ABSTRACT
Multidrug resistance (MDR), virulence and transferable elements potentiate Pseudomonas aeruginosa's role as an opportunistic pathogen creating a high risk for public health. In this study, we evaluated the possible association of multidrug resistance, virulence factors and integrons with intrahospital P. aeruginosa strains isolated from patients at Cumana hospital, Venezuela. Relevant clinical-epidemiological data were collected to study 176 strains (2009-2016) isolated from different hospital units. Bacterial resistance was classified as susceptible, low-level resistant (LDR), multidrug resistant (MDR) and extensively drug-resistant (XDR). Most strains produced pyoverdine, DNase, gelatinase and hemolysin. Around 73% of the strains showed some type of movement. MDR and XDR strains increased from 2009 (24.2% and 4.8%, respectively) to 2016 (53.1% and 18.8%); while LDR decreased from 64.5% to 6.3%. The exoU and exoS genes were found in a significant number of strains (38.1 and 7.4%, respectively). Class I integrons were detected in 35.8% of the strains and the frequency was associated with resistance (42.9, 22.4, 41.4 and 61.9%, for susceptible, LDR, MDR and XDR, respectively). The MDR/XDR strains were positively associated with hemolysins and exoU, but negatively associated with bacterial twitching. MDR/XDR phenotypes were also associated with the Intensive Care Unit (ICU), septicemia, bronchial infection and diabetic foot ulcers, as well as long hospital stay (≥10 days) and previous antimicrobial treatment. High frequency of MDR/XDR strains and their association with class I integrons and virulence factors can increase the infection potential, as well as morbidity and mortality of patients attending this hospital and could spread infection to the community, creating a health risk for the region.
KEYWORDS: Pseudomonas aeruginosa, Virulence factors, MDR, XDR, Integrons, Type III Secretion Systems
INTRODUCTION
P. aeruginosa is a ubiquitous environmental bacterium that causes numerous opportunistic human infections, which are clinically and epidemiologically relevant due to the difficulty associated with the treatment of these infections and the transmission of resistance mechanisms. They can remain in the environment, producing repeated relapses in inpatients, increasing their morbidity and mortality, as well as health care costs1.
The pathogenicity of Pseudomonas is associated with numerous virulence factors, such as the presence of lipopolysaccharides, pili and flagella, which are all part of the cell structure, as well as other factors synthesized by the cell and excreted into the surrounding tissue, where the infection develops, such as toxins, proteolytic enzymes, DNases and pigments2. Biofilm formation is another virulence factor due to the biofilm's inherent resistance to antimicrobial agents and the selection of phenotypic variants. P. aeruginosa biofilm presents different resistance mechanisms that make it clinically responsible for many chronic infections, controlled by quorum sensor systems that regulate the transformation from planktonic cells into sessile ones3.
It has also been shown that, depending on the type of infection, P. aeruginosa can adapt by changing its iron uptake system using pyoverdine or pyochelin, characterized by their high and low affinity for iron, respectively, which in turn activate the production of acute virulence factors. The success of P. aeruginosa in causing infections is determined by a complex interaction between pathogenicity, antimicrobial resistance and regulation networks interconnecting resistance and virulence4 and these interactions are currently the subject of numerous studies.
Multidrug resistant (MDR) P. aeruginosa strains are capable of colonizing environments and multiplying in conditions in which the rest of bacterial species are unable, granting them an evolutionary advantage and a better ability to cause epidemic outbreaks. In a hospital setting, a pathogen must be virulent and resistant to antimicrobials for an infection to persist, leading to increased patient morbidity5. In this sense, integrons, which are mobile genetic elements that can carry bacterial genes, have also been considered responsible for the spread of resistance to many antimicrobial agents; they are not essential to bacteria, but can provide additional genes that allow them to better adapt to their environment6.
In Venezuela, antimicrobial agents are frequently prescribed to the population, and currently, hospitals have a limited supply of drugs for their patients’ therapies, resulting in an increasing frequency of MDR P. aeruginosa due to the lack of timely and adequate treatments7. Since 1989, the presence of P. aeruginosa has been reported in intrahospital infections in Venezuela, with different resistances to beta-lactam antibiotics, aminoglycosides and quinolones8–10. Reports of MDR P. aeruginosa strains began with metallo-beta-lactamase positive strains reported in 200411 up to MDR strains found in several hospitals12. However, there are no studies evaluating the simultaneous presence of virulence factors, resistance genes and mobile genetic elements in this species, even though they are essential to understand the pathogen's behavior and to design adequate therapeutic and control strategies to fight these organisms. Therefore, the objective of the present study was to evaluate how the virulence factors, resistance genes and integrons are affecting the epidemiological outcome of P. aeruginosa strains isolated from patients in the general hospital of Cumana, Venezuela.
MATERIALS AND METHODS
Study population and bioethical standards
We evaluated 176 P. aeruginosa strains isolated from patients diagnosed with clinical infections and indications of culture and antibiogram performed in the Antonio Patricio de Alcala University Hospital (HUAPA) in Cumana, collected from June to December 2009 (62 strains), January to December 2010 (42 strains), February to December 2012 (40 strains), and January to June 2016 (32 strains). Clinical and epidemiological relevant data were collected from patients, after the signature of an informed consent form. Each strain was isolated from a different individual, and they were independent of any epidemic outbreak. Biological samples and data collection were carried out following the guidelines established by the IIBCA-UDO biosecurity and bioethics committee (CoBioBios). The presence of infection was defined using criteria established by the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC)13, which classifies an infection as a healthcare-associated infection (HAI) if the date on which the specific site of the infection is detected takes place at least 48 h after the patient's admission14.
Toxin production
Hemolysins were produced in blood agar (8.0%) cultures. A transparent halo around the colonies indicated the presence of P. aeruginosa's beta-hemolysin and the absence of a hemolysis halo was interpreted as a negative result15. Pigment production was evaluated in cetrimide agar cultures; targeted pigments were pyocyanin (blue-green), fluorescein (yellow), pyoverdine (green), pyorubin (red) and pyomelanin (brown)16.
Biofilm production
Bacteria were cultured in LB media in 96-well plates to quantify the production of biofilm, using P. aeruginosa PA01 strain as a positive control17. A sterile broth was used as the negative control to ensure sterility and the absence of non- media-specific components. Strains were considered as non-biofilm producers when their optical densities were ≤ 0.108 (≤ Average of Negative Control + 2σ) and biofilm producers when densities were > 0.108 (> ANC + 2σ).
Motility tests
Motility tests were carried out in sterile Petri dishes with Luria-Bertani agar (LB) using different concentrations of agar to evaluate swarming (0.5%), swimming (0.3%) and twitching (1.2%) as the different types of motility found in P. aeruginosa18 strains. Ten microliters of the media containing overnight cultured strains were inoculated on the agar surface for the swarming testing; while for swimming, 10 μL of the strains were inoculated into a thick agar by puncture; and for twitching, 10 μL of cultures were inoculated at the bottom of the agar. A well characterized Proteus mirabilis (LGM7881), swarming producer strain, and another well characterized immobile Klebsiella pneumoniae LGM1589 strain were used as positive and negative controls. All plates were incubated at 37 °C for 24 h. To assess the twitching motility, the agar was removed from the plate and 25 mL of 1% crystal violet dye were added, incubated for 30 min, then the stain was poured off the plate and air dried. Finally, the average diameters of the growing areas of the different types of motility were measured with a graduated ruler and expressed in millimeters (mm).
