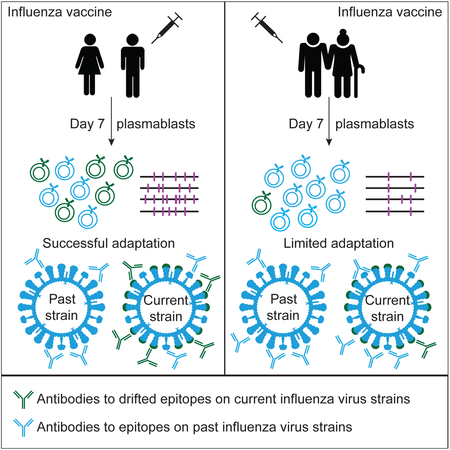
SUMMARY
Influenza is a leading cause of death in the elderly, and the vaccine protects only a fraction of this population. A key aspect of antibody-mediated anti-influenza virus immunity is adaptation to antigenically distinct epitopes on emerging strains. We examined factors contributing to reduced influenza vaccine efficacy in the elderly and uncovered a dramatic reduction in the accumulation of de novo immunoglobulin gene somatic mutations upon vaccination. This reduction is associated with a significant decrease in the capacity of antibodies to target the viral glycoprotein, hemagglutinin (HA), and critical protective epitopes surrounding the HA receptor-binding domain. Immune escape by antigenic drift, in which viruses generate mutations in key antigenic epitopes, becomes highly exaggerated. Due to this reduced adaptability, most B cells activated in the elderly cohort target highly conserved but less potent epitopes. Given these findings, vaccines driving immunoglobulin gene somatic hypermutation should be a priority to protect elderly individuals.
Graphical Abstract
eTOC blurb
Influenza virus vaccination elicits poor efficacy in elderly individuals. Henry et al. find that elderly adults have a reduced accumulation of de novo immunoglobulin gene somatic mutations and are unable to adapt their antibody responses upon influenza virus vaccination. These results should be considered when designing vaccines for elderly populations.
INTRODUCTION
The detrimental effect of aging on the immune system or “immunosenescence” is thought to be a major cause of morbidity and mortality in elderly adults by increasing susceptibility to bacterial, fungal and viral infections (Chen et al., 2009; Frasca and Blomberg, 2014; Marrie, 2000). The great majority of influenza deaths occur within populations older than 65 years, and aged individuals have a significantly reduced antibody response to influenza vaccination (Goodwin et al., 2006; Sasaki et al., 2011; Thompson et al., 2003). A critical component of antibody-mediated immunity to influenza virus is adaptation to antigenically distinct epitopes on emerging drifted and shifted strains. Immunoglobulin gene somatic hypermutation is predicted to be critical for this adaptation. While the mechanism of V(D)J recombination diversifies the initial variable gene repertoire, B cells undergo affinity maturation following antigen exposure in germinal centers (GCs) through the process of somatic hypermutation (SHM) (Eisen, 2014). In mice, there is a reduction in SHM with age (Miller and Kelsoe, 1995; Yang et al., 1996) and a reduction of the size of GCs (Zheng et al., 1997). In humans, conflicting results have been published to date (Chong et al., 2003; Rosner et al., 2001; Troutaud et al., 1999), though older adults exhibited restricted clonal diversity, signifying a reduced substrate for mounting novel responses and decreased fine-tuning of B-cell receptor (BCR) specificities by SHM (de Bourcy et al., 2017; Jiang et al., 2013). Functional pathways and B cell differentiation associated with SHM against influenza virus antigens have also been shown to be altered in various contexts (reviewed in (Cancro et al., 2009; Frasca and Blomberg, 2014)). This substantial published evidence of immune decline suggests that aged subjects may have a limited capacity to undergo critical adaptations of their antibody response by SHM.
Plasmablasts are a transient population of B cells activated upon antigen exposure, reflecting the ongoing immune response (Wrammert et al., 2008). We used the degree by which clonal plasmablasts, derived from the same progenitor with the same V(D)J rearrangements, have differentially mutated their antibody variable genes as a measure of recent mutation after influenza vaccination. Here we report that elderly individuals have a reduced accumulation of de novo mutations in their plasmablast immunoglobulin variable genes (IgV) associated with a decreased adaptability of their antibody responses to influenza virus.
RESULTS
Influenza-reactive plasmablasts from elderly individuals have reduced de novo mutations
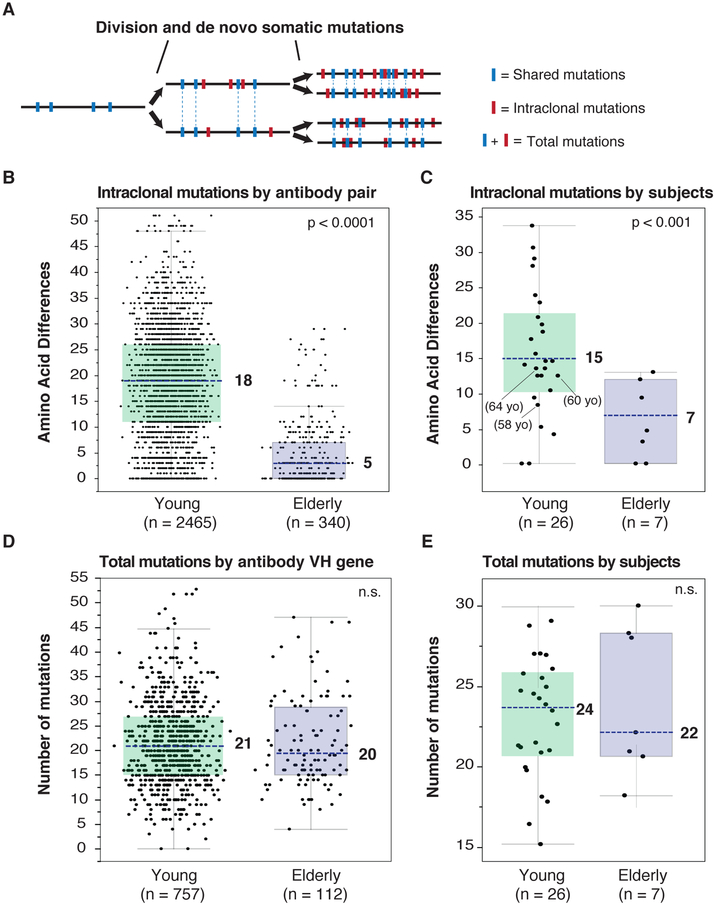
Monoclonal antibodies (mAbs) were generated from the plasmablasts that arose specifically against the administered influenza vaccine (Smith et al., 2009) from 13 elderly individuals (71-89 years old) and 26 younger adults (22-64 years old) at day 7 post-immunization. Individuals were recruited between 2006 and 2011 and received either a trivalent seasonal vaccine (Fluzone or Fluvirin) or the monovalent 2009 pandemic H1N1 vaccine (all vaccines were inactivated influenza virus vaccines) (Tables S1 and S2). To distinguish recent from preexisting mutations, we analyzed the frequency of unique amino acids between paired clonotypes from verified influenza-reactive IgV genes for 2,465 clonal pairs from the young- and 340 pairs from elderly-subjects (Figure 1A and figure S1A). We observed that elderly individuals had significantly reduced accumulation of de novo VH somatic mutations (Figure 1B and 1C). While B-cell clones from young adults had 18 amino acid differences per clonal IgV pair or 15 differences by subject, the elderly subjects had only five amino acid differences by IgV pair and seven by subject. The same observation was true for the light chain genes, though with less accumulation of mutations on the whole (Figure S1B). Because age-related changes in immune responses might develop progressively, we compared the intraclonal mutations by subject’s age 50 or lower, between the ages of 50 and 70 years, and over 70 years old. While there is a decrease in de novo mutations in the 50-70 year old group compared to subjects less than 50, there were not statistically less intraclonal mutations than the younger cohort. Interestingly, the 50 to 70 year old subjects had antibody genes that were also not significantly more diversified than the most aged cohort (Figure S1C), and thus appear to have an intermediate phenotype. Gender did not affect the results (Figure S1D). The overall mutation frequency did not differ between elderly and young adults, either by IgV gene (Figure 1D) or by subject (Figure 1E), nor did the frequency of non- and synonymous mutations (data not shown). Thus, influenza immunization activates apparent memory B cells with highly mutated IgV genes from both elderly and young individuals. However, the young cohort showed a continued recent accumulation of mutations, whereas the elderly cohort appeared to have an essentially fixed B-cell repertoire, lacking recent adaptation by somatic mutation.
