Abstract
Background
Multiplex molecular panels are relentlessly replacing conventional methods for the detection of enteric pathogens from stool samples in clinical and research laboratories. Here we evaluated four commercial multiplex real-time PCR assays for the detection of Cryptosporidium hominis/parvum, Giardia duodenalis and Entamoeba histolytica.
Methods
The diagnostic performance of the Gastroenteritis/Parasite Panel I (Diagenode), the RIDAGENE Parasitic Stool Panel (R-Biopharm), the Allplex Gastrointestinal Parasite Panel 4 (Seegene) and the FTD Stool Parasites (Fast Track) real-time PCR methods was assessed against a reference panel of 126 well-characterized DNA samples including Cryptosporidium hominis (n = 29), Cryptosporidium parvum (n = 3), Giardia duodenalis (n = 47), Entamoeba histolytica (n = 3), other parasite species (n = 20), and apparently healthy subjects (n = 24).
Principal findings
Obtained diagnostic sensitivities ranged from 53–88% for Cryptosporidium hominis/parvum, and from 68–100% for G. duodenalis. The R-Biopharm method achieved the best performance for the detection of Cryptosporidium hominis/parvum both in terms of diagnostic sensitivity (87.5%) and detection limit (a 100-fold increase compared to other tests). The Fast Track method was particularly suited for the detection of G. duodenalis, achieving a 100% sensitivity and a detection limit at least 10-fold superior. Detection of E. histolytica was similarly achieved by all compared methods except Diagenode.
Conclusions
Diagnostic performance varied largely depending on the method used and the targeted pathogen species. Factors including test sensitivity/specificity, cost, patient population surveyed, laboratory workflow, and diagnostic algorithm should be carefully considered when choosing the most appropriate multiplex PCR platform.
Introduction
Enteric Giardia duodenalis, Cryptosporidium spp., and Entamoeba histolytica are the most important diarrhoea-causing protozoa globally. Infections by these parasites cause significant morbidity and mortality primarily among children living in resource-poor settings in developing countries [1], but are also a significant public health concern in developed nations [2]. Indeed, these three protozoan species account for up to 70% of the gastrointestinal parasites diagnosed every year at hospital-based microbiology laboratories in Europe [3]. Additionally, both G. duodenalis and Cryptosporidium spp. are increasingly recognized as important waterborne and foodborne pathogens all over the world [4–6].
Light microscopy stands as the preferred routine diagnosis method for enteric protozoan parasites in most clinical settings. Although this technique is labour-intensive, lacks sensitivity, and requires skilled technicians, its simplicity and low cost outweighs the above mentioned limitations and makes microscopy suited for resource-limited laboratories particularly in endemic, high-prevalence areas. However, in high-income countries where parasite prevalence rates and burden are typically low and diagnostic sensitivity become an issue, a different diagnostic approach is clearly needed [7,8]. Other pressing issues include growing costs of labour, increased sample testing, a desire for improved throughput, and optimized laboratory workflows. All together, these facts explain why microscopy is being progressively replaced by highly sensitive DNA-based assays as first-line routine diagnostic methods for intestinal parasites in many clinical settings in western countries, mainly in Europe [8,9].
In recent years a wide diversity of in-house real-time PCR (qPCR) assays have been developed for detecting enteric viral, bacterial and parasitic, diarrhoea-causing, agents, with the trend fast moving from single pathogen detection to a multiplex approach allowing simultaneous identification of multiple pathogens [10,11]. An additional advantage of this technology is that it can be easily adapted to specific diagnostic requirements (e.g. pathogen combinations) depending on the population or patient group under study. Currently, several multiplex gastrointestinal pathogen panel tests are commercially available, including fully integrated robotic systems incorporating DNA extraction, amplification, detection, and analysis directly from stool samples [9,12]. A number of these methods (e.g. BD MAX Enteric Parasite Panel, Dickinson and Company, USA; Luminex xTAG Gastrointestinal Pathogen Panel, Luminex Corporation, Canada; NanoCHIP GIP, Savyon Diagnostics Ltd, Israel) have received clearance from public health agencies like the U.S. Food and Drug Administration (USA) and are being progressively incorporated in the diagnostic algorithms of modern clinical laboratories, particularly those attending large populations of paediatric, immunocompromised, or returning traveller populations [9]. Of notice, most of the studies conducted to evaluate the diagnostic performance of these and similar methods were based on prospectively and/or retrospectively collected stool samples with a previous diagnosis by microscopy examination [13–19]. In addition, a recent inter-laboratory external quality assessment scheme has evidenced the need of harmonization of molecular-based protocols and procedures for the detection of enteric protozoa within the European Union [20].
