Abstract
Only a tiny fraction of the nanomedicine-design space has been explored, owing to the structural complexity of nanomedicines and the lack of relevant high-throughput synthesis and analysis methods. Here, we report a methodology for determining structure–activity relationships and design rules for spherical nucleic acids (SNAs) functioning as cancer-vaccine candidates. First, we identified ~1,000 candidate SNAs on the basis of reasonable ranges for 11 design parameters that can be systematically and independently varied to optimize SNA performance. Second, we developed a high-throughput method for making SNAs at the picomolar scale in a 384-well format, and used a mass spectrometry assay to rapidly measure SNA immune activation. Third, we used machine learning to quantitatively model SNA immune activation and identify the minimum number of SNAs needed to capture optimum structure–activity relationships for a given SNA library. Our methodology is general, can reduce the number of nanoparticles that need to be tested by an order of magnitude, and could serve as a screening tool for the development of nanoparticle therapeutics.
Nanotechnology is beginning to play a major role in the development of new therapeutic modalities. Currently, over 100 drugs based on nanomaterials are in clinical trials or approved for therapeutic use1. These structures are promising because of their multifunctionality, which directly relates to their relatively large size and often complex architectures when compared with conventional small molecules or biologics. However, due to this complexity, little attention has been paid to how structural changes inform biological activity. Consider, for example, spherical nucleic acids (SNAs), which are made by chemically arranging short sequences of DNA or RNA around a nanoparticle core (Fig. 1a)2,3. SNAs exhibit properties that are substantively different from the short, linear oligonucleotides that comprise them, including the ability to actively cross mammalian cell membranes without the need for transfection reagents, a resistance to nuclease degradation, and the ability to carry large and complex cargo (such as oligonucleotides and peptides) into many cell types4–7.
Fig. 1 |. SNA architecture and design properties.
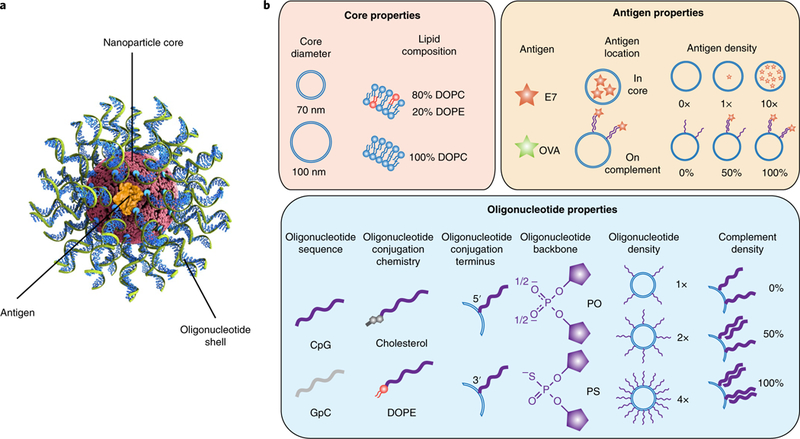
a, The three components of immunostimulatory SNAs. b, The parameters investigated for each of the SNA design properties, organized by core, antigen and oligonucleotide property categories. The potential design space has 3,072 variants. PO, phosphodiester; PS, phosphorothioate.
These properties make SNAs an attractive candidate in cancer immunotherapy, as structures with dual functionality can be rapidly prepared from lipids, oligonucleotide adjuvants and peptide antigens. When delivered to antigen presenting cells (APCs), SNAs activate the immune system and, in a lymphoma model, show superior activity compared with the same free antigen and linear oligonucleotides5. However, the modularity of an SNA allows for a large number of possible designs and compositions, and identifying the nanoparticle architectures best for inducing multiple aspects of cellular immune responses, such as potency, selectivity and efficacy, remains a challenge. Furthermore, understanding how variations in SNA structure influence any individual step in generating immune responses at the cellular level (for example, Toll-like receptor (TLR) activation and antigen presentation) is also challenging, particularly if the dependence on activity is nonlinear across multiple variables.
Here, we describe a new approach for synthesizing a library of SNAs that are qualitatively similar but structurally distinct, in conjunction with a mass spectrometry-based screening protocol that can rapidly and quantitatively determine the ability of an SNA structure to activate the TLR9 pathway. First, we show how this methodology can be used to make and screen ~1,000 SNA architectures (800 of which are unique). In addition, we describe how machine learning models can be trained with this data and subsequently used to accurately predict the TLR9 stimulatory activity of SNAs based on structural features. Significantly, these models provide a ranking of the order of importance of 11 structural parameters, as well as SNA drug concentration. The library screen and analysis by machine learning revealed several non-intuitive and nonlinear consequences of structural variation on TLR9 activation; identification of these relationships was made possible only by the parallel examination of multiple variables. Collectively, these insights have important implications in the design of SNA-based therapeutics. Additionally, since this methodology can be extended to other nanotherapeutics, this work points towards a new way of designing and optimizing nanomedicines for a wide variety of uses.
Results and discussion
Modular design of SNAs.
Immunostimulatory SNAs consist of three modular components—the nanoparticle core, oligonucleotide shell and peptide antigen—each of which can be arranged in a variety of configurations5. To establish an appropriate library for high-throughput evaluation, we focused on 11 properties across these components (Fig. 1b). We used 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) to form liposomes that are biocompatible, straightforward to synthesize and capable of encapsulating the antigen8. We focused on two liposome core sizes with average diameters of ~70 and ~100 nm that were made from DOPC or a mixture of 80% DOPC and 20% DOPE, respectively. The size of the SNA can influence its rate of cellular uptake, and inclusion of DOPE in the liposomes is believed to affect the peptide release rate and endosomal escape, which is important for peptide processing9,10.
The oligonucleotide shell serves two roles. It facilitates cellular uptake and serves as the adjuvant, which activates the innate immune system in a sequence-specific manner5. The oligonucleotides used in the design of SNAs in the library varied in five ways: sequence, backbone chemistry, conjugation chemistry to the liposome, site of lipid functionalization and surface density of presentation by SNAs (albeit over a narrow range). We chose a CpG DNA oligonucleotide (ODN1826) known to activate mouse TLR9, as well as an inactive control where the CpG motif is inverted to GpC11,12. TLR9 is an endosomal protein that recognizes unmethylated CpG oligonucleotides associated with bacteria and viruses13. To explore the importance of backbone composition, we synthesized linear oligonucleotides with either phosphodiester (PO) or phosphorothioate (PS) backbones, since phosphorothioate oligonucleotides are known to induce higher immune activation, but SNAs comprising phosphodiester backbones present activities comparable to phosphorothioate structures5,14. We evaluated distinct strategies for conjugating oligonucleotides to the nanoparticles by preparing structures with cholesterol or DOPE, both of which insert into the liposomal cores and can be chemically attached to the 3′ or 5′ ends of the oligonucleotides. Finally, since oligonucleotide density is known to influence cellular uptake and protein binding of SNAs, we evaluated the oligonucleotide surface density at 0.5, 1 and 2 pmol cm−2 (referred to as 1×, 2× and 4×, respectively)15,16. The 4× structure represents the upper limit of what is synthetically viable via our high-throughput procedures at present.
