Abstract
The Polycomb Repressive Complex 2 (PRC2) is a chromatin-associated methyltransferase catalyzing mono-, di- and trimethylation of lysine 27 on histone H3 (H3K27). This activity is required for normal organismal development and maintenance of gene expression patterns to uphold cell identity. PRC2 function is often deregulated in disease and is a promising candidate for therapeutic targeting in cancer. In this review, we discuss the molecular mechanisms proposed to take part in modulating PRC2 recruitment and shaping H3K27 methylation patterns across the genome. This includes consideration of factors influencing PRC2 residence time on chromatin and PRC2 catalytic activity with a focus on the mechanisms giving rise to regional preferences and differential deposition of H3K27 methylation. We further discuss existing evidence for functional diversity between distinct subsets of PRC2 complexes with the aim of extracting key concepts and highlighting major open questions towards a more complete understanding of PRC2 function.
50-word ‘blurb’
PRC2 function is crucial for the specification and maintenance of cellular identity during normal development and is often deregulated in disease. Laugesen et al. discuss the orchestration of PRC2 recruitment and deposition of H3K27 methylation by factors influencing PRC2 residence time at specific chromatin regions and modulating its catalytic activity.
Introduction
Specification and preservation of cellular identity require intricate regulation of gene expression and tight control of transcriptional states over cell generations. Gene expression patterns are established by networks of transcription factors (TFs) with chromatin-associated proteins contributing an additional layer of transcriptional control. Polycomb group (PcG) proteins are important chromatin-associated factors contributing to the maintenance of transcriptional repression (Schuettengruber et al., 2017).
In line with their crucial role in stabilizing transcriptional patterns and maintaining cell fate, many PcG proteins are essential for normal cellular functions and organismal development (Laugesen and Helin, 2014; Margueron and Reinberg, 2011). In recent years, genes encoding PRC2 subunits have been found mutated or deregulated in diseases including cancer, and small-molecule inhibitors abolishing PRC2 function have entered clinical trials for the treatment of several cancer types (Laugesen et al., 2016). Comprehensive insight into the molecular mechanisms governing PRC2 function is therefore crucial for a better understanding of both normal biology and human disease and to guide the development of novel therapeutic strategies.
Here, we discuss the recent surge of interesting data breaking new ground for our understanding of the molecular mechanisms underlying PRC2 recruitment to chromatin and deposition of its catalytic products.
Polycomb Repressive Complexes
PcG proteins assemble in large multimeric protein complexes with distinct catalytic activities and cellular functions. The most widely studied are the Polycomb Repressive Complexes 1 and 2 (PRC1 and PRC2). While PRC1 mono-ubiquitylates lysine 119 on histone H2A (H2AK119ub1) through its catalytic subunit, the RING1A or RING1B ubiquitin ligase, PRC2 catalyzes mono-, di- and tri-methylation on lysine 27 on histone H3 (H3K27me1, H3K27me2 and H3K27me3). Both PRC1 and PRC2 exist in different forms with distinct subunit compositions potentially influencing the structure and mechanistic properties of the complexes (Chittock et al., 2017).
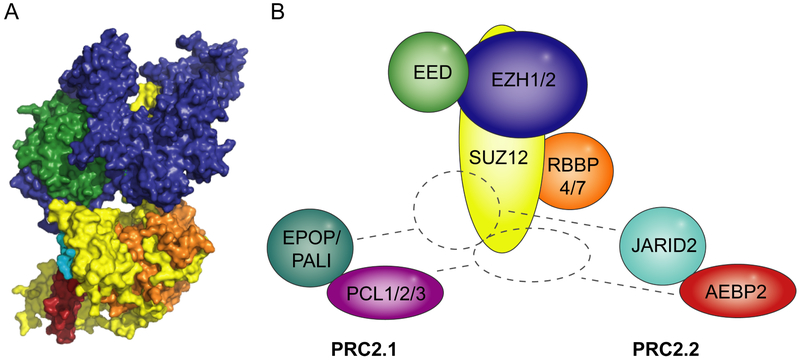
The core PRC2 subunits, SUZ12, EED, and the methyltransferase EZH2 or its closely related homolog EZH1, associate with equimolar stoichiometries and are all required for the catalytic activity of the complex (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Margueron et al., 2008; Muller et al., 2002; Shen et al., 2008; Smits et al., 2013). Along with a histone-binding protein, RBBP4 or RBBP7, the PRC2 core complex forms distinct subcomplexes by incorporating different combinations of substoichiometric interaction partners with suggested roles in PRC2 recruitment or regulation of PRC2 activity. Protein interaction data shows segregation into two main subtypes of PRC2 termed PRC2.1 (containing a PCL homolog (PCL1-3) along with EPOP (C17ORF96) or PALI (C10ORF12)) and PRC2.2 (containing JARID2 and AEBP2) (Hauri et al., 2016) (Figure 1).
Figure 1: Structure of PRC2 with associated non-core subunits.
A. Crystal structures, PDB ID: 5WAI (Chen et al., 2018) and PDB ID: 5LS6 (Vaswani et al., 2016), superimposed onto Cryo-EM structure, PDB ID: 6C23 (Kasinath et al., 2018a), showing core PRC2 (SUZ12 (yellow), EED (green), EZH2 (blue)) with fragments of AEBP2 (red) and JARID2 (turquoise). B. Schematic drawing of PRC2 with indication of core subunits and potential for incorporation of distinct sets of non-core subunits to form PRC2.1 and PRC2.2 ((Hauri et al., 2016)).
H3K27 methylation
PRC2 is the only identified methyltransferase with activity towards H3K27 and is responsible for all H3K27 methylation in mouse embryonic stem cells (mESCs) (Hojfeldt et al., 2018). The different degrees of H3K27 methylation (H3K27me1/me2/me3) show distinct genomic distributions: H3K27me1 is found on 5-10 % of all histone H3 in a cell and enriched within gene bodies of actively transcribed genes, H3K27me2 is very abundant, marking 50-70% of total histone H3 and covering inter- and intragenic regions with suggested roles in prevention of inappropriate promoter or enhancer activity, and H3K27me3 (present on 5-10 % of histone H3) is strongly enriched at sites overlapping with PRC2 binding, but also found at lower levels in other genomic regions (Ferrari et al., 2014; Hojfeldt et al., 2018; Jung et al., 2010) (Figure 2).
Figure 2: Overview of the genomic distribution of H3K27 methylation.