Here we aimed to evaluate the diagnostic sensitivity and specificity of four commercial multiplex qPCR assay for the specific detection of Cryptosporidium hominis/parvum, Giardia duodenalis and Entamoeba histolytica. The study was conducted against a reference panel of DNA samples extracted from stool specimens.
Materials and methods
Ethics statement
The study design and consent procedures involved in this survey have been approved by the Research Ethics Committee of the Carlos III Health Institute under reference number CEI PI 17_2017-v3. Written informed consent was not required for this study because the stool samples used were exclusively intended for routine clinical diagnostic procedures. All samples were anonymized using a unique laboratory identifier code to guarantee the anonymity and confidentiality of the patients.
DNA reference panel
A total of 126 well-characterised DNA samples extracted and purified from stool specimens of clinically confirmed patients were obtained from a previously published study by our laboratory [21]. These included PCR-positive samples for Cryptosporidium hominis (n = 29), C. parvum (n = 3), Giardia duodenalis (n = 47), and Entamoeba histolytica (n = 3). No Cryptosporidium species other than C. hominis and C. parvum were assessed. Potential cross-reactivity was assessed against DNA samples positive for E. dispar (n = 10), Leishmania infantum (n = 2), Trypanosoma cruzi (n = 2), Toxoplasma gondii (n = 2), Ascaris lumbricoides (n = 3), Strongyloides stercoralis (n = 1), and apparently healthy subjects (n = 24) with a negative result for Cryptosporidium hominis/parvum, G. duodenalis and E. histolytica/dispar by PCR. Four aliquots of each individual DNA sample were prepared and stored at –20°C until tested with each assay to prevent degradation by freeze-thawing. The full dataset showing the characteristics of the DNA sample panel used and the diagnostic results obtained with each method is shown in S1 Table.
Multiplex real-time PCR assays
The four commercial multiplex real-time PCR (qPCR) assays compared here were the Gastroenteritis/Parasite Panel I (Diagenode, Seraing, Belgium), the RIDAGENE Parasitic Stool Panel (R-Biopharm, Darmstadt, Germany), the Allplex Gastrointestinal Parasite Panel 4 (Seegene, Seoul, Korea), and the FTD Stool Parasites (Fast Track Diagnostics, Luxembourgh). The main features of these assays, including previously reported diagnostic sensitivity and specificity values, are shown in Table 1.
Table 1. Main features and reported diagnostic performance of the four commercial multiplex real-time PCR assays compared in the present study for the detection of Cryptosporidium hominis/parvum, Giardia duodenalis, and Entamoeba histolytica from clinical DNA samples.
| Method | Manufacturer | Automated DNA extraction | Sensitivity (%) | Specificity (%) | References |
|---|---|---|---|---|---|
| Gastroenteritis/Parasite Panel I | Diagenode | No | 92–100 | 100 | [3,15] |
| RIDAGENE Parasitic Stool Panel | R-Biopharm | No | 95–100a | 99–100a | – |
| Allplex Gastrointestinal Parasite Panel 4 | Seegene | Yes | NS | NS | – |
| FTD Stool Parasites | Fast Track | No | NS | NS | [13] |
Notes: the RIDAGENE Parasitic Stool Panel (R-Biopharm) is designed to also detect Dientamoeba fragilis. The Allplex Gastrointestinal Parasite Panel 4 (Seegene) is designed to also detect Blastocystis hominis, Dientamoeba fragilis, and Cyclospora cayetanensis.
NS: not specified.
a As reported by the manufacturer.