Enzymes production
DNAse activity was studied using the DNAse test to rule out the presence of deoxyribonuclease19. A transparent halo around the colonies indicated nuclease activity, whereas the lack of a halo indicated a negative result. A Serratia marcescens strain (ATCC 1388) was used as the control. Pseudomonas's capacity to produce proteolytic enzymes (gelatinases) was determined by using the puncture method in tubes containing 4% gelatin-agar. Total or partial medium hydrolysis attested the production of the enzyme (positive result), whereas a lack of hydrolysis was interpreted as a negative result20.
Resistance phenotypes
The agar diffusion method was used to evaluate resistance to the following antimicrobials: piperacillin/tazobactam (100/10 µg), aztreonam (30 µg), ceftazidime (30 µg), cefepime (30 µg), meropenem (10 µg), imipenem (10 µg), colistin (10 µg), polymyxin B (300 U), amikacin (30 µg), gentamicin (10 µg), tobramycin (10 µg), netilmicin (10 µg), levofloxacin (5 µg), norfloxacin (5 µg) and ciprofloxacin (5 µg). All essays were conducted using the criteria established by Clinical and Laboratory Standards Institute. Resistance phenotypes were defined using the guidelines established for P. aeruginosa21, and strains were characterized as low-level resistant (LDR, <3 antimicrobial categories), multidrug resistant (MDR, non-susceptible to ≥3 categories), extensively drug-resistant (XDR, non-susceptible to all but ≤2 categories), and pandrug-resistant (PDR, non-susceptible to all antimicrobial categories) after testing all categories of microbial agents used in Venezuela, except for those for which this bacterium is intrinsically resistant. The quality of antimicrobial discs and biochemical tests was verified using Escherichia coli (ATTC 25922) and P. aeruginosa (ATTC 27853) as control strains.
Detection of genes
DNA extraction was carried out using pure P. aeruginosa cultures. A Wizard Genomic DNA purification kit (Promega, Wisconsin, USA) was used for extraction according to the manufacturer's specifications. The presence of the type 3 secretion systems (SST3) exoS and exoU genes were detected by PCR using oligonucleotides ExoS-F/R (1587 pb) and ExoU-F/R (761 pb) according to Ajayi et al.22. Strains PA01 and PA103 were assayed as positive controls for exoS and exoU, respectively. The detection of integrases int1-F/R (1202 pb), int2-F/R (973 pb) and int3-F/R (239 pb) genes was conducted based on the protocol described by Fonseca et al.23 Amplified products were run in 2% agarose gels and stained with GelRed Nucleic Acid (Biotium, California, USA) using TBE 1X buffer for 30 min at 100 V cm. The size of the fragments was estimated using a molecular marker (100 pb from Promega, and Gene Ruler 1Kb plus from Thermo Fisher Scientific, Massachussets, USA). The resulting bands were visualized by transillumination with ultraviolet light, and all gels were photographically documented.
Analysis of data
Results were presented in tables and figures. The relationships between MDR and the presence of virulence factors, clinical and epidemiological factors as well as integrons were evaluated by a binary logistic regression test, with a 95% confidence interval, using the SPSS statistical software, version 18. (IBM, New York, USA).
RESULTS
Identification of resistant phenotypes
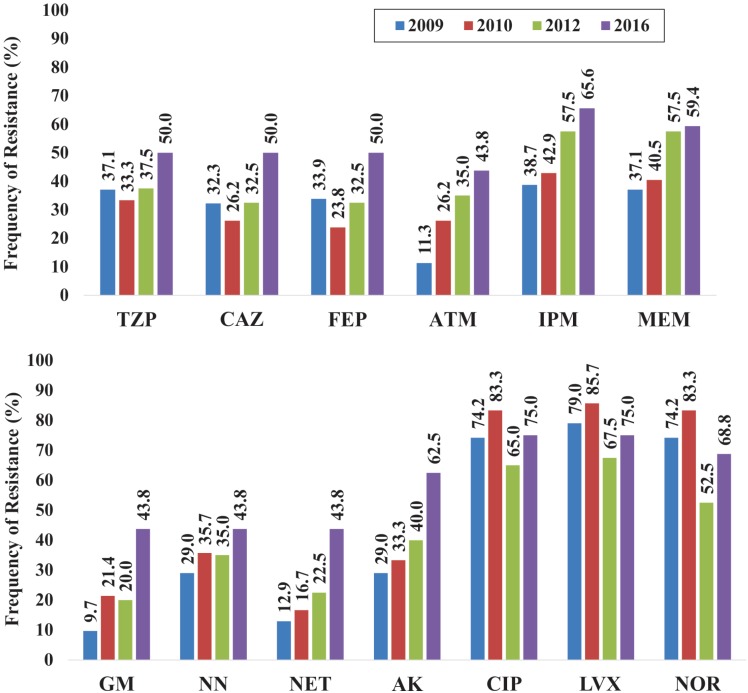
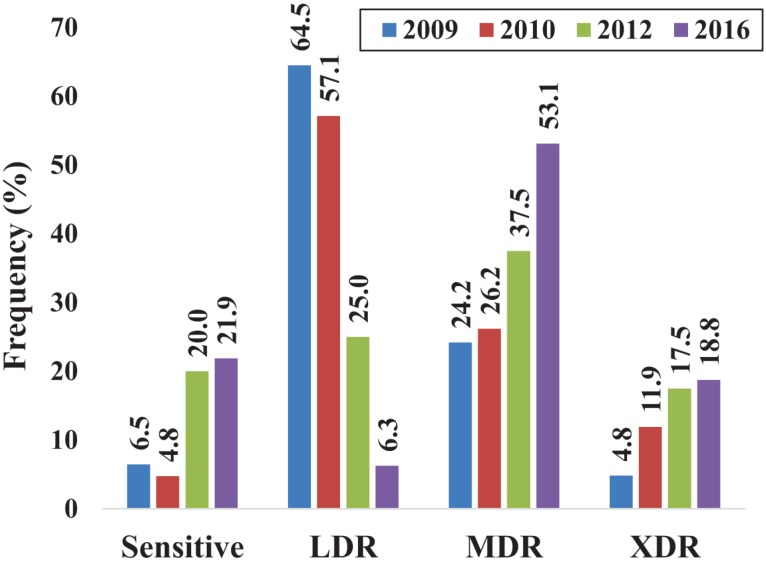
The present study demonstrated an increase in the frequency of resistant strains to beta-lactam antibiotics from 2009 to 2016, especially to carbapenems (Figure 1). Strains resistant to aminoglycosides have also increased, and even though the resistance to all fluoroquinolones remained ≥ 52.0% throughout the evaluated years and it has slightly decreased in the strains isolated in 2012 and 2016. All strains were susceptible to polymyxin B and colistin. The frequency of Pseudomonas aeruginosa MDR and XDR phenotypes strains from HUAPA (Figure 2) increased from 2009 to 2016, at the expense of the decrease of the LDR phenotype, showing a clear trend of resistance gain to more antibiotic categories.
Figure 1. Frequency of P. aeruginosa strains resistant to betalactamics (top) as well as aminoglycosides and quinolones (bottom). TZP: piperacillin/tazobactam, CAZ: ceftazidime, FEP: cefepime, ATM: aztreonam, IPM: imipenem, MEM: meropenem, GM: gentamicin, NN: tobramycin, NET: netilmicin, AK: amikacin, CIP: ciprofloxacin, LVX: levofloxacin; NOR: norfloxacin.
Figure 2. Distribution of P. aeruginosa strains resistant phenotypes throughout the years. Low-level resistant (LDR, <3 antimicrobial categories), Multidrug resistant (MDR, non-susceptible to ≥3), extensively drug-resistant strains (XDR, non-susceptible to all but ≤2 antimicrobial categories).