A. Average ChIP-seq signals for H3K27me1 (light blue), H3K27me2 (medium blue), and H3K27me3 (dark blue) over active and inactive genes (relative gene lengths +/− 20 kb) based on tracing of plotted ChIP-seq data from (Hojfeldt et al., 2018). B. Stylized representation of genomic snapshots of plotted ChIP-seq tracks of H3K27 methylation and SUZ12 (red) illustrating overlapping patterns of SUZ12 and H3K27me3 by an inactive gene, enrichment of H3K27me1 over an active gene and intergenic H3K27me2. TSS: Transcription start site, TTS: Transcription termination site, CGI: CpG island, kb: kilobases.
Most studies probing PRC2 function focus on H3K27me3, which is considered a hallmark of PRC2-mediated gene repression. While data from Drosophila (Muller et al., 2002; Pengelly et al., 2013) clearly support a biological role of H3K27 methylation, the functional significance of each degree of H3K27 methylation and the molecular basis for their differential genomic deposition remain incompletely understood.
PRC2 occupancy and differential deposition of H3K27 methylation
As evidenced by many ChIP-seq studies, detectable PRC2 binding is observed at a relatively small part of the genome, primarily overlapping with the H3K27me3-positive CpG island (CGI) promoters of silent genes (Ku et al., 2008; Tanay et al., 2007), while its catalytic products are found on 70-80 % of all histone H3 in mESCs (Jung et al., 2010; Peters et al., 2003). With PRC2 being responsible for all H3K27 methylation in mESCs (Hojfeldt et al., 2018) and bulk methylation of nascent histones occurring after DNA replication (Reveron-Gomez et al., 2018), it is clear that PRC2 must interact with chromatin throughout most of the genome.
Thus, the widely dispersed H3K27me2 and intragenic H3K27me1 are products of PRC2 interactions occurring along the genome, where PRC2 cannot be detected in ChIP-seq studies. These interactions are most likely transient with very short residence times, but their ‘ChIP-invisibility’ could also reflect that they occur with minimally involved interaction surfaces such as just the PRC2 catalytic site engaged with the histone H3 tail, which could further hamper efficient cross-link capture. In accordance with this, single-molecule microscopy studies of PRC2 dynamics confirm that ∼80% of PRC2 is highly mobile, while the remaining PRC2 is stably bound with long residence time (Youmans et al., 2018). The stably bound PRC2 molecules likely represent the pool of PRC2 that is CGI-bound and captured in ChIP studies.
Interestingly, PRC2 is most efficient at catalyzing the initial methylation steps to produce H3K27me1 and H3K27me2, while conversion to H3K27me3 is more time-consuming (Sneeringer et al., 2010; Zee et al., 2012). This is reflected in rapid restoration of global levels of H3K27me1, followed by H3K27me2, and finally H3K27me3 after DNA replication (Alabert et al., 2014) or upon release from PRC2 inhibition (Hojfeldt et al., 2018). Genomic mapping of the rapidly occurring H3K27me1/me2 reveal that it is initially produced at sites ultimately proceeding to become H3K27me3-positive, demonstrating that strongly PRC2-bound sites have the highest catalytic rates (Hojfeldt et al., 2018; Reveron-Gomez et al., 2018). The strong overlap of H3K27me3 with sites identified as PRC2-bound in ChIP analyses could therefore indicate a requirement for stable PRC2 binding in order to allow the more time-consuming catalytic conversion of H3K27me2 to H3K27me3 (Sneeringer et al., 2010; Zee et al., 2012), while the more rapidly produced H3K27me1 and H3K27me2 may arise through more transient interactions. Importantly, however, H3K27me3 is also detected at lower levels in regions without stably associated PRC2. In this context, it is interesting to consider that a minimal, catalytically competent, truncated PRC2 (consisting of just EZH2, EED and the C-terminal VEFS domain of SUZ12) is able to catalyze normal global levels of H3K27 methylation, despite having undetectable chromatin binding as measured by ChIP analysis (Hojfeldt et al., 2018). This surprising observation demonstrates that stable PRC2 binding is not a requirement for the formation of high global levels of H3K27me3, and stabilization of PRC2 at CGIs could therefore be viewed as a way of focusing the H3K27me3 distribution.
On the term ‘recruitment’
The molecular mechanisms that facilitate PRC2 occupancy at CGIs are colloquially, and in this review, termed ‘recruitment’. The term implies a mechanism similar to what in gene regulation is mediated by TFs, which target genomic regions through sequence-specific DNA-binding domains and recruit cofactors through protein-protein interaction domains. However, no such recruiter is known for PRC2. Sequence-specific factors are likely not involved in direct recruitment, as CGIs lack recurring motifs and PRC2 binding can be achieved by synthetic or non-evolved sequences (Lynch et al., 2012; Mendenhall et al., 2010; Wachter et al., 2014). Instead, PRC2-bound CGIs are characterized by high CpG content, unique DNA conformations, lack of transcription and co-occupancy of PRC1 and H2AK119ub1. Thus, PRC2 subunits or interactors utilizing these features to support long residence time at CGIs might be considered as PRC2 recruiters.
In addition to such recruiters, factors directly influencing the catalytic activity of PRC2 may modulate the local deposition of H3K27 methylation. Patterns of H3K27 methylation might be further modulated through active demethylation by the KDM6 family of demethylases. While dispensable for early embryonic development and their loss not leading to global increases in H3K27 methylation (Shpargel et al., 2014), in vitro studies suggest that they may be required for gene activation in response to specific signaling events (reviewed in (Kooistra and Helin, 2012)). Moreover, recent results indicate that histone turnover could be an important mechanism contributing to the establishment of H3K27 methylation patterns (Chory et al., 2019).
Factors impacting PRC2 recruitment and activity
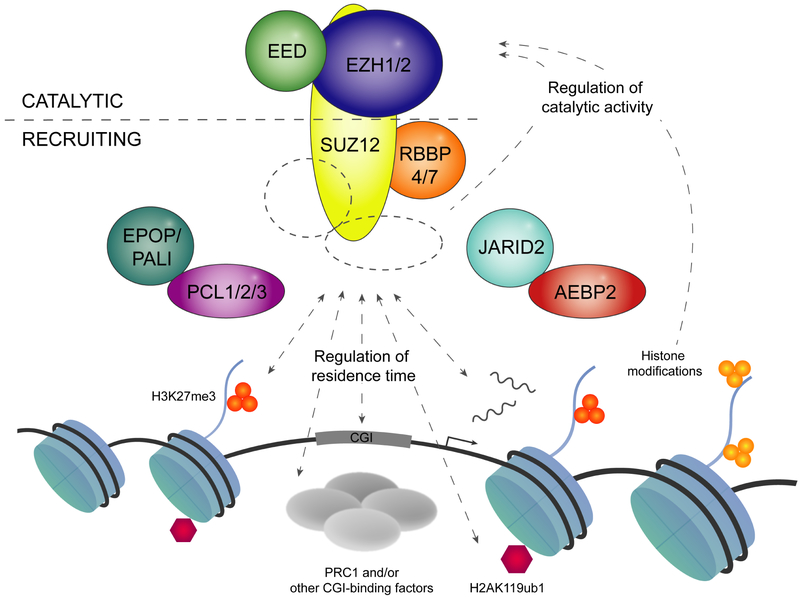
Many different factors have been proposed to modulate PRC2 recruitment and catalytic activity. These include direct interaction of core PRC2 members and a set of non-core PRC2 subunits with DNA and histones and further modulation of residence time and catalytic activity by local chromatin context, RNA, PRC1-mediated H2AK119ub1 and other histone modifications (summarized in Figure 3 and Table 1). In the following, we will review the potential impact of each of these factors on PRC2 recruitment and their role in shaping H3K27 methylation patterns.
