Abstract
Methanogenesis is an ancient metabolism of key ecological relevance, with direct impact on the evolution of Earth’s climate. Recent results suggest that the diversity of methane metabolisms and their derivations have probably been vastly underestimated. Here, by probing thousands of publicly available metagenomes for homologues of methyl-coenzyme M reductase complex (MCR), we have obtained ten metagenome-assembled genomes (MAGs) belonging to potential methanogenic, anaerobic methanotrophic and short-chain alkane oxidizing archaea. Five of these MAGs represent under-sampled (e.g., Verstraetearchaeota, Methanonatronarchaeia, ANME-1) or previously genomically undescribed (ANME-2c) archaeal lineages. The remaining five MAGs correspond to lineages that are only distantly related to previously known methanogens and span the entire archaeal phylogeny. Comprehensive comparative annotation significantly expands the metabolic diversity and energy conservation systems of MCR-bearing archaea. It also suggests the potential existence of a yet uncharacterized type of methanogenesis linked to short-chain alkane/fatty acid oxidation in a previously undescribed class of archaea (‘Ca. Methanoliparia’). We redefine a common core of marker genes specific to methanogenic, anaerobic methanotrophic and short-chain alkane-oxidizing archaea, and propose a possible scenario for the evolutionary and functional transitions that led to the emergence of such metabolic diversity.
Methanogenesis is an archaeal-specific metabolism of key relevance in the anaerobic degradation of organic matter and biogas production1,2. It is considered one of the most ancient energetic metabolisms3,4 with direct impact on the evolution of the Earth’s climate system5. Methanogens have been detected in virtually all types of anaerobic environments. Until recently, all methanogens were thought to belong to two euryarchaeal clades, named Class I and Class II methanogens6. The majority of Class I/II methanogens can grow by reducing CO2 into methane using H2 as an electron donor7. Several representatives of the Methanosarcinales (Class II methanogens) use additional energetic substrates, including acetate and methylated compounds8. Methanosphaera spp. (Class I methanogens) are restricted to the reduction of methanol with H29. Regardless of the encoded methanogenic pathway, all members of Class I/II methanogens possess the H4MPT methyl-branch of the Wood-Ljungdahl pathway (m-WL), the N5-Methyltetrahydromethanopterin:coenzyme M methyltransferase complex (MtrABCDEFGH or MTR), and the methyl-coenzyme M reductase complex (McrABG or MCR)10. The same enzymes are present in ANaerobic MEthanotrophic archaea (ANME) and are used in reverse to oxidize methane11–13.
Our understanding of the diversity and metabolic versatility of methanogenic archaea is undergoing a rapid transformation with the availability of additional isolates and metagenome-assembled genomes (MAGs). This has revealed additional lineages only distantly related to Class I/II methanogens14–16, including Methanomassiliicoccales10, Methanofastidiosales17, Methanonatronarchaeia18 and Verstraetearchaeota19. A striking characteristic of these recently described methanogens is the absence of the MTR complex, a partial or missing m-WL pathway, and the presence of specific methyltransferases for the utilization of methylated compounds. Accordingly, they are predicted to be limited to reduce methylated compounds with H2 for methanogenesis, which was experimentally validated for Methanomassiliicoccales20 and Methanonatronarchaeia18. Interestingly, the implication of a divergent McrABG-like complex in the oxidation of short-chain alkanes (butane, propane) has been demonstrated in two representatives of a recently described euryarchaeal order, the ‘Ca. Syntrophoarchaeales’21. Divergent MCR sequences were also found in two members of the Bathyarchaeota22 (TACK superphylum), in the GoM-Arc1 (a lineage within the Methanosarcinales)23, and from environmental samples24. Altogether, this suggests that methanogens, anaerobic methanotrophs and short-chain alkane oxidizers may have an even wider phylogenetic and environmental distribution than previously anticipated, provoking new questions on the diversity and evolution of these metabolisms.
Results and Discussion
Additional lineages of archaea with an MCR or MCR-like complex
To identify previously undescribed lineages of potential methanogens, anaerobic methanotrophs and short-chain alkane oxidizers, we probed available metagenomes from the JGI/IMG database for McrA homologues and identified sequences distantly related to well characterized lineages (Methods). Ten MAGs were reconstructed from the corresponding metagenomes sourced from a wide range of anoxic environments including an inland petroleum reservoir from Brazil, oil seeps from USA25, soda lake sediments from Russia, and hot-springs from China and USA (Table 1). Nine of the ten MAGs had an estimated completeness ranging from 78.4 to 94.4%, and one was only 51.5% complete. Estimated contamination (without strain heterogeneity) ranged from 0 to 3.3%.
Table 1. General information on the ten MAGs obtained in this study.
| Genome | Origin | Scaffold (nbr) | Size (Mb) | Genes (nbr) | GC (%) | Compl. (%) | Cont. (%) | Strain hetero.* | Cont. exd. strain hetero. (%) |
|---|---|---|---|---|---|---|---|---|---|
| NM1a | Enrichment culture (50°C) from petroleum sample, Brazil | 12 | 1.26 | 1388 | 35.7 | 92.5 | 1.3 | 0 | 1.3 |
| NM1b | Santa Barbara Channel oil seeps, USA | 183 | 1.66 | 1860 | 43.8 | 90.2 | 3.6 | 66.7 | 1.2 |
| NM2 | Santa Barbara sediments, USA | 210 | 1.03 | 1254 | 41.8 | 51.5 | 2 | 0 | 2 |
| NM3 | Enrichment culture (40°C) from petroleum sample, Brazil | 26 | 1.49 | 1578 | 55.2 | 85.5 | 2.9 | 0 | 2.9 |
| NM4 | Yellowstone sulfidic hot spring, USA | 122 | 1.42 | 1603 | 43.4 | 85.5 | 1.8 | 66.7 | 0.6 |
| Verst-YHS | Yellowstone sulfidic Hot Spring, USA | 46 | 1.05 | 1220 | 28.4 | 94.4 | 0 | 0 | 0 |
| Mnatro-ASL | Altai Soda Lake sediments, Russia | 73 | 1.34 | 1451 | 42.2 | 93.3 | 1.3 | 0 | 1.3 |
| ANME-1-THS | Tibetan Hot Spring sediment, China | 181 | 2.03 | 2155 | 48.7 | 78.4 | 4.9 | 33 | 3.3 |
| GoM-Arc1-GOS | Gulf of Mexico natural Oil Seep, USA | 119 | 1.46 | 1623 | 41.0 | 91.2 | 0 | 0 | 0 |
| ANME-2c | Gulf of Mexico natural oil seep, USA | 249 | 2.66 | 2867 | 48.5 | 92 | 2 | 0 | 2 |
Genome ID, origin, number of scaffolds, number of protein-coding genes, guanine-cytosine (GC) content, estimated completeness (Compl.), estimated contamination (Cont.), strain heterogeneity (Strain) and contamination excluding strain heterogeneity (Cont. excl. strain hetero.) are shown.
percentage of contamination that can be due to binning of contigs from closely related strains.
Four MAGs represent three previously undescribed lineages only distantly related to known methanogenic/methanotrophic archaea (Fig. 1): NM1 (NM1a and NM1b MAGs) branches within the Methanotecta superclass15, between Archaeoglobales and the clade formed by ‘Ca. Syntrophoarchaeales’ and ‘Ca. Methanophagales’15 (ANME-1); NM3 branches within the Acherontia superclass15, at the base of the clade formed by the non-methanogenic Theionarchaea26 and ‘Ca. Methanofastidiosa’ (former WSA2/Arc1); NM4 branches within the TACK superphylum and is related to Korarchaeum cryptofilum. In the NM4 MAG, the markers for methane metabolism are present on two contigs with a lower coverage than the other contigs and contain few genes related to Korarchaeum. However, an independent study supports that these contigs belong to the same organism, provisionally named “Candidatus Methanodesulfokores washburnensis” for its methane- and sulfur-cycling capacities (McKay et al., submitted). One additional MAG (NM2) was too partial to assess its phylogenetic placement.