As our test case, we chose the OVA257–264 peptide from ovalbumin—a well-studied model antigen. Since peptide properties can vary dramatically with amino acid composition, we also tested a peptide antigen from the E7 protein of human papillomavirus17. To study how the release rate of the antigen influences nuclear factor kappa light chain enhancer of activated B cells (NF-κB) activation, we evaluated SNAs wherein the antigen was either encapsulated within the SNA architecture or chemically conjugated to oligonucleotides complementary to the CpG oligonucleotides and associated with SNAs through nucleic acid hybridization. As a control, we investigated how the addition of a complement affects TLR9 stimulation.
We synthesized and tested three subsets of SNAs (OVA-encapsulated SNAs, E7-encapsulated SNAs and surface-presented OVA SNAs) representing the key possible combinations of the parameters, with a few synthesis-limited exceptions noted below regarding lipid composition, oligonucleotide surface density and surface-conjugated peptide antigen (see Methods and Table 1). Variation across the 11 structural features—spanning the nanoparticle core, oligonucleotide chemistry, surface presentation of oligonucleotides and incorporation of antigen—led to the design of a library with 960 total SNAs, 800 of which are unique.
Table. 1 |.
SNA design space: the total design space investigated in this study, divided into three subsets
| SNA property | Encapsulated oVA subset (336 SNAs) | Encapsulated E7 subset (336 SNAs) | Conjugated oVA subset (288 SNAs) | ||
|---|---|---|---|---|---|
| Oligonucleotide conjugation chemistry | Cholesterol | DOPE | Cholesterol | DOPE | DOPE |
| Antigen | OVA | OVA | E7 | E7 | OVA |
| Antigen location | Core | Core | Core | Core | Complement |
| Core diameter (nm) | 70 and 100 | 70 and 100 | 70 and 100 | 70 and 100 | 70 and 100 |
| Lipid composition (%DOPC) | 80 and 100% | 100% | 80 and 100% | 100% | 100% |
| Antigen density | 0×, 1× and 10× | 0×, 1× and 10× | 0×, 1× and 10× | 0×, 1× and 10× | 0, 50 and 100% |
| Oligonucleotide sequence | CpG and GpC | CpG and GpC | CpG and GpC | CpG and GpC | CpG and GpC |
| Oligonucleotide conjugation terminus | 5′ and 3′ | 5′ and 3′ | 5′ and 3′ | 5′ and 3′ | 5′ and 3′ |
| Oligonucleotide backbone | PO and PS | PO and PS | PO and PS | PO and PS | PO and PS |
| Oligonucleotide density | 1× and 2× | 1×, 2× and 4× | 1× and 2× | 1×, 2× and 4× | 1×, 2× and 4× |
| Complement density | NA | NA | NA | NA | 0, 50 and 100% |
NA, not applicable; PO, phosphodiester; PS, phosphorothioate.
High-throughput screening of SNA libraries.
To enable the screening of SNA libraries, we developed a high-throughput assay for the rapid and quantitative measurement of cellular responses to the SNAs (Fig. 2a). We cultured RAW-Blue macrophages in 384-well plates and treated each well with a distinct SNA at 4 oligonucleotide concentrations between 1 nM and 1 μM (each separated by a factor of 10). RAW-Blue cells are engineered to secrete secreted embryonic alkaline phosphatase (SEAP) on activation of NF-κB (a major transcription factor that is activated by TLR9 signalling), as well as other signals, to regulate the immune response. We collected the culture media and determined the concentration of SEAP using SAMDI (self-assembled monolayers for MALDI, where MALDI stands for matrix-assisted laser desorption/ionization) mass spectrometry—a label-free assay for high-throughput, quantitative analysis of enzymatic activity18–21. SAMDI uses monolayers presenting a selective capture chemistry against a background of non-binding tri(ethylene glycol) groups to isolate substrates and products from a complex mixture21,22. Subsequent analysis of the monolayers by MALDI mass spectrometry (MALDI-MS) quantitates the amount of substrate and product, which is a direct measure of the enzyme concentration (Fig. 2b,c). Here, we mixed the media containing SEAP with a phosphorylated peptide substrate, captured the substrate and dephosphorylated product on monolayers and then analysed the samples by SAMDI (see Methods for experimental details). We chose this technology for its ability to quantify enzyme activities at high throughput, without dependence on the common optical methods, which can be negatively affected by the light scattering and absorbance of the nanoparticles. These artefacts are difficult to correct because of their dependence on nanoparticle properties such as size, concentration and aggregation. Furthermore, SAMDI is compatible with small sample volumes for analysis, thereby reducing the amounts of SNAs, cells and reagents necessary for evaluation (by around sixfold compared with the amounts used in optical assays).
Fig. 2 |. SAMdi assay workflow.
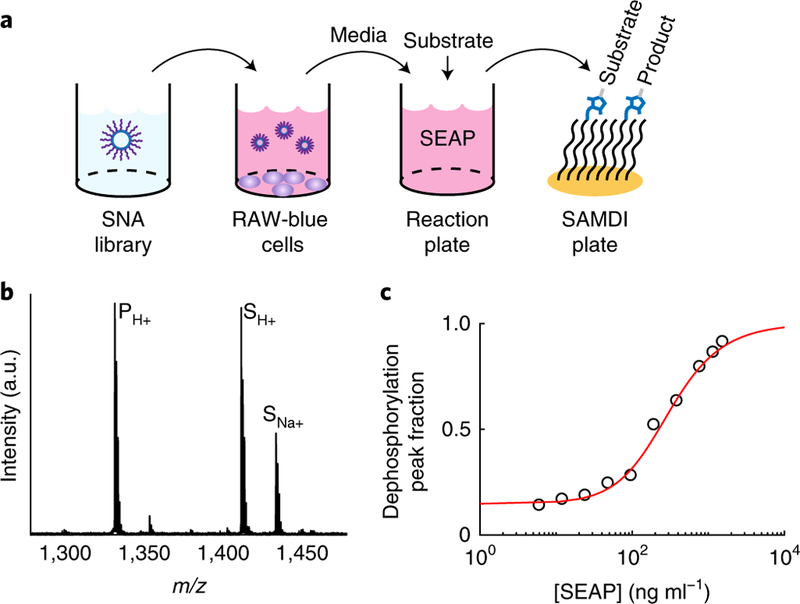
a, Assay used to evaluate the structure– activity relationships between SNA properties and TLR9 activation of APCs. Libraries of SNAs are incubated with RAW-Blue macrophages in 384-well plates. The macrophages have been engineered to secrete SEAP into the media. After ~16 h, the media is transferred, processed and mixed with a phosphorylated substrate. The solution is transferred to SAMDI plates with 1,536 spot arrays of monolayers presenting maleimides to selectively capture the substrate and product by a maleimide–thiol reaction. b, An example SAMDI spectrum showing the immobilized substrate and product. Performing MALDI-MS on the self-assembled monolayers (that is, SAMDI) results in mass spectra containing quantitative information on the relative amounts of substrate and product (that is, the extent of dephosphorylation). c, An example standard curve used to convert the SAMDI spectral data for the library into SEAP concentration.
With this assay, we measured the responses to 960 SNAs at 4 concentrations and with 2 biological replicates, and acquired 2 SAMDI spectra for each sample. Along with standards and controls, more than 8,500 cell culture wells were used, and more than 17,000 SAMDI spectra were analysed. These data revealed many insights into the importance of each structural feature, and how the combinations of features impact immune activation. Below, we highlight some of the most prominent trends.
SNAs induce higher immune activation than linear oligonucleotides.