Figure 3: Schematic representation of factors potentially influencing PRC2 recruitment or catalytic activity.
SUZ12 acts as scaffold for the complex, containing a C-terminal VEFS domain binding to EED and EZH2/1 and a N-terminal domain with associated non-core subunits. Chromatin binding is dependent on the N-terminal part of SUZ12, and CGI residence time may be modulated by non-core subunits, transcriptional activity and colocalizing factors including PRC1, H2AK119ub1 and H3K27me3. The catalytic activity of PRC2 may be influenced by non-core subunits and post-translational modifications of histones (H3K27me3, H3K4me3 and H3K36me2/me3).
Table 1.
Mammalian a PRC2 subunits and their potential roles in PRC2 function
| Chromatin binding | Catalytic activity | References | |
|---|---|---|---|
| Core PRC2 subunits | |||
| EZH1/2 | H3 tail (substrate) via SET domain. Nucleosome-binding by EZH1. | Catalytic subunit. EZH2 activity > EZH1 activity. | (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Lee et al., 2018a; Muller et al., 2002; Son et al., 2013) |
| EED | H3K27me3 (aromatic cage). | Required for catalytic activity. Allosteric activation via binding of H3K27me3 and potentially JARID2K116me3. | (Cao and Zhang, 2004; Lee et al., 2018a; Lee et al., 2018b; Margueron et al., 2009) |
| SUZ12 | N-terminal SUZ12 required for chromatin binding in vivo. | VEFS-domain required for catalytic activity in vitro and in vivo. Stimulation of activity by dense chromatin. | (Choi et al., 2017; Hojfeldt et al., 2018; Yuan et al., 2012) |
| Non-core PRC2 subunits | |||
| RBBP4/7 | Core histone binding. | Stimulation of in vitro activity. | (Cao and Zhang, 2004; Murzina et al., 2008; Schmitges et al., 2011) |
| PRC2.1 | |||
| EPOP | Stimulation of in vitro activity. | (Zhang et al., 2011) | |
| PALI | Stimulation of in vitro activity. | (Conway et al., 2018) | |
| PCL1-3 | CGI-binding (EH domain). | Stimulation of in vitro activity. | (Cao et al., 2008; Li et al., 2017; Sarma et al., 2008; Wang et al., 2017) |
| PRC2.2 | |||
| AEBP2 | Stimulation of in vitro activity. | (Cao and Zhang, 2004; Lee et al., 2018a) | |
| JARID2 | H2AK119ub1-binding (N-terminal ubiquitin-interaction motif). | Allosteric activation through EED-based recognition of EZH2-deposited JARID2K116me3. | (Blackledge et al., 2014; Cooper et al., 2014; Cooper et al., 2016; Kalb et al., 2014; Sanulli et al., 2015; Son et al., 2013) |
While Table 1 summarizes the properties of mammalian PRC2 subunits and their roles in PRC2 function, some cited references include data from other species, in particular Drosophila.
Physical properties of PRC2-bound regions
PcG proteins were originally identified in Drosophila and PRC recruitment has been studied extensively in this context. Here, they are recruited to Polycomb Response Elements (PREs), which are long DNA stretches with binding sites for TFs that contribute to the recruitment of PRCs to chromatin. Most of the TFs involved in PRC recruitment in Drosophila are not conserved in mammals, and while several other TFs have been shown to interact with mammalian PRCs, they do not confer a general mechanism for PRC recruitment in mammals (Schuettengruber et al., 2017).
Mammalian PRC1 and PRC2 display largely overlapping genomic binding patterns (Boyer et al., 2006; Bracken et al., 2006), primarily enriched near the transcription start site in CpG-rich promoters of non-transcribed genes (Ku et al., 2008; Tanay et al., 2007), implicating CGIs as the mammalian cis element required for PRC recruitment. Ectopic, unmethylated CGI-like DNA is able to recruit PRC2, when incorporated into transcriptionally inactive genomic regions (Jermann et al., 2014; Lynch et al., 2012; Mendenhall et al., 2010; Wachter et al., 2014) and PRC2 is recruited to CpG-rich promoters of previously active genes upon excision of activating motifs (Hosogane et al., 2016; Mendenhall et al., 2010) or drug-induced blockage of transcription (Riising et al., 2014).
It has been speculated whether cis-regulatory PRC2-binding elements might be conserved that are not defined by sequence motifs recognized by TFs, but rather DNA compositions that impart physical properties such as nucleosome spacing, DNA shape or overall chromatin conformation. In this context, the PCL proteins, a family of non-core PRC2 subunits, were recently shown to bind CpG-rich DNA through a mechanism suggested to include recognition of specific DNA shape (Li et al., 2017; Perino et al., 2018), but the significance of DNA shape as a determinant of PRC2 recruitment is unclear. Interestingly, both core PRC2 and PCL-containing PRC2 have been shown to interact most strongly with naked DNA, indicating that nucleosome-depleted regions with extended linker DNA observed at PRC2-bound CGIs and Drosophila PREs (Deal et al., 2010; Mito et al., 2007; Riising et al., 2014) may directly contribute to PRC2 binding (Choi et al., 2017; Wang et al., 2017).
While extended linker DNA promotes increased residence time and deposition of H3K27 methylation, dense chromatin has been shown to stimulate PRC2 activity in a SUZ12-dependent manner (Yuan et al., 2012). The functional implication of this observation, and whether it might influence deposition of H3K27me3 in heterochromatic non-CGI contexts remains to be explored.
