.Bayliss R, Harris R, Coutte L, Monier A, Fronzes R, Christie PJ, Driscoll PC, Waksman G. 2007. NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc Natl Acad Sci U S A 104:1673–1678. 10.1073/pnas.0609535104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walldén K, Williams R, Yan J, Lian PW, Wang L, Thalassinos K, Orlova EV, Waksman G. 2012. Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc Natl Acad Sci U S A 109:11348–11353. 10.1073/pnas.1201428109. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peña A, Matilla I, Martín-Benito J, Valpuesta JM, Carrascosa JL, de la Cruz F, Cabezón E, Arechaga I. 2012. The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J Biol Chem 287:39925–39932. 10.1074/jbc.M112.413849. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prevost MS, Waksman G. 2018. X-ray crystal structures of the type IVb secretion system DotB ATPases. Protein Sci 27:1464–1475. 10.1002/pro.3439. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gordon JE, Costa TRD, Patel RS, Gonzalez-Rivera C, Sarkar MK, Orlova EV, Waksman G, Christie PJ. 2017. Use of chimeric type IV secretion systems to define contributions of outer membrane subassemblies for contact-dependent translocation. Mol Microbiol 105:273–293. 10.1111/mmi.13700. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kubori T, Koike M, Bui XT, Higaki S, Aizawa S, Nagai H. 2014. Native structure of a type IV secretion system core complex essential for Legionella pathogenesis. Proc Natl Acad Sci U S A 111:11804–11809. 10.1073/pnas.1404506111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frick-Cheng AE, Pyburn TM, Voss BJ, McDonald WH, Ohi MD, Cover TL. 2016. Molecular and structural analysis of the Helicobacter pyloricag type IV secretion system core complex. mBio 7:e02001-15. 10.1128/mBio.02001-15. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sgro GG, Costa TR, Cenens W, Souza DP, Cassago A, Coutinho de Oliveira L, Salinas RK, Portugal RV, Farah CS, Waksman G. 2018. CryoEM structure of the core complex of a bacterial killing type IV secretion system. Nat Microbiol 3:1429–1440. 10.1038/s41564-018-0262-z. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cascales E, Christie PJ. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci U S A 101:17228–17233. 10.1073/pnas.0405843101. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. 2014. Structure of a type IV secretion system. Nature 508:550–553. 10.1038/nature13081. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Redzej A, Ukleja M, Connery S, Trokter M, Felisberto-Rodrigues C, Cryar A, Thalassinos K, Hayward RD, Orlova EV, Waksman G. 2017. Structure of a VirD4 coupling protein bound to a VirB type IV secretion machinery. EMBO J 36:3080–3095. 10.15252/embj.201796629. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cascales E, Christie PJ. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170–1173. 10.1126/science.1095211. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Atmakuri K, Cascales E, Christie PJ. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol 54:1199–1211. 10.1111/j.1365-2958.2004.04345.x. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J Mol Biol 341:961–977. 10.1016/j.jmb.2004.06.052. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jakubowski SJ, Cascales E, Krishnamoorthy V, Christie PJ. 2005. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J Bacteriol 187:3486–3495. 10.1128/JB.187.10.3486-3495.2005. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li F, Alvarez-Martinez C, Chen Y, Choi KJ, Yeo HJ, Christie PJ. 2012. Enterococcus faecalis PrgJ, a VirB4-like ATPase, mediates pCF10 conjugative transfer through substrate binding. J Bacteriol 194:4041–4051. 10.1128/JB.00648-12. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Christie PJ, Vogel JP. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol 8:354–360. 10.1016/S0966-842X(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kubori T, Nagai H. 2016. The type IVB secretion system: an enigmatic chimera. Curr Opin Microbiol 29:22–29. 10.1016/j.mib.2015.10.001. [PubMed] [DOI] [PubMed] [Google Scholar]