Varying the design parameters of SNAs induced a broad range of immune activation (Figs. 3a,d shows the encapsulated OVA subset with the active CpG oligonucleotide sequence, and Supplementary Fig. 1 shows the encapsulated E7 subset). Almost all of the SNAs with the active oligonucleotide sequence outperformed the linear phosphodiester oligonucleotide. Additionally, many SNAs, including those with a phosphodiester backbone, were more potent than the linear oligonucleotide with the phosphorothioate backbone.
Fig. 3 |. trends in immune activation due to changes in the oligonucleotide properties of SNAs.
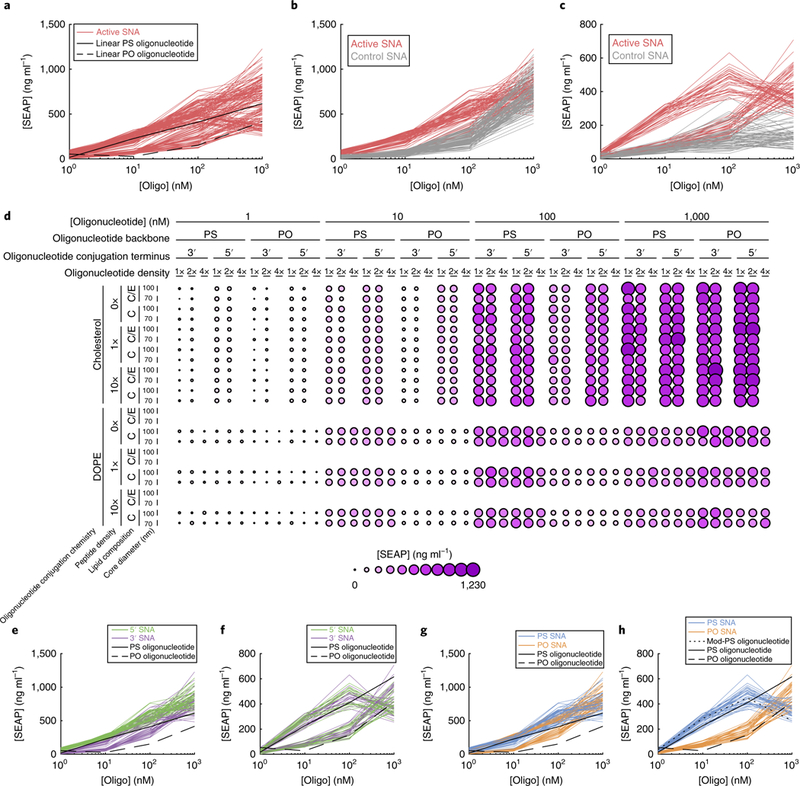
a, SEAP concentrations observed for all of the active-sequence SNAs in the encapsulated OVA subset (all data in this figure are from this subset), compared with the PO and PS versions of linear oligonucleotides with the same active sequence. b,c, Comparison of SNAs with the active and control sequences, for groups of SNAs with cholesterol-conjugated oligonucleotides (b) and those with DOPE-conjugated oligonucleotides (c). d, Dimension stacking plot of the active-sequence SNAs, showing the SEAP concentration for each combination of design properties. Larger and darker circles indicate greater SEAP concentration. For core lipid composition: C, 100% DOPC lipid composition; C/E, mixture of 80% DOPC and 20% DOPE. e,f, Comparison of 5′ and 3′ conjugation termini of SNAs with active sequences, grouped by conjugation chemistry (cholesterol conjugation, e; DOPE conjugation, f). g,h, Comparison of PO and PS backbones of SNAs with active sequences, grouped by conjugation chemistry (cholesterol conjugation, g; DOPE conjugation, h). Mod-PS oligonucleotide, DOPE-conjugated PS oligonucleotide.
Conjugation chemistry of oligonucleotide–liposome association significantly affects immune activation by SNAs.
With 11 design parameters under investigation, we sought to identify the relative importance of design choices on immune activation. Multifactor analysis of variance (ANOVA) (Supplementary Table 1) revealed, unsurprisingly, that oligonucleotide concentration and oligonucleotide sequence (that is, active or control) heavily influenced activation. After sequence, the feature that had the greatest impact on immune activation was the lipid moiety conjugated to the oligonucleotide for liposome attachment. Cholesterol conjugation resulted in higher levels of immune activation than DOPE conjugation (P = 4.8 × 10−16). However, SNAs with cholesterol-conjugated oligonucleotides without CpG motifs also induced similarly high levels of activation at the 1 μM oligonucleotide concentration ([SEAP]: 798 and 747 ng ml−1 for active and inactive, respectively; Fig. 3b), indicating a sequence-independent activation of TLR9. The linear oligonucleotide does not activate TLR9; therefore, these results indicate that these SNAs may activate NF-κB via another mechanism. One possible explanation is that cholesterol groups delivered to cells on the SNA induce additional activation. Our cholesterol conjugation chemistry utilizes carbamates, which can be cleaved by esterases, including sterol O-acyltransferases23. Any potentially released cholesterol, which is known to activate the unfolded protein response pathway in macrophages, may also induce NF-κB activation24.
In contrast, SNAs without CpG-containing oligonucleotides conjugated to DOPE (instead of cholesterol) lead to dramatically lower secretion of SEAP compared with their cholesterol-conjugated counterparts (P < 1 × 10−16; Fig. 3c). We conclude that DOPE conjugation provides a way to synthesize SNAs that trigger an innate immune response exclusively through activation of TLR9. However, the combination of TLR9 stimulation and non-specific activation by SNAs with cholesterol-conjugated oligonucleotides may be advantageous for inducing a greater overall immune response.
The conjugation terminus of the oligonucleotide influences the immune activation in a conjugation chemistry-dependent manner.
Because of the dominant effects of conjugation chemistry, we analysed the remaining SNA properties separately for SNAs with cholesterol- and DOPE-conjugated oligonucleotides. Interestingly, we observed differences in the preferred conjugation terminus when different conjugation chemistries were used (Fig. 3e,f). With cholesterol conjugation, 5′-conjugated SNAs showed significantly higher activity than 3′-conjugated SNAs (OVA subset: P < 2.2 × 10−16 for all concentrations; mean [SEAP]: 566 and 439 ng ml−1 at 100 nM for 5′ and 3′ conjugation, respectively); however, DOPE-conjugated SNAs did not show a difference with conjugation terminus (OVA subset: P = 1 for all concentrations; mean [SEAP]: 324 and 330 ng ml−1 at 100 nM for 5′ and 3′ conjugation, respectively). Furthermore, conjugation from the 5′ terminus did not lead to loss of immune activation for either conjugation chemistry, which contradicts reports that modifications at the 5′ end inactivate the TLR9 activity of linear CpG oligonucleotides25,26.
The phosphorothioate oligonucleotide backbone increases immune activation compared with the phosphodiester backbone.
Similar to well-known trends with linear oligonucleotides, the oligonucleotide backbone also influenced the immunostimulatory activity of the SNAs (Supplementary Table 1 and Fig. 3g,h)14. SNAs with phosphorothioate backbones generally outperformed their phosphodiester counterparts (P = 5 × 10−9 for DOPE- and P = 2.7 × 10−4 for cholesterol-conjugated SNAs). However, a more pronounced dependence on oligonucleotide backbone was observed with DOPE-conjugated SNAs than with cholesterol-conjugated SNAs. For DOPE-conjugated SNAs, the mean SEAP concentrations were 191 and 463 ng ml−1 for phosphodiester and phosphorothioate backbones, respectively, whereas for cholesterol-conjugated SNAs, they were 431 and 573 ng ml−1 (all at 100 nM).