Proposed roles of RNA in modulating PRC2 recruitment
There have been many reports of RNAs interacting with PRC2, and some have been proposed to guide PRC2 to specific loci. Examples of PRC2-associated ncRNAs include the cis-acting XIST-RepA transcript proposed to guide PRC2 recruitment to the X chromosome during X chromosome inactivation (Kohlmaier et al., 2004), and the ncRNA HOTAIR transcribed from the HOXC locus and proposed to drive PRC2 recruitment and silencing in trans to the HOXD locus (Rinn et al., 2007). However, while RepA is required for X inactivation, it is dispensable for PRC2 binding (da Rocha et al., 2014), and HOTAIR-mediated repression of the HOXD locus has been shown to be independent of PRC2 (Portoso et al., 2017). Therefore, it still remains uncertain whether long ncRNAs play a direct role in recruiting PRC2 to chromatin.
In addition to the long ncRNAs with proposed roles in PRC2 recruitment, several RNA immunoprecipitation experiments have shown massive ‘non-specific’ RNA binding by EZH2 or SUZ12 (Davidovich et al., 2013; Kaneko et al., 2014; Kaneko et al., 2013; Zhao et al., 2010). Some have considered that short transcripts from PRC2-bound genes might play a role in ‘tethering’ PRC2 to it target gene promoters (Kanhere et al., 2010), but the functional relevance of this model remains unclear. Others have proposed that RNA precludes PRC2 from binding to actively transcribed regions through competition or interference with chromatin binding interfaces (Beltran et al., 2016). This is supported by findings that RNA competes with DNA binding to PRC2, while the PRC2 autocatalytic activity is unaffected by the presence of RNA (Wang et al., 2017).
Thus, while a general role of sequence-specific ncRNAs in targeting PRC2 seems tenuous, competitive displacement of PRC2 by small RNAs preventing PRC2 binding in transcribed regions may explain how PRC2 ‘senses’ transcriptional activity.
Reciprocal relationship between PRC1 and PRC2
The coinciding binding patterns of PRC1 and PRC2 along with observations that PRC1-binding is abolished in E(z) (the Drosophila ortholog of EZH1/2) mutant flies (Rastelli et al., 1993), and the preference for H3K27me2/me3-binding by chromodomains in the CBX-components of PRC1 (Cao et al., 2002) led to the proposal of the classical, hierarchical model of PRC recruitment. This model proposes that initial PRC2 binding and methylation of H3K27 directs the recruitment of PRC1, which in turn facilitates chromatin compaction and stable repression of non-transcribed genes (Francis et al., 2004). Accordingly, PRC1 recruitment was considered secondary to PRC2 binding and much focus was put on elucidation of the mechanisms governing PRC2 recruitment. However, RING1A/B form distinct subcomplexes (termed PRC1.1-1.6), of which only the ‘canonical’ PRC1 complexes, PRC1.2 and PRC1.4, contain a CBX subunit (Blackledge et al., 2015; Gao et al., 2012; Lagarou et al., 2008). These findings correlate well with the observation that PRC2 depletion has relatively mild effects on global H2AK119ub1 levels (Leeb et al., 2010), indicating that at least a subset of PRC1 complexes, catalyzing a substantial fraction of total H2AK119ub1, relies on PRC2- and H3K27me3-independent recruitment mechanisms (Tavares et al., 2012). One such mechanism was identified by findings that KDM2B mediates recruitment of PRC1.1 through its CXXC-domain, which binds unmethylated CpG-rich DNA (Farcas et al., 2012; He et al., 2013; Wu et al., 2013). Along with indications that H2AK119ub1 modulates binding and/or catalytic activity of PRC2 through interaction with JARID2 (Blackledge et al., 2014; Cooper et al., 2014; Cooper et al., 2016; Kalb et al., 2014), this points to an, as of yet incompletely understood, reciprocal relationship between PRC1 and PRC2, which may contribute synergistically to the development of strongly bound and modified Polycomb domains (reviewed in (Blackledge et al., 2015)).
Histone modifications influencing PRC2 binding or activity
In addition to the above-mentioned interaction of H2AK119ub1 with JARID2-containing PRC2 complexes, several other histone modifications may influence PRC2 binding or catalytic activity, including the PRC2 product H3K27me3 as well as H3K36me2/me3 and H3K4me3.
The methyltransferase activity of EZH2 can be allosterically activated by binding of H3K27me3 by an aromatic cage in the core PRC2 subunit EED, located proximally to the PRC2 active site. This binding has been suggested to be required for effective deposition and spreading of H3K27me3 in PRC2- bound regions (Hansen et al., 2008; Margueron et al., 2009; Oksuz et al., 2018). Structural studies show that a single PRC2 complex may indeed bind between two nucleosomes, allowing H3K27me3 from one nucleosome to contact EED, while the active site of EZH2 contacts the neighboring nucleosome (Poepsel et al., 2018). The allosteric effect is proposed to arise via subtle conformational changes reaching from the EED-EZH2 interface to the active site in the EZH2 SET domain (Jiao and Liu, 2015, 2016; Lee et al., 2018b) and may contribute to the enrichment of H3K27me3 domains in regions that already contain the modification. While EED-mediated H3K27me3-binding may well modulate PRC2 activity to facilitate ‘spreading’ of H3K27me3, it is certainly dispensable for PRC2 recruitment, since SUZ12 binds independently of PRC2 complex formation and H3K27 methylation (Hojfeldt et al., 2018), and H3K27me3-binding by EED is also not sufficient to facilitate recruitment and maintenance of H3K27me3 domains (Coleman and Struhl, 2017; Laprell et al., 2017).
The Tudor domains of PCL proteins have been shown to interact with H3K36me3, and the interaction of PCLs with H3K36me3 has been proposed to promote PRC2 recruitment, deposition of H3K27me3 and transcriptional repression (Ballare et al., 2012; Brien et al., 2012; Cai et al., 2013). Conversely, findings that H3K36me2/me3 and H3K4me3 inhibit PRC2 activity and that H3K4me3 inhibits PRC2 binding in vitro have given rise to a putative role of these modifications in exclusion of PRC2 activity from transcribed regions (Schmitges et al., 2011). While PCL1-mediated recognition of H3K36me3 has been confirmed to inhibit the catalytic activity of PRC2 both in vitro and in vivo, chromatin binding remains unperturbed (Li et al., 2017; Musselman et al., 2012; Yuan et al., 2011). Furthermore, a recent study found that PRC2 binds nucleosomes carrying different histone modifications with similar affinities, and the affinities of PRC2 to histones are much lower than those observed between PRC2 and DNA (Wang et al., 2017). Thus, it seems likely that PCL proteins promote stabilization of PRC2-binding to DNA, while any local interactions with H3K36me2/me3 or H3K4me3 might limit the catalytic efficiencies rather than affecting chromatin binding.