In an attempt to normalise initial experimental conditions among assays, DNA samples (5 μL for all methods excepting the FTD Stool Parasites method, for which 10 μL were used following the manufacturer´s recommendation) were tested undiluted in a 25 μL final volume. No sample duplicates were carried out. In the case of PCR inhibition, the sample was diluted 10-fold and retested. Experiments were performed on a Corbett Rotor-Gene 6000 qPCR cycler (Qiagen Corbett, Hilden, Germany) except otherwise indicated. All four methods included appropriate negative, positive, and qPCR inhibition controls. Multiplex qPCR protocols were conducted in strict accordance with the manufacturer´s instructions with few modifications as follows:
RIDAGENE Parasitic Stool Panel (R-Biopharm). Amplification reactions were conducted on an Mx3005P qPCR instrument (Agilent Technologies, Waldbronn, Germany) provided by the manufacturer as the method was not yet fully validated for the Rotor-Gene 6000 qPCR system at the time of the analyses.
Allplex Gastrointestinal Parasite Panel 4 (Seegene). This method includes an automated DNA extraction system that was not used in the present study for comparative reasons. Amplification reactions were carried out on the CFX96 qPCR instrument (Bio-Rad Laboratories, CA, USA) provided by the manufacturer.
Simulated mixed infections
To mimic natural co-infections involving double (Cryptosporidium + Giardia) and triple (Cryptosporidium + Giardia + E. histolytica or E. dispar) pathogen combinations a total of 10 different simulated mixes were artificially generated. Each combination was prepared by mixing equal amounts of individual DNA samples from the reference panel (S1 Table). Simulated mixed infections were also used to indirectly assess diagnostic sensitivities and specificities of each method.
Relative detection limit
Ten-fold serial dilutions of individual Cryptosporidium (undiluted to 10−3) and Giardia (10−1 to 10−4) positive DNA samples from the reference panel were tested in each compared method for detection limit determination based on cycle threshold (Ct) values obtained during qPCR. This assessment was not conducted for E. histolytica due to the low number of samples available and the high Ct values associated to them.
Data analyses
The Shapiro-Wilk's test was used to assess the normality of distribution of the Ct values obtained in Cryptosporidium- and Giardia-positive samples during qPCR analyses with each method evaluated. Once normality was demonstrated, an analysis of variance (ANOVA) for simultaneous comparison of methods was conducted. A probability (P) value < 0.05 was considered evidence of statistical significance. Statistical analyses were performed using the software package SPSS version 21.0 (IBM Corp., USA).
Results
The diagnostic performance results of the four multiplex qPCR methods compared here are summarized in Table 2. The R-Biopharm method was the most sensitive (87.5%) assay for the detection of Cryptosporidium hominis/parvum, with the Fast Track assay performing poorly (53.1%). All four methods detected the three C. parvum DNA samples assessed. When tested undiluted, Cryptosporidium-positive DNA samples generated an elevated number of inhibitory reactions mostly resolved when re-tested in a 1:10 dilution. This was particularly true for the R-Biopharm and the Diagenode methods (of note, the former manufacturer specifically recommends diluting faecal suspensions 1:3 prior to DNA extraction). This issue was observed neither for Giardia- nor Entamoeba-positive DNA samples. One-way ANOVA test results showed significant (P < 0.05) variations in the distribution of obtained Ct values among the four multiplex qPCR methods assessed here, although no one-by-one direct comparison of methods was attempted due to unsurmountable differences in equipment (e.g. thermocycler used) features and assay (e.g. initial volume of DNA tested) procedures. In line with the findings mentioned above, the R-Biopharm method produced lower mean Ct values either in unpaired (mean: 30.1; 95% CI: 28.6–31.5) (Fig 1A) and paired (mean: 28.2; 95% CI: 26.9–29.6) (Fig 1B) Cryptosporidium-positive DNA samples than the other three methods tested.