In contrast, at the highest concentration of 1 μM, SNAs with phosphodiester oligonucleotides outperformed their phosphorothioate counterparts. Notably, the activity induced by DOPE-conjugated SNAs with phosphorothioate oligonucleotides consistently decreased when the oligonucleotide concentration increased from 100 nM to 1 μM. The DOPE-conjugated phosphorothioate linear oligonucleotide, but not the phosphodiester backbone, showed a similar reduction in activity at 1 μM (Fig. 3h), suggesting that this behaviour is due to the specific stimulatory properties of the DOPE-conjugated oligonucleotide.
These results lead us to conclude that DOPE-conjugated oligonucleotides with phosphorothioate backbones provide an advantage if greater potency is desired. Phosphorothioate backbones have the added benefit of resistance to nuclease degradation in vivo27. However, these results also show that SNAs with oligonucleotides composed of phosphodiester backbones can achieve similar levels of activation when present at higher concentrations. While class B CpG oligonucleotides are less effective with phosphodiester backbones, using SNAs with phosphodiester oligonucleotides may be worth the loss in potency because of the reduction in toxicity and cost, since the SNA structure may provide sufficient resistance to nuclease activity28–30.
Oligonucleotide density on the surface of the nanoparticle has a small and variable impact on immune activation.
Surprisingly, there was not a strong or consistent trend in how oligonucleotide density affected activity, with neither the highest nor lowest densities showing the best activity. In previous studies, SNAs with higher oligonucleotide densities led to higher biological activity in cellular uptake and RNAse H-mediated degradation of messenger RNA; however, the nanoparticle designs in those studies were limited to gold cores, and used different core sizes and oligonucleotide densities compared with this study15,16. From these observations, we conclude that the choice of oligonucleotide density for these constructs over this narrow density range should be based on other considerations, such as stability in vivo, which is inextricably linked to potency.
Core diameter and lipid composition influence the immune activation of SNAs in an encapsulated peptide-specific manner.
In both encapsulated SNA subsets, the lipid composition generally did not have a significant impact on activity, as determined by ANOVA (Supplementary Table 1), except in one particular context discussed below. Additionally, core diameter was not a significant parameter in the encapsulated OVA subset, whereas it had a significant impact with encapsulated E7 group.
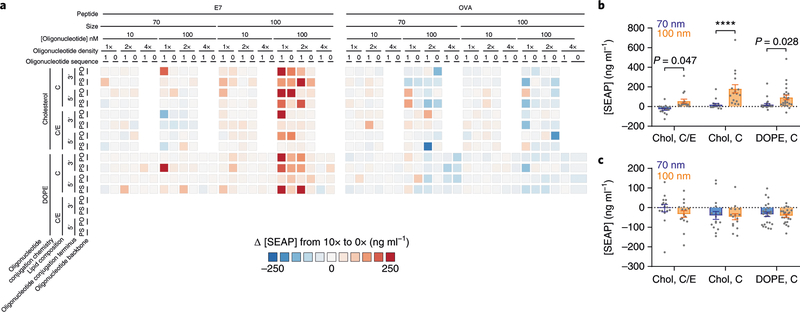
Since all combinations of parameters evaluated were both with and without peptide, we were able to isolate the effects of peptide encapsulation by comparing pairs of SNAs with identical properties except for the amount of peptide encapsulation. We subtracted the SEAP concentration of the SNA without peptide from the SNA with the highest peptide concentration (Fig. 4a). This analysis revealed that core diameter and lipid composition were influential when E7, but not OVA, was encapsulated. Specifically, for the E7 subset, SNAs with 100 nm cores containing peptide induced higher levels of NF-κB activation (P = 5.7 × 10−5), and the magnitude of this effect also depended on lipid composition (Fig. 4b). Within the subset of SNAs with cholesterol-conjugated oligonucleotides on 100 nm cores, the SNAs with 100% DOPC cores showed higher immune activation than those with 80% DOPC and 20% DOPE cores (P = 0.0011; Fig. 4b). We observed no dependence between the presence of antigen and immune activation when the antigen was OVA (Fig. 4c).
Fig. 4 |. trends in immune activation due to changes in the peptide encapsulation of SNAs.
a, Difference in SEAP concentration between each SNA with the highest peptide concentration (10×) and the corresponding SNA without any peptide (0×) for the two encapsulated peptide subsets. b,c, Average difference in SEAP concentration between SNAs with 10× and 0× E7 peptide concentrations, grouped by core diameter and the combination of conjugation chemistry (cholesterol (Chol) versus DOPE) and lipid composition, for the E7 (b) and OVA subsets (c), at 100 nM oligonucleotide concentration (n = 16 for Chol, C/E and Chol, C; n = 24 for DOPE, C). Bar heights and error bars represent means ± s.e.m. P values were calculated by two-way ANOVA with Sidak’s post-hoc test. All differences between means with P < 0.05 are indicated (****P < 0.0001).
These results clearly illustrate that peptide encapsulation can impact the ability of SNAs to activate TLR9 and reveal crosstalk between the molecular components of SNAs intended to induce innate or adaptive immunity. Unlike oligonucleotides, the physicochemical properties of peptides vary dramatically with sequence, which can affect their interaction with the rest of the SNA structure. For example, the differences in the isoelectric points of the peptides, which are 5.7 and 8.8 for the E7 and OVA peptides, respectively, result in different net charges for the peptides, which could affect their interaction with the positively charged liposome core. We conclude that the interactions between liposomes and peptides must be taken into account when designing and evaluating nanomedicines, as they can lead to large shifts in the immune activation of SNAs, especially at high levels of peptide encapsulation.
Effects of hybridization of complementary strands onto SNAs.
The versatility of the SNA architecture allows for alternative methods of incorporating the antigen into the structure, apart from loading in the lipid core. We investigated one such alternative—conjugation of the antigen to a complementary oligonucleotide, which is then hybridized to a lipid-anchored oligonucleotide. As a control, we also synthesized SNAs with the complementary oligonucleotide but without peptide conjugation. In these SNAs, the CpG-containing oligonucleotide is double stranded, and thus is differentiated from SNAs with only single-stranded oligonucleotides. In this conjugated OVA subset, we used DOPE-conjugated oligonucleotides to prevent the non-specific NF-κB activation by cholesterol-conjugated SNAs described above.
Our results show that SNAs synthesized with this strategy shared some trends with their single-stranded counterparts. After oligonucleotide sequence, the most influential property on immune activation was backbone chemistry, with phosphorothioate backbones outperforming phosphodiester versions (Fig. 5a). Again, we found that the core properties of lipid composition and core diameter were not significant.
Fig. 5 |. trends in immune activation due to hybridization.