Collectively, the functional implication of histone modifications in regulating PRC2 binding or activity are incompletely understood and may well be influenced by the exact molecular composition of PRC2 according to the incorporation of different non-core subunits with distinct properties. Importantly, however, none of the described modifications explain the specific binding of PRC2 to CGIs, and co-occurring histone modifications appear to primarily influence PRC2 function through modulation of its catalytic activity, rather than through orchestration of binding patterns.
Properties of PRC2 subunits
Both core and non-core PRC2 subunits contain several putative DNA- or histone-binding domains (reviewed in (Margueron and Reinberg, 2011)), but no single factor has been shown to be solely responsible for establishing PRC2 binding at CGIs. The potential role of each domain and subunit in mediating PRC2 recruitment has been addressed through loss-of-function studies, mutational analyses and assessment of in vitro or in vivo PRC2 binding. In vitro binding assays with recombinant PRC2 show that core PRC2 has intrinsic affinity for chromatin with the highest affinities observed for linker DNA (Choi et al., 2017; Wang et al., 2017). As described in more detail below, this interaction can be modulated by the presence of different non-core PRC2 subunits.
The SUZ12 N-terminal is required for chromatin-binding
It was recently found that the N-terminal part of SUZ12 (lacking the VEFS domain) localizes to PRC2 target genes in the absence of PRC2 complex formation and H3K27 methylation (Hojfeldt et al., 2018). In accordance with this, single-molecule tracing in live-cell imaging experiments shows that the fraction of chromatin-bound N-terminal SUZ12 corresponds to that of holo-PRC2, while VEFS-PRC2 (comprised of the C-terminal SUZ12 VEFS domain bound to EED and EZH2) displays increased cellular dynamics lacking chromatin interactions with long residence time. Interestingly, the chromatin-bound fraction is also reduced in cells expressing specific N-terminal mutations in SUZ12 disrupting the interaction with PCL2 and/or AEBP2, suggesting that these subunits contribute to the binding of PRC2 to chromatin (Youmans et al., 2018).
Thus, the interactions with H3 tails with various modifications mediated by EED and EZH2 are not sufficient to mediate strong chromatin binding, while the N-terminal part of SUZ12 is, either in itself or through interaction with other factors, mediating chromatin binding in a manner independent from the rest of the core PRC2 complex. In line with this, a host of recent structural studies illustrate that the non-core PRC2 subunits, suggested to contribute to PRC2 recruitment, bind to the N-terminal part of SUZ12 (reviewed in (Kasinath et al., 2018b)) and may thus contribute to the observed chromatin localization of both N-terminal SUZ12 and PRC2 as a whole.
Non-core PRC2 subunits regulating PRC2 recruitment and activity
RBBP4/7 is present in both PRC2.1 and PRC2.2 complexes and binds PRC2 through the N-terminal part of SUZ12 (Schmitges et al., 2011). Its incorporation increases the intrinsic activity of core PRC2 on oligonucleosome substrates (Cao and Zhang, 2004). The importance of histone-binding by RBBP4/7 for PRC2 recruitment is difficult to test experimentally, as they are subunits in several other chromatin-associated complexes and essential for cell proliferation. RBBP4 has been proposed to guide some of these complexes to genomic regions through recognition of unmethylated lysine 4 on histone H3 (H3K4me0) (Schmitges et al., 2011). However, pull-down assays with H3K4me0-peptides recover components of the NuRD deacetylase complex, but not PRC2, illustrating that incorporation of RBBP4 into PRC2 changes its H3K4me0-binding (Chen et al., 2018). Structural analyses further reveal that both SUZ12 and the non-core subunit AEBP2 can interact with RBBP4 via its known H3 tail binding surface (Chen et al., 2018; Kasinath et al., 2018a), which would preclude such binding. Thus, it appears unlikely that histone-binding by RBBP4/7 is involved in guiding PRC2 chromatin binding.
The PCL proteins, part of PRC2.1, have recently been shown to bind CpG-rich DNA through their winged helix DNA-binding domains in their N-terminal extended homology (EH) region. Supporting the role of PCLs in PRC2 recruitment, knockout of Pcl2/Mtf2, leads to reduced SUZ12-binding at CGIs. While one data set shows a strong concomitant depletion of H3K27me3 (Perino et al., 2018), in a separate study, H3K27me3 patterns were only modestly affected upon Pcl2 knockout (Li et al., 2017). The remaining H3K27me3 has been suggested to stem from redundancy from the other PCL homologs. In support of this notion, PCL3/PHF19 has also been suggested to promote PRC2 recruitment to a subset of Polycomb target genes (Ballare et al., 2012; Brien et al., 2012). The role of PCLs in PRC2 recruitment was further substantiated by findings that PCL1/PHF1-containing PRC2 complexes associate two-to-three-fold more stably with nucleosome arrays compared to core PRC2 alone. The stabilization is dependent on the N-terminal part of PCL1/PHF1 and yields increased deposition of H3K27me3 (Choi et al., 2017). This study confirms other observations that PRC2 binds linker DNA with affinities several orders of magnitude higher than those observed for PRC2-histone interactions mediated by EED and RBBP4/7 (Cao et al., 2002; Choi et al., 2017; Lee et al., 2018a; Margueron et al., 2009). These recent studies on the PCL proteins suggest an important role for these proteins in PRC2 chromatin binding and potentially provide an interaction that contribute to CGI binding strength and preference of PRC2.1.
The PRC2.1 component EPOP has been demonstrated to interact with CGIs regardless of their transcriptional status. Despite described roles in stimulating PRC2 activity (Zhang et al., 2011), depletion of EPOP has been shown to promote increased PRC2 binding and H3K27me3 at PRC2 targets (Beringer et al., 2016; Liefke and Shi, 2015), and its role in modulating PRC2 binding or H3K27 methylation remains incompletely understood. In PRC2.1, EPOP is mutually exclusive with PALI1/2 (Alekseyenko et al., 2014; Hauri et al., 2016), and Pali1/2 knockout was found to yield slightly reduced H3K27me3 at some PRC2 target genes (Conway et al., 2018), suggesting that the specific incorporation of either subunit might further differentiate PRC2.1 function.
Addition of the PRC2.2 component AEBP2 to RBBP4-PRC2 further potentiates the methylation readout of in vitro studies (Cao and Zhang, 2004). A recent study confirmed the stimulatory effect, showing that AEBP2 promotes both increased oligonucleosome-binding and stimulation of catalytic activity of both EZH1-PRC2 and EZH2-PRC2 in a mechanism distinct from the allosteric stimulation of the H3K27me3-EED interaction (Lee et al., 2018a).