Figure 1.
Placement of nine MAGs described in this study in the reference phylogeny of Archaea. NM2 was not included due to low completeness. Bayesian phylogeny (PhyloBayes, CAT+GTR+ Γ4) based on concatenation of 40 conserved phylogenetic markers (8,564 amino acid positions) and 156 genomes/MAGs (see Supplementary Table 5 and Supplementary Table 6 for detail). Node supports refer to posterior probabilities, and for reasons of readability only values above 0.8 are shown. The tree is rooted according to Raymann et al.52. The scale bar represents the average number of substitutions per site. Black arrows point to the 9 obtained MAGs and accolated pie charts indicate their estimated completeness. Colors indicate that genomes of these lineages encode an MCR/MCR-like complex, Class I/II methanogens are in green, methyl-dependent hydrogenotrophic lineages are in red, methanotrophs are in orange (some being within Class II), potential or validated shot-chain alkane users are in in blue. NM1 could also have a methane metabolism (see text for discussion).
Finally, five MAGs correspond to currently under-sampled archaeal lineages: a deep-branching Verstraetearchaeota, a third Methanonatronarchaeia, a second ‘Ca. Methanophagales’ (ANME-1), a second GoM-Arc1 (showing a close relationship with the methanotrophic Methanoperedens), and the first representative of ANME-2c (Fig. 1).
Phylogeny and functional inference of MCR/MCR-like complexes
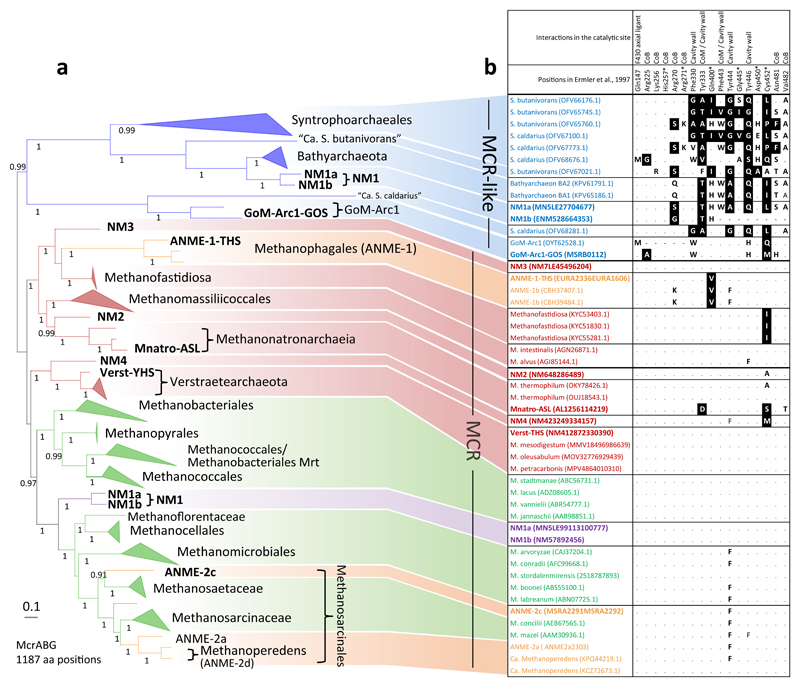
To investigate the evolutionary relationships and characteristics of the MCR complexes identified in this study, we built a phylogeny based on a concatenated alignment of McrABG subunits. This phylogeny is in overall agreement with recently published ones16,18. Three MAGs (NM1a, NM1b and GoM-Arc1-GOS) encode alternative McrABG-like complexes that cluster with those of ‘Ca. Syntrophoarchaeum’ and Bathyarchaeota (Fig. 2A, in blue). The presence of an MCR-like complex and the absence of a canonical MCR in GoM-Arc1-GOS are consistent with the recent description of a MAG of this lineage23, and represent so far a unique feature within the Methanosarcinales.
Figure 2.
Phylogeny of the MCR/MCR-like complex and conservation of important positions in the catalytic site. A) Unrooted Bayesian phylogeny (CAT+GTR+Γ4) based on a concatenation of McrABG/McrABG-like subunits (1,187 amino acid positions) from 109 genomes/MAGs (see Supplementary Table 6 for details). Node supports refer to posterior probabilities, and for reasons of readability only values above 0.8 are shown. The scale bar represents the average number of substitutions per site. The color code is similar to that in Fig. 1 with the exception of NM1 which have both an MCR-like (in blue) and a canonical MCR (in purple) (see text for discussion). B) Conservation of 17 residues previously described to interact with CoM, CoB, F430 cofactors, making part of the substrate cavity wall, or having post-translational modifications27,47,75. Replacement of conserved amino acids associated to a negative value in the Blosum45 matrix are indicated by white on black background, those with a null or positive value in the Blosum45 matrix are in bold, “.” indicate conserved positions and “-“ indicate missing positions in the sequence due to sequencing incompleteness.
The remaining MAGs harbour canonical MCR complexes (Fig. 2A), branching next to their closest MCR-bearing neighbors in the reference archaeal phylogeny (Fig. 1), suggesting no recent horizontal gene transfers (HGT). The clustering of NM3 with Methanofastidiosa supports an early presence of methanogenesis in the Acherontia. The clustering of NM4 with Verstraetearchaeota, support that it is a genuine methanogenic/methanotrophic representative of the TACK. The separate branching of ANME-2c from the other ANME-2 lineages suggests that anaerobic methane oxidation in Methanosarcinales emerged multiple times independently from methanogenic ancestors. Interestingly, both NM1a and NM1b encode, in addition to the McrABG-like complex, a canonical MCR complex branching at the base of Class II methanogens, consistently with the reference phylogeny. The coexistence of MCR and MCR-like complexes in the same archaeon has never been observed before and brings into question the metabolism of this lineage (see below).
It is striking to observe that most of the predicted or experimentally proven methyl-dependent hydrogenotrophic methanogens are closely related in the MCR tree (Fig. 2A, in red), irrespective of their placement in the reference phylogeny (Fig. 1). This might be the consequence of ancient exchanges of the MCR complex among these lineages, whose direction is hard to define. Nevertheless, some more recent transfers may be identified. For example, ’Ca. Methanophagales’ (ANME-1) MCRs branch far from their Methanotecta relatives, and might have acquired their MCR complex from a methanogenic member of the Acherontia.
The clustering of MCR-like homologues belonging to distantly related lineages (Fig. 2A, in blue) is also puzzling. This might be due to HGT and/or tree reconstruction artefacts linked to their high sequence divergence with respect to canonical MCRs, exemplified by their longer-than-average branches. Such divergence is probably related to a change in function, as MCR-like complexes are involved in activating short-chain alkanes (butane and propane) in ‘Ca. Syntrophoarchaeum’21. Accordingly, several residues playing an important role in canonical MCR, either by interacting with cofactors, forming the catalytic site cavity wall or being post-translationally modified, are not conserved in ‘Ca. Syntrophoarchaeum’ sequences (Fig 2B). The replacement of large aromatic residues (e.g. Phe330, Tyr333, Tyr444, Tyr446) present in the cavity wall of canonical MCR27 by smaller ones in ‘Ca. Syntrophoarchaeum’ MCR-like complexes could have occurred to accommodate butane/propane (larger substrate than methane) in the catalytic site (Fig. 2B). The presence of smaller amino acids at these positions in NM1 and Bathyarchaeota MCR-like complex suggest a similar function in short-chain alkane oxidation. Finally, the MCR-like sequences of GoM-Arc1 show fewer modifications at these sites, suggesting the utilization of a smaller alkane, possibly ethane or methane.