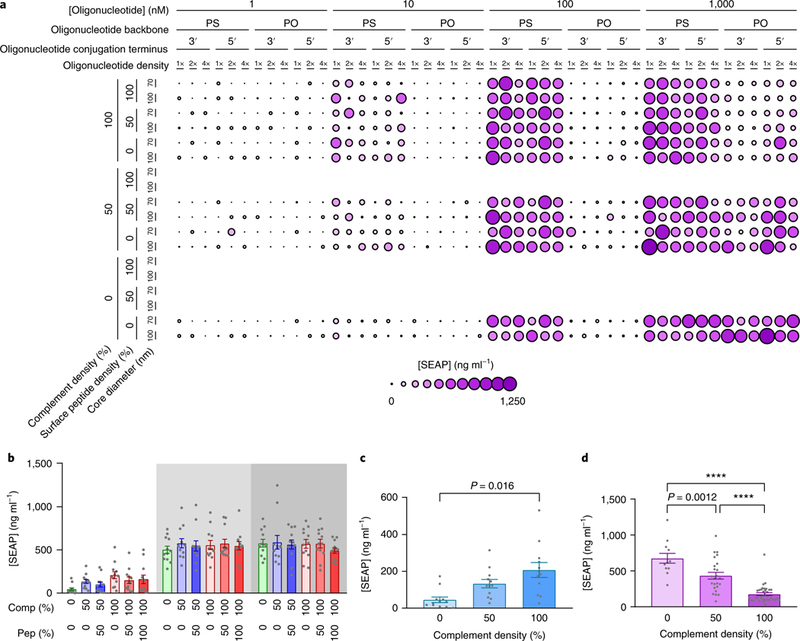
a, SEAP concentrations for all active-sequence SNAs in the surface-presented OVA subset. b, Mean SEAP concentration of PS-backbone, active-sequence SNAs, grouped by the combinations of complement density (Comp.) and surface antigen density (Pep.) (n = 12). Shaded areas indicate the response at 10 nM, 100 nM and 1,000 nM oligonucleotide concentration, from left to right. c, Mean SEAP concentration of SNAs with a phosphorothioate backbone, 0% peptide and active sequence, as a function of complement density, at a 10 nM oligonucleotide concentration (n = 12). d, Mean SEAP concentration of SNAs with a phosphodiester backbone and active sequence, as a function of complement density, at a 1 μM oligonucleotide concentration (100%: n = 36, 50%: n = 24, 0%: n = 12). Bar heights and error bars represent means ± s.e.m. P values were calculated by one-way ANOVA with Tukey’s post-hoc test. All differences between means with P < 0.05 are indicated (****P < 0.0001).
Interestingly, for the SNAs with phosphorothioate oligonucleotides, addition of the complement oligonucleotide (either to half or all of the anchored oligonucleotides) did not change immune activation at concentrations of 100 nM or 1 μM, respectively (Fig. 5b). Furthermore, there was no difference between SNAs composed of the complement with and without conjugated peptide. However, at low concentrations (10 nM), higher complement densities led to higher immune activation (Fig. 5c). This effect may be a function of SNA uptake, where higher complement densities create higher charge densities on the surface and increase the uptake of SNAs, which in turn leads to higher immune activation. In contrast, complementation strongly reduced the activity of phosphodiester-backbone SNAs at the highest concentration tested (Fig. 5d). A possible explanation for the decreased activity in duplexed SNAs is that the duplexing interferes with the oligonucleotide interaction with TLR9; however, it is not clear why the interaction with TLR9 would be different with phosphodiester and phosphorothioate backbones. These results suggest that the strategy of including antigens by duplexing antigen-conjugated complementary oligonucleotides is effective with phosphorothioate SNAs, without concern for losing activation of TLR9.
Supervised machine learning captures nonlinearity of property interactions and confirms trends in the biological importance of properties.
Because many of the parameters studied were interdependent, we utilized supervised machine learning models and evaluated their performance to better understand SNA properties. We applied supervised models to automatically predict immune activity from SNA properties with the expectation that properties relevant to immune activation would improve a model’s predictive capability31,32. These models differ from traditional data analysis in that instead of explicitly programming them with formulas, they ‘learn’ from the data on their own. Specifically, we employed multiple linear regression, logistic regression and nonlinear XGBoost to fit training data, and cross-validation of test data was conducted using the Q2 statistic. Q2 quantifies the accuracy of the predicted SEAP concentrations against measured values, and ranges from −∞ to 1, where 0 indicates no predictive power (equivalent to predicting the mean) and 1 indicates perfect prediction33,34.
We trained each model with all combinations of properties (that is, two properties at a time, three properties at a time, and so on) and analysed their Q2 performance. As additional properties were added to the models, the Q2 performance increased, plateauing for most models and decreasing in the XGBoost model for the surface- presented OVA subset (Fig. 6a,b). Since clear nonlinear trends were observed in the data, as described above, the model performance increased with the nonlinearity of the model in both subsets (mean increase from 0.53 for the linear model to 0.83 for XGBoost). Analysis of the most predictive SNA property combinations demonstrates that highly predictive properties remain significant and informative as more properties are introduced into the model (Supplementary Fig. 2a,b). In addition, the order of importance of the properties was largely consistent between the encapsulated OVA and surface-conjugated OVA subsets, suggesting that the ordering is robust regardless of peptide localization. To ensure that these trends are not artefacts, we repeated this analysis with randomized data. Q2 values for all of these models were zero or below, indicating that the predictions are specific to our data (Supplementary Fig. 3). In addition, we calculated the standard deviations of the error between the predicted and actual values for the XGBoost to further validate the model (Supplementary Fig. 4). The standard deviations were in the range of 10–30 ng ml−1, which is very small compared with the range of activities that exceeds 1,000 ng ml−1.
Fig. 6 |. Analysis by machine learning.
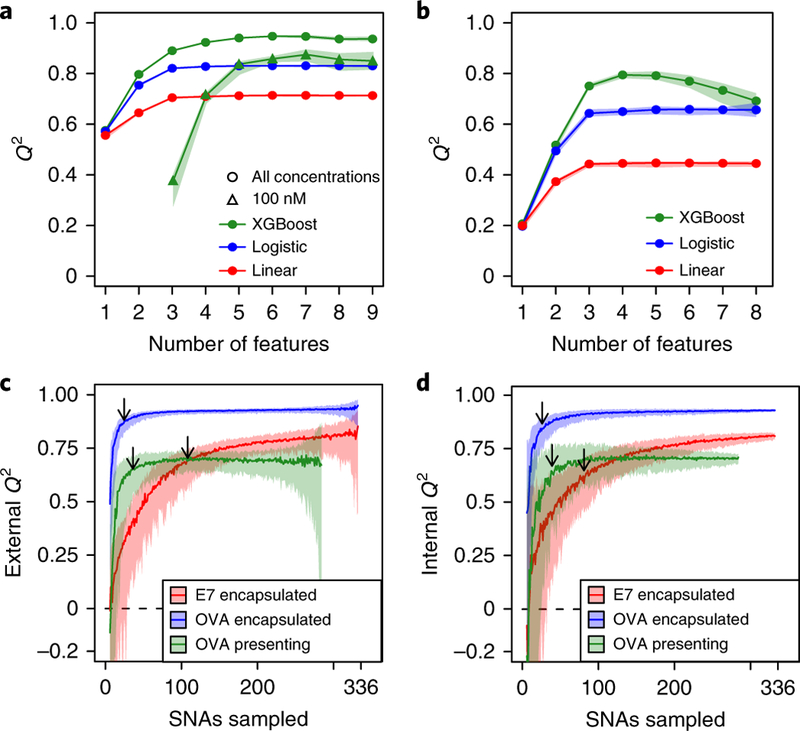
a,b, Q2 values of the highest-performing SNA property combinations are shown across different numbers of properties for encapsulated OVA (a) and surface-presented OVA subsets (b). In a, for the XGBoost model, the active sequence and 100 nM subset is shown, in addition to both active and inactive sequences for all concentrations. c,d, XGBoost Q2 performance when selecting and training on a random SNA subsample and testing predictions on the unselected SNAs (c) or cross-validating within the selected subsample (d). The shading represents 90% confidence intervals. The arrows point to the points of diminishing returns.