Virulence factors
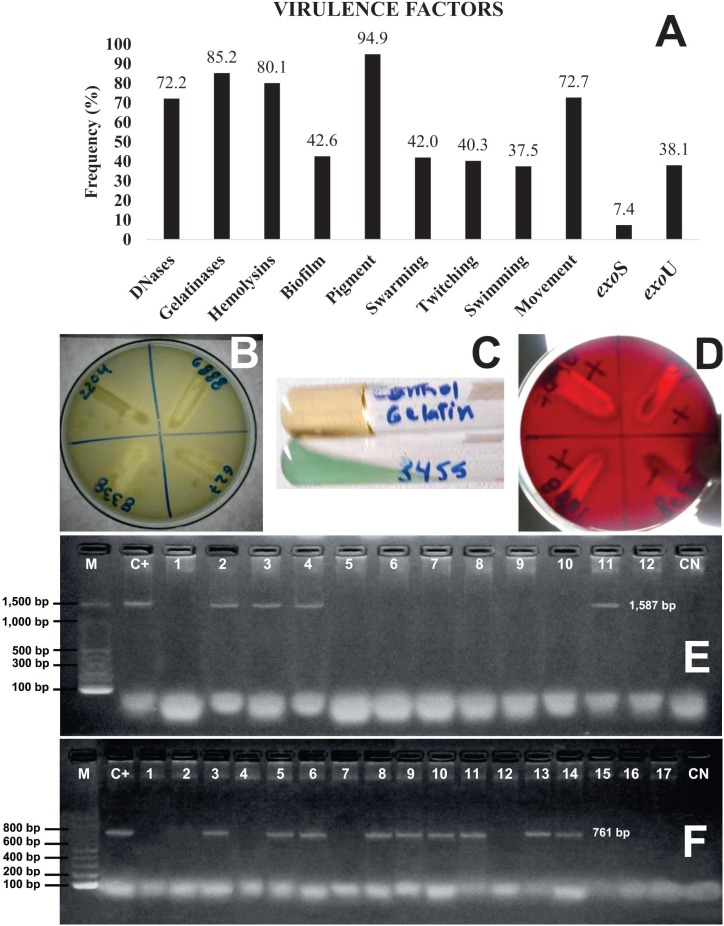
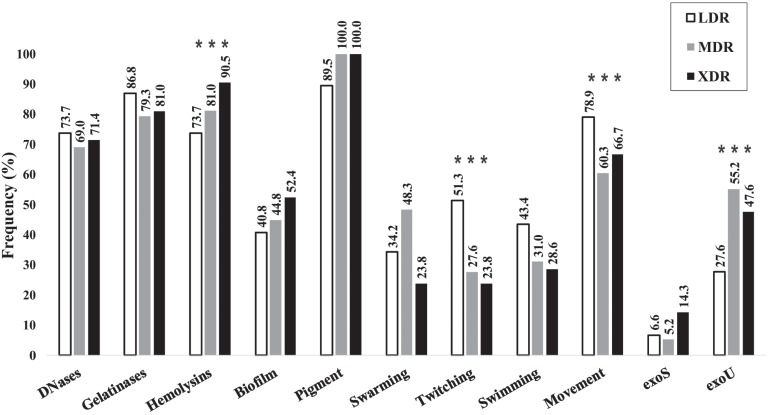
Pigment was the most common virulence factor produced by Pseudomonas aeruginosa strains (Figure 3A), followed by gelatinases, hemolysins and DNases (Figures 3B, 3C and 3D). About 40% of the strains showed biofilm and the different types of movement. The detection of exoS and exoU genes showed the latter to be more frequent (Figures 3A, 3E and 3F) and the simultaneous presence of exoS+exoU genes was found in only 1.7% of the strains.
Figure 3. A) Frequency of the different virulence factors detected; example of the tests for the production of DNases (B), gelatinases (C), hemolysins (D), PCR amplification for P. aeruginosa exoS (E) and exoU (F) genes. bp: base pairs, M: molecular weight markers 100 bp ladder, C+: positive Control, C-: negative Control.
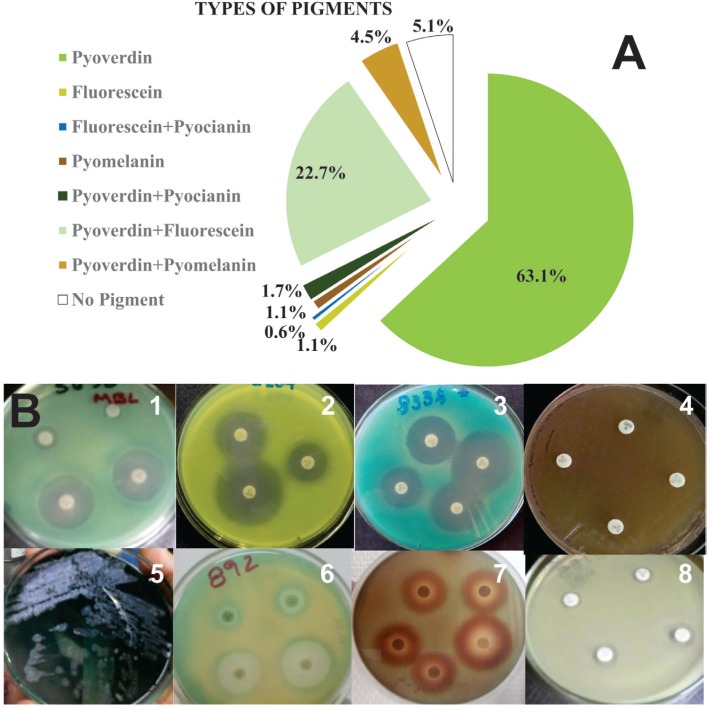
The production of pyoverdine alone was the most frequently found bacterial phenotype (Figure 4A and 4B1), followed by pyoverdine+fluorescein (Figure 4B6), no pigment (Figue 4B8) and pyoverdine+pyomelanin (Figure 4B7). Simultaneous pigment production was more evident when strains were subjected to antimicrobial susceptibility tests, which showed a halo of one of the pigments around the antimicrobial disc (Figure 4B1-7) and the other pigment spread across the plate. Additionally, the metallic sheen characteristic of P. aeruginosa (Figure 4B5) has also been observed in other three strains.
Figure 4. Relative frequency of types of pigments produced by P. aeruginosa strains: A) Examples of different pigments and pigment combinations produced in Mueller-Hinton agar with antibiotics; B) 1: pyoverdine, 2: fluorescein, 3: fluorescein + pyocyanin, 4: pyomelanin, 5: pyoverdine + pyocyanin, 6: pyoverdine + fluorescein, 7: pyoverdine + pyomelanin, and 8: no pigment.
Association of factors
Production of DNases, gelatinase, biofilm and pigment was not statistically associated with the level of resistance shown by the strains (Table 1, Figure 5), whereas hemolysins production was directly related to resistance. In addition, the presence of the exoS gene was not statistically related to resistance, while exoU gene was significantly more frequent in MDR and XDR strains, compared to LDR and susceptible strains.