Figure 1. VH mutation analysis of influenza-reactive plasmablast antibody genes.
(A) To analyze intraclonal diversity, de novo mutations of antibody genes from the same clonal pool were compared (i.e., intraclonal mutations in red). Shared mutations (in blue) were not considered for intraclonal diversity but both shared and de novo mutations were considered for total VH number of mutations.
(B-C) Antibody VH genes intraclonal mutations represented as variable distribution with quartiles (blue dotted line: median). Statistical significance: Mann–Whitney U test. (B) Amino-acid differences by antibody pair in each group (young adults n = 2465; elderly adults n = 340). (C) Intraclonal mutations by subject in each group (young adults n = 26; elderly adults n = 7).
(D-E) Total number of mutations for antibody VH genes represented as variable distribution (blue dotted line: median). Statistical significance: Mann–Whitney U test. (D) Total number of mutations by antibody VH gene in each group (young adults n = 757 influenza VH sequences; elderly n = 112). (E) Total number of mutations by subject in each group (young adults n = 26; elderly adults n = 7). n.s. not significant.
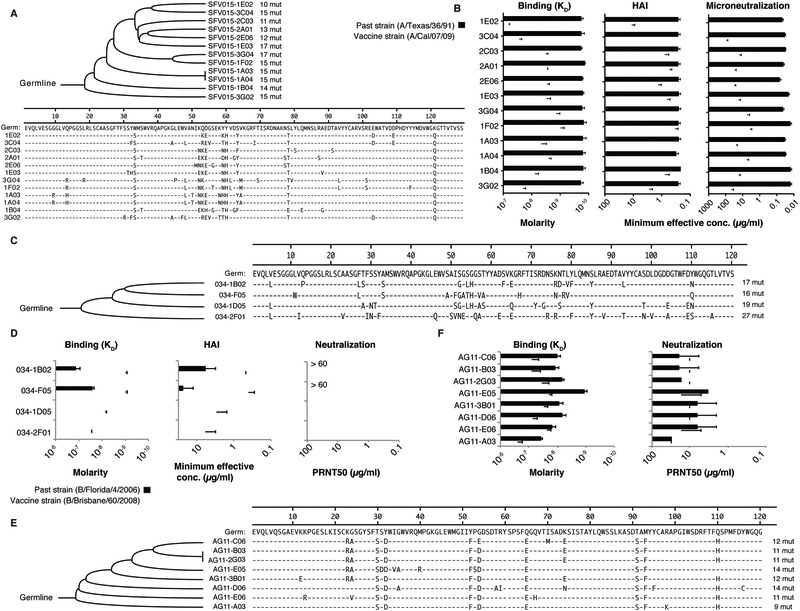
Intraclonal somatic mutations should allow the evolution of B cells to divergent influenza virus strains. We observed that members of a single plasmablast clonotype induced by the pH1N1 monovalent A/California/07/2009 vaccine strain were more reactive with the historical influenza strain A/Texas/36/1991. Mutationally distinct antibodies had up to 100-fold differences in binding avidity and widely variant HAI and microneutralization capacities against A/California/07/2009 (Figure 2A and 2B). Thus, it appears that the precursor B-cell clone was originally induced by a virus related to the A/Texas/36/1991 strain. We then compared clonotypes from young and elderly individuals when plasmablasts were activated by a seasonal influenza virus strain. The various antibodies from the young clonotype bound with higher avidity to the current vaccine strain (B/Brisbane/60/2008) than a past strain (B/Florida/4/2006) and had higher HAI and neutralization capacities against B/Brisbane/60/2008 (Figure 2C and 2D). Additionally two of the clonotype members did not display any binding to the past strain. In contrast, antibodies isolated from an elderly individual had higher binding avidity and neutralization capacity to the past strain (B/Florida/4/2006) compared to the vaccine strain (B/Brisbane/60/2008) and all the clonotype members had a similar binding/neutralizing pattern (Figure 2E and 2F). These analyses suggest that antibody responses decline with age in part because of a loss of efficient somatic hypermutation.
Figure 2. Newly introduced mutations confer the capacity to adapt to drifted and shifted influenza viruses.
(A-B) Plasmablast clonal lineage (12 mAbs) induced after the monovalent 2009 pandemic influenza vaccine. (A) Phylogenetic tree depicting the relationship between the variable gene sequences of the clonal lineage. (B Approximated binding KD determined by ELISA, HAI titers, and neutralization titers to H1N1 A/Texas/36/1991 and A/California/07/2009 virus strains.
(C-D) Plasmablast clonal lineage (4 mAbs) induced after TIV seasonal vaccine in a young adult. (C) Phylogenetic tree depicting the relationship between the variable gene sequences of the clonal lineage (D) Approximated binding KD to rHA determined by ELISA, HAI titers, and neutralization titers to B/Florida/4/2006 (past strain) and B/Brisbane/60/2008 (vaccine strain) viruses.
(E-F) Plasmablast clonal lineage (8 mAbs) induced after TIV seasonal vaccine in an elderly individual. (E) Phylogenetic tree depicting the relationship between the variable gene sequences of the clonal lineage. (F) Approximated binding KD to rNA determined by ELISA and neutralization titers to B/Florida/4/2006 (past strain) and B/Brisbane/60/2008 (vaccine strain) viruses. The assays were performed in duplicate 2-3 times for each antibody; in B, D and F. shown are mean +/− SD.
Influenza-reactive antibodies from elderly individuals predominantly target epitopes outside of the HA receptor binding site and have reduced potency
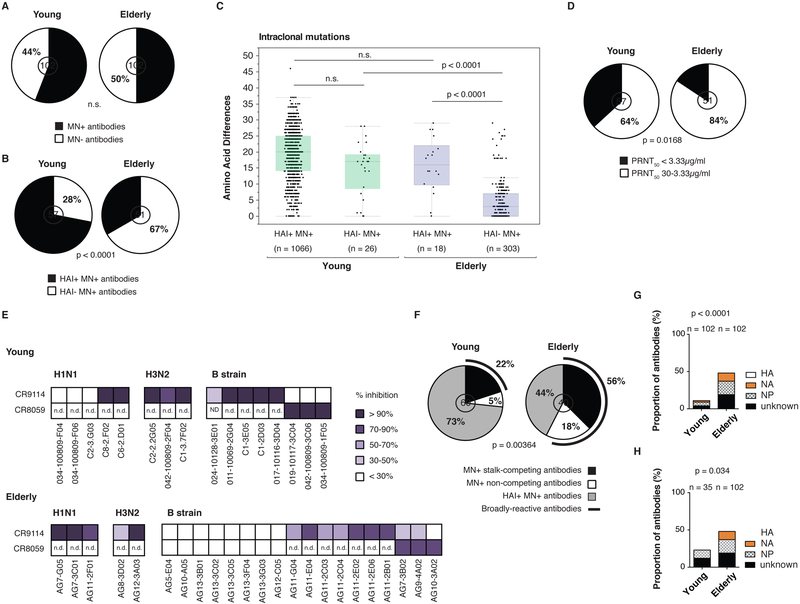
Having a reduced capacity to adapt to recent influenza strains by somatic mutation should alter the protective capacity of induced antibodies. We analyzed 102 influenza virus-reactive mAbs from the 13 elderly individuals and 18 of the young adults for their influenza strain-matched virus reactivity and protective capacity (Table S1). Although the proportion of total neutralizing mAbs did not differ between the elderly and young cohorts (Figure 3A), the proportion of HAI positive mAbs was substantially reduced in the elderly individuals (33% for elderly versus 72% for young adults, p < 0.0001) (Figure 3B). Consistently, after vaccination there was also less of an increase in HAI antibody titer observed in the serum of our elderly cohort compared to the young cohort (Figure S1E and (Sasaki et al., 2011) while the virus ELISA antibody titers did not differ (Figure S1F). To distinguish between the inability to adapt to any epitope by SHM versus a greater use of memory cells accounting for the epitope shift in the elderly cohort, we analyzed the accumulation of intraclonal mutations by epitope (Figure 3C). The young subjects had high de novo mutation frequencies for genes encoding antibodies binding either the RBD (HAI+MN+) or more conserved non-RBD (HAI−MN+) epitopes on HA or other viral proteins. However, the elderly cohort showed much less adaptation to the non-RBD epitopes than the young cohort (p < 0.0001). The few antibodies that were able to bind the RBD from the elderly cohort were derived from IgV genes with evidence of increased recent mutation, as well as a minority of the HAI−MN+ antibodies. We also independently compared the frequency of unique amino acid differences in response to the 2009 pH1N1 strain by young individuals and found no difference in de novo mutation frequency (Figure S1G). Potently neutralizing epitopes are predicted to be the most robustly counter-selected as influenza virus adapts to herd immunity. We observed that the frequency of antibodies that potently neutralize below the median PRNT50 of 3.33 μg/ml was significantly reduced with age (16% for elderly versus 36% for young adults, p = 0.0168) (Figure 3D). Additionally, elderly subjects had lower MN titers in the serum post-vaccination compared to young adults as shown by lower MN fold increases between d0 and d28 (Figure S1H). In conclusion it appears that there is a reduction with age in the ability to adapt the immunoglobulin (Ig) repertoire to highly potent epitopes and drifting epitopes on the HA head.