Another PRC2.2 subunit, JARID2, has been suggested as a potential PRC2 recruiter and modulator of PRC2 activity based on its interaction with PRC2, significant overlap in genomic target sites and slight affinity for GC-rich DNA through its ARID domain. While loss of Jarid2 in mESCs leads to differentiation defects, it yields only modest effects on PRC2 binding and H3K27me3 patterns (Landeira et al., 2010; Li et al., 2010; Pasini et al., 2010; Peng et al., 2009; Shen et al., 2009). Interestingly, JARID2 has been directly linked to PRC1-facilitated PRC2 recruitment by studies showing that forced deposition of H2AK119ub1 promotes PRC2 recruitment in a manner dependent on a ubiquitin-interaction motif in the JARID2 N-terminal, and in vitro binding preference of JARID2 with H2AK119ub1-modified nucleosomes over unmodified nucleosomes (Cooper et al., 2016). Conversely, JARID2 does not enhance nucleosome-binding beyond five-component PRC2 in the absence of H2AK119ub1, but its inclusion in methyltransferase assays leads to increased catalytic output (Kalb et al., 2014; Son et al., 2013; Wang et al., 2017). This may stem from an autostimulatory feedback loop involving trimethylation of JARID2K116 by PRC2, which allosterically activates PRC2 via recognition by EED in a manner analogous to allosteric stimulation by H3K27me3 (Justin et al., 2016; Sanulli et al., 2015). As a consequence of H2AK119ub1 enrichment at CGIs, JARID2-mediated H2AK119ub1-binding could contribute to the CGI-preference of PRC2.2. The modest loss of H3K27me3 globally and locally at CGIs in cells lacking JARID2, however, does not support a critical role of the proposed stimulatory role of JARID2K116me3, and any potential role in conferring CGI preference through H2AK119ub1-binding would at least be covered by redundancy from other mechanisms.
Functional interplay between PRC2.1 and PRC2.2?
Based on observations from mass spectrometry data that non-core PRC2 subunits segregate into two distinct classes of complexes (PRC2.1 and PRC2.2) (Hauri et al., 2016), it is interesting to speculate about potential functional distinctions of these subcomplexes. Comprehensive comparisons of genomic binding sites of each subcomplex remain to be seen. As of yet, however, distinct sets of PRC2 target genes dependent on PRC2.1 or PRC2.2, respectively, have not been demonstrated. ChIP-seq studies of the different non-core subunits show large overlaps with core PRC2 (Brien et al., 2012; Grijzenhout et al., 2016; Li et al., 2017; Pasini et al., 2010), and it therefore seems likely that they support PRC2 recruitment to overlapping sites rather than each subcomplex orchestrating PRC2 recruitment to distinct sets of target sites.
In mESCs, knockout of either Pali1/2 or Aebp2 leads to increased chromatin association of the other factor and subtle effects on expression levels of other non-core subunits. The observed effects on H3K27me3 are modest, but the study highlights how loss of individual subunits may, in addition to removing the targeted factor, skew the ‘balance’ of PRC2 subcomplexes with potentially different catalytic competences or binding affinities (Conway et al., 2018). Studies of de novo PRC2 recruitment and H3K27 methylation upon re-expression of EED in Eed knockout mESCs show that combined knockout of Jarid2 and Pcl2 exacerbate the phenotype observed upon single knockout of Pcl2 (Oksuz et al., 2018), indicating that PRC2.1 and PRC2.2 both contribute to the establishment of PRC2 binding. It is worth noting that knockout of these non-core subunits appears to influence the expression levels of both core and non-core PRC2 components to different degrees. Thus, these recent studies illustrate the difficulty in assigning effects on PRC2 binding or catalytic activities to single factors in an intricately regulated network of interaction partners with partially redundant functions.
A range of in vivo studies illustrate that knockout of non-core PRC2 subunits yield milder phenotypes with later embryonic lethality compared with knockout of core PRC2 (Brien et al., 2012; Conway et al., 2018; Grijzenhout et al., 2016; Laugesen and Helin, 2014; Li et al., 2017; Liefke and Shi, 2015; Pasini et al., 2010), indicating that the non-core subunits collaborate and compensate for each other in guiding PRC2 recruitment and H3K27 methylation. In addition to such partial molecular redundancy providing biological robustness, the different features of each non-core subunit in regulating chromatin binding and catalytic activity of PRC2 could yield flexibility to PRC2 function. While the studied subunits are all expressed in mESCs, differential expression patterns during development or variation in expression levels over the cell cycle or in response to intrinsic or extrinsic stimuli could influence PRC2 function. Modulation of the exact composition of PRC2 complexes may thus provide plasticity and responsiveness to distinct molecular cues in different cellular contexts.
Concluding remarks
The recent surge of data probing PRC2 structure and function highlights that control of PRC2 function and specification of H3K27 methylation patterns encompass an intricate interplay of factors modulating PRC2 residence time in specific chromatin regions and factors directly impacting the catalytic activity of PRC2. Collectively, the emerging picture is that core PRC2 is capable of binding linker DNA (Choi et al., 2017; Wang et al., 2017), while CGI-binding is stabilized by PCL proteins (Choi et al., 2017; Li et al., 2017). Other subunits seem to contribute to shaping H3K27 methylation patterns by further modulating binding patterns and/or influencing PRC2 catalytic rate. Negative regulation includes potential preclusion of PRC2 binding by RNA transcripts (Beltran et al., 2016; Wang et al., 2017) and prevention of aberrant deposition of H3K27me3 in transcribed regions through inhibition of catalytic activity by H3K4me3 and H3K36me2/me3 (Li et al., 2017; Schmitges et al., 2011). Conversely, other factors colocalizing with PRC2 at CGIs, including non-core PRC2 subunits, H3K27me3, and PRC1-deposited H2AK119ub1, may further stimulate activity in these regions (Cooper et al., 2016; Lee et al., 2018b; Margueron et al., 2009; Sanulli et al., 2015).
The definition of PRC2.1 and PRC2.2 together with recent structural insight and functional studies provide a very strong framework for understanding how the PRC2 complex is bound to specific sites of the genome. However, a number of open questions remain. In our view, some of the most pertinent challenges to undertake include studies to address the role of nucleosome density and nucleosome exchange in regulating H3K27 methylation patterns and PRC2 activity, studies to understand what role, if any, RNA plays in regulating PRC2 chromatin binding or catalytic activity, comprehensive studies addressing the individual and overlapping roles of non-core PRC2 subunits, and structural studies demonstrating how the N-terminal part of SUZ12, in association with non-core subunits and/or other factors, interacts with CpG-containing DNA.
Further dissection of PRC2 function and the role of its catalytic products will provide important insight to the biological processes regulated by this complex and help guide the design of novel therapeutic strategies targeting PRC2 activity.