Expanded diversity of methyl-dependent hydrogenotrophic methanogens
The NM3 and NM4 MAGs share several similarities with the recently discovered order-level lineages of methanogens that were proposed or experimentally proven to perform methyl-dependent hydrogenotrophic methanogenesis10,17–19 (Fig. 3, Supplementary Table 1). First, these relatively complete MAGs (85,5% completeness) lack at least 24 genes coding for the MTR complex, H4MPT biosynthesis, and the H4MPT methyl-branch of the WL pathway, otherwise present in all Class I/II methanogens (Supplementary Table 1). Second, they encode [Ni-Fe] hydrogenases and methyltransferases with the potential to support methanogenesis from methanol (MtaABC) in NM3 and NM4 and methanethiol (MtsAB) in NM3 (Fig. 3; Supplementary Table 1). Interestingly, energy conservation complexes of NM3 are mostly similar to Methanofastidiosales17 (Supplementary Fig. 1), their closest related methanogens in the reference phylogeny (Fig. 1). Altogether, this suggests that NM3 and NM4 rely on methyl-dependent hydrogenotrophic methanogenesis (Fig. 3; Supplementary Discussion for details on energy conservation in NM3 and NM4).
Figure 3.
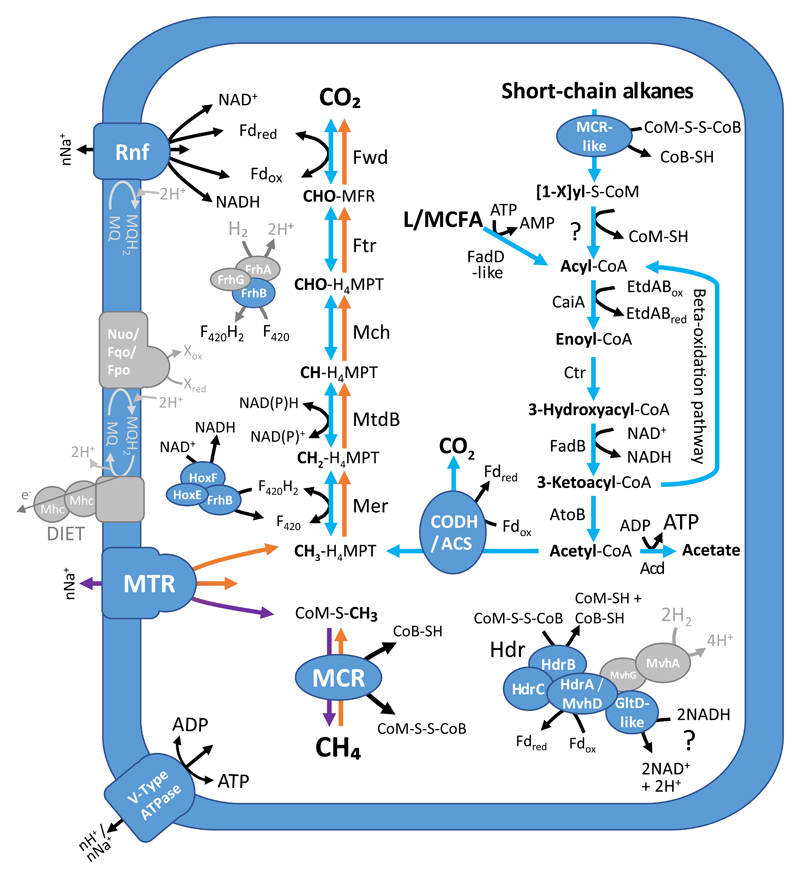
Predicted methane and short-chain alkane metabolism of the MAGs described in this study, with the exception of NM1, which is presented in Fig. 4, and NM2, which has a low completeness. Colored arrows correspond to reactions modifying or transferring the C1 carbon group of the substrate. Details on the annotation of the enzymes are presented in Supplementary Table 1. MFR, methanofuran; H4MPT, tetrahydromethanopterin; Fd, ferredoxin; F420, coenzyme F420; LCFA, Long Chain Fatty Acids; MQ, menaquinone; Mp, methanophenazine; Mhc, c-type multiheme cytochromes; DIET, Direct Interspecies Electron Transfer. Grey color indicates the absence of the enzyme, complex, reaction or compound. Comparisons of with other methane-cycling or short-chain alkane oxidizers, which are discussed in the text, are presented in Supplementary Figs 1 and 3. The percentages between brackets indicate the estimated completeness of the corresponding MAGs.
The predicted methanogenesis pathway in Verst-YHS (Verstraetearchaeota) and Mnatro-ASL (Methanonatronarchaeia) MAGs also supports methyl-dependent hydrogenotrophic methanogenesis (Fig. 3, Supplementary Table 1), as described in the first genomic assemblies for these lineages18,19. However, comparison of the energy conservation enzymes in the seven currently available Verstraetearchaeota (order Methanomethyliales) suggests an alternative model than previously described19 (Fig. 3, Supplementary Table 1). Indeed, we found that all Methanomethyliales MAGs (95% average completeness) lack the HdrA/MvhD and possibly MvhAG subunits of the electron-bifurcating complex HdrABC/MvhADG, suggesting that this complex is absent in these archaea. In contrast, we identified in these genomes a gene cluster encoding a potential complex composed of a membrane-bound hydrogenase and of HdrBC (tentatively named Energy-converting Hydrogenase D or Ehd; Supplementary Fig. 2). We propose that this complex could be involved in a previously unreported mode of energy conservation associated with methanogenesis (Fig. 3; Supplementary Discussion).
Insights into methane and short-chain alkane oxidizers
GoM-Arc1-GOS, ANME-1-THS and ANME-2c MAGs possess a WL pathway and lack the methyltransferases and [Ni-Fe] hydrogenases required for methylotrophic and hydrogenotrophic methanogenesis, respectively (Fig. 3; Supplementary Table 1), similar to all available MAGs of methanotrophs and short-chain alkane oxidizer (Supplementary Fig. 3). Although they encode an AMP-producing acetyl-CoA synthetase (Acs) which is used for aceticlastic methanogenesis in Methanosaeta spp., they could rather use it for acetate assimilation11. Comparison with methanotrophs and short-chain alkane oxidizers also reveals a common core of enzymes for energy conservation, comprising the F420H2:quinone (or phenazine) oxidoreductase (Fqo/Fpo) and a potential electron confurcating complex (HdrABC/MvhD/FdhB28) coded by a conserved gene cluster (Supplementary Fig. 4). ANME-2c and GoM-Arc1-GOS encode 17 and 10 multiheme c-type cytochromes respectively, supporting the importance of direct electron transfer to syntrophic partners in anaerobic methane29,30 and short-chain alkane oxidation21 metabolisms (Supplementary Fig. 3; Supplementary Table 1).
ANME-1-THS MAG is the first sequenced representative of a “Land clade” within the ‘Ca. Methanophagales’ (Supplementary Fig. 5), suggesting different adaptations to environmental conditions than members of the ANME-1b clade, which are mainly from marine methane seeps. ANME-1-THS differs from the ANME-1b MAG31 by the presence of a bacterial-like Rnf complex that could couple the NAD:ferredoxin oxidoreduction with chemiosmotic gradient generation/utilisation (Fig. 3; Supplementary Fig. 6; Supplementary Discussion). If these genes are not in the missing region of this MAG, ANME-1-THS might also differ from the other ANMEs by the lack of multiheme c-type cytochromes to transfer electrons from methane oxidation to a syntrophic partner (Fig. 3; Supplementary Fig. 3). Alternatively, two PsrABC-like complexes, including a molybdenum/selenocysteine-containing dehydrogenase subunit, could be involved in the reduction of inorganic compounds such as polysulfide/elemental sulfur32,33 (Fig. 3). This contrasts with ANME-1b MAG which misses the membrane integral (PsrC-like) subunit needed to transfer electrons from membrane-associated electron transporters (Supplementary Fig. 3). These characteristics might indicate growth of ANME-1-THS without bacterial syntrophs.