Figure 3. Most influenza virus-reactive antibodies from elderly individuals are low potency and target conserved epitopes on HA and on other viral proteins.
(A-B) HAI and in vitro neutralization by plaque reduction neutralization assay (n = 102 mAbs in each group). Statistical significance: Fisher’s exact test. The assays were performed 2-3 times for each antibody. The number in the middle of the pie charts represents the total number of mAbs tested. (A) Proportion of neutralizing antibodies in each group. (B) Proportion of HAI+ neutralizing antibodies in each group.
(C) Intraclonal mutations based on HAI status. Amino acid differences by antibody pair (young adults HAI+MN+ n = 1066, HAI−MN+ n = 26; elderly adults HAI+MN+ n = 18, HAI−MN+ n = 303) represented as variable distribution with quartiles. Statistical significance: Mann–Whitney test. n.s. not significant.
(D) Proportion of highly potent neutralizing antibodies. The concentration 3.33 μg/ml was used as a cut-off value based on the median PRNT50 of all neutralizing antibodies. Statistical significance: Fisher’s exact test. The number in the middle of the pie charts represents the total number of mAbs tested.
(E) Competition for conserved epitopes on HA. Biotinylated mAbs CR9114 (stalk-reactive, MN+) and CR8059 (head HAI−MN+) were tested for binding to rHA protein by ELISA with or without the presence of a competitor mAb. Only HA-reactive mAbs were used as competitors. The experiment was done in duplicate 2-3 times. The average percentage of competition was calculated. Represented here are different degrees of inhibition (> 90%, 70%-90%, 50%-70%, 30%-50% or < 30%). An inhibition greater than 50% was considered a positive competition. “n.d.”, not determined.
(F) Proportion of antibodies targeting conserved epitopes on HA in each group. HA-reactive MN+ antibodies were divided into HAI+MN+, HAI−MN+ competing and HAI−MN+ non-competing mAbs. The number in the middle of the pie charts represents the total number of mAbs tested. Statistical significance: chi-square test.
(G-H) Proportion of HA-, NA- and NP-reactive antibodies. (G) All mAbs in each group (n = 102 for each group) were compared. (H) Antibodies induced by the Fluzone vaccine (n = 35 for the young cohort and n = 102 for the elderly cohort) were compared. Statistical significance: Fisher’s exact test.
Influenza-reactive antibodies from elderly individuals target conserved epitopes on HA and on other viral proteins
Past serological studies suggest that there will be selection in the elderly subjects for previously experienced B cells targeting predominantly conserved epitopes (Nachbagauer et al., 2016; Rajendran et al., 2017). We tested the possibility that HA-reactive mAbs target conserved epitopes (stalk and non-RBD head) (Figure 3E). 38% (15/40) of HA-reactive mAbs from the elderly individuals were found to neutralize via known conserved epitopes on HA, compared to 22% (12/60) of mAbs from the young individuals (p = 0.00364) (Figure 3F). Interestingly, 18% (7/40) of the B strain-reactive neutralizing mAbs from the elderly individuals bound novel conserved epitopes (non-competing). Inactivated influenza vaccines can induce rare B cells to NA, NP and other non-HA proteins (Wrammert et al., 2008). A surprisingly high frequency of mAbs from the elderly versus the young cohorts targeted NA, NP or other non-HA proteins (p < 0.0001) (Figure 3G). Although two different seasonal trivalent influenza vaccines were used in the young individual cohort (Fluzone, split virus vaccine or Fluvirin, subunit vaccine), so that the amount of the various antigens present in the vaccine might differ, responses to the non-HA proteins occurred independently of vaccine composition (Figure 3H). Additionally, we tested the antibody serum reactivity to the HA-stalk, NA and NP proteins pre- and post- vaccination in a group of our young (n=16) and elderly (n=9) individuals. We found increased antibody titers against the group 1 stalk, H1N1 NA and NP at day 0 in the elderly compared to the young, as well as increased antibody titers against the H1N1 NA in the elderly at d28 (Figure S2B, S2C and S2D). We did not observe any statistically significant serological differences to the HA stalk or non-HA proteins for the H3N2 or B influenza strains. We also tested an additional 38 young and 33 elderly individuals that received matched vaccines in the 2009-2010, 2012-2013 or 2016-2017 influenza seasons (Figure S2E, S2F and S2G). The elderly individuals in this cohort had higher serum antibody titers against the group 1 HA-stalk and the H1N1 NP protein. These results were consistent with the serology from the previous cohort with the exception of the H1N1 NA protein. These results are also consistent with previous work that had shown that individuals born before 1957 were imprinted by H1N1 influenza viruses during their childhood (Gostic et al., 2016). While serology cannot resolve current from historical responses, on the whole, the serological analyses revealed similarities but also differences compared to the plasmablast analysis.
NP is more conserved than the HA head domain (Air, 2012; Babar and Zaidi, 2015) (Figure S3). Indeed, some mAbs isolated cross-reacted to NP from both H1N1 and H3N2 influenza strains (Figure S4A) and we observed a trend towards higher antibody titers to H3N2 NP at d28 in the serum of elderly. Moreover, all the NA-reactive antibodies isolated were cross-reactive within subtypes and inhibited NA activity in the NA-Star assay, suggesting that the epitope targeted overlaps with the enzymatic active site that is highly conserved on the head of NA (Figure S3, S4B and S4C). Finally the elderly immunoglobulin repertoire had several biases, including preferential use of the VH1 IgV genes such as VH1-69, a feature of HA stalk-reactive antibodies. The other biases were due to specific clonal expansions in some of the individuals and cannot be generalized to the elderly population (Figure S4D). No differences were noted in JH gene usage or the light chain repertoire between the young and elderly cohorts (Figure S4E and S4F). Thus antibodies from elderly individuals target predominantly conserved epitopes on HA and other influenza virus proteins.