Table 1. Statistical correlation of different virulence factors with P. aeruginosa strains MDR/XDR phenotypes.
| Virulence Factor | MDR/XDR | Total | P | OR(95% CI) | ||
|---|---|---|---|---|---|---|
| – | + | |||||
| DNAase | – | 24 | 25 | 49 | 0.343 | |
| + | 70 | 57 | 127 | |||
| Gelatinase | – | 10 | 16 | 26 | 0.106 | |
| + | 84 | 66 | 150 | |||
| Hemolysin | – | 22 | 13 | 35 | 0.044 | 2.3(1.0-5.3) |
| + | 72 | 69 | 141 | |||
| Pyoverdine | – | 11 | 3 | 14 | 0.062 | |
| + | 83 | 79 | 162 | |||
| Fluorescein | – | 69 | 64 | 133 | 0.479 | |
| + | 25 | 18 | 43 | |||
| Pyomelanin | – | 89 | 77 | 166 | 0.725 | |
| + | 5 | 5 | 10 | |||
| Swarming | – | 50 | 44 | 94 | 0.996 | |
| + | 41 | 33 | 74 | |||
| Twitching | – | 43 | 54 | 97 | 0.005 | 0.4(0.2-0.8) |
| + | 48 | 23 | 71 | |||
| Swimming | – | 51 | 51 | 102 | 0.271 | |
| + | 40 | 26 | 66 | |||
| Movement | – | 17 | 31 | 48 | 0.004 | 0.4(0.2-0.7) |
| + | 77 | 51 | 128 | |||
| Biofilm | – | 56 | 45 | 101 | 0.776 | |
| + | 38 | 37 | 75 | |||
| exoS | – | 87 | 76 | 163 | 0.974 | |
| + | 7 | 6 | 13 | |||
| exoU | – | 71 | 38 | 109 | <0.001 | 3.6(1.9-6.8) |
| + | 23 | 44 | 67 | |||
| Integrase I | – | 70 | 43 | 113 | 0.003 | 2.7(1.4-5.0) |
| + | 24 | 39 | 63 | |||
P: probability, OR: Odds Ratio, CI: confidence interval.
Figure 5. Frequency of P. aeruginosa strains that produce virulence factors according to the resistance phenotype described in Figure 2. Factors associated to MDR/XDR are highlighted with three asterisks.
When analyzing the types of movement shown by the strains, swarming but not swimming was directly correlated, while twitching was inversely correlated to the level of resistance (Table 1, Figure 5). When comparing the presence of any type of movement versus non-motile strains, we have also found an inverse correlation with the level of resistance.
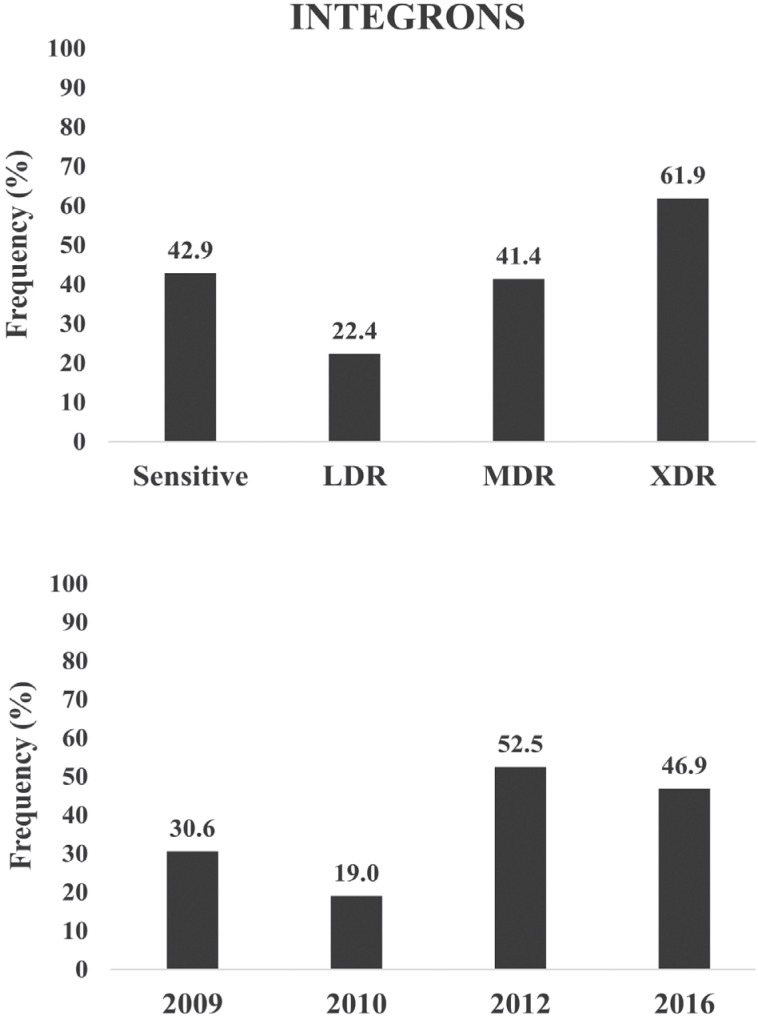
An increase in the frequency of class I integrons has been observed for the years 2012 and 2016, compared to previous years (Figure 6). Additionally, a higher frequency of these sequences was found in XDR and MDR strains when compared to LDR ones, although sensitive strains have shown the same frequency as MDRs.
Figure 6. Frequency of P. aeruginosa strains containing class I integrons according to the resistance phenotype shown (top) and throughout the years (bottom). Resistant phenotypes as described in Figure 2.
The analysis of virulence factors as risk factors for MDR/XDR resistance has shown that the presence of hemolysin, exoU gene and class I integrons has increased the risk of strains to present an MDR/XDR level of resistance 2.3, 3.6 and 2.7 times higher, respectively (Table 1), while the presence of twitching reduced the risk of MDR/XDR resistance by 2.6 times.
Epidemiological clinical factors
When MDR was analyzed with respect to the presence of epidemiological clinical factors such as age, sex, hospital unit, type of sample, length of hospital stay and previous antimicrobial treatment, a statistically significant association (P <0.05) was found for some of these factors (Table 2). In this sense, patients treated at the Intensive Care Unit were 3.4 times more likely to become infected by an MDR/XDR P. aeruginosa strain than a LDR or a susceptible strain. In addition, patients with blood stream infection (septicemia), bronchial infection or diabetic foot ulcers were 7.9, 6.7 and 9.9 times more likely to be infected by an MDR/XDR P. aeruginosa strain, while patients with other types of infection (urine, sputum, ear swabs, eye and vagina) were 5 times less likely to be infected by an MDR/XDR strain.
Table 2. Statistical correlations of different patients’ clinical and epidemiological factors, (from whom P. aeruginosa was isolated), with MDR/XDR phenotypes.
| Factor | MDR/XDR | P | OR(95% CI) | |||
|---|---|---|---|---|---|---|
| – | + | Total | ||||
| Units | Medical | 38 | 27 | 65 | 0.871 | |
| ICU | 10 | 26 | 36 | 0.030 | 3.4(1.2-10.2) | |
| Surgery | 18 | 9 | 27 | 0.462 | ||
| Outpatient | 13 | 10 | 23 | 0.748 | ||
| Pediatric/Neonatal | 15 | 10 | 25 | 0.807 | ||
| Sample | Blood | 7 | 12 | 19 | 0.003 | 7.9(2.1-30.2) |
| Bronchial secretion | 11 | 16 | 27 | 0.003 | 6.7(2.0-23.0) | |
| Diabetic foot ulcer | 13 | 28 | 41 | <0.001 | 9.9(3.1-31.9) | |
| Skin secretion | 40 | 21 | 61 | 0.117 | ||
| Other a | 23 | 5 | 28 | 0.002 | 0.2(0.1-0.6) | |
| Stay | Short (<10 days) | 70 | 43 | 113 | 0.019 | 2.3(1.1-4.) |
| Long (≥10 days) | 21 | 29 | 50 | |||
| Sex | Male | 53 | 55 | 108 | 0.355 | |
| Female | 35 | 27 | 62 | |||
| Age | <10 years | 13 | 11 | 24 | 0.964 | |
| 10-60 years | 52 | 41 | 93 | 0.721 | ||
| >60 years | 16 | 18 | 34 | 0.573 | ||
| Previous antimicrobial treatment | No | 78 | 44 | 122 | <0.001 | 4.2(2.1-8.4) |
| Yes | 16 | 38 | 54 | |||
P: probability, OR: Odds Ratio, CI: confidence interval.