The gene content of GoM-Arc1-GOS is consistent with the recent description of the first member of the GoM-Arc1 lineage23. While GoM-Arc1 members encode an MCR-like complex possibly involved in short-chain alkane oxidation (Fig. 3), they lack the beta-oxidation pathway proposed to be involved in butane/propane utilization in ‘Ca. Syntrophoarchaeales’21 (Supplementary Fig. 3). If GoM-Arc1 members are capable of oxidizing ethane (CH3CH3), as suggested by the fewer modifications observed in the catalytic site of its MCR-like complex relative to canonical MCRs (Fig. 2), the oxidation of the ethyl-group would lead to an acetyl-group that could directly enter the oxidative WL pathway, making the beta-oxidation pathway unnecessary (Fig. 3; Supplementary Discussion). With the presence of Fqo, HdrABC/MvhD/FdhB, multiheme c-type cytochromes, and HdrDE (Supplementary Table 1), the energy conservation system associated with this potential ethane-oxidation metabolism in GoM-Arc1 would mostly resemble that associated with methanotrophy in their closely related ANME-2 lineages (Supplementary Fig. 3). The question remains whether the MCR-like homologs of GoM-Arc1 could also be capable of methane oxidation.
A previously uncharacterised type of methanogenesis?
The two NM1 MAGs represent the first archaea predicted to encode both an MCR and an MCR-like complex (Fig. 2), suggesting that they might be potentially capable of both methane and short-chain alkane metabolisms (Fig. 4; Supplementary Table 1). Interestingly, while both NM1 MAGs encode the MTR and the m-WL pathway similarly to Class I/II methanogens, they lack the [Ni-Fe] hydrogenases (MvhA and FrhA) and methyltransferases needed for hydrogenotrophic and methylotrophic methanogenesis, respectively. They also diverge from Class I/II methanogens by the replacement of the F420 dependent methylene-tetrahydromethanopterin dehydrogenase (Mtd) by MtdB, which relies on NAD(P) redox cofactor in Methylobacterium extorquens34.
Figure 4.
Predicted methane, short-chain alkane and long/medium-chain fatty acid metabolism of the two MAGs NM1a (“Ca. Methanoliparum thermophilum” completeness of 92.5%) and NM1b (“Ca. Methanolliviera hydrocarbonicum” completeness of 90.2%) belonging to the candidate class “Ca. Methanoliparia”. Colored arrows correspond to reactions present in both MAGs which modify or transfer the carbon group(s) of the substrate. Predicted possible metabolisms are the utilisation of short-chain alkanes and long/medium-chain fatty acids (L/MCFA) (blue), methanotrophy (orange) and methanogenesis from short-chain alkanes or L/MCFA (purple). Details on the annotation of the enzymes are provided in Supplementary Table 1. MFR, methanofuran; H4MPT, tetrahydromethanopterin; Fd, ferredoxin; F420, coenzyme F420; L/MCFA, Long/medium chain fatty acids; MQ, menaquinone; Mhc, c-type multiheme cytochromes; DIET, Direct Interspecies Electron Transfer. Grey color indicates the absence of the corresponding enzyme, complex, reaction or compound in both MAGs.
Beyond the presence of an MCR-like complex, the potential ability of NM1 for short-chain alkane oxidation is also suggested by the presence of a complete beta-oxidation pathway with several gene copies per step, and a complete WL pathway (including CODH/ACS) as in ‘Ca. Syntrophoarchaeales’21. In addition, NM1 encode multiple long-chain fatty acid acyl-CoA synthases (FadD-like), not present in ‘Ca. Syntrophoarchaeales’. Long chain fatty acids (LCFA) activated with these enzymes can enter the beta-oxidation pathway. NM1a and NM1b also encode multiple AMP-forming acetyl-CoA synthetase (Acd) to generate ATP from LCFA degradation. These enzymatic redundancies suggest a versatility toward substrates, as previously proposed for Syntrophus aciditrophicus35 and Archaeoglobus fulgidus36. Consistently, analysis of the environmental distribution of NM1 (Supplementary Fig. 7) reveals their common association with anoxic hydrocarbon-rich environments including methane seeps and oil-rich environments, where short-chain alkanes and long-chain carboxylic acids can be present in substantial concentrations37,38. In particular, NM1a and NM1b originate from an enrichment culture based on petroleum fluids and from a natural oil seep.
In addition to this potential wide substrate range, NM1 also contrast with ‘Ca. Syntrophoarchaeales’ in terms of energy conservation by lacking homologues of the NADH/F420H2:quinone oxidoreductase (Nuo/Fqo) and multiheme c-type cytochromes (Fig. 4; Supplementary Table 1). Also, NM1 contain an Rnf complex potentially using NAD instead of menaquinone for ferredoxin oxidoreduction, similarly to ANME-1-THS (Supplementary Fig. 6; Supplementary Discussion). In the absence of membrane-bound enzymes involved in oxidoreduction of lipid-soluble electron carriers, of multiheme c-type cytochromes for direct interspecies electron transfer, of confurcating [Fe]-hydrogenase for interspecies H2 transfer39, and of enzymes involved in dissimilatory reduction of inorganic compounds, the nature of the terminal electron acceptor coupled to alkane/LCFA oxidation remains elusive. Although both MAGs are mostly complete (~90%), it cannot be excluded that some of these enzymes are coded in their missing regions, or that an alternative way to transfer electrons to a terminal acceptor exists (e.g. utilisation of the assimilatory-type sulfite reductase present in both MAGs for dissimilatory reduction of sulfite, direct electron transfer not relying on cytochromes, or utilization of cytochromes produced by a syntrophic partner). Alternatively, we speculate that in NM1, methanogenesis involving the canonical MCR complex could act as a sink for the electrons produced during alkane and LCFA oxidation. Several electron-bifurcating/confurcating complexes encoded in the two NM1 MAGs (Supplementary Fig. 8) together with the Rnf complex could be involved in this metabolism. The conversion of alkane and LCFA into CH4 and acetate is thermodynamically feasible but was only reported to occur through syntrophic partnerships between a bacterium (performing the beta-oxidation) and a H2-consuming methanogen40,41, and it thus remains to be proven experimentally whether this can occur in a single organism.
Based on the presence of methane and short-chain alkane/fatty acid-related enzymes and the preferential association with hydrocarbon-rich environments, we propose the provisional class ‘Candidatus Methanoliparia’, with ‘Candidatus Methanoliparum thermophilum’ for NM1a and ‘Candidatus Methanolliviera hydrocarbonicum’ for NM1b (see Supplementary Discussion for full taxonomy and nomenclature).
A core of markers related to methane and short chain-alkane metabolisms
A group of 38 genes present in most methanogens and absent from most other organisms, generally referred to as “methanogenesis core markers”, was previously defined from Class I/II methanogen genomes42,43 (Supplementary Table 2). Half of them have an unknown function. The others correspond to MCR and MTR subunits, enzymes for biosynthesis and activation of the F430 prosthetic group of MCR, and post-translational modifications in the McrA catalytic site44,45. We reassessed the occurrence of these markers in the ten assembled MAGs as well as reference genomes covering all recently discovered lineages of methanogens, methanotrophs and short-chain alkane oxidizers (Table 2).
Table 2. Occurrence in methanogens, methanotrophs and short-chain alkane users of 38 genes previously suggested as methanogenesis markers.