Influenza-reactive antibodies from aged individuals arise from cross-reactive memory B cells generated early in life
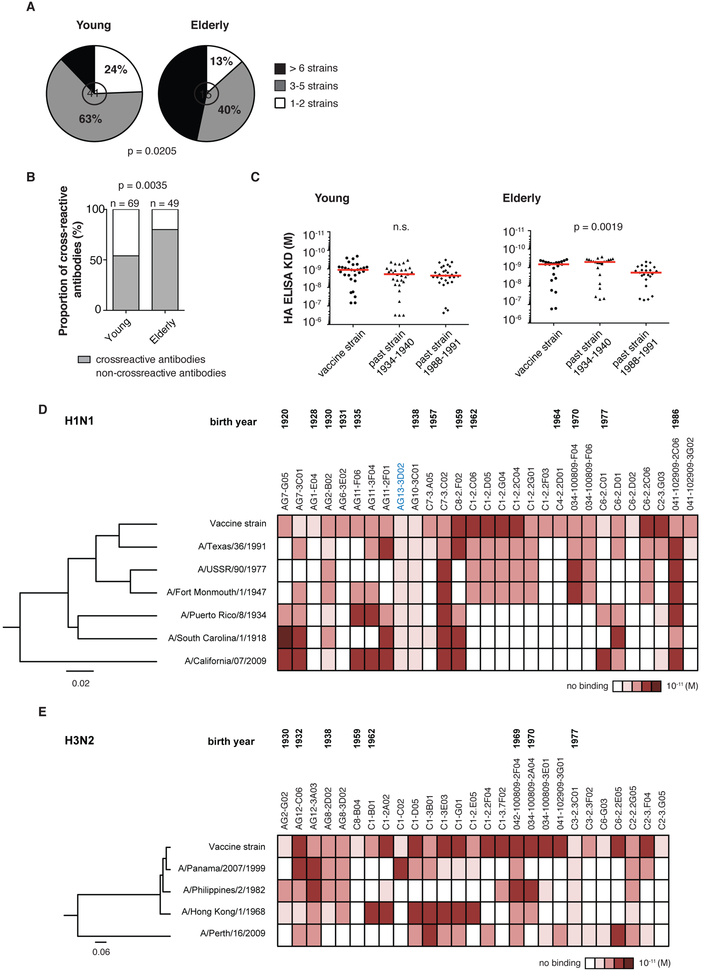
The binding of sites outside of the HA RBD predicts that the antibodies from the elderly cohort will be more cross-reactive. Indeed, 47% of the influenza A HA-reactive mAbs generated from the elderly individuals bound to six or more strains compared to only 12% for the young adults (Figure 4A). Reactivity to the B strains did not differ (Figure S2A) likely because the B strains in these vaccines exhibit particularly low rates of evolutionary change (Figure S3) (Chen and Holmes, 2008; Ferguson et al., 2003). In addition, the mAbs from the elderly individuals were more cross-reactive to two historical HA strains than those from young adults (p = 0.0035) (Figure 4B). Unlike for the young cohort, a subset of antibodies from the elderly individuals had higher affinity to historical HA strains (circulating during their childhood) compared to the vaccine strains, and lower affinity to more contemporary strains (Figure 4C, 4D and 4E). These findings suggest that upon influenza immunization, elderly individuals recall highly cross-reactive memory B cells generated earlier in life. Thus, original antigenic sin becomes exaggerated with age because of reduced adaptation of the Ig repertoire to recent influenza virus strains
Figure 4. Elderly individuals rely on cross-reactive memory B cells generated early in life.
(A) Proportion of cross-reactive antibodies capable of binding to the number of rHAs listed in the legend. MAbs reactive to H1N1 vaccine strains were tested against six other H1N1 rHAs and one H5N1 rHA; mAbs reactive to H3N2 vaccine strains were tested against five other H3N2 rHAs. Statistical significance: chi-square test.
(B) Proportion of cross-reactive antibodies capable of binding to HA of historic strains (A/Puerto Rico/8/1934 for H1N1 and B/Lee/1940 for B strain). All H1N1 and B strains HA-reactive antibodies were included. We did not include H3N2 strains as the first strain appeared in 1968 when a majority of the young subjects were already born. Statistical significance: Fisher’s exact test.
(C) Approximated KD values for cross-reactive antibodies by rHA ELISA. Vaccine strains: A/Solomon Islands/6/2006 and A/Brisbane/59/2007 (H1N1) and B/Brisbane/60/2008 and B/Malaysia/2506/2004. Historic strains were A/Puerto Rico/8/1934 (H1N1) and B/Lee/1940. The more “contemporary” past strains were A/Texas/36/1991 (H1N1) and B/Yamagata/16/1988. The assays were performed in duplicate three times for each antibody and shown are representative KD values. Statistical significance: paired Friedman test.
(D-E) Cross-reactivity to past influenza H1N1 and H3N2 rHAs tested by ELISA. The assays were performed in duplicate three times for each antibody. ELISA binding affinities represented by KD (M) were plotted as a heatmap. HAs clustered by amino acid sequence phylogeny, multiple alignments performed using CLUSTALW algorithm. Rooted tree constructed using neighbor-joining method and visualized using FigTree v1.4.0 software. The antibody in blue cross-reacts to influenza A and B strains. (D) H1N1 vaccine strains: A/Brisbane/59/2007 and A/Solomon Islands/6/2006. (E) H3N2 vaccine strains: A/Wisconsin/57/2005 and A/Uruguay/716/2007.
DISCUSSION
In this study, we observed a dramatic reduction with age in the ability to adapt the immunoglobulin repertoire to drifting epitopes, likely due to the reduced accumulation of IgV gene mutations (fixed repertoire). However a greater reliance on immune memory with age likely may also contributes to this inflexibility, as a more extensive immune history might activate memory cells not requiring further mutations. Additionally elderly individuals may recruit fewer memory B cells to be reactivated following antigen re-exposure resulting in a more frequent reliance on single expanded B cell clones. High preexisting serum antibody titers to influenza strains to which one has been previously exposed has been suggested to impede current responses due to rapid immunogen clearance or epitope masking (reviewed in (Henry et al., 2018)). One possibility is that the more extensive immune history of the elderly cohort studied herein might induce a similar response, impeding de novo germinal center induction and antibody diversification. However, we do not believe this is the case as the analyses of baseline titers before vaccination showed that the aged cohort did not contain more virus specific antibody, and that what antibody that was present tended to bind non-HA proteins to which the elderly cohort had an increased response. Further, such masking responses are known to occur only on recent, high-titer responses to the same influenza strain (i.e., from the vaccine of the previous year), but not to de novo strains or after a period without vaccination (Andrews et al., 2015b). When this masking-effect occurs, there are few specific plasmablasts to the recent strain activated at day 7 post-vaccination. The ready detection of plasmablasts from the elderly cohort in this study does not support an effect from epitope masking or clearance by preexisting antibody.
Previous studies using high-throughput sequencing of antibody transcripts from peripheral blood B cells revealed a reduced number of B cell lineages and a reduction in intralineage entropy (diversification) with age (de Bourcy et al., 2017; Jiang et al., 2013). One of the major drawbacks of these studies is the lack of link between the antibody repertoire sequencing data and the influenza-reactivity status of the clones. A more recent study combined high-throughput repertoire sequencing to the expression of a few mAbs (Ju et al., 2018), with the caveat that the repertoire sequencing was done on total plasmablasts, not antigen-reactive plasmablasts. An increased breadth of binding across influenza strains was noted with several antibodies from elderly individuals using HA1 peptides. This study was suggestive and corroborative but ultimately limited and not generalizable based on the limited numbers of antibodies tested. Further, the results herein provide important insight into the functional consequences of reduced diversity in the anti-influenza antibody repertoire with age.
Our study focused mainly on antibodies produced by plasmablasts, a transient population of Ig-secreting cells that reflects predominantly the re-activation of memory B cells. It is notable that in the case of the group 1 stalk, the plasmablast response was reflective of both preexisting serological responses as well as serology from 28 days post-vaccination that reflects contributions of newly induced naïve B cells, GC reactions, and newly established long-lived plasma cells. Differences between plasmablast and serology analyzes were to be expected as the plasmablast population mirrors the ongoing immune response (antibodies from currently activated B cells) while serology represents antibody derived from a subject’s entire history of exposure to influenza over a lifespan.
An interesting corollary is the response of a young cohort to the 2009 pandemic H1N1 strain upon first exposure only: because this strain was highly divergent, the subjects had memory cells only to conserved epitopes (mostly on the HA stalk) that were targeted by early activated plasmablasts (Li et al., 2012; Wrammert et al., 2011). Upon subsequent exposures, specific epitopes on the immunodominant HA globular head were targeted instead because they could now adapt (Andrews et al., 2015a). The elderly cohort is essentially stalled with the capacity to only target these conserved epitopes for any influenza strain with memory cells because they can no longer adapt (modeled in Figure 5).
Figure 5. Mechanistic model showing the first response of young adults to the pandemic 2009 H1N1 strain compared to the general response of elderly individuals to any influenza strain.
In young adults, first exposure to the 2009 H1N1 pandemic strain resulted in a biased response towards highly conserved epitopes on the stalk or head domains of HA (Wrammert et al., 2011; Andrews et al., 2015). Upon subsequent exposure to this strain, the biased response to these conserved epitopes was lost, and the divergent globular head was then predominantly targeted, similar to the past responses to H1N1 strains prior to 2009 and to all H3N2 and B strains. This biased response to conserved epitopes upon first exposure to that novel strain and the mechanism driving this response represents an interesting corollary to that of the aged cohort to any strain. That is, young adults in 2009 had memory B cells that could only target the conserved epitopes, simply based on a lack of immune history. Subsequently they could adapt to the novel epitopes and since have a typical response biased for the globular head of the 2009 H1 strain and all strains. Because the elderly population has lost the capacity to adapt by somatic hypermutation in recent years, their response becomes analogous to first exposure to that highly divergent pandemic strain, but now indefinitely, and to all recent influenza variants arising.