For the encapsulated OVA and surface-presented OVA subsets, the Q2 value stopped increasing beyond five and four properties, respectively (Fig. 6a). At first glance, one might conclude that only these highly predictive properties are relevant; however, when repeating this analysis with fixed values for sequence and concentration (the two features with the greatest impact), the Q2 values stopped increasing after another five properties were added (Fig. 6a), indicating that formerly seemingly non-predictive properties do, in fact, influence immune activation (Supplementary Fig. 2c). Taken together, these properties, which appear non-influential in a global context, become impactful in a restricted design space.
Capturing the maximum structure–activity relationship with minimum SNA synthesis and evaluation.
Next, we investigated whether a similar Q2 level is attainable with fewer, randomly selected SNA designs. This question is particularly relevant when synthesis and evaluation of full libraries are impractical, but when exploration of a large design space is desired. In this case, one could synthesize a random subset that would capture the most important trends and then suggest additional candidates to evaluate. To this end, we simulated this process by training an XGBoost model on a random selection of SNAs and testing predictions on the remaining, unselected SNAs within the three subsets (Fig. 6c). We identified the points of diminishing returns, which balance the minimum number of SNAs with maximum Q2, by calculating the sample size closest to training size 1 and Q2 = 1. This point is 90, 20 and 31 SNAs (out of 336, 336 and 288 SNAs) with Q2 = 0.67, 0.88 and 0.66 for the encapsulated E7, encapsulated OVA and surface-presented OVA subsets, respectively. These points represent a mean of 16% of the total number of SNAs, suggesting that a small number of randomly selected SNAs can predict structure–activity relationships of a relatively large SNA library. In practice, this external Q2 (prediction of non-synthesized SNAs) cannot be measured with a randomized subsample, but an internal Q2 can be measured by cross-validating within the randomized subsample. We show that the internal and external Q2 values are highly correlated (Fig. 6d and Supplementary Fig. 5), suggesting that we can identify points of diminishing returns as we continually synthesize random SNAs from an arbitrary library size. Combined with the high-throughput SNA synthesis and characterization approach described above, the machine learning analysis shows that a combined experimental and computational method can probe and predict the structure–activity relationships of tens of thousands of SNAs with a much smaller subset (order of thousands) of structures.
Outlook.
This work, as well as other approaches35,36, makes clear the need to consider the full range of structure–activity relationships when designing nanomedicines by high-throughput processes. Although high-throughput techniques are industry standards in the combinatorial screening of small-molecule drugs, such approaches are just beginning to be implemented to define structure–activity relationships for therapeutic nanoconstructs. The data presented here show that such properties can be strongly interrelated in non-obvious ways, and emphasize the risks of using limited datasets to make global conclusions about one structural consideration being more critical than others. Our results show that predictions about SNA activity simply based on what is known about the individual components of an SNA (that is, CpG, OVA, E7 and phospholipids) are inaccurate in many cases. This interdependence and non-linearity are underscored when applying the nonlinear machine learning models, as opposed to linear ones, in predicting the biological response of SNAs. Indeed, to realize rational approaches to vaccinology, this work makes a strong case for the combination of high-throughput experimentation and computational analysis in determining the structure–activity relationships of nanomedicines in general and SNAs in particular. Note that this study did not directly pursue the identification or optimization of a candidate immunotherapeutic for a specific disease. Rather, this effort has examined how a single key biochemical step (TLR9 activation) in the generation of an immune response can be activated by nearly 1,000 variations in SNA structure.
A key finding from our use of supervised machine learning to analyse the data generated for ~1,000 SNA structures is the accuracy of predicting activities when using data obtained only from relatively small sublibraries. However, a broader use of this approach will require the careful design of structures in the random subsets to ensure the validity of predictions, and follow-up with experimental confirmation of predicted activities. In our study, we applied the machine learning independently for libraries defined by the selection of antigen and position of the antigen, with the benefit of knowing that the selection of antigen influences activity. This type of approach to structure–activity relationships is limited to predictions based on properties included in the training set. In addition to the unavoidable potential of missing unique combinatorial effects that are not captured in the sampled space, the accuracy and scalability of prediction that can be accomplished by supervised machine learning may differ for libraries consisting of other types of nanostructures.
Our constraint to a single readout and its dependence on a wide range of variables has led to a lesson in the design and development of a type of nanomedicine that could not have been extracted by the alternative and more conventional approach of evaluating a small number of candidate structures and analysing multiple immune system readouts (for example, cytokine expression or cellular proliferation) in vivo; such an approach is severely limited in throughput by the small number of structures that can be prepared and evaluated in parallel. Conversely, we note that TLR9 activation in a model cell line reports on one key step among many involved in raising cellular immune responses (for example, biodistribution, antigen delivery and presentation by different types of APCs), and that structures optimized for a single output may not be the best therapeutic candidates when examined in vivo. The pursuit of identifying and ultimately arriving at SNA (or other nanoparticle) immunotherapeutic agents will be well served by the combination of using high-throughput library screens in cellular assays, and in vivo examination of structures whose selection is informed by the library screen.
Methods
Materials.
DOPE and DOPC were purchased from Avanti Polar Lipids. Phosphoramidites for DNA synthesis were purchased from Glen Research. Peptide antigens were custom ordered from GenScript. 2,2’-dipyridyldisulfide, hexadecylphosphonic acid, tris(2-carboxyethyl)phosphine hydrochloride, maleimide and 2,4,6-trihydroxyacetophenone were purchased from Sigma– Aldrich. Monolayer disulfides were purchased from Chemtos. Peptide synthesis reagents were purchased from AnaSpec and MilliporeSigma.
DNA synthesis.
DNA was synthesized with a MerMade 12 synthesizer. Cholesterol modification was done on the column in the synthesizer using 3′-Cholesteryl-TEG CPG (Glen Research) for 3′ modifications and Cholesteryl-TEG Phosphoramidite (Glen Research) for 5′ modifications. For DOPE-modified oligonucleotides, a thiol-modified oligonucleotide was synthesized. DNA sequences are shown in Supplementary Table 2.
Synthesis of DOPE-SMPB.
To 1 mol equivalent of succinimidyl 4-(p-maleimidophenyl)butyrate (SMPB; Thermo Fisher Scientific) and 1 mol equivalent of N,N-diisopropylethylamine was added 1 ml of DOPE as received from Avanti Polar Lipids (25 mg ml−1 in chloroform). The reaction was incubated for 24 h at room temperature. The reaction was checked for completion with thin-layer chromatography using 20% methanol in dichloromethane as the mobile phase. On disappearance of the DOPE band in thin-layer chromatography, the reaction was washed three times with water, and the organic phase was dried under vacuum.
DOPE modification of oligonucleotides.
The thiol-modified oligonucleotide was reduced with 200 mM dithiothreitol (DTT) in 100 mM phosphate buffer (pH 8.0) for 2 h at 40 °C. The oligonucleotide was purified away from DTT with NAP-10 columns using water as the mobile phase (GE Healthcare). The reduced oligonucleotide was immediately reacted with DOPE-SMPB as follows: DOPE-SMPB (50 mol equivalents) was dissolved in ethanol in the same volume as the oligonucleotide. The two solutions were mixed together and incubated at room temperature for 48 h. The reaction mixture was washed with chloroform three times to remove excess lipid. The interface and the aqueous phase was lyophilized. The reaction yield and purity were determined by 20% denaturing polyacrylamide gel electrophoresis gels. Typically, yields were greater than 90% and no further cleanup was performed.