| MCR | “6 marker cluster” | F430 bioS and activation | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nbr of genomes / lineage | Avg genome completness (%) | m1_mcrA | m2_mcrB | m3_mcrG | m4_Predicted rotamase | m5_DUF2102 | m6_DUF2112 | m7_YjiL-like | m8_DUF2113 | m9_arCOG03226 | m10_atwA | m11_mcrC | m12_mcrD | m13_cfbD | m14_cfbE | ||
| Class I / II | Methanobacteriales | 24 | 99 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Methanopyrales | 2 | 100 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Methanococcales | 15 | 100 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Methanosarcinales* | 39 | 99 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0,97 | |
| Methanomicrobiales | 16 | 99 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0,94 | 1 | |
| Methanocellales | 3 | 100 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Methanoflorentaceae | 1 | 97 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| SCAO & AOM | Msar_ANME-2a | 1 | 99 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Msar_ANME-2c | 1 | 92 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Msar_ANME-2d | 2 | 98 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Methanophagales (incl. ANME-1-THS) | 2 | 79 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | |
| NM1§‡ | 2 | 91 | 1 | 1 | 1 | 1 | 1 | 0,5 | 0,5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Msar_GoM-Arc1 (incl. GoM-Arc1-GOS)§ | 2 | 78 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | |
| Syntrophoarchaeales | 2 | 93 | 1 | 1 | 1 | 1 | 0,5 | 1 | 1 | 1 | 1 | 1 | 0,5 | 1 | 1 | 1 | |
| CH3-dep hydro | Methanomethyliales (incl. Verst-YHS)§ | 7 | 95 | 1 | 1 | 1 | 1 | 0,86 | 1 | 0,86 | 1 | 1 | 1 | 1 | 1 | 0,86 | 0,71 |
| Methanomassiliicoccales | 11 | 99 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0,91 | 0,91 | |
| Methanofastidiosa§ | 8 | 86 | 1 | 1 | 1 | 0,75 | 1 | 1 | 1 | 0,88 | 1 | 1 | 1 | 1 | 1 | 1 | |
| NM3§ | 1 | 86 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | |
| NM4§ | 1 | 86 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Methanonatronarchaeia (incl. Mnat-ASL) | 3 | 97 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| NM2§ | 1 | 52 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | |
| Bathyarchaeota BA1/BA2 | 2 | 92 | 1 | 1 | 1 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 1 | 0,5 | 0,5 | 0 | 0 | |
| Essentiality index in M. maripaludis | 0 | 0 | 0 | 0 | 3 | 2,5 | 0,5 | 0,5 | 3,5 | 0 | 0 | 0 | 0 | 0 | |||
| MTR | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| m15_mtxX | m16_AIR synthase-related | m17_Zn-ribbon protein | m18_DUF2099 | m19_DUF2117 | m20_Gly-thioamidation (McrA PTM) | m21_DUF2124 | m22_DUF2098 | m23_Soluble P-type ATPase | m24_DUF2111 | m25_arCOG04853 | m26_DUF2114 | m27_mtrA | m28_mtrB | m29_mtrC | m30_mtrD | m31_mtrE | m32_arCOG04885 | m33_Arg-methyltransf. (McrA PTM) | m34_DUF1894 | m35_DUF1890 | m36_DUF2115 | m37_DUF2119 | m38_DUF2121 |
| 1 | 1 | 1 | 1 | 0,95 | 0,95 | 0,68 | 1 | 1 | 0,59 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 1 | 1 | 1 | 0,87 | 1 | 1 | 0,73 | 1 | 0,8 | 0,93 | 1 | 1 | 0,93 | 1 | 1 | 1 | 1 | 1 | 1 | 0,87 | 0,87 | 1 | 0,47 | 1 |
| 0,97 | 1 | 1 | 1 | 0,92 | 1 | 0,87 | 1 | 1 | 0,97 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0,97 | 1 | 1 | 0,67 | 0,21 | 0,9 | 0,67 |
| 1 | 1 | 1 | 1 | 0,44 | 1 | 0,81 | 1 | 0,94 | 0,81 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0,31 |
| 1 | 1 | 1 | 1 | 0,67 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0,5 |
| 0 | 0,5 | 0 | 1 | 1 | 1 | 1 | 0 | 0,5 | 0,5 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0,5 | 0 | 0,5 | 0,5 | 0 | 1 | 0 |
| 0 | 1 | 0 | 0,5 | 1 | 0,5 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 |
| 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 0,5 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0,5 | 1 | 1 | 1 | 0 | 0 |
| 1 | 1 | 1 | 1 | 1 | 0 | 0,5 | 0,5 | 0 | 0 | 0,5 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0,50 | 0 |
| 0,71 | 1 | 0,71 | 1 | 0,86 | 0,86 | 1 | 1 | 1 | 0,14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 1 | 0,91 | 0,27 | 1 | 0,09 | 1 | 0 | 1 | 0 | 0 | 0,09 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 0,63 | 0,63 | 0,88 | 1 | 0,88 | 0 | 1 | 0 | 0,5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 1 | 1 | 1 | 0,67 | 0,33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0 | 0,5 | 0 | 1 | 0,5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 0 | 9,5 | 0,5 | 3 | 0,5 | 1 | 5,5 | 16 | 10,5 | 11,5 | 23 | 6 | 0 | 0 | 0,5 | 0 | 0 | 0 | 13 | 5 | 6 | 3 | 3 | 2 |
The numbers in the cells colored in red/green indicate the proportion of genomes of one lineage that have a given marker (all = 1, none = 0). It is possible that some of the markers are present in the missing regions of the incomplete MAGs/genomes. For gene essentiality in Methanococcus maripaludis S2: predicted essential genes (Essentiality index ≤ 2) are highlighted in yellow and predicted none essential gene (Essentiality index ≥ 5) are highlighted in grey60. “m1”, “m2”, etc means “marker 1”, “marker 2” etc; Msar, Methanosarcinales; AOM, Anaerobic Oxidation of Methane; SCAO, short-chain alkane oxidation; CH3-dep hydro, methyl-dependent hydrogenotrophic methanogenesis; MCR, methyl-coenzyme M reductase complex; MTR, N5-Methyltetrahydromethanopterin: coenzyme M methyltransferase complex; PTM, involved in post-translational modification of McrA residues; F430 prosthetic group of MCR.
Excluding ANME-2 and GoM-Arc1 genomes.
NM1 MAGs encode for a SCAO metabolism and possibly for methanogenesis or methanotrophy.
Metabolic classification relying only on genomic predictions. The numbering and ordering of the marker was chosen to reflect their occurrence in methanogens, methanotrophs and short-chain alkane oxidizers (i.e. from the most occurrent to the less occurrent) with few modifications in the order to gather markers involved in same functions or being part of same enzymatic complexes.
Our analysis shows that some markers are no longer universal in Class I/II methanogens (e.g. m37, 38). Also, many marker genes shared by all or most Class I/II methanogens were predicted to be nonessential in Methanococcus maripaludis S246 (Table 2). These non-universal and nonessential genes could possibly be involved in fine-tuning of methanogenesis (e.g. post-translational modification of MCR47) or in its regulation under specific environmental conditions that are not encountered by all methanogens. For example, m21 and m24 are missing in several methanogens from nutrient-rich environments, such as Methanobrevibacter, Methanosphaera and Methanocorpusculum, and could be involved in regulatory processes related to changes in substrate/nutrient availability.
All the lineages of predicted and experimentally proven methyl-dependent hydrogenotrophic methanogens17–19 lack numerous markers (Table 2), similarly to what was previously noted in Methanomassiliicoccales48. These markers correspond to MTR complex subunits (m27-31), an McrA post-translational modification enzyme (m33)45 and several uncharacterized markers that are mostly nonessential in M. maripaludis46 (Table 2). The existence of the same pattern in NM3 and NM4 supports our inference of a potential methyl-dependent hydrogenotrophic methanogenesis. Finally, Bathyarchaeota BA1 and BA222 which were described as methyl-dependent hydrogenotrophic methanogens22 but possess an MCR-like complex instead of the canonical MCR (Fig. 2), lack almost all methanogenesis markers (Table 2), questioning their actual metabolism.
Several homologues of the methanogenesis markers are also known to be present in non-methanogenic archaea. This is the case of the MCR/MCR-like (m1-3) and MTR (m27-31) complexes in archaeal methanotrophs7 and GoM-Arc123, as well as the MCR-like complex in ‘Ca. Syntrophoarchaeales’21. Based on our analysis, archaeal methanotrophs and short-chain alkane oxidizers also appear to possess numerous markers previously exclusively associated with methanogenesis (Table 2), supporting the common origin and functional links of these metabolisms.
In addition to the MCR/MCR-like complex subunits, the most specific and conserved markers in all lineages of methanogens, methanotrophs and short-chain alkane oxidizers appear to be the genes involved in the biosynthesis (nflD/cfbD, murD/cfbE and possibly mcrD49) and activation (atwA and possibly mcrC50) of the F430 prosthetic group of MCR, along with six genes encoding uncharacterized proteins (m4 to m9) (Table 2). These six genes are co-localized in most genomes (Supplementary Fig. 9) and are among those that were predicted to be co-transcribed in Methanolobus psychrophilus R1551, suggesting they operate in a common process. These six marker enzymes do not co-purify with MCR50. However, their phylogeny (Supplementary Fig. 10) and their restriction to archaea having MCR or MCR-like complexes strongly suggest they are involved in essential aspects of the regulation, folding and/or function of the respective holoenzymes (Supplementary Discussion).