Finally our study suggests that a primary goal for improving influenza immunity in elderly populations would be to improve the induction of germinal centers and thereby enhance antibody affinity maturation. In that context, correlates to measure novel vaccine efficacy in elderly populations should include the accumulation of intra-clonal Ig gene mutations and improved targeting of the HA RBD. There are now vaccines designed specifically for people 65 years and older with evidence of greater efficacy (DiazGranados et al., 2014; DiazGranados et al., 2016; Domnich et al., 2017; McElhaney et al., 2013). It will be important to determine if these more robust vaccine compositions improve the deficiencies reported here.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to the Lead contact, Patrick C. Wilson (wilsonp@uchicago.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study Cohort
Healthy volunteers receiving an influenza immunization were enrolled from 2006 to 2010 (younger adults, 22-64 years of age and elderly adults, 70-89 years of age). The institutional review boards of Stanford University, The University of Chicago, and Oklahoma Medical Research Foundation approved the study. Informed consent was obtained from all participants. Blood samples were collected from each participant and plasmablasts were isolated at day 7 after immunization.
Exclusion criteria included history of immune deficiency, known or suspected impairment of immunologic function, recent or current use of immunosuppressive medication (including glucocorticoids) autoimmune disease (including rheumatoid arthritis treated with immunosuppressive medication), use of any anti-coagulation medication, history of Guillain–Barré Syndrome, known allergies to eggs or vaccine components and any condition which, in the opinion of the investigator, might interfere with volunteer safety, study objectives or the ability of the participant to understand or comply with the study protocol.
Study cohort included individuals of both gender identity (58% female and 42% male) and gender did not affect the results of the study. Additional information about the subjects and the vaccine they received is presented in Table S1 and Table S2. For full monoclonal antibody characterization (n = 102 per cohort), we focused on individuals who received the seasonal inactivated influenza vaccine (TIV, Fluzone®, Sanofi Pasteur, split vaccine or Fluvirin®, Novartis, subunit vaccine depending on the years) because none of the aged individuals received the monovalent pandemic 2009 vaccine. We also focused on matching influenza virus strains between the two cohorts and analyzed responses to two different H1N1, two different H3N2, and two different B strains (Table S1).
Additional serum samples pre- and post-vaccination were obtained from Stanford University (Fluzone 2009-2010, young 18-51, elderly >70) (Sasaki et al., 2011), the University of Rochester (Fluarix 2016-2017, young 18-51, elderly >65) as well as from Duke University Medical Center in 2012-2013 (Cobey et al., 2018).
Cells, viruses, and recombinant proteins
Human Embryonic Kidney HEK293T (female, # CRL-11268) and Madin Darby Canine Kidney MDCK (female, # CCL-34, NBL-2) cells were purchased and authenticated by the American Type Culture Collection (ATCC). The MDCK London Line (female, IRR #FR-58) were purchased and authenticated by IRR. All cell lines were maintained under a humidified atmosphere of 5% CO2 at 37°C. HEK293T cells were maintained in Advanced-DMEM supplemented with 2% ultra-low IgG Fetal Bovine Serum (FBS) (Invitrogen), 1% L-Glutamine (Invitrogen) and 1% antibiotic-antimycotic (Invitrogen). Both MDCK cell lines were maintained in DMEM supplemented with 10% FBS (Invitrogen), 1% L-Glutamine (Invitrogen) and 1% Penicillin-Streptomycin (Invitrogen). All cell lines were used at low-passages. All influenza virus stocks used for the assays were freshly grown in specific pathogen-free eggs, purified and titered. Recombinant HA proteins derived from influenza virus strains A/Solomon Islands/6/2006 (H1N1), A/Brisbane/59/2007 (H1N1), A/Wisconsin/57/2005 (H3N2), A/Uruguay/716/2007 (H3N2), A/Perth/16/2009 (H3N2), A/Victoria/361/2011 (H3N2), B/Malaysia/2506/2004, B/Florida/4/2006 and B/Brisbane/60/2008 were obtained from BEI resources. Recombinant NA proteins derived from influenza virus strains A/Brisbane/59/2007 and B/Florida/4/2006 were obtained from BEI resources. All the other HA and NA proteins were expressed in the baculovirus expression system as described before (Margine et al., 2013). All recombinant NP proteins were obtained from Sino Biological.
METHOD DETAILS
Monoclonal antibody production
Monoclonal antibodies were generated as previously described (Smith et al., 2009) in accordance with the University of Chicago Institutional Review Board. Briefly, plasmablasts were single cell sorted in 96-well plates and the BCR heavy and light chain variable regions were amplified and cloned into expression vectors. HEK293T cells were co-transfected with the plasmids using polyethylenimine (PEI, Polysciences) and supernatant containing the secreted antibody was collected 4-5 days after transfection and purified using protein A beads (Pierce). Multiple batches of the same antibody were produced and tested to ensure replication of the results.
ELISA assays
Plates were coated with recombinant proteins at various concentrations depending on the protein (from 0.5-2 μg/ml) in phosphate buffered saline (PBS, pH 7.4) overnight at 4ΰC. For viruses, plates were coated with eight he magglutination units (HAU) of virus in carbonate buffer overnight at 4°C. After blocking, antibodies were incubated (starting concentration 10 μg/ml) for 1 h at 37°C. Horseradis h peroxidase (HRP)-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch) was used to detect binding of the mAbs, followed by development with Super Aquablue ELISA substrate (eBiosciences). Absorbance was measured at 405 nm on a microplate spectrophotometer (BioRad). To standardize the assays, antibodies with known binding characteristics were included on each plate and the plates were developed when the absorbance of these controls reached 3.0 ± 0.1 OD units. An antibody was considered positive (strain-reactive) if the OD at 405 nm was > 0.5 after background subtraction when the positive control had reached an OD of 3. KD values of antibody binding were determined by Scatchard analysis using nonlinear regression (one site binding model) on GraphPad Prism software. All ELISAs were done in duplicates 2-3 times.
For serum antibody reactivity, serum samples were serially diluted 3-fold seven times, starting at a 1:100 dilution. To standardize the results, all plates coated with the same virus strain or recombinant protein were allowed to develop for the same amount of time. Serum antibody 50% effective concentration (EC50) levels were determined by nonlinear regression analysis of the serially diluted serum OD values, as calculated by GraphPad Prism. The EC50 takes into account both the concentration (maximum binding) and affinity of Abs (slope of the binding curve) and is therefore an accurate measure of total binding and generally correlates with the endpoint titer (Andrews et al., 2015b). Due to the limited amount of serum available, all serum ELISAs were done in duplicates twice.
Competition ELISA assay
Competition ELISAs were performed by inhibition of binding of each biotinylated antibody of interest at the half-maximal binding concentration with a ten-fold molar excess of competitor antibody. HRP-conjugated streptavidin (Southern Biotech) was used for detection. The absorbance value of each antibody against itself is scored at 100% inhibition and comparison of different antibodies was done as a percentage of this 100% inhibition. All competition ELISAs were done in duplicates 2-3 times.
Hemagglutination inhibition assay
Viruses diluted to eight HA units/50 μl and 25 μl were combined in duplicate wells with an equal volume of antibody serially diluted in PBS and incubated at room temperature (RT) for 45 minutes. Fifty microliters of 0.5% Turkey red blood cells (Lampire Biological) was then added and incubated for 1 h at RT. Minimum effective concentrations were read based on the final dilution for which haemagglutination was observed. The assay was performed in duplicate 2-3 times.
Serum samples were inactivated by treatment with receptor-destroying enzyme (RDE; Denka Seiken) for 18 h at 37°C. Sodium citrate was added, and RDE was inactivated by heating the solution to 56°C for 30 min. Inactivated serum samples were added at a final dilution of 1:10 in PBS and then diluted 2-fold down a U-bottom 96-well plate. The minimum serum dilution, which inhibited hemagglutination, was recorded. The assay was performed in duplicate twice.
Plaque assay and PRNT50 assay
Plaque assays were performed as previously described (Wrammert et al., 2011) with the exception that cells were incubated for 48 hours with the agar overlay. MDCK London Line cells were used. Plaques were counted, and the final concentration of antibody that reduced plaque numbers to 50% (PRNT50) was determined using GraphPad Prism software. The assay was performed in duplicate 2-3 times.