Other types of samples: urine, sputum, ear swabs, eye and vagina.
Furthermore, patients with longer hospital stay had a 2.3 times higher risk to be infected by an MDR/XDR strain, while those with a previous antimicrobial treatment showed a 4.2 times higher risk of being infected by these strains.
DISCUSSION
The presence of MDR/XDR P. aeruginosa strains in the HUAPA hospital has increased over the past years. Teixeira et al.24 evaluated strains between 2010 and 2011 and reported resistance to one or more antimicrobial agents in 65% of the analyzed strains. Data reported here on MDR and XDR strains have shown the severity of this medical issue for the patients attending this hospital due to the higher risk of morbidity and mortality and also the risk of spreading of resistant strains to the population living in this region. The increase of resistant strains of this species has a greater impact because of the natural resistance mechanisms that this bacterial species has.
Although most beta-lactam antibiotics are faced with lower resistance than carbapenems, this high resistance to carbapenems should be highlighted and it is probably due to the indiscriminate use of carbapenems in this particular hospital or to the lack of a controlled use of carbapenems in intrahospital infections, leading to the emergence of MDR strains. Other studies have shown that P. aeruginosa strains isolated from patients subjected to prolonged antimicrobial treatments are more prone to adapt to antimicrobial agents and to become MDR strains; this selection phenomenon is the reason behind the difficulty to control infections caused by MDR strains25,26.
The percentages of MDR and XDR strains in the present study were higher than those reported by Vega and Dowzicky27 who studied antimicrobial susceptibility among gram-positive and gram-negative bacteria from Latin American between 2004 and 2015; a percentage of 26.7% of MDR P. aeruginosa was found and the countries presenting the highest percentages of MDR P. aeruginosa isolates were Guatemala, Venezuela, Honduras and Brazil (43.8, 32.6, 31.9 and 31.5%, respectively).
Pigment production was the most frequent virulence factor in HUAPA strains throughout the evaluated periods, both in susceptible and LDR strains. Even though pigment production was somewhat higher in MDR and XDR strains, the difference was not statistically significant. The simultaneous production of pigments by P. aeruginosa strains from HUAPA was more evident when MDR and XDR strains were subjected to antimicrobial susceptibility tests. The presence of pigments is the distinctive characteristic of the fluorescens group of Pseudomonas species, and it is also an evidence of the increased virulence of strains found at this hospital. Pigments are necessary for infection to occur in patients. Moreover, pyoverdine and pyomelanin facilitate virulence, biofilm development allowing species to establish chronic infections4,28. Pigment combinations have been associated with the status or severity of infections caused by P. aeruginosa26.
Another important aspect of pyoverdine synthesis in P. aeruginosa is its quorum sensor (QS) regulation mechanism, which modulates the existence of other virulence factors such as proteases, exotoxins and biofilm4.
Among the other virulence factors identified in HUAPA strains, such as gelatinases, hemolysins and DNases, only hemolysins were associated with MDR, but production of these factors is variable in P. aeruginosa strains. Georgescu et al.18 reported that the most frequently expressed soluble virulence factors are beta-hemolysins (83.3%) and gelatinases (66.7%) and less frequently DNases (16.7%), whereas Holban et al.29 reported an increase in DNases and a decrease in gelatinases in wound secretion isolates.
In HUAPA strains, beta-hemolysin production plays an important role in spreading the infection and extending wounds, since these enzymes (which form pores on cell membranes) facilitate the invasion of Pseudomonas into eukaryotic cells, protecting the bacterium from the host's defense mechanisms and the antimicrobial treatment14.
For their part, DNases have important implications for virulence because they degrade extracellular DNA that will be used as a source of nitrogen, carbon and phosphate; they support P. aeruginosa's biofilm matrix maintenance and their role in cell signaling to induce the expression of genes essential for resistance to antibiotics have been evidenced, leading to evasion of the host immune system30.
Gelatinase production in HUAPA strains was also an important virulence factor, since proteases degrade a variety of proteins, including matrix components of different cells. Galdino et al.31, studying clinical strains of P. aeruginosa from Brazil, observed that gelatinase and elastase activities and the level of production varied depending on the bacteria's site of isolation and antimicrobial susceptibility.
Another virulence factor evaluated in the present study was SST3 (Secretion System Type 3) because of the detection of the exoS and exoU genes in HUAPA strains. The exoU gene was associated with MDR, and its highest frequency was found in wound and bronchial secretions and blood samples from HUAPA patients, which has also been reported by Holban et al.32, who detected higher proportions of positive results for exoS than for the exoU enzyme. These genes are traits of clinical P. aeruginosa strains that have exoU secretion, higher impact on mortality, persistence of chronic diseases and dissemination of P. aeruginosa infections in comparison the exoS strain secretors, but it can vary depending on the site and the characteristics of the disease33,34.
According to Feltman et al.33, exoS and exoU distribution is not random and the authors found that most isolates carrying exoS sequences lack exoU, and vice versa, which was confirmed by the results of 115 P. aeruginosa clinical and environmental isolates among which 82 contained exoS and 31 contained exoU, and only one isolate contained both genes.
Production of biofilms by P. aeruginosa has been associated with an increase on antimicrobial resistance35,36, by means of resistance genes accumulation and also to the fact that biofilms matrix penetration is difficult for antimicrobial agents.
Resistance to antipseudomonal beta-lactam antibiotics is apparently proportional to the increment of infection chronicity, which is in turn associated with biofilm formation and pyoverdine production26. More studies on antimicrobial resistance and simultaneous production of virulence factors are essential to understand the behavior of pathogenenic bacteria and to develop effective therapeutic strategies6.
Results of the present study showed that susceptible strains with class I integrons began to appear during the 2012 period, possibly acquired by horizontal transference of MDR or XDR P. aeruginosa intrahospital strains and increased in number as well as integron acquisition. Integrons have resistance cassettes that may favor the presence of MDR strains, which has been reported in P. aeruginosa strains with class I integrons37–40. Integrons were found to be associated with MDR strains of Pseudomonas, however, Fonseca et al.23 found no correlations between the presence of class I integron and MDR in clinical P. aeruginosa strains. Other studies have shown that P. aeruginosa isolates from patients with a history of long antimicrobial treatment developed MDR/XDR; long hospital stay and readmission to the Intensive Care Unit were associated with MDR in patients with bacteremia caused by this species41,42.
Results of the present study demonstrated that HUAPA strains increased their MDR throughout the different sampling periods; these strains were also characterized by the presence of different virulence factors (pigments, biofilms, among others) favoring infections by P. aeruginosa. However, only hemolysins, exoU genes and class I integrons were associated with MDR/XDR.
The present study demonstrated that P. aeruginosa employs various mechanisms to maintain infection in patients admitted to the studied hospital; in addition to risk factors identified by the study (attending the Intensive Care Unit, previous antimicrobial treatment, long hospital stay), these conditions predispose patients to P. aeruginosa MDR strains infections, which require infection-specific therapeutic, isolation and control measures to decrease both, the infection rate and the dissemination of bacterial resistance mechanisms in hospitals.
Funding Statement
The present study was financed by the Ministerio del Poder Popular para la Ciencia, Tecnologia e Innovacion (G-2012000780) and the Comision de Investigacion, Universidad de Oriente (CI-02-040102-1781-12), Venezuela.