Synthesis of liposomes.
DOPC (25 mg in chloroform) was transferred to a glass vial and dried overnight into a thin film, first under an N2 stream followed by high vacuum. For DOPC-DOPE mixture liposomes, 20 mol% DOPE was added to the 25 mg of DOPC before drying. The lipid film was rehydrated with 1 ml of 1× phosphate buffered saline (PBS) and vortexed until no more clumps were visible. For encapsulated peptides, the peptide was dissolved into the PBS at 0.1 and 1 mg ml−1. The lipid suspensions were frozen in liquid nitrogen and thawed in a bath sonicator with sonication. The freeze–thaw was repeated three times. The solution was then extruded through 200, 100, 80 and 50 nm filters. Two filters were used for each extrusion, and the solution was passed through these filters 11 times. The liposomes were split into 2 after the 80 nm extrusion. Half of the solution was saved, and the remainder was extruded through a 50 nm filter. The liposomes were dialysed against 1× PBS overnight to remove non-encapsulated peptide. The liposomes were characterized by dynamic light scattering for size (z-average reported), and phosphatidylcholine assay for concentration (MilliporeSigma). The liposomes extruded through the 50 and 80 nm filters had z-averages of ~70 and ~100 nm, respectively. For DOPC-DOPE mixture SNAs, DOPE did not interfere with the phosphatidylcholine assay, so we assumed that the DOPC-to-DOPE ratio remained 80:20. The liposome concentrations were calculated from the diameter and the lipid concentration, as described by Banga et al.37.
Synthesis of complementary oligonucleotides with peptide.
The complementary oligonucleotides were reduced with DTT as described above and mixed with 55 equivalents of 2,2’-dipyridyldisulfide in 100 mM phosphate buffer (pH 8.0). The reaction was incubated at 40 °C for 24 h. The reaction process was monitored by absorption of pyridinethione at 343 nm. On completion, the modified oligonucleotide was washed 3 times with water in a 3K MWCO spin filter. The oligonucleotide was then mixed with 1 equivalent of cysteine-modified OVA (CSIINFEKL) and incubated at 40 °C overnight. The process was again monitored at 343 nm and washed with a spin filter as described above.
Duplex formation.
The purified peptide–oligonucleotide conjugate and 1 equivalent of the lipid-conjugated oligonucleotide were mixed in duplex buffer (30 mM HEPES (pH 7.4), 100 mM potassium acetate and 2 mM magnesium acetate). The mixture was heated to 65 °C for 10 min and slow-cooled to room temperature by turning off the heat block and allowing the temperature to equilibrate.
SNA synthesis.
Lipid-modified oligonucleotides or duplexes were mixed with liposomes in a 384-well plate in a 40 µl final volume. The final concentration of lipid-modified oligonucleotide or duplex in each well was 10 µM. The concentration of liposomes was adjusted to accommodate SNAs of various oligonucleotide densities. After mixing, the plate was sealed and incubated at room temperature for 24 h.
Synthesis of the peptide substrate.
The CRpY-NH2 peptide substrate was synthesized using standard fluorenylmethoxycarbonyl solid-phase peptide synthesis methods on a Rink Amide resin. The amino terminus was acetylated. The peptide was purified by reverse-phase high-performance liquid chromatography on a C-18 column in a gradient from water to acetonitrile, and fractions were checked for the correct mass by MALDI-MS. The peptide was lyophilized and stored as a solid until use.
SAMDI plate and monolayer preparation.
Stainless steel plates custom designed for use in MALDI instruments were cleaned and used to evaporate a 1,536-spot pattern of 5 nm titanium (0.02 nm s−1), then 35 nm gold (0.05 nm s−1), using an aluminium mask. The gold array plates were incubated overnight at 4 °C in an ethanolic solution containing a 1:4 ratio of an asymmetric disulfide terminated with a maleimide group and a tri(ethylene glycol) group, and a symmetric disulfide terminated with tri(ethylene glycol) groups, with a 0.5 mM total disulfide concentration. The plates were then rinsed with ethanol, dried and placed in a solution of 10 mM hexadecylphosphonic acid in ethanol for 10 min at room temperature. Plates were then rinsed with ethanol, dried and then used for the SEAP assay.
SEAP assay.
RAW-Blue cells (InvivoGen) were cultured as described by the manufacturer. The cells were collected and suspended at 550,000 cells ml−1, and 17,000 cells were distributed into 384-well culture plates with a Thermo Fisher Scientific Multidrop Combi. Next, 10× SNA solutions were added to the cell culture plates with a Tecan liquid handler, then cultured at 37 °C under 5% CO2. After ~16 h, the cell culture plates were centrifuged at 300 rcf for 1 min, then 10 μl of media was transferred to a 384-well reaction plate. Recombinant SEAP (0–1,600 ng ml−1) was prepared in media from untreated cells and then added to empty wells. This was used as the standard curve. To minimize free thiol in the media, which competes with substrate immobilization, 1 μl of 11 mM tris(2-carboxyethyl)phosphine hydrochloride in water was added to the plates and they were incubated for 15 min at 60 °C to first reduce cystine to cysteine. The 60 °C incubation also inactivates any potential phosphatases other than SEAP, which is stable at 60 °C. Next, 1 μl of 12 mM maleimide was added to react with free cysteines for 1 h at 37 °C. Some 8 μl of 75 μM CRpY peptide substrate in reaction buffer (300 mM Tris, pH 8.5 and 2.5 mM MgCl2) was added to the reaction plate, then incubated for 1 h at 37 °C. To this, 2 μl of 11 mM pridoxal 5′-phosphate hydrate in reaction buffer was added. Next, 0.75 μl of the reaction solutions were transferred to 1,536-spot SAMDI array plates and incubated for 1 h at 37 °C. The plates were rinsed with water and ethanol, then dried with air. Matrix (15 mg ml−1 2,4,6-trihydroxyacetophenone in acetone) was applied to the SAMDI plates and they were analysed by MALDI using an Applied Biosystems SCIEX TOF/TOF 5800 MALDI instrument in positive reflector mode. The spectra were analysed by calculating the areas under the curves for the [M + H]+ and [M + Na]+ disulfide peaks corresponding to the substrate and product masses. Each SNA subset was tested in two wells (biological replicates) and each sample was tested on two SAMDI spots (technical replicates). Technical replicates with subthreshold signal-to-noise ratios were excluded from the analysis.
Quantitative structure–activity relationship model.
We trained quantitative structure–activity relationship models to predict immune activation from SNA properties. The training data contained 336 SNA rows with 9 property columns for datasets 1 and 2, and 288 SNA rows with 8 property columns for dataset 3. The response vector—also called the predicted variable—is the immune activation measured via the SEAP concentration. We used cross-validation, where a sample of data are left out for model testing, to calculate the predictive power Q2 metric:
In this formulation, yi is the immune activation for test SNA i, is the predicted immune activation, is the immune activation of the training set, and n is the number of cross-validated test SNAs. The Q2 metric can take on values from −∞ to 1, where 1 is perfect prediction, 0 is equivalent to random performance (such as predicting the mean immune activation of all SNAs in the training set), and negative values represent worse than random performance. We use fivefold cross-validation, where a random 80% of the data are selected for training, with the remaining 20% as validation. We selected three models to test for linear relationships among SNA properties and immune activation: linear regression, logistic regression and the nonlinear model XGBoost. If all relationships were linear, Q2 would be similar for all models. Similarly, logistic regression can fit trends that are more complex than linear regression, but it still treats multiple properties as linearly related and is still a linear model. For all models, we created an explicit null model by randomizing the data values before model training.