Microneutralization assay
Serum samples were inactivated by treatment with receptor-destroying enzyme (RDE; Denka Seiken) for 18 h at 37°C. Sodium citrate was added, and RDE was inactivated by heating the solution to 56°C for 30 min. Inactivate d serum samples were 2-fold diluted in minimal essential medium with tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (infection medium) at a concentration of 1 μg/ml in 96-well cell culture plates. Viruses were diluted to a concentration of 100 PFU per 50 μl in infection medium. Fifty microliters of diluted sera was incubated with 50 μl of virus for 1 h at room temperature. MDCK cells (ATCC) were washed once with PBS, and 100 μl of serum-virus mixture was added onto cells. Cells were incubated at 37°C for 1 h, washed once with PBS, and 50 μl of diluted serum and 50 μl of infection medium were added to each well. Infected cells were incubated for 48 h. Fifty microliters of supernatants were collected and 50μl of 0.5% Turkey red blood cells (Lampire Biological) was then added and incubated for 1 h at RT. The minimum serum dilution, which inhibited hemagglutination, was recorded. Due to the limited amount of serum available, all serum ELISAs were done in duplicates twice.
NA-Star assay
The NA-Star influenza neuraminidase inhibitor resistance detection kit (Applied Biosystems) was used and manufacturer's instructions were followed. Briefly, 25 μl of each antibody in serial two-fold dilutions in NA-Star assay buffer (starting concentration 100 μg/ml) was mixed with 25 μl of virus at the determined 4× EC50 (half maximum effective concentration) and incubated at 37°C for 20 min. After adding 10 μl of 1000× diluted NA-Star substrate, the plates were incubated for 30 min. Reaction was stopped by adding 60 μl of NA-Star accelerator. The chemiluminescence was determined using the DTX 880 plate reader. IC50 values were determined using Prism software. The assay was performed in duplicate 2-3 times.
Immunoglobulin repertoire analysis
By utilizing antibody variable genes from single plasmablast cells, we could both ensure influenza virus binding and know with certainty that clonal B cells, even those with identical variable genes (no mutational differences), were derived from different cells. The V, D, and J gene segments and regions of all Ig sequences were determined using the stand-alone NCBI IgBlast search engine (https://www.ncbi.nlm.nih.gov/igblast/) and the IMGT sequence database (http://www.imgt.org). Variable gene clones were determined by aligning all sequences sharing identical progenitor sequence as predicted by IgBLAST (the same V, D and J genes and CDR3 lengths) with Clustal Omega to provide hierarchical comparisons. Frequencies of somatic mutation were calculated for the CDR1 and CDR2 regions of each paired sequence in the hierarchical alignments. Clones were identified as those sequences with the same predicted precursor sequences and that had mutational frequencies in the untemplated regions (n and p additions) of the CDR3s not exceeding the frequency of base-pair differences calculated to occur based on the CDR1 and CDR2 plus the standard deviation. As verification, clonal lineages were verified and essentially identical using the Clonalyst software such that the results of the clonal analyses do not differ by either clone-calling method. Rate of intraclonal mutations were determined from the amino acid sequence alignments from all clonal influenza-reactive sequences on a pairwise basis. A total of 869 influenza virus-reactive antibody heavy chain (VH) sequences (young adults n = 757; elderly individuals n = 112) were cloned and we identified 2,465 clonal pairs of VH genes from all of the young subjects and 340 pairs from seven of the elderly subjects (Table S2). The 2,465 pairs derive from the pairwise comparison of the clones, which multiplies depending on the number of related sequences within each clonal expansion.
Evolutionary analysis of influenza proteins
To compare rates of evolutionary change among different influenza antigens, complete protein-coding nucleotide sequences for HA, NA and NP of influenza A/H3N2, A/H1N1 and B were downloaded from the NCBI Influenza Virus Database. Influenza B isolates were assigned to the Victoria or Yamagata lineages by blasting HA sequences against reference strains B/Victoria/2/87 and B/Yamagata/16/1988 (NA and NP sequences of influenza B isolates for which HA sequences were not available were ignored). Validation of this assignment method using HA sequences available on GISAID (for which lineage identity is typically available) suggests it is accurate in 98.8% of cases (11,340/11,481 isolates; data not shown). Sequences were aligned using MAFFT v. 7.310 (Katoh et al., 2002) with variable gap opening penalties for different subtypes/lineages but identical gap opening penalties for different segments of the same subtype/lineage: 3 for A/H3N2, 1.53 (the default value) for A/H1N1, 8 for B/Victoria and 12 for Yamagata. We calculated the amino acid divergence between each sequence and the corresponding sequence from a reference isolate for each subtype/lineage (A/Hong Kong/01/1968 for A/H3N2, A/Puerto Rico/8-1/1934 for A/H1N1, B/Victoria/02/1987 for B/Victoria and B/Yamagata/16/1988 for B/Yamagata). Amino acid divergence was calculated as the number of amino acid differences between each sequence and the reference sequence divided by the number of sites without gaps or ambiguous amino acids in either sequence. Amino acid divergence was computed across the entire sequence of each segment and separately for the head HA1 and HA2 domains of HA and the head, stem (including the transmembrane domain) and active site regions of NA. Start and end positions for the HA1 and HA2 domains were based on A/Hong Kong/01/1968 (accession number AP039310) for A/H3N2, A/Puerto Rico/8-1/1934 for A/H1N1 (accn. ACV49545) and A/Brisbane/60/2008 (a Yamagata strain; PDB:4FQM) for both B/Yamagata and B/Victoria. Start and end positions for the NA regions were based on (Air, 2012). Positions were visually adjusted to account for indels in the alignments obtained in this study. In addition to amino acid divergence, the number of potentially N-linked glycosylated sites was estimated as the number of N-X[ST]-X motifs, where X is any amino acid except proline. The discontinuity in the B/Victoria NA corresponds to acquisition of a Yamagata-lineage NA by Victoria viruses through reassortment in the early 2000s (Langat et al., 2017). Data and code implementing the analysis are available at github.com/cobeylab/flu_response_aging.
QUANTIFICATION AND STATISTICAL METHODS
All immunoglobulin repertoire mutation graphing were performed using JMP10. The exact value of n, representing the number of antibody VH genes or the number of subjects depending on the analysis, was indicated in the figure legend. Statistical significance was determined using GraphPad Prism two-tailed nonparametric unpaired Mann–Whitney U test.
All other statistical analyses were performed using GraphPad Prism. The exact value of n, representing the number of monoclonal antibodies or the number of subjects depending on the analysis, was indicated in the figure legend. Statistical significance was determined using two-tailed nonparametric unpaired Mann–Whitney U test, Fisher’s exact test, chi-square test or paired Friedman test, as detailed in the figure legends. Nonparametric tests were used because of non-normal distribution for the majority of the data, as determined by GraphPad Prism using the D’Agostino and Pearson omnibus normality test. P-values equal to or lower than 0.05 were considered significant.
DATA AND SOFTWARE AVAILABILITY
All antibody sequences have been deposited to GenBank and are available under accession numbers MH205200-MH205603.