Footnotes
FUNDING
The present study was financed by the Ministerio del Poder Popular para la Ciencia, Tecnologia e Innovacion (G-2012000780) and the Comision de Investigacion, Universidad de Oriente (CI-02-040102-1781-12), Venezuela.
REFERENCES
- 1.López-Pueyo MJ, Barcenilla-Gaite F, Amaya-Villar R, Garnacho-Montero J. Multirresistencia antibiótica en unidades de críticos. Med Intensiva. 2011;35:41–53. doi: 10.1016/j.medin.2010.07.011. [DOI] [PubMed] [Google Scholar]; 1. López-Pueyo MJ, Barcenilla-Gaite F, Amaya-Villar R, Garnacho-Montero J. Multirresistencia antibiótica en unidades de críticos. Med Intensiva. 2011;35:41-53. [DOI] [PubMed]
- 2.Jácome PR, Alves LR, Cabral AB, Lopes AC, Maciel MA. Phenotypic and molecular characterization of antimicrobial resistance and virulence factors in Pseudomonas aeruginosa clinical isolates from Recife, State of Pernambuco, Brazil. Rev Soc Bras Med Trop. 2012;45:707–712. doi: 10.1590/s0037-86822012000600010. [DOI] [PubMed] [Google Scholar]; 2. Jácome PR, Alves LR, Cabral AB, Lopes AC, Maciel MA. Phenotypic and molecular characterization of antimicrobial resistance and virulence factors in Pseudomonas aeruginosa clinical isolates from Recife, State of Pernambuco, Brazil. Rev Soc Bras Med Trop. 2012;45:707-12. [DOI] [PubMed]
- 3.Wei Q, Ma LZ. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci. 2013;14:20983–21005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]; 3. Wei Q, Ma LZ. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci. 2013;14:20983-1005. [DOI] [PMC free article] [PubMed]
- 4.Cornelis P, Dingemans J. Pseudomonas aeruginosa adapts it iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol. 2013;3:75–75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]; 4. Cornelis P, Dingemans J. Pseudomonas aeruginosa adapts it iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol. 2013;3:75. [DOI] [PMC free article] [PubMed]
- 5.Martínez JL, Baquero F. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin Microbiol Rev. 2002;15:647–679. doi: 10.1128/CMR.15.4.647-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; 5. Martínez JL, Baquero F. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin Microbiol Rev. 2002;15:647-79. [DOI] [PMC free article] [PubMed]
- 6.Beceiro A, Tomás M, Bou G. Resistencia a los antimicrobianos y virulencia, ¿una asociación beneficiosa para el mundo microbiano? Enferm Infecc Microbiol Clin. 2012;30:492–499. doi: 10.1016/j.eimc.2012.01.011. [DOI] [PubMed] [Google Scholar]; 6. Beceiro A, Tomás M, Bou G. Resistencia a los antimicrobianos y virulencia, ¿una asociación beneficiosa para el mundo microbiano? Enferm Infecc Microbiol Clin. 2012;30:492-9. [DOI] [PubMed]
- 7.Sociedad Venezolana de Infectología emitió un pronunciamiento ante la crisis humanitaria. [[cited 2018 Dec 5]]. Available from: http://revistasic.gumilla.org/2016/sociedad-venezolana-de-infectologia-emitio-un-pronunciamiento-ante-la-crisis-humanitaria/; 7. Sociedad Venezolana de Infectología emitió un pronunciamiento ante la crisis humanitaria. [cited 2018 Dec 5]. Available from: http://revistasic.gumilla.org/2016/sociedad-venezolana-de-infectologia-emitio-un-pronunciamiento-ante-la-crisis-humanitaria/
- 8.Pitteloud JJ, De Rodríguez I, Navarro P, De González M, Pérez E, Apollonia B. Infecciones nosocomiales en el Hospital Universitario de Caracas. Rev Argent Microbiol. 1989;21:25–30. [PubMed] [Google Scholar]; 8. Pitteloud JJ, De Rodríguez I, Navarro P, De González M, Pérez E, Apollonia B. Infecciones nosocomiales en el Hospital Universitario de Caracas. Rev Argent Microbiol. 1989;21:25-30. [PubMed]
- 9.Araque-Granadillo MC, Nieves-Blanco BM. Estudio microbiologico de infecciones pulmonares de origen intrahospitalario. Rev Latinoam Microbiol. 1993;35:147–152. [PubMed] [Google Scholar]; 9. Araque-Granadillo MC, Nieves-Blanco BM. Estudio microbiologico de infecciones pulmonares de origen intrahospitalario. Rev Latinoam Microbiol. 1993;35:147-52. [PubMed]
- 10.Rodríguez AJ, Samaniego DR, Soskin A, Rodríguez CN, Canese J, de Canese JO, et al. Comparative study of antimicrobial resistance of Pseudomonas aeruginosa strains isolated from patients of Caracas and Asunción in a 4-year-period. Chemotherapy. 2002;48:164–167. doi: 10.1159/000063867. [DOI] [PubMed] [Google Scholar]; 10. Rodríguez AJ, Samaniego DR, Soskin A, Rodríguez CN, Canese J, de Canese JO, et al. Comparative study of antimicrobial resistance of Pseudomonas aeruginosa strains isolated from patients of Caracas and Asunción in a 4-year-period. Chemotherapy. 2002;48:164-7. [DOI] [PubMed]
- 11.Mendes RE, Castanheira M, Garcia P, Guzman M, Toleman MA, Walsh TR, et al. First isolation of blaVIM-2 in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother. 2004;48:1433–1434. doi: 10.1128/AAC.48.4.1433-1434.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; 11. Mendes RE, Castanheira M, Garcia P, Guzman M, Toleman MA, Walsh TR, et al. First isolation of blaVIM-2 in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother. 2004;48:1433-4. [DOI] [PMC free article] [PubMed]
- 12.Guevara A, de Waard J, Araque M. Detección del gen blaVIM-2 en cepas de Pseudomonas aeruginosa productoras de metalo b-lactamasa aisladas en una unidad de cuidados intensivos en Ciudad Bolívar, Venezuela. Rev Chilena Infectol. 2009;26:336–341. [PubMed] [Google Scholar]; 12. Guevara A, de Waard J, Araque M. Detección del gen blaVIM-2 en cepas de Pseudomonas aeruginosa productoras de metalo b-lactamasa aisladas en una unidad de cuidados intensivos en Ciudad Bolívar, Venezuela. Rev Chilena Infectol. 2009;26:336-41. [PubMed]
- 13.Pujol M, Limón E. Epidemiología general de las infecciones nosocomiales. Sistemas y programas de vigilancia. Enferm Infecc Microbiol Clin. 2013;31:108–113. doi: 10.1016/j.eimc.2013.01.001. [DOI] [PubMed] [Google Scholar]; 13. Pujol M, Limón E. Epidemiología general de las infecciones nosocomiales. Sistemas y programas de vigilancia. Enferm Infecc Microbiol Clin. 2013;31:108-13. [DOI] [PubMed]
- 14.Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, et al. Health care-associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med. 2002;137:791–797. doi: 10.7326/0003-4819-137-10-200211190-00007. [DOI] [PubMed] [Google Scholar]; 14. Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, et al. Health care-associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med. 2002;137:791–7. [DOI] [PubMed]
- 15.Georgescu M, Gheorghe I, Curutiu C, Lazar V, Bleotu C, Chifiriuc MC. Virulence and resistance features of Pseudomonas aeruginosa strains isolated from chronic leg ulcers. BMC Infect Dis. 2016;16(Suppl 1):92–92. doi: 10.1186/s12879-016-1396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]; 15. Georgescu M, Gheorghe I, Curutiu C, Lazar V, Bleotu C, Chifiriuc MC. Virulence and resistance features of Pseudomonas aeruginosa strains isolated from chronic leg ulcers. BMC Infect Dis. 2016;16 Suppl 1:92. [DOI] [PMC free article] [PubMed]
- 16.Merino LA. Pseudomonas aeruginosa: una bacteria con personalidades múltiples. Rev Argent Microbiol. 2007;39:143–143. [PubMed] [Google Scholar]; 16. Merino LA. Pseudomonas aeruginosa: una bacteria con personalidades múltiples. Rev Argent Microbiol. 2007;39:143. [PubMed]