Finally, several markers are present in archaeal lineages without MCR/MCR-like complexes (Supplementary Table 3) and are possibly remnants of an ancestral methane-metabolism (Supplementary Fig. 11-13; Supplementary Discussion).
Taken together, these observations indicate that none of the previously defined methanogenesis markers are unique to methanogens but are rather more generally indicative of metabolisms involving MCR or MRC-like complexes, including methanogenesis, methanotrophy, and short-chain alkane oxidation. Elucidating the roles of these markers (MCR-Associated Markers or MAM) will be essential not only for understanding methanogenesis, but also anaerobic methanotrophy and short-chain alkane oxidation in archaea.
Evolution of methane and short-chain alkane metabolisms
Our results significantly extend recent data by highlighting the overwhelming presence of lineages with an MCR or MCR-like complex in the Archaea (Fig. 1). This supports an early origin of methanogenesis in this domain of life, and multiple losses of this metabolism during archaeal diversification4,14,18,52.
The sharing of a common set of genes (Table 2) clearly indicates that methanogens, anaerobic methanotrophs and short-chain alkane oxidizers are evolutionarily linked. However, it remains unclear which type of metabolism is the most ancient, and what evolutionary and functional transitions led to such diversity14. The antiquity of the WL pathway53,54, and the recent proposal that the root of the archaeal tree might lie in between Class I and II methanogens52, would suggest that CO2-dependent hydrogenotrophic methanogenesis is the ancestral type of methanogenesis. Nevertheless, the growing diversity of methyl-dependent hydrogenotrophic methanogens, including this work (Fig. 1 in red), indicates that this metabolism has been largely overlooked. Its origin and evolutionary relationship with CO2-dependent hydrogenotrophic methanogenesis remain unclear. The fact that it is a simpler metabolism, requiring fewer genes than CO2-dependent hydrogenotrophic methanogenesis might suggest its earliest origin. However, it may also signify that it could have emerged later through loss of the WL pathway and/or HGT, as suggested by the grouping of most archaea sharing this metabolism in the phylogenies of MCR (Fig. 2A) and of m4 to m9 markers (Supplementary Fig. 10). Also, the clustering of NM4 with Verstraetearchaeota on a separate and well-supported clade in the MCR tree (Fig. 2A) is compatible with a possible inheritance of this metabolism from the last archaeal common ancestor, even under the classical root in between Euryarchaeota and the TACK. However, the possibility of an acquisition through ancient HGT cannot be excluded at present. More insights into the ancestral type of methanogenesis might also be gained from re-examination of the root of the archaeal tree52 including all recently discovered archaeal lineages.
The phylogenetic placement of the ANME lineages (Fig. 1), strongly suggests that the capabilities for anaerobic methanotrophy emerged multiple times independently during archaeal diversification. In the Methanosarcinales this could have occurred relatively recently and repeatedly by reversal of methanogenesis, possibly through switch of function of a resident canonical MCR, leading to the different ANME-2 (Fig. 2A) and possibly ANME-3 lineages. The pool of genes associated with energy conservation in methanogenic and methanotrophic Methanosarcinales is in fact relatively similar55 (Fig. 3 and Supplementary Fig. 3) and some methanogenic Methanosarcinales encode c-type multiheme cytochromes11 providing the necessary background for electron transfer in AOM archaea.
The identification and experimental demonstration of the capacity for oxidation of short-chain alkanes (butane, propane) by a divergent MCR-like complex in the Synthrophoarchaeales21 is among the most interesting findings of the recent years in the field of environmental microbiology. Our results extend the distribution of these MCR-like complexes in the archaea (Fig. 2A), and therefore also of potential short-chain alkane oxidation capabilities (Figs. 3 and 4). The rapid evolutionary rates of MCR-like homologues coupled to the change of key residues (Fig. 2B) suggest that these complexes might have arisen from canonical MCRs through modifications in the catalytic site to accommodate larger hydrocarbons than methane. Transitions between anaerobic methanotrophy and short-chain alkane utilisation could have occurred in both directions as suggested by i) the close phylogenetic relationships between ‘Ca. Methanophagales’ and ‘Ca. Syntrophoarchaeales’ and the position of GoM-Arc1 within a clade comprising ANME-2a/ANME-2d (Fig. 1), ii) the proposed mechanism of alkane activation in their MCR/MCR-like complexes21, iii) their very similar modes of energy conservation (Supplementary Fig. 3), and iv) their numerous shared markers (Table 2). If GoM-Arc1 is a short-chain alkane oxidizer, as suggested by its MCR-like complex, this capacity could have emerged from methanotrophy. Conversely, ‘Ca. Methanophagales’ (ANME-1) might have shifted from short-chain alkane oxidation to methanotrophy after acquisition of their MCR through HGT (Fig. 2). Finally, the first report of co-existence of an MCR and an MCR-like complex in members of the “Ca. Methanoliparia” class opens up the possibility of an additional type of methanogenesis associated with alkane and/or LCFA oxidation.
Further exploration of archaeal lineages with an MCR/MCR-like complex and their experimental characterization will lead to a more complete understanding of methane metabolisms and their derivations, as well as their environmental impact.
Methods
Metagenomic database probing and contig binning
Contigs of 6108 metagenomes publicly available on the IMG/JGI database in April 2017 were screened for the presence of COG4058, corresponding to McrA, using search tools of the database. 819 contigs containing an McrA sequence with a minimal length of 750 bp were downloaded. The McrA sequences present on these contigs were aligned on those of 188 published genomes by using Mafft56 (mafft-linsi) and were trimmed with BMGE57 (BLOSUM30). A maximum likelihood (ML) phylogeny was calculated in IQTree58 with the TEST option for best model selection and 100 bootstrap replicates. Metagenomes containing one or several contigs coding for an McrA homologue that was only distantly related to know lineages or belonged to undersampled lineages were downloaded from the IMG database. These metagenomes were assembled with MetaSPAdes59, IDBA-UD60 and Newbler (Roche) (see Supplementary Table 4 for details). The contigs were binned with ESOM, MetaBAT61, ABAWACA 1.07 (http://ggkbase.berkeley.edu/), MaxBin 2.062 and CONCOT63 (Supplementary Table 4). An in-house pipeline (Let-it-bin, https://github.com/QuentinLetourneur/Let-it-bin) was used for read trimming, assembly, and contig binning. Two of the MAGs were refined using DAS_Tool64. Completeness and contamination of the assembled MAGs were estimated with CheckM65.
Phylogenomic analyses
A reference archaeal phylogeny was built from a concatenation of 40 phylogenetic markers corresponding to the 36 proteins of the Phylosift dataset66, plus the alpha and beta subunits of the RNA polymerase and two universal ribosomal proteins (L30, S4) (Supplementary Table 5). We used a subset of the genomes available for each order/class/phylum level lineages (Supplementary Table 6) to minimize biased associated with uneven distribution of taxa among them (e.g. >100 taxa in Halobacteriales vs. 3 taxa in Methanocellales). The 147 genomes were chosen because they were the most complete and the most distant to each other within each lineage. Two phylogenies were built from a concatenation of McrABG and of six co-localized markers (m4 to m9) specific to genomes encoding an MCR/MCR-like complex. Sequences used for these trees were searched by HMM in the ten MAGs obtained in this study and in genomes present in the NCBI or IMG-databases, aligned with Mafft56 (mafft-linsi), trimmed with BMGE57 (BLOSUM30) and concatenated with an in-house script. Before concatenation, the genes of the two datasets (McrABG and m4 to m9) were tested for congruence using the Internode Certainty (IC)67 test in RaxML68. Maximum likelihood phylogenies for each gene and blind concatenations were calculated in IQTree58 with the TEST option for best model selection and 100 bootstrap replicates. Sequences causing strong incongruences (with a bootstrap>=80%) at high taxonomic ranks (order to phylum as applicable) were removed, and the procedure was repeated until no further incongruence was found. Bayesian phylogenies were constructed in PhyloBayes69 under the CAT+GTR+Γ4 model. Four independent Markov chain Monte Carlo chains were run until convergence and checked by sampling ever two cycles with a 25% burn-in. Support at nodes was evaluated by posterior probability values. ML phylogenies were constructed in IQ-TREE58 under the LG+C60 model.