Acknowledgments
We thank members of the Helin laboratory for discussions. The work in the Helin laboratory is supported by the Danish Cancer Society (R167-A10877), the Danish National Research Foundation (DNRF82), the Independent Research Fund Denmark (6153-000005; 7016-00067; 8020-00044), The Neye Foundation, the Novo Nordisk Foundation (NNF; NNF16OC0023234), through a center grant from the NNF to the NNF Center for Stem Cell Biology (NNF17CC0027852), and through the Memorial Sloan Kettering Cancer Center Support Grant (NIH P30 CA008748).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, and Groth A (2014). Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 16, 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Gorchakov AA, Kharchenko PV, and Kuroda MI (2014). Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc Natl Acad Sci U S A 111, 2488–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballare C, Lange M, Lapinaite A, Martin GM, Morey L, Pascual G, Liefke R, Simon B, Shi Y, Gozani O, et al. (2012). Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat Struct Mol Biol 19, 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, Fisher CL, Sibley CR, Foster BM, Bartke T, et al. (2016). The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res 26, 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer M, Pisano P, Di Carlo V, Blanco E, Chammas P, Vizan P, Gutierrez A, Aranda S, Payer B, Wierer M, et al. (2016). EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol Cell 64, 645–658. [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, et al. (2014). Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Rose NR, and Klose RJ (2015). Targeting Polycomb systems to regulate gene expression: modifications to a complex story. Nat Rev Mol Cell Biol 16, 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, and Helin K (2006). Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20, 1123–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brien GL, Gambero G, O'Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, et al. (2012). Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat Struct Mol Biol 19, 1273–1281. [DOI] [PubMed] [Google Scholar]
- Cai L, Rothbart SB, Lu R, Xu B, Chen WY, Tripathy A, Rockowitz S, Zheng D, Patel DJ, Allis CD, et al. (2013). An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell 49, 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, and Zhang Y (2008). Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol 28, 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, and Zhang Y (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043. [DOI] [PubMed] [Google Scholar]
- Cao R, and Zhang Y (2004). SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 15, 57–67. [DOI] [PubMed] [Google Scholar]
- Chen S, Jiao L, Shubbar M, Yang X, and Liu X (2018). Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding. Mol Cell 69, 840–852 e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittock EC, Latwiel S, Miller TC, and Muller CW (2017). Molecular architecture of polycomb repressive complexes. Biochem Soc Trans 45, 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Bachmann AL, Tauscher K, Benda C, Fierz B, and Muller J (2017). DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation. Nat Struct Mol Biol 24, 1039–1047. [DOI] [PubMed] [Google Scholar]
- Chory EJ, Calarco JP, Hathaway NA, Bell O, Neel DS, and Crabtree GR (2019). Nucleosome Turnover Regulates Histone Methylation Patterns over the Genome. Mol Cell 73, 61–72 e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RT, and Struhl G (2017). Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway E, Jerman E, Healy E, Ito S, Holoch D, Oliviero G, Deevy O, Glancy E, Fitzpatrick DJ, Mucha M, et al. (2018). A Family of Vertebrate-Specific Polycombs Encoded by the LCOR/LCORL Genes Balance PRC2 Subtype Activities. Mol Cell 70, 408–421 e408. [DOI] [PubMed] [Google Scholar]
- Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, et al. (2014). Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep 7, 1456–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Grijzenhout A, Underwood E, Ancelin K, Zhang T, Nesterova TB, Anil-Kirmizitas B, Bassett A, Kooistra SM, Agger K, et al. (2016). Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat Commun 7, 13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, and Pirrotta V (2002). Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111, 185–196. [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, et al. (2014). Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol Cell 53, 301–316. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, and Cech TR (2013). Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol 20, 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, and Henikoff S (2010). Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328, 1161–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, et al. (2012). KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife 1, e00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stutzer A, Fischle W, Bonaldi T, and Pasini D (2014). Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol Cell 53, 49–62. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, and Woodcock CL (2004). Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577. [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, and Reinberg D (2012). PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45, 344–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grijzenhout A, Godwin J, Koseki H, Gdula MR, Szumska D, McGouran JF, Bhattacharya S, Kessler BM, Brockdorff N, and Cooper S (2016). Functional analysis of AEBP2, a PRC2 Polycomb protein, reveals a Trithorax phenotype in embryonic development and in ESCs. Development 143, 2716–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, and Helin K (2008). A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10, 1291–1300. [DOI] [PubMed] [Google Scholar]
- Hauri S, Comoglio F, Seimiya M, Gerstung M, Glatter T, Hansen K, Aebersold R, Paro R, Gstaiger M, and Beisel C (2016). A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep 17, 583–595. [DOI] [PubMed] [Google Scholar]
- He J, Shen L, Wan M, Taranova O, Wu H, and Zhang Y (2013). Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol 15, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojfeldt JW, Laugesen A, Willumsen BM, Damhofer H, Hedehus L, Tvardovskiy A, Mohammad F, Jensen ON, and Helin K (2018). Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat Struct Mol Biol 25, 225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosogane M, Funayama R, Shirota M, and Nakayama K (2016). Lack of Transcription Triggers H3K27me3 Accumulation in the Gene Body. Cell Rep 16, 696–706. [DOI] [PubMed] [Google Scholar]
- Jermann P, Hoerner L, Burger L, and Schubeler D (2014). Short sequences can efficiently recruit histone H3 lysine 27 trimethylation in the absence of enhancer activity and DNA methylation. Proc Natl Acad Sci U S A 111, E3415–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L, and Liu X (2015). Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350, aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L, and Liu X (2016). Structural analysis of an active fungal PRC2. Nucleus 7, 284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HR, Pasini D, Helin K, and Jensen ON (2010). Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol Cell Proteomics 9, 838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, De Marco V, Haire LF, Walker PA, Reinberg D, et al. (2016). Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun 7, 11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, and Muller J (2014). Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 21, 569–571. [DOI] [PubMed] [Google Scholar]
- Kaneko S, Son J, Bonasio R, Shen SS, and Reinberg D (2014). Nascent RNA interaction keeps PRC2 activity poised and in check. Genes Dev 28, 1983–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, and Bonasio R (2013). PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol 20, 1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. (2010). Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 38, 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinath V, Faini M, Poepsel S, Reif D, Feng XA, Stjepanovic G, Aebersold R, and Nogales E (2018a). Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359, 940–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinath V, Poepsel S, and Nogales E (2018b). Recent structural insights into PRC2 regulation and substrate binding. Biochemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, and Wutz A (2004). A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2, E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra SM, and Helin K (2012). Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 13, 297–311. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. (2008). Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4, e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, and Reinberg D (2002). Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16, 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, and Verrijzer CP (2008). dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 22, 2799–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jorgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. (2010). Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol 12, 618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprell F, Finkl K, and Muller J (2017). Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 356, 85–88. [DOI] [PubMed] [Google Scholar]