- 17.Gómez J, Gómez-Lus ML, Bas P, Ramos C, Cafini F, Maestre JR, et al. ¿Es la cuantificación del biofilm un elemento diferenciador en la patogenia de bacilos gramnegativos? Rev Esp Quimioter. 2013;26:97–102. [PubMed] [Google Scholar]; 17. Gómez J, Gómez-Lus ML, Bas P, Ramos C, Cafini F, Maestre JR, et al. ¿Es la cuantificación del biofilm un elemento diferenciador en la patogenia de bacilos gramnegativos? Rev Esp Quimioter. 2013;26:97-102. [PubMed]
- 18.Wang S, Yu S, Zhang Z, Wei Q, Yan L, Ai G, et al. Coordination of swarming motility, biosurfant synthesis, and biolm matrix exopolysacharide production in Pseudomonas aeruginosa. Appl Environ Microbiol. 2014;80:6724–6732. doi: 10.1128/AEM.01237-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; 18. Wang S, Yu S, Zhang Z, Wei Q, Yan L, Ai G, et al. Coordination of swarming motility, biosurfant synthesis, and biolm matrix exopolysacharide production in Pseudomonas aeruginosa. Appl Environ Microbiol. 2014;80:6724-32. [DOI] [PMC free article] [PubMed]
- 19.Jeffries CD, Holtman DF, Guse DG. Rapid method for determining the activity of microorganisms on nucleic acids. J Bacteriol. 1957;73:590–591. doi: 10.1128/jb.73.4.590-591.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]; 19. Jeffries CD, Holtman DF, Guse DG. Rapid method for determining the activity of microorganisms on nucleic acids. J Bacteriol. 1957;73:590-1. [DOI] [PMC free article] [PubMed]
- 20.MacFaddin JF. Pruebas bioquímicas para la identificación de bacterias de importancia clínica. 3ᵃ ed. Buenos Aires: Editorial Medica Panamericana; 2003. [Google Scholar]; 20. MacFaddin JF. Pruebas bioquímicas para la identificación de bacterias de importancia clínica. 3ᵃ ed. Buenos Aires: Editorial Medica Panamericana; 2003.
- 21.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]; 21. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268-81. [DOI] [PubMed]
- 22.Ajayi T, Allmond LR, Sawa T, Wiener-Kronish JP. Single-nucleotide-polymorphism mapping of the Pseudomonas aeruginosa type III secretion toxins for development of a diagnostic multiplex PCR system. J Clin Microbiol. 2003;41:3526–3531. doi: 10.1128/JCM.41.8.3526-3531.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; 22. Ajayi T, Allmond LR, Sawa T, Wiener-Kronish JP. Single-nucleotide-polymorphism mapping of the Pseudomonas aeruginosa type III secretion toxins for development of a diagnostic multiplex PCR system. J Clin Microbiol. 2003;41:3526-31. [DOI] [PMC free article] [PubMed]
- 23.Fonseca EL, Vieira VV, Cipriano R, Vicente AC. Class 1 integrons in Pseudomonas aeruginosa isolates from clinical settings in Amazon region, Brazil. FEMS Immunol Med Microbiol. 2005;44:303–309. doi: 10.1016/j.femsim.2005.01.004. [DOI] [PubMed] [Google Scholar]; 23. Fonseca EL, Vieira VV, Cipriano R, Vicente AC. Class 1 integrons in Pseudomonas aeruginosa isolates from clinical settings in Amazon region, Brazil. FEMS Immunol Med Microbiol. 2005;44:303-9. [DOI] [PubMed]
- 24.Teixeira B, Rodulfo H, Carreño N, Guzmán M, Salazar E, De Donato M. Aminoglycoside resistance genes in Pseudomonas aeruginosa isolates from Cumana, Venezuela. Rev Inst Med Trop Sao Paulo. 2016;58:13–13. doi: 10.1590/S1678-9946201658013. [DOI] [PMC free article] [PubMed] [Google Scholar]; 24. Teixeira B, Rodulfo H, Carreño N, Guzmán M, Salazar E, De Donato M. Aminoglycoside resistance genes in Pseudomonas aeruginosa isolates from Cumana, Venezuela. Rev Inst Med Trop Sao Paulo. 2016;58:13. [DOI] [PMC free article] [PubMed]
- 25.Casellas JM. Resistencia a los antibacterianos en América Latina: consecuencias para la infectología. Rev Panam Salud Publica. 2011;30:519–528. [PubMed] [Google Scholar]; 25. Casellas JM. Resistencia a los antibacterianos en América Latina: consecuencias para la infectología. Rev Panam Salud Publica. 2011;30:519-28. [PubMed]
- 26.Mayer-Hamblett N, Rosenfeld M, Gibson RL, Ramsey BW, Kulasekara HD, Retsch-Bogart GZ, et al. Pseudomonas aeruginosa in vitro phenotypes distinguish cystic fibrosis infection stages and outcomes. Am J Respir Crit Care Med. 2014;190:289–297. doi: 10.1164/rccm.201404-0681OC. [DOI] [PMC free article] [PubMed] [Google Scholar]; 26. Mayer-Hamblett N, Rosenfeld M, Gibson RL, Ramsey BW, Kulasekara HD, Retsch-Bogart GZ, et al. Pseudomonas aeruginosa in vitro phenotypes distinguish cystic fibrosis infection stages and outcomes. Am J Respir Crit Care Med. 2014;190:289-97. [DOI] [PMC free article] [PubMed]
- 27.Vega S, Dowzicky MJ. Antimicrobial susceptibility among Gram-positive and Gram-negative organisms collected from the Latin American region between 2004 and 2015 as part of the Tigecycline Evaluation and Surveillance Trial. Ann Clin Microbiol Antimicrob. 2017;16:50–50. doi: 10.1186/s12941-017-0222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; 27. Vega S, Dowzicky MJ. Antimicrobial susceptibility among Gram-positive and Gram-negative organisms collected from the Latin American region between 2004 and 2015 as part of the Tigecycline Evaluation and Surveillance Trial. Ann Clin Microbiol Antimicrob. 2017;16:50. [DOI] [PMC free article] [PubMed]
- 28.Rodríguez-Rojas A, Mena A, Martín S, Borrell N, Oliver A, Blazquez J. Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology. 2009;155:1050–1057. doi: 10.1099/mic.0.024745-0. [DOI] [PubMed] [Google Scholar]; 28. Rodríguez-Rojas A, Mena A, Martín S, Borrell N, Oliver A, Blazquez J. Inactivation of the hmgA gene of Pseudomonas aeruginosa leads to pyomelanin hyperproduction, stress resistance and increased persistence in chronic lung infection. Microbiology. 2009;155:1050-7. [DOI] [PubMed]
- 29.Holban AM, Bleotu C, Chifiriuc MC, Bezirtzoglou E, Lazar V. Role of Pseudomonas aeruginosa quorum sensing (QS) molecules on the viability and cytokine profile of human mesenchymal stem cells. Virulence. 2014;5:303–310. doi: 10.4161/viru.27571. [DOI] [PMC free article] [PubMed] [Google Scholar]; 29. Holban AM, Bleotu C, Chifiriuc MC, Bezirtzoglou E, Lazar V. Role of Pseudomonas aeruginosa quorum sensing (QS) molecules on the viability and cytokine profile of human mesenchymal stem cells. Virulence. 2014;5:303-10. [DOI] [PMC free article] [PubMed]
- 30.Mulcahy H, Charron-Mazenod L, Lewenza S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ Microbiol. 2010;12:1621–1629. doi: 10.1111/j.1462-2920.2010.02208.x. [DOI] [PubMed] [Google Scholar]; 30. Mulcahy H, Charron-Mazenod L, Lewenza, S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ Microbiol. 2010;12:1621-9. [DOI] [PubMed]
- 31.Galdino AC, Viganor L, Ziccardi M, Nunes AP, Dos Santos KR, Branquinha MH, et al. Heterogeneous production of proteases from Brazilian clinical isolates of Pseudomonas aeruginosa. Enferm Infecc Microbiol Clin. 2017;35:630–637. doi: 10.1016/j.eimc.2016.06.015. [DOI] [PubMed] [Google Scholar]; 31. Galdino AC, Viganor L, Ziccardi M, Nunes AP, Dos Santos KR, Branquinha MH, et al. Heterogeneous production of proteases from Brazilian clinical isolates of Pseudomonas aeruginosa. Enferm Infecc Microbiol Clin. 2017;35:630-7. [DOI] [PubMed]
- 32.Holban AM, Chifiriuc MC, Cotar AI, Bleotu C, Grumezescu AM, Banu O, et al. Virulence markers in Pseudomonas aeruginosa isolates from hospital acquired infections occurred in patients with underlying cardiovascular disease. Rom Biotech Lett. 2013;18:8843–8854. [Google Scholar]; 32. Holban AM, Chifiriuc MC, Cotar AI, Bleotu C, Grumezescu AM, Banu O, et al. Virulence markers in Pseudomonas aeruginosa isolates from hospital acquired infections occurred in patients with underlying cardiovascular disease. Rom Biotech Lett. 2013;18:8843-54.