Table 2. Diagnostic performance of the four commercial multiplex real-time PCR assays compared in the present study for the detection of Cryptosporidium hominis/parvum, Giardia duodenalis, and Entamoeba histolytica using a reference panel of well-characterized DNA samples (n = 126) from clinical specimens and healthy subjects.
| Diagenode | R-Biopharm | Seegene | Fast Track | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Protozoan pathogen | Samples (n) | + | % | + | % | + | % | + | % |
| Cryptosporidium spp. | 32 | 24 | 75.0 | 28 | 87.5 | 25 | 78.1 | 17 | 53.1 |
| C. hominis | 29 | 21 | 72.4 | 25 | 86.2 | 22 | 75.9 | 14 | 48.3 |
| C. parvum | 3 | 3 | 100 | 3 | 100 | 3 | 100 | 3 | 100 |
| Giardia duodenalis | 47 | 32 | 68.1 | 37 | 78.7 | 43 | 91.5 | 47 | 100 |
| Entamoeba histolytica | 3 | 0 | 0.0 | 2 | 66.7 | 3 | 100 | 2 | 66.7 |
| Other parasite species | 20 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Entamoeba dispar | 10 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Leishmania infantum | 2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Trypanosoma cruzi | 2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Toxoplasma gondii | 2 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Ascaris lumbricoides | 3 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Strongyloides stercoralis | 1 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 |
| Healthy subjects | 24 | 0a | 0.0 | 1b | 4.2 | 1c | 4.2 | ND | ND |
Notes: ´+´ refers to positive detection, ND stands for ´not determined´.
a Only 13/24 samples tested.
b Sample positive to G. duodenalis with a Ct value = 37.73.
c Sample positive to Cryptosporidium hominis/parvum. with a Ct value = 40.20.
Fig 1. Dotplot showing the distribution of Ct values for Cryptosporidium-positive DNA samples obtained with each of the four commercial multiplex real-time PCR assays evaluated in this study.

Mean values and standard deviation ranges for each group are represented by large and short horizontal bars, respectively. Unpaired (panel A) and paired (panel B) groups were represented for comparative purposes.
Regarding G. duodenalis, the Fast Track method showed a 100% sensitivity, closely followed by the Seegene assay (91.5%). The latter method also detected all three E. histolytica positive samples, whereas the R-Biopharm and Fast Track produced a positive result for two out of these three samples. When Ct values for G. duodenalis-positive samples were plotted, the Fast Track method generated the lowest figures either in unpaired (mean: 26.6; 95% CI: 25.5–27.8) (Fig 2A) and paired (mean: 25.2; 95% CI: 23.7–26.8) (Fig 2B) sample groups. Very similar results were obtained with the Diagenode assay. No obvious association between Giardia-negative samples by the Diagenode, R-Biopharm, and Seegene methods and the magnitude of the qPCR Ct values at initial diagnosis (ranging from 20.7 to 32.8) was observed (S1 Table).
Fig 2. Dotplot showing the distribution of Ct values for G. duodenalis-positive DNA samples obtained with each of the four commercial multiplex real-time PCR assays evaluated in this study.
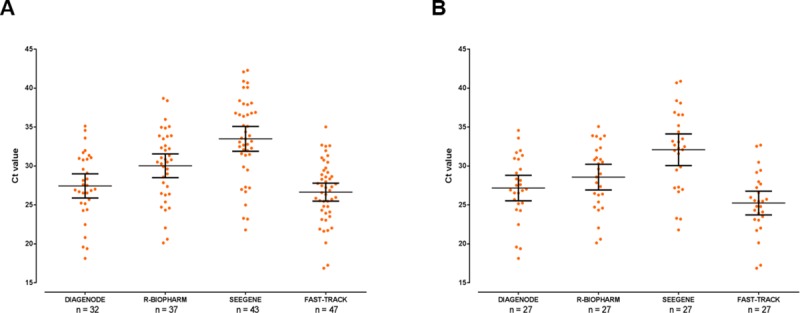
Mean values and standard deviation ranges for each group are represented by large and short horizontal bars, respectively. Unpaired (panel A) and paired (panel B) groups were represented for comparative purposes.
No non-specific amplification/cross-reactivity was seen in any of the four multiplex qPCR methods when DNA samples from other parasitic/commensal species (E. dispar, L. infantum, T. cruzi, T. gondii, A. lumbricoides and S. stercoralis) were tested. All 24 DNA samples from apparently healthy subjects tested negative, excepting one sample that was G. duodenalis-positive by R-Biopharm (Ct = 37.7) and a sample that was Cryptosporidium-positive by Seegene (Ct = 40.2). A number of previously unnoticed co-infections were observed within the panels of selected DNA samples positive for Cryptosporidium hominis/parvum, G. duodenalis and E. histolytica/dispar, most of them associated with high (≥ 35) Ct values (S1 Table). This finding is most likely due to the comparatively higher detection sensitivity of the multiplex qPCR methods with those of the conventional PCRs used in the primary diagnosis of the samples, although the occurrence of false-positive results could not be completely ruled out.