- Laugesen A, and Helin K (2014). Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14, 735–751. [DOI] [PubMed] [Google Scholar]
- Laugesen A, Hojfeldt JW, and Helin K (2016). Role of the Polycomb Repressive Complex 2 (PRC2) in Transcriptional Regulation and Cancer. Cold Spring Harb Perspect Med 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Holder M, Grau D, Saldana-Meyer R, Yu JR, Ganai RA, Zhang J, Wang M, LeRoy G, Dobenecker MW, et al. (2018a). Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2. Mol Cell 70, 435–448 e435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Yu JR, Kumar S, Jin Y, LeRoy G, Bhanu N, Kaneko S, Garcia BA, Hamilton AD, and Reinberg D (2018b). Allosteric Activation Dictates PRC2 Activity Independent of Its Recruitment to Chromatin. Mol Cell 70, 422–434 e426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, and Wutz A (2010). Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 24, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, and Reinberg D (2010). Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24, 368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liefke R, Jiang J, Kurland JV, Tian W, Deng P, Zhang W, He Q, Patel DJ, Bulyk ML, et al. (2017). Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 549, 287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefke R, and Shi Y (2015). The PRC2-associated factor C17orf96 is a novel CpG island regulator in mouse ES cells. Cell Discov 1, 15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MD, Smith AJ, De Gobbi M, Flenley M, Hughes JR, Vernimmen D, Ayyub H, Sharpe JA, Sloane-Stanley JA, Sutherland L, et al. (2012). An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J 31, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. (2009). Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, and Reinberg D (2008). Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32, 503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, and Reinberg D (2011). The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, and Bernstein BE (2010). GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6, e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, and Henikoff S (2007). Histone replacement marks the boundaries of cis-regulatory domains. Science 315, 1408–1411. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, and Simon JA (2002). Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111, 197–208. [DOI] [PubMed] [Google Scholar]
- Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, et al. (2008). Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16, 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde ME, Hong Z, Allen C, Roy S, Nunez JK, Nickoloff J, et al. (2012). Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1. Nat Struct Mol Biol 19, 1266–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksuz O, Narendra V, Lee CH, Descostes N, LeRoy G, Raviram R, Blumenberg L, Karch K, Rocha PP, Garcia BA, et al. (2018). Capturing the Onset of PRC2-Mediated Repressive Domain Formation. Mol Cell 70, 1149–1162 e1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, and Helin K (2010). JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464, 306–310. [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, and Wysocka J (2009). Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139, 1290–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly AR, Copur O, Jackle H, Herzig A, and Muller J (2013). A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339, 698–699. [DOI] [PubMed] [Google Scholar]
- Perino M, van Mierlo G, Karemaker ID, van Genesen S, Vermeulen M, Marks H, van Heeringen SJ, and Veenstra GJC (2018). MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat Genet 50, 1002–1010. [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. (2003). Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12, 1577–1589. [DOI] [PubMed] [Google Scholar]
- Poepsel S, Kasinath V, and Nogales E (2018). Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat Struct Mol Biol 25, 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portoso M, Ragazzini R, Brencic Z, Moiani A, Michaud A, Vassilev I, Wassef M, Servant N, Sargueil B, and Margueron R (2017). PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J 36, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli L, Chan CS, and Pirrotta V (1993). Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J 12, 1513–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveron-Gomez N, Gonzalez-Aguilera C, Stewart-Morgan KR, Petryk N, Flury V, Graziano S, Johansen JV, Jakobsen JS, Alabert C, and Groth A (2018). Accurate Recycling of Parental Histones Reproduces the Histone Modification Landscape during DNA Replication. Mol Cell 72, 239–249 e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising EM, Comet I, Leblanc B, Wu X, Johansen JV, and Helin K (2014). Gene silencing triggers polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol Cell 55, 347–360. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanulli S, Justin N, Teissandier A, Ancelin K, Portoso M, Caron M, Michaud A, Lombard B, da Rocha ST, Offer J, et al. (2015). Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol Cell 57, 769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, and Reinberg D (2008). Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol 28, 2718–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. (2011). Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42, 330–341. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Bourbon HM, Di Croce L, and Cavalli G (2017). Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 171, 34–57. [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, and Orkin SH (2009). Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139, 1303–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, and Orkin SH (2008). EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32, 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel KB, Starmer J, Yee D, Pohlers M, and Magnuson T (2014). KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet 10, e1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits AH, Jansen PW, Poser I, Hyman AA, and Vermeulen M (2013). Stoichiometry of chromatin-associated protein complexes revealed by label-free quantitative mass spectrometry-based proteomics. Nucleic Acids Res 41, e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, and Copeland RA (2010). Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A 107, 20980–20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Shen SS, Margueron R, and Reinberg D (2013). Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev 27, 2663–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, O'Donnell AH, Damelin M, and Bestor TH (2007). Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci U S A 104, 5521–5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. (2012). RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaswani RG, Gehling VS, Dakin LA, Cook AS, Nasveschuk CG, Duplessis M, Iyer P, Balasubramanian S, Zhao F, Good AC, et al. (2016). Identification of (R)-N-((4-Methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1 -(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a Potent and Selective Inhibitor of Histone Methyltransferase EZH2, Suitable for Phase I Clinical Trials for B-Cell Lymphomas. J Med Chem 59, 9928–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter E, Quante T, Merusi C, Arczewska A, Stewart F, Webb S, and Bird A (2014). Synthetic CpG islands reveal DNA sequence determinants of chromatin structure. Elife 3, e03397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Paucek RD, Gooding AR, Brown ZZ, Ge EJ, Muir TW, and Cech TR (2017). Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol 24, 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Johansen JV, and Helin K (2013). Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell 49, 1134–1146. [DOI] [PubMed] [Google Scholar]
- Youmans DT, Schmidt JC, and Cech TR (2018). Live-cell imaging reveals the dynamics of PRC2 and recruitment to chromatin by SUZ12-associated subunits. Genes Dev 32, 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. (2012). Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337, 971–975. [DOI] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, and Zhu B (2011). H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286, 7983–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zee BM, Britton LM, Wolle D, Haberman DM, and Garcia BA (2012). Origins and formation of histone methylation across the human cell cycle. Mol Cell Biol 32, 2503–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jones A, Sun CW, Li C, Chang CW, Joo HY, Dai Q, Mysliwiec MR, Wu LC, Guo Y, et al. (2011). PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells 29, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, and Lee JT (2010). Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40, 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]