Statistics.
Multi-way ANOVA was performed on each SNA subset using MATLAB software. Statistical comparisons of paired data were made using the two-tailed Wilcoxon test. Unpaired data were compared with a two-tailed t-test for single comparison. Finally, ANOVA was chosen for multiple comparisons using R and Graphpad Prism software.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number U54CA199091. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work made use of the IMSERC at Northwestern University, which has received support from Northwestern University and the State of Illinois.
Footnotes
Code availability
The custom codes used to generate the results reported in this manuscript are available from the corresponding authors upon reasonable request.
Data availability
The data that support the findings of this study are available within the paper and its Supplementary Information. All data generated in this study are available from the corresponding authors upon reasonable request.
Competing interests
C.A.M. and M.M. own stock from Exicure, which has licensed the SNA technology. M.M. owns stock in SAMDI Tech—the company that has licensed the SAMDI technology.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41551–019-0351–1.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bobo D, Robinson KJ, Islam J, Thurecht KJ & Corrie SR Nanoparticle-based medicines: a review of FDA-approved materials and clinical trials to date. Pharm. Res 33, 2373–2387 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Mirkin CA, Letsinger RL, Mucic RC & Storhoff JJ A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 382, 607–609 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Cutler JI, Auyeung E & Mirkin CA Spherical nucleic acids. J. Am. Chem. Soc 134, 1376–1391 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Choi CHJ, Hao L, Narayan SP, Auyeung E & Mirkin CA Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl Acad. Sci. USA 110, 7625–7630 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radovic-Moreno AF et al. Immunomodulatory spherical nucleic acids. Proc. Natl Acad. Sci. USA 112, 3892–3897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosi NL et al. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 312, 1027–1030 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Seferos DS, Prigodich AE, Giljohann DA, Patel PC & Mirkin CA Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano Lett 9, 308–311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J et al. A review on phospholipids and their main applications in drug delivery systems. Asian J. Pharm 10, 81–98 (2015). [Google Scholar]
- 9.Schroit AJ, Madsen J & Nayar R Liposome–cell interactions: in vitro discrimination of uptake mechanism and in vivo targeting strategies to mononuclear phagocytes. Chem. Phys. Lipids 40, 373–393 (1986). [DOI] [PubMed] [Google Scholar]
- 10.Simoes S, Slepushkin V, Duzgunes N & Pedroso de Lima M. C. On the mechanisms of internalization and intracellular delivery mediated by pH-sensitive liposomes. Biochim. Biophys. Acta 1515, 23–37 (2001). [DOI] [PubMed] [Google Scholar]
- 11.McCluskie MJ & Davis HL CpG DNA as mucosal adjuvant. Vaccine 18, 231–237 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Krieg AM et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374, 546–549 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Hemmi H et al. A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Zhao Q, Temsamani J, Iadarola PL, Jiang Z & Agrawal S Effect of different chemically modified oligodeoxynucleotides on immune stimulation. Biochem. Pharmacol 51, 173–182 (1996). [DOI] [PubMed] [Google Scholar]
- 15.Giljohann DA et al. Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett 7, 3818–3821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prigodich AE, Alhasan AH & Mirkin CA Selective enhancement of nucleases by polyvalent DNA-functionalized gold nanoparticles. J. Am. Chem. Soc 133, 2120–2123 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gendron KB, Rodriguez A & Sewell DA Vaccination with human papillomavirus type 16 E7 peptide with CpG oligonucleotides for prevention of tumor growth in mice. Arch. Otolaryngol. Head Neck Surg 132, 327–332 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Berns EJ, Cabezas MD & Mrksich M Cellular assays with a molecular endpoint measured by SAMDI mass spectrometry. Small 12, 3811–3818 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Min DH, Tang WJ & Mrksich M Chemical screening by mass spectrometry to identify inhibitors of anthrax lethal factor. Nat. Biotechnol 22, 717–723 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Mrksich M Mass spectrometry of self-assembled monolayers: a new tool for molecular surface science. ACS Nano 2, 7–18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su J & Mrksich M Using mass spectrometry to characterize self-assembled monolayers presenting peptides, proteins, and carbohydrates. Angew. Chem. Int. Ed. Engl 41, 4715–4718 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Su J, Rajapaksha TW, Peter ME & Mrksich M Assays of endogenous caspase activities: a comparison of mass spectrometry and fluorescence formats. Anal. Chem 78, 4945–4951 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Humerickhouse R, Lohrbach K, Li L, Bosron WF & Dolan ME Characterization of CPT-11 hydrolysis by human liver carboxylesterase isoforms hCE-1 and hCE-2. Cancer Res 60, 1189–1192 (2000). [PubMed] [Google Scholar]
- 24.Li Y et al. Free cholesterol-loaded macrophages are an abundant source of tumor necrosis factor-α and interleukin-6: model of NF-κB and MAP kinase-dependent inflammation in advanced atherosclerosis. J. Biol. Chem 280, 21763–21772 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Yu D, Zhao Q, Kandimalla ER & Agrawal S Accessible 5′-end of CpG-containing phosphorothioate oligodeoxynucleotides is essential for immunostimulatory activity. Bioorg. Med. Chem. Lett 10, 2585–2588 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Kandimalla ER et al. Conjugation of ligands at the 5′-end of CpG DNA affects immunostimulatory activity. Bioconjug. Chem 13, 966–974 (2002). [DOI] [PubMed] [Google Scholar]
- 27.De Clercq E, Eckstein E & Merigan TC Interferon induction increased through chemical modification of a synthetic polyribonucleotide. Science 165, 1137–1139 (1969). [DOI] [PubMed] [Google Scholar]
- 28.Roberts TL, Sweet MJ, Hume DA & Stacey KJ Cutting edge: species-specific TLR9-mediated recognition of CpG and non-CpG phosphorothioate-modified oligonucleotides. J. Immunol 174, 605–608 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Flierl U et al. Phosphorothioate backbone modifications of nucleotide-based drugs are potent platelet activators. J. Exp. Med 212, 129–137 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henry SP et al. Complement activation is responsible for acute toxicities in rhesus monkeys treated with a phosphorothioate oligodeoxynucleotide. Int. Immunopharmacol 2, 1657–1666 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Chen T & Guestrin C XGBoost: a scalable tree boosting system In Proc. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (ACM, 2016). [Google Scholar]
- 32.Menard S Applied Logistic Regression Analysis Vol. 106 (Sage, Thousand Oaks, 2002). [Google Scholar]
- 33.Schuurmann G, Ebert RU, Chen J, Wang B & Kuhne R External validation and prediction employing the predictive squared correlation coefficient test set activity mean vs training set activity mean. J. Chem. Inf. Model 48, 2140–2145 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Golbraikh A & Tropsha A Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. J. Comput. Aided. Mol. Des 16, 357–369 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Akinc A et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol 26, 561–569 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson DG, Lynn DM & Langer R Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angew. Chem. Int. Ed. Engl 42, 3153–3158 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Banga RJ, Chernyak N, Narayan SP, Nguyen ST & Mirkin CA Liposomal spherical nucleic acids. J. Am. Chem. Soc 136, 9866–9869 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.