- 87.Ghosal D, Chang YW, Jeong KC, Vogel JP, Jensen GJ. 2017. In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep 18:726–732. 10.15252/embr.201643598. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jeong KC, Ghosal D, Chang YW, Jensen GJ, Vogel JP. 2017. Polar delivery of Legionella type IV secretion system substrates is essential for virulence. Proc Natl Acad Sci U S A 114:8077–8082. 10.1073/pnas.1621438114. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clarke M, Maddera L, Harris RL, Silverman PM. 2008. F-pili dynamics by live-cell imaging. Proc Natl Acad Sci U S A 105:17978–17981. 10.1073/pnas.0806786105. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bradley DE. 1980. Morphological and serological relationships of conjugative pili. Plasmid 4:155–169. 10.1016/0147-619X(80)90005-0. [DOI] [PubMed] [Google Scholar]
- 91.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, König W, Backert S. 2007. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449:862–866. 10.1038/nature06187. [PubMed] [DOI] [PubMed] [Google Scholar]
- 92.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. 2011. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog 7:e1002237. 10.1371/journal.ppat.1002237. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. 2014. Genes required for assembly of pili associated with the Helicobacter pyloricag type IV secretion system. Infect Immun 82:3457–3470. 10.1128/IAI.01640-14. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rohde M, Püls J, Buhrdorf R, Fischer W, Haas R. 2003. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol 49:219–234. 10.1046/j.1365-2958.2003.03549.x. [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.Chang YW, Shaffer CL, Rettberg LA, Ghosal D, Jensen GJ. 2018. In vivo structures of the Helicobacter pyloricag type IV secretion system. Cell Rep 23:673–681. 10.1016/j.celrep.2018.03.085. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Babic A, Lindner AB, Vulic M, Stewart EJ, Radman M. 2008. Direct visualization of horizontal gene transfer. Science 319:1533–1536. 10.1126/science.1153498. [PubMed] [DOI] [PubMed] [Google Scholar]
- 97.Costa TRD, Ilangovan A, Ukleja M, Redzej A, Santini JM, Smith TK, Egelman EH, Waksman G. 2016. Structure of the bacterial sex F pilus reveals an assembly of a stoichiometric protein-phospholipid complex. Cell 166:1436–1444.e10. 10.1016/j.cell.2016.08.025. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sagulenko E, Sagulenko V, Chen J, Christie PJ. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J Bacteriol 183:5813–5825. 10.1128/JB.183.20.5813-5825.2001. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bhatty M, Cruz MR, Frank KL, Gomez JA, Andrade F, Garsin DA, Dunny GM, Kaplan HB, Christie PJ. 2015. Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance) and PrgC contribute to plasmid transfer, biofilm formation and virulence. Mol Microbiol 95:660–677. 10.1111/mmi.12893. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schmitt A, Jiang K, Camacho MI, Jonna VR, Hofer A, Westerlund F, Christie PJ, Berntsson RP. 2018. PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction. Mol Microbiol 109:291–305. 10.1111/mmi.13980. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.González-Rivera C, Khara P, Awad D, Patel R, Li YG, Bogisch M, Christie PJ. 2019. Two pKM101-encoded proteins, the pilus-tip protein TraC and Pep, assemble on the Escherichia coli cell surface as adhesins required for efficient conjugative DNA transfer. Mol Microbiol 111:96–117. 10.1111/mmi.14141. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Guzmán-Herrador DL, Steiner S, Alperi A, González-Prieto C, Roy CR, Llosa M. 2017. DNA delivery and genomic integration into mammalian target cells through type IV A and B secretion systems of human pathogens. Front Microbiol 8:1503. 10.3389/fmicb.2017.01503. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith MA, Coinçon M, Paschos A, Jolicoeur B, Lavallée P, Sygusch J, Baron C. 2012. Identification of the binding site of Brucella VirB8 interaction inhibitors. Chem Biol 19:1041–1048. 10.1016/j.chembiol.2012.07.007. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Shaffer CL, Good JA, Kumar S, Krishnan KS, Gaddy JA, Loh JT, Chappell J, Almqvist F, Cover TL, Hadjifrangiskou M. 2016. Peptidomimetic small molecules disrupt type IV secretion system activity in diverse bacterial pathogens. mBio 7:e00221-16. 10.1128/mBio.00221-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