Testing of artificially-prepared mixed infections showed that, overall, the R-Biopharm method delivered the most consistent diagnostic results (Table 3). This method detected Cryptosporidium hominis/parvum in all simulated mixed infections except one, with lower Ct values than those obtained by the other three assays. Both the R-Biopharm and the Diagenode methods were also the only procedures able to detect G. duodenalis in all 10 mixed combinations assessed, even considering that the Fast Track system typically generated lower Ct values for this particular species. Additionally, detection of E. histolytica was only achieved by the R-Biopharm and the Fast Track methods in two out of the three samples tested, the latter assay showing the lowest Ct values.
Table 3. Diagnostic performance of the four commercial multiplex real-time PCR assays compared in the present study for the detection of Cryptosporidium hominis, Giardia duodenalis, and Entamoeba histolytica in artificially prepared DNA samples mimicking mixed infections.
Cycle-threshold values are shown.
| Cryptosporidium hominis | Giardia duodenalis | Entamoeba histolytica | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Combination | Diagenode | R-Biopharm | Seegene | Fast Track | Diagenode | R-Biopharm | Seegene | Fast Track | Diagenode | R-Biopharm | Seegene | Fast Track |
| 1: Cr + G + Eh | 28.9 | 28.9 | 30.3 | 33.5 | 32.0 | 38.7 | 39.1 | 30.2 | Neg. | 41.8 | Neg. | 30.4 |
| 2: Cr + G + Eh | 38.6 | 31.3 | 34.4 | Neg. | 20.9 | 25.7 | 25.0 | 19.4 | Neg. | 38.7 | Neg. | 33.4 |
| 3: Cr + G + Eh | Neg. | Neg. | Neg. | Neg. | 24.9 | 29.5 | 28.2 | 23.7 | Neg. | Neg. | Neg. | Neg. |
| 4: Cr + G + Ed | 37.6 | 29.1 | 33.4 | Neg. | 28.4 | 31.8 | 33.9 | Neg. | – | – | – | – |
| 5: Cr + G | 26.7 | 25.9 | 27.0 | 34.2 | 23.8 | 27.8 | 27.7 | 29.4 | – | – | – | – |
| 6: Cr + G | 28.0 | 27.5 | 28.6 | Neg. | 25.5 | 29.8 | 29.8 | 19.4 | – | – | – | – |
| 7: Cr + G | 40.9 | 30.5 | 34.3 | Neg. | 31.5 | 32.6 | Neg. | 23.7 | – | – | – | – |
| 8: Cr + G | Neg. | 37.7 | Neg. | Neg. | 26.8 | 29.6 | 30.1 | 26.8 | – | – | – | – |
| 9: Cr + G | Neg. | 29.7 | 32.6 | Neg. | 32.5 | 39.1 | 38.3 | 21.9 | – | – | – | – |
| 10: Cr + G | 39.9 | 30.6 | 34.0 | Neg. | 27.6 | 32.3 | 31.9 | 24.9 | – | – | – | – |
Notes: ´Neg.´ refers to a negative detection in the presence of parasitic DNA; ´–´ refers to a negative detection in the absence of parasitic DNA; Cr, Cryptosporidium hominis; G, Giardia duodenalis; Eh, Entamoeba histolytica; Ed, Entamoeba dispar.
Confirming previous diagnostic findings, relative detection limit analyses based on serial dilutions of one sample positive for Cryptosporidium and one sample positive for G. duodenalis demonstrated that the R-Biopharm method was two order of magnitude more sensitive for the detection of Cryptosporidium than the other three assays evaluated (Fig 3A). For the detection of G. duodenalis, the best performance was achieved by the Fast Track method, at least one order of magnitude more sensitive than that of the other assays (Fig 3B).
Fig 3.