Supplementary Material
Data S1, Summary information table of the 204 fully characterized monoclonal antibodies included in this study, Related to Figures 3 and 4.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CD19 PE-AF610 conjugate | Invitrogen/Thermo Scientific | Cat# MHCD1922 RRID: AB_10373379 |
| Anti-human CD27 R-PE conjugate | Invitrogen/Thermo Scientific | Cat# MHC2704 RRID: AB_10392393 |
| Anti-human CD38 APC-Cy5.5 conjugate | Invitrogen/Thermo Scientific | Cat# MHCD3819 RRID: AB_10371760 |
| Anti-human CD3 FITC conjugate | Invitrogen/Thermo Scientific | Cat# MHCD0301 RRID: AB_10376003 |
| Anti-human CD20 FITC conjugate | Invitrogen/Thermo Scientific | Cat# MHCD2001 RRID: AB_10373690 |
| HRP-conjugated goat anti-human IgG antibody | Jackson Immuno Research | Cat# 109-035-098 RRID:AB_2337586 |
| Streptavidin-HRP | Southern Biotech | Cat#7100-05 |
| Bacterial and Virus Strains | ||
| NEB® 5-alpha Competent E. coli | NEB | Cat# C2988J |
| A/Solomon Islands/6/2006 (H1N1) | Patrick Wilson’s laboratory stock | N/A |
| A/Brisbane/59/2007(H1N1) | Patrick Wilson’s laboratory stock | N/A |
| A/California/07/2009 (H1N1) | Patrick Wilson’s laboratory stock | N/A |
| A/Wisconsin/57/2005 (H3N2) | Patrick Wilson’s laboratory stock | N/A |
| A/Uruguay/716/2007 (H3N2) | Patrick Wilson’s laboratory stock | N/A |
| B/Malaysia/2506/2004 | Patrick Wilson’s laboratory stock | N/A |
| B/Florida/4/2006 | Patrick Wilson’s laboratory stock | N/A |
| B/Brisbane/60/2008 | Patrick Wilson’s laboratory stock | N/A |
| Biological Samples | ||
| Human PBMC | This study | N/A |
| Turkey red blood cells | Lampire Biological Laboratories | Cat# 7209403 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| A/Solomon Islands/6/2006 (H1N1) HA | BEI RESOURCES | NR-15170 |
| A/Brisbane/59/2007 (H1N1) HA | BEI RESOURCES | NR-28607 |
| A/Wisconsin/57/2005 (H3N2) HA | BEI RESOURCES | NR-49237 |
| A/Uruguay/716/2007 (H3N2) HA | BEI RESOURCES | NR-15168 |
| B/Malaysia/2506/2004 HA | BEI RESOURCES | NR-15172 |
| B/Florida/4/2006 HA | BEI RESOURCES | NR-15169 |
| B/Brisbane/60/2008 HA | BEI RESOURCES | NR-19239 |
| A/Brisbane/10/2007 (H1N1) NA | BEI RESOURCES | NR-43785 |
| A/Wisconsin/67/2005 (H3N2) NA | BEI RESOURCES | NR-19237 |
| A/Perth/16/2009 (H3N2) HA | IRR | FR-472 |
| A/Puerto Rico/8/1934 (H1N1) NP | Sino Biological | 11675-V08B |
| A/Aichi/2/1968 (H3N2) NP | Sino Biological | 40207-V08B |
| B/Florida/4/2006 NP | Sino Biological | 40438-V08B |
| A/California/04/2009 (H1N1) HA | Florian Krammer’s laboratory stock | N/A |
| A/California/04/2009 (H1N1) NA | Florian Krammer’s laboratory stock | N/A |
| A/Vietnam/1203/2004 (H5N1) NA | Florian Krammer’s laboratory stock | N/A |
| A/Texas/36/1991 (H1N1) HA | Florian Krammer’s laboratory stock | N/A |
| A/Texas/36/1991 (H1N1) NA | Florian Krammer’s laboratory stock | N/A |
| A/USSR/90/1977 HA (H1N1) HA | Florian Krammer’s laboratory stock | N/A |
| A/Fort Monmouth/1/1947 HA (H1N1) HA | Florian Krammer’s laboratory stock | N/A |
| A/Puerto Rico/8/1934 (H1N1) HA | Florian Krammer’s laboratory stock | N/A |
| A/South Carolina/1/1918 (H1N1) HA | Florian Krammer’s laboratory stock | N/A |
| A/Hong Kong/1/1968 (H3N2) HA | Florian Krammer’s laboratory stock | N/A |
| A/Hong Kong/1/1968 (H3N2) NA | Florian Krammer’s laboratory stock | N/A |
| A/Philippines/2/1982 (H3N2) HA | Florian Krammer’s laboratory stock | N/A |
| A/Panama/2007/1999 (H3N2) HA | Florian Krammer’s laboratory stock | N/A |
| A/Hong Kong/485197/2014 (H3N2) NA | Florian Krammer’s laboratory stock | N/A |
| B/Brisbane/60/2008 NA | Florian Krammer’s laboratory stock | N/A |
| B/Yamagata/16/1988 HA | Florian Krammer’s laboratory stock | N/A |
| B/Yamagata/16/1988 NA | Florian Krammer’s laboratory stock | N/A |
| B/Florida/4/2006 NA | Florian Krammer’s laboratory stock | N/A |
| B/Lee/1940 HA | Florian Krammer’s laboratory stock | N/A |
| Chimeric H6/1 HA (H6 head from A/mallard/Sweden/81/02 combined with H1 stalk from A/California/04/2009) | Florian Krammer’s laboratory stock | N/A |
| Chimeric H6/1 PR8 HA (H6 head from A/mallard/Sweden/81/02 combined with H1 stalk from A/Puerto Rico/8/1934) | Florian Krammer’s laboratory stock | N/A |
| Chimeric H5/3 HA (H5 head from A/mallard/Sweden/24/2002 combined with H3 stalk from A/Perth/16/2009) | Florian Krammer’s laboratory stock | N/A |
| Chimeric H8/B HA (H8 head from A/Vietnam/1203/2004 combined with B stalk from B/Florida/4/2006) | Florian Krammer’s laboratory stock | N/A |
| PEI 25K, Transfection Grade | Polysciences | Cat# 23966-2 |
| Super Aquablue ELISA substrate | Thermo Scientific | Cat# 00-4203-58 |
| EZ-link Sulfo-NHS-Biotin | Thermo Scientific | Cat# 21217 |
| RDE II by Denka Seiken | Hardy Diagnostics | Cat# 370013 |
| Trypsin, TPCK treated | Sigma-Aldrich | Cat# T8802 |
| Crystal violet solution | Sigma-Aldrich | Cat# V5265 |
| Pierce™ Protein A agarose | Thermo Scientific | Cat# 20334 |
| Critical Commercial Assays | ||
| NA-Star® Influenza Neuraminidase Inhibitor Resistance Detection Kit | Thermo Scientific | Cat# 437C4422 |
| Experimental Models: Cell Lines | ||
| MDCK cells, London line (plaque assay) | IRR | Cat# FR-58 |
| MDCK cells (microneutralization assay) | ATCC | Cat# CCL-34 |
| HEK293T Cell Line | ATCC | Cat# CRL-11268 |
| Experimental Models: Organisms/Strains | ||
| Recombinant DNA | ||
| IgG-AbVec | Patrick Wilson’s laboratory stock | N/A |
| Igκ-AbVec | Patrick Wilson’s laboratory stock | N/A |
| Igλ-AbVec | Patrick Wilson’s laboratory stock | N/A |
| Software and Algorithms | ||
| GraphPad Prism (version 7.0) | GraphPad Software Inc |
http://www.graphpad.com RRID: SCR_002798 |
| Jmp (version 10.0) | SAS |
https://www.jmp.com/en_us/software.html RRID: SCR_014242 |
| IMGT/V-QUEST | Immunogenetics, Marie-Paule Lefranc |
http://www.imgt.org/IMGT_vquest/share/textes/ RRID: SCR_010749 |
| IgBlast | NCBI |
http://www.ncbi.nlm.nih.gov/igblast/ RRID: SCR_002873 |
| FigTree v1.4.0 | Andrew Rambaut’s laboratory |
http://tree.bio.ed.ac.uk/software/figtree RRID: SCR_008515 |
| Clustal Omega | EMBL-EBI |
http://www.ebi.ac.uk/Tools/msa/clustalo/ RRID: SCR_001591 |
| Clonalyst software | Laboratory of Computational Immunology, Boston University | https://www.bu.edu/computationalimmunology/research/software/ |
| MAFFT v. 7.310 | CBRC |
http://mafft.cbrc.jp/alignment/server/ RRID:SCR_011811 |
| Deposited data | ||
| Antibody sequences, Genbank accession numbers MH205200-MH205603 | Genbank, NCBI |
https://www.ncbi.nlm.nih.gov/genbank/ |
Highlights.
Elderly adults have less de novo somatic hypermutations in immunoglobulin variable genes
Elderly individuals have less adaptability in their antibody responses to influenza
Antibodies from the elderly target conserved, often less potent hemagglutinin epitopes
ACKNOWLEDGMENTS
We thank Y. Huang, A. Hirsh, Shirin Strohmeier, Daniel Stadlbauer, Raffael Nachbagauer, Andres Javier, Parnavi Desai and F. Amanat for technical assistance. We thank K. Neu and J. Guthmiller for critical comments. This study was supported in part by the National Institute of Allergy and Infectious Disease; National Institutes of Health grant numbers U19AI082724 (P.C.W.), P01AI097092 (P.C.W.), U19AI109946 (P.C.W.), U19AI057266 (P.C.W.), HHSN272201400005C (P.C.W.). H.G. and X-S.H. were supported by AI057229 (Stanford). F.K. was supported by R01 AI117287 and NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400008C. S.E.H. was supported by 1R01AI113047, 1R01AI108686 and CEIRS HHSN272201400005C. M.C.V and S.C were supported by DP2AI117921, CEIRS HHSN272201400005C and the James S. McDonnell Complex Systems Scholar Award. K.E.S received support from the NIA, Duke Pepper Older Americans Independence Center, P30AG028716. J.H was supported by UL1TR002001 and CEIRS HHSN272201400005C.