- 33.Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology. 2001;147:2659–2669. doi: 10.1099/00221287-147-10-2659. [DOI] [PubMed] [Google Scholar]; 33. Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology. 2001;147:2659-69. [DOI] [PubMed]
- 34.Shaver CM, Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun. 2004;72:6969–6977. doi: 10.1128/IAI.72.12.6969-6977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; 34. Shaver CM, Hauser AR. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun. 2004;72:6969-77. [DOI] [PMC free article] [PubMed]
- 35.Perez LR, Antunes AL, Freitas AL, Barth AL. When the resistance gets clingy: Pseudomonas aeruginosa harboring metallo-β-lactamase gene shows high ability to produce biofilm. Eur J Clin Microbiol Infect Dis. 2012;31:711–714. doi: 10.1007/s10096-011-1363-2. [DOI] [PubMed] [Google Scholar]; 35. Perez LR, Antunes AL, Freitas AL, Barth AL. When the resistance gets clingy: Pseudomonas aeruginosa harboring metallo-β-lactamase gene shows high ability to produce biofilm. Eur J Clin Microbiol Infect Dis. 2012;31:711-4. [DOI] [PubMed]
- 36.Gonçalves I, Dantas RC, Ferreira ML, Batistão DW, Gontijo-Filho PP, Ribas RM. Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz J Microbiol. 2017;48:211–217. doi: 10.1016/j.bjm.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; 36. Gonçalves I, Dantas RC, Ferreira ML, Batistão DW, Gontijo-Filho PP, Ribas RM. Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz J Microbiol. 2017;48:211-7. [DOI] [PMC free article] [PubMed]
- 37.Yan H, Li L, Zong M, Alam MJ, Shinoda S, Shi L. Occurrence and characteristics of Class 1 and 2 integrons in clinical bacterial isolates from patients in south China. J Health Sci. 2010;56:442–450. [Google Scholar]; 37. Yan H, Li L, Zong M, Alam MJ, Shinoda S, Shi L. Occurrence and characteristics of Class 1 and 2 integrons in clinical bacterial isolates from patients in south China. J Health Sci. 2010;56:442-50.
- 38.Ruiz-Martínez L, López-Jiménez L, Fusté E, Vinuesa T, Martínez JP, Viñas M. Class 1 integrons in environmental and clinical isolates of Pseudomonas aeruginosa. Int J Antimicrob Agents. 2011;38:398–402. doi: 10.1016/j.ijantimicag.2011.06.016. [DOI] [PubMed] [Google Scholar]; 38. Ruiz-Martínez L, López-Jiménez L, Fusté E, Vinuesa T, Martínez JP, Viñas M. Class 1 integrons in environmental and clinical isolates of Pseudomonas aeruginosa. Int J Antimicrob Agents. 2011;38:398-402. [DOI] [PubMed]
- 39.Budak F, Kasap M, Kolayli F, Karadenizli A, Vahaboglu MH. Integron-associated resistance genes among multidrug-resistant Pseudomonas aeruginosa isolated from clinical specimens. Turk J Med Sci. 2012;42:149–156. [Google Scholar]; 39. Budak F, Kasap M, Kolayli F, Karadenizli A, Vahaboglu MH. Integron-associated resistance genes among multidrug-resistant Pseudomonas aeruginosa isolated from clinical specimens. Turk J Med Sci. 2012;42:149-56.
- 40.Odumosu BT, Adeniyi BA, Chandra R. Analysis of integrons and associated gene cassettes in clinical isolates of multidrug resistant Pseudomonas aeruginosa from Southwest Nigeria. Ann Clin Microbiol Antimicrob. 2013;12:29–29. doi: 10.1186/1476-0711-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]; 40. Odumosu BT, Adeniyi BA, Chandra R. Analysis of integrons and associated gene cassettes in clinical isolates of multidrug resistant Pseudomonas aeruginosa from Southwest Nigeria. Ann Clin Microbiol Antimicrob. 2013;12:29. [DOI] [PMC free article] [PubMed]
- 41.Lye DC, Earnest A, Ling ML, Lee TE, Yong HC, Fisher DA, et al. The impact of multidrug resistance in healthcare-associated and nosocomial Gram-negative bacteraemia on mortality and length of stay: cohort study. Clin Microbiol Infect. 2012;18:502–508. doi: 10.1111/j.1469-0691.2011.03606.x. [DOI] [PubMed] [Google Scholar]; 41. Lye DC, Earnest A, Ling ML, Lee TE, Yong HC, Fisher DA, et al. The impact of multidrug resistance in healthcare-associated and nosocomial Gram-negative bacteraemia on mortality and length of stay: cohort study. Clin Microbiol Infect. 2012;18:502-8. [DOI] [PubMed]
- 42.Valderrama SL, González PF, Caro MA, Ardila N, Ariza B, Gil F, et al. Factores de riesgo para bacteriemia por Pseudomonas aeruginosa resistente a carbapenémicos adquirida en un hospital colombiano. Biomedica. 2016;36:69–77. doi: 10.7705/biomedica.v36i2.2784. [DOI] [PubMed] [Google Scholar]; 42. Valderrama SL, González PF, Caro MA, Ardila N, Ariza B, Gil F, et al. Factores de riesgo para bacteriemia por Pseudomonas aeruginosa resistente a carbapenémicos adquirida en un hospital colombiano. Biomedica. 2016;36:69-77. [DOI] [PubMed]