Metabolic prediction
Gene prediction was performed using Prodigal70. All metabolic genes were identified using hidden Markov models (HMMs) searches with PFAM, TIGR and custom HMM profiles. Annotation of proteins displayed in Supplementary Table 1 were improved by inspecting the genomic context of the metabolic genes using RAST71 and SyntTax72 (http://archaea.u-psud.fr/synttax/), by phylogenetic analyses including the sequences of characterized enzymes and by identifying their conserved domains using CD-search batch73 (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). Genomic context was also inspected to identify conserved patterns among multiple genomes. Identification of energy conservation systems was performed with MacSyFinder74, by defining specific and sensitive models for each system followed by manual curation.
Supplementary Material
Acknowledgements
We thank Rudolf Thauer for feedback on an earlier version of the manuscript. G.B. acknowledges support from the Institut Pasteur through a Roux-Cantarini fellowship. P.S.A. is supported by a PhD fellowship from Paris Diderot University and by funds from the PhD Program “Frontières du Vivant (FdV)-Programme Bettencourt”. S.G. acknowledges funding from the French National Agency for Research Grant ArchEvol (ANR-16-CE02-0005-01). This work used the computational and storage services (TARS cluster) provided by the IT department at Institut Pasteur, Paris. S.J.H. acknowledges support from the US Department of Energy (DOE) JGI supported by the Office of Science of US DOE Contract DE-AC02-05CH11231, the Natural Sciences and Engineering Research Council (NSERC) of Canada, Genome British Columbia, Genome Canada, Canada Foundation for Innovation (CFI), and the Tula Foundation. I.N.S.G. and V.M.O. are grateful to São Paulo Research Foundation - FAPESP (process numbers 2011/14501-6 and 2013/20436-8) and Petrobras for financial support and to Dr. Neil Gray and Dr. Ian Head from the School of Civil Engineering and Geosciences at Newcastle University for lab facilities. W-J.L. was supported by Key Projects of Ministry of Science and Technology (MOST) (Nos. 2013DFA31980, 2015FY110100). G.M. was supported by the ERC Advanced Grant PARASOL (No. 322551). L.J.M. appreciates funding from the NASA Postdoctoral Program through the NASA Astrobiology Institute and W.P.I. was supported by the Montana Agricultural Experiment Station (Project 911300).
Footnotes
Data availability
MAG sequences are available in the BioProject PRJNA472146 and Biosamples SAMN10387997, SAMN10390728, SAMN10390732, SAMN10390733, SAMN10390735, SAMN10390736, SAMN10390737, SAMN10390738, SAMN10390739. NM2 sequences corresponding to markers reported in Table 2 are deposited under MK202738 to MK202758.
The data that support the findings of this study are available from the corresponding author upon request.
Author contributions
G.B. and S.G. conceived the study. L.J.M., L-X.C., I.N.S.-G., C.M.K.S., G.L.A., W.-J.L., S.J. H., G.M., V.M.d.O., W.P.I., and J.F.B. sequenced and assembled the metagenomes. G.B. screened the IMG database for McrA and identified these metagenomes. Q. L., A. G. and G. B. developed the pipeline Let-it-bin. G.B. performed the contig binning of NM1a, NM1b, NM2, NM3, NM4, Verst-YHS, and Mnatro-ASL MAGs. L-X.C. carried out the contig binning of ANME-1-THS MAG and C.M.K.S. those of GoM-Arc1-GOS and ANME-2c MAGs. G.B. inferred the metabolism associated to each MAG and performed all phylogenetic analyses. P.A. performed the congruence tests. G.B. and S.G. wrote the manuscript. All authors read and commented on the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Demirel B, Scherer P. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: A review. Rev Environ Sci Biotechnol. 2008;7:173–190. [Google Scholar]
- 2.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueno Y, Yamada K, Yoshida N, Maruyama S, Isozaki Y. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature. 2006;440:516–519. doi: 10.1038/nature04584. [DOI] [PubMed] [Google Scholar]
- 4.Sousa FL, et al. Early bioenergetic evolution. Philosophical Transactions of the Royal Society B: Biological Sciences. 2013;368 doi: 10.1098/rstb.2013.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasting JF, Siefert JL. Life and the evolution of Earth’s atmosphere. Science. 2002;296:1066–1068. doi: 10.1126/science.1071184. [DOI] [PubMed] [Google Scholar]
- 6.Bapteste E, Brochier C, Boucher Y. Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea. 2005;1:353–363. doi: 10.1155/2005/859728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitman WB, Bowen TL, Boone DR. The Methanogenic Bacteria. Prokaryotes. 2006;3 doi: 10.1007/0-387-30743-5_9. [DOI] [Google Scholar]
- 8.Kendall MM, Boone DR. The Prokaryotes. 2006. pp. 244–256. [DOI] [Google Scholar]
- 9.Oren A. The Prokaryotes: Other Major Lineages of Bacteria and The Archaea. 2014. pp. 165–193. 9783642389. [Google Scholar]
- 10.Borrel G, et al. Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol Evol. 2013;5:1769–1780. doi: 10.1093/gbe/evt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timmers PHA, et al. Reverse Methanogenesis and Respiration in Methanotrophic Archaea. Archaea. 2017;2017 doi: 10.1155/2017/1654237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheller S, Ermler U, Shima S. Anaerobic Utilization of Hydrocarbons, Oils, and Lipids. 2017. pp. 1–29. [DOI] [Google Scholar]
- 13.Hallam SJ, et al. Reverse methanogenesis: Testing the hypothesis with environmental genomics. Science. 2004;305:1457–1462. doi: 10.1126/science.1100025. [DOI] [PubMed] [Google Scholar]
- 14.Borrel G, Adam PS, Gribaldo S. Methanogenesis and the Wood-Ljungdahl Pathway: An Ancient, Versatile, and Fragile Association. Genome Biol Evol. 2016;8:1706–1711. doi: 10.1093/gbe/evw114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adam PS, Borrel G, Brochier-Armanet C, Gribaldo S. The growing tree of Archaea: New perspectives on their diversity, evolution and ecology. ISME J. 2017;11:2407–2425. doi: 10.1038/ismej.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spang A, Caceres EF, Ettema TJG. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science. 2017;357 doi: 10.1126/science.aaf3883. [DOI] [PubMed] [Google Scholar]
- 17.Nobu MK, Narihiro T, Kuroda K, Mei R, Liu W-T. Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J. 2016;10:2478–2487. doi: 10.1038/ismej.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorokin DY, et al. Discovery of extremely halophilic, methyl-reducing euryarchaea provides insights into the evolutionary origin of methanogenesis. Nat Microbiol. 2017;2:17081. doi: 10.1038/nmicrobiol.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanwonterghem I, et al. Methylotrophic methanogenesis discovered in the novel archaeal phylum Verstraetearchaeota. Nat Microbiol. 2016;1:16170. doi: 10.1038/nmicrobiol.2016.170. [DOI] [PubMed] [Google Scholar]
- 20.Brugère JF, et al. Archaebiotics: Proposed therapeutic use of archaea to prevent trimethylaminuria and cardiovascular disease. Gut Microbes. 2014;5:5–10. doi: 10.4161/gmic.26749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laso-Pérez R, et al. Thermophilic archaea activate butane via alkyl-coenzyme M formation. Nature. 2016;539:396–401. doi: 10.1038/nature20152. [DOI] [PubMed] [Google Scholar]
- 22.Evans PN, et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science. 2015;350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- 23.Dombrowski N, Seitz KW, Teske AP, Baker BJ. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome. 2017;5 doi: 10.1186/s40168-017-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKay LJ, Hatzenpichler R, Inskeep WP, Fields MW. Occurrence and expression of novel methyl-coenzyme M reductase gene (mcrA) variants in hot spring sediments. Sci Rep. 2017;7 doi: 10.1038/s41598-017-07354-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawley ER, et al. Metagenomes from two microbial consortia associated with Santa Barbara seep oil. Mar Genomics. 2014;18:97–99. doi: 10.1016/j.margen.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Seitz KW, Lazar CS, Hinrichs K-U, Teske AP, Baker BJ. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J. 2016;10:1696–1705. doi: 10.1038/ismej.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science. 1997;278:1457–1462. doi: 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]