Footnotes
DECLARATION OF INTERESTS
The authors declare that they have no competing interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Air GM (2012). Influenza neuraminidase. Influenza Other Respir Viruses 6, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, et al. (2015a). Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med 7, 316ra192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SF, Kaur K, Pauli NT, Huang M, Huang Y, and Wilson PC (2015b). High preexisting serological antibody levels correlate with diversification of the influenza vaccine response. J Virol 89, 3308–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babar MM, and Zaidi NU (2015). Protein sequence conservation and stable molecular evolution reveals influenza virus nucleoprotein as a universal druggable target. Infect Genet Evol 34, 200–210. [DOI] [PubMed] [Google Scholar]
- Cancro MP, Hao Y, Scholz JL, Riley RL, Frasca D, Dunn-Walters DK, and Blomberg BB (2009). B cells and aging: molecules and mechanisms. Trends Immunol 30, 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, and Holmes EC (2008). The evolutionary dynamics of human influenza B virus. J Mol Evol 66, 655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Kozlovsky BF, Effros RB, Grubeck-Loebenstein B, Edelman R, and Sztein MB (2009). Vaccination in the elderly: an immunological perspective. Trends Immunol 30, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Y, Ikematsu H, Yamaji K, Nishimura M, Kashiwagi S, and Hayashi J (2003). Age-related accumulation of Ig V(H) gene somatic mutations in peripheral B cells from aged humans. Clin Exp Immunol 133, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobey S, Gouma S, Parkhouse K, Chambers BS, Ertl HC, Schmader KE, Halpin RA, Lin X, Stockwell TB, Das SR, et al. (2018). Poor Immunogenicity, Not Vaccine Strain Egg Adaptation, May Explain the Low H3N2 Influenza Vaccine Effectiveness in 2012-2013. Clin Infect Dis 67, 327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bourcy CF, Angel CJ, Vollmers C, Dekker CL, Davis MM, and Quake SR (2017). Phylogenetic analysis of the human antibody repertoire reveals quantitative signatures of immune senescence and aging. Proc Natl Acad Sci U S A 114, 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiazGranados CA, Dunning AJ, Kimmel M, Kirby D, Treanor J, Collins A, Poliak R, Christoff J, Earl J, Landolfi V, et al. (2014). Efficacy of high-dose versus standard-dose influenza vaccine in older adults. N Engl J Med 371, 635–645. [DOI] [PubMed] [Google Scholar]
- DiazGranados CA, Dunning AJ, Robertson CA, Talbot HK, Landolfi V, and Greenberg DP (2016). Effect of Previous-Year Vaccination on the Efficacy, Immunogenicity, and Safety of High-Dose Inactivated Influenza Vaccine in Older Adults. Clin Infect Dis 62, 1092–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domnich A, Arata L, Amicizia D, Puig-Barbera J, Gasparini R, and Panatto D (2017). Effectiveness of MF59-adjuvanted seasonal influenza vaccine in the elderly: A systematic review and meta-analysis. Vaccine 35, 513–520. [DOI] [PubMed] [Google Scholar]
- Eisen HN (2014). Affinity enhancement of antibodies: how low-affinity antibodies produced early in immune responses are followed by high-affinity antibodies later and in memory B-cell responses. Cancer Immunol Res 2, 381–392. [DOI] [PubMed] [Google Scholar]
- Ferguson NM, Galvani AP, and Bush RM (2003). Ecological and immunological determinants of influenza evolution. Nature 422, 428–433. [DOI] [PubMed] [Google Scholar]
- Frasca D, and Blomberg BB (2014). B cell function and influenza vaccine responses in healthy aging and disease. Curr Opin Immunol 29, 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin K, Viboud C, and Simonsen L (2006). Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine 24, 1159–1169. [DOI] [PubMed] [Google Scholar]
- Gostic KM, Ambrose M, Worobey M, and Lloyd-Smith JO (2016). Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354, 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton NS, Sachs D, Chen CJ, Hai R, and Palese P (2013). Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc Natl Acad Sci U S A 110, 20248–20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C, Palm AE, Krammer F, and Wilson PC (2018). From Original Antigenic Sin to the Universal Influenza Virus Vaccine. Trends Immunol 39, 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, He J, Weinstein JA, Penland L, Sasaki S, He XS, Dekker CL, Zheng NY, Huang M, Sullivan M, et al. (2013). Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci Transl Med 5, 171ra119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju CH, Blum LK, Kongpachith S, Lingampalli N, Mao R, Brodin P, Dekker CL, Davis MM, and Robinson WH (2018). Plasmablast antibody repertoires in elderly influenza vaccine responders exhibit restricted diversity but increased breadth of binding across influenza strains. Clin Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, and Miyata T (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langat P, Raghwani J, Dudas G, Bowden TA, Edwards S, Gall A, Bedford T, Rambaut A, Daniels RS, Russell CA, et al. (2017). Genome-wide evolutionary dynamics of influenza B viruses on a global scale. PLoS Pathog 13, e1006749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, et al. (2012). Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A 109, 9047–9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margine I, Palese P, and Krammer F (2013). Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J Vis Exp, e51112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrie TJ (2000). Community-acquired pneumonia in the elderly. Clin Infect Dis 31, 1066–1078. [DOI] [PubMed] [Google Scholar]
- McElhaney JE, Beran J, Devaster JM, Esen M, Launay O, Leroux-Roels G, Ruiz-Palacios GM, van Essen GA, Caplanusi A, Claeys C, et al. (2013). AS03-adjuvanted versus non-adjuvanted inactivated trivalent influenza vaccine against seasonal influenza in elderly people: a phase 3 randomised trial. Lancet Infect Dis 13, 485–496. [DOI] [PubMed] [Google Scholar]
- Miller C, and Kelsoe G (1995). Ig VH hypermutation is absent in the germinal centers of aged mice. J Immunol 155, 3377–3384. [PubMed] [Google Scholar]
- Nachbagauer R, Choi A, Izikson R, Cox MM, Palese P, and Krammer F (2016). Age Dependence and Isotype Specificity of Influenza Virus Hemagglutinin Stalk-Reactive Antibodies in Humans. MBio 7, e01996–01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran M, Nachbagauer R, Ermler ME, Bunduc P, Amanat F, Izikson R, Cox M, Palese P, Eichelberger M, and Krammer F (2017). Analysis of Anti-Influenza Virus Neuraminidase Antibodies in Children, Adults, and the Elderly by ELISA and Enzyme Inhibition: Evidence for Original Antigenic Sin. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner K, Winter DB, Kasmer C, Skovgaard GL, Tarone RE, Bohr VA, and Gearhart PJ (2001). Impact of age on hypermutation of immunoglobulin variable genes in humans. J Clin Immunol 21, 102–115. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Sullivan M, Narvaez CF, Holmes TH, Furman D, Zheng NY, Nishtala M, Wrammert J, Smith K, James JA, et al. (2011). Limited efficacy of inactivated influenza vaccine in elderly individuals is associated with decreased production of vaccine-specific antibodies. J Clin Invest 121, 3109–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Garman L, Wrammert J, Zheng NY, Capra JD, Ahmed R, and Wilson PC (2009). Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc 4, 372–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, and Fukuda K (2003). Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289, 179–186. [DOI] [PubMed] [Google Scholar]
- Troutaud D, Drouet M, Decourt C, Le Morvan C, and Cogne M (1999). Age-related alterations of somatic hypermutation and CDR3 lengths in human Vkappa4-expressing B lymphocytes. Immunology 97, 197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. (2011). Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 208, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, et al. (2008). Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Stedra J, and Cerny J (1996). Relative contribution of T and B cells to hypermutation and selection of the antibody repertoire in germinal centers of aged mice. J Exp Med 183, 959–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Han S, Takahashi Y, and Kelsoe G (1997). Immunosenescence and germinal center reaction. Immunol Rev 160, 63–77. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1, Summary information table of the 204 fully characterized monoclonal antibodies included in this study, Related to Figures 3 and 4.
Data Availability Statement
All antibody sequences have been deposited to GenBank and are available under accession numbers MH205200-MH205603.