- 28.Arshad A, et al. A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front Microbiol. 2015;6 doi: 10.3389/fmicb.2015.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wegener G, Krukenberg V, Riedel D, Tegetmeyer HE, Boetius A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature. 2015;526:587–590. doi: 10.1038/nature15733. [DOI] [PubMed] [Google Scholar]
- 30.McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature. 2015;526:531–535. doi: 10.1038/nature15512. [DOI] [PubMed] [Google Scholar]
- 31.Meyerdierks A, et al. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ Microbiol. 2010;12:422–439. doi: 10.1111/j.1462-2920.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- 32.Grimaldi S, Schoepp-Cothenet B, Ceccaldi P, Guigliarelli B, Magalon A. The prokaryotic Mo/W-bisPGD enzymes family: A catalytic workhorse in bioenergetic. Biochim Biophys Acta - Bioenerg. 2013;1827:1048–1085. doi: 10.1016/j.bbabio.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 33.Jormakka M, et al. Molecular mechanism of energy conservation in polysulfide respiration. Nat Struct Mol Biol. 2008;15:730–737. doi: 10.1038/nsmb.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hagemeier CH, Chistoserdova L, Lidstrom ME, Thauer RK, Vorholt JA. Characterization of a second methylene tetrahydromethanopterin dehydrogenase from Methylobacterium extorquens AM1. Eur J Biochem. 2000;267:3762–3769. doi: 10.1046/j.1432-1327.2000.01413.x. [DOI] [PubMed] [Google Scholar]
- 35.McInerney MJ, et al. The genome of Syntrophus aciditrophicus: Life at the thermodynamic limit of microbial growth. Proc Natl Acad Sci. 2007 doi: 10.1073/pnas.0610456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klenk H-P, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 37.Milkov AV. Molecular and stable isotope compositions of natural gas hydrates: A revised global dataset and basic interpretations in the context of geological settings. Org Geochem. 2005;36:681–702. [Google Scholar]
- 38.Meredith W, Kelland SJ, Jones DM. Influence of biodegradation on crude oil acidity and carboxylic acid composition. Org Geochem. 2000;31:1059–1073. [Google Scholar]
- 39.Sieber JR, McInerney MJ, Gunsalus RP. Genomic Insights into Syntrophy: The Paradigm for Anaerobic Metabolic Cooperation. Annu Rev Microbiol. 2012 doi: 10.1146/annurev-micro-090110-102844. [DOI] [PubMed] [Google Scholar]
- 40.Zengler K, Richnow HH, Rosselló-Mora R, Michaelis W, Widdel F. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature. 1999 doi: 10.1038/45777. [DOI] [PubMed] [Google Scholar]
- 41.Stams AJM, Sousa DZ, Kleerebezem R, Plugge CM. Role of syntrophic microbial communities in high-rate methanogenic bioreactors. Water Sci Technol. 2012 doi: 10.2166/wst.2012.192. [DOI] [PubMed] [Google Scholar]
- 42.Gao B, Gupta RS. Phylogenomic analysis of proteins that are distinctive of Archaea and its main subgroups and the origin of methanogenesis. BMC Genomics. 2007;8:86. doi: 10.1186/1471-2164-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaster AK, et al. More than 200 genes required for methane formation from H2 and CO2 and energy conservation are present in Methanothermobacter marburgensis and Methanothermobacter thermautotrophicus. Archaea. 2011;2011 doi: 10.1155/2011/973848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nayak DD, Mahanta N, Mitchell DA, Metcalf WW. Post-translational thioamidation of methyl-coenzyme M reductase, a key enzyme in methanogenic and methanotrophic archaea. Elife. 2017;6:e29218. doi: 10.7554/eLife.29218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lyu Z, et al. Mmp10 is required for post-translational methylation of arginine at the active site of methyl-coenzyme M reductase. BioRxiv. 2017 211441. [Google Scholar]
- 46.Sarmiento F, Mrázek J, Whitman WB. Genome-scale analysis of gene function in the hydrogenotrophic methanogenic archaeon Methanococcus maripaludis. Proc Natl Acad Sci U S A. 2013;110:4726–4731. doi: 10.1073/pnas.1220225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner T, Kahnt J, Ermler U, Shima S. Didehydroaspartate Modification in Methyl-Coenzyme M Reductase Catalyzing Methane Formation. Angew Chemie - Int Ed. 2016;55:10630–10633. doi: 10.1002/anie.201603882. [DOI] [PubMed] [Google Scholar]
- 48.Borrel G, et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics. 2014;15:679. doi: 10.1186/1471-2164-15-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng K, Ngo PD, Owens VL, Yang X, Mansoorabadi SO. The biosynthetic pathway of coenzyme F430 in methanogenic and methanotrophic archaea. Science. 2016;354:339–342. doi: 10.1126/science.aag2947. [DOI] [PubMed] [Google Scholar]
- 50.Prakash D, Wu Y, Suh SJ, Duin EC. Elucidating the process of activation of methyl-coenzyme M reductase. J Bacteriol. 2014;196:2491–2498. doi: 10.1128/JB.01658-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J, et al. Global mapping transcriptional start sites revealed both transcriptional and post-transcriptional regulation of cold adaptation in the methanogenic archaeon Methanolobus psychrophilus. Sci Rep. 2015;5:9209. doi: 10.1038/srep09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raymann K, Brochier-Armanet C, Gribaldo S. The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci. 2015;112:6670–6675. doi: 10.1073/pnas.1420858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss MC, et al. The physiology and habitat of the last universal common ancestor. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.116. [DOI] [PubMed] [Google Scholar]
- 54.Adam PS, Borrel G, Gribaldo S. Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc Natl Acad Sci. 2018;115:E5837. doi: 10.1073/pnas.1716667115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGlynn SE. Energy Metabolism during Anaerobic Methane Oxidation in ANME Archaea. Microbes Environ. 2017;32:5–13. doi: 10.1264/jsme2.ME16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peng Y, Leung HCM, Yiu S-M, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 61.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu YW, Simmons BA, Singer SW. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2015;32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 63.Alneberg J, et al. Binning metagenomic contigs by coverage and composition. Nat Methods. 2014;11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- 64.Sieber CMK, et al. Recovery of genomes from metagenomes via a dereplication, aggregation, and scoring strategy. BioRxiv. 2017 doi: 10.1101/107789. 107789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Darling AE, et al. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014;2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kobert K, Salichos L, Rokas A, Stamatakis A. Computing the Internode Certainty and Related Measures from Partial Gene Trees. Mol Biol Evol. 2016;33:1606–1617. doi: 10.1093/molbev/msw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 69.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 70.Hyatt D, et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aziz RK, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oberto J. SyntTax: A web server linking synteny to prokaryotic taxonomy. BMC Bioinformatics. 2013;14:4. doi: 10.1186/1471-2105-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abby SS, Néron B, Ménager H, Touchon M, Rocha EPC. MacSyFinder: A program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS One. 2014;9 doi: 10.1371/journal.pone.0110726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kahnt J, et al. Post-translational modifications in the active site region of methyl-coenzyme M reductase from methanogenic and methanotrophic archaea. FEBS J. 2007;274:4913–4921. doi: 10.1111/j.1742-4658.2007.06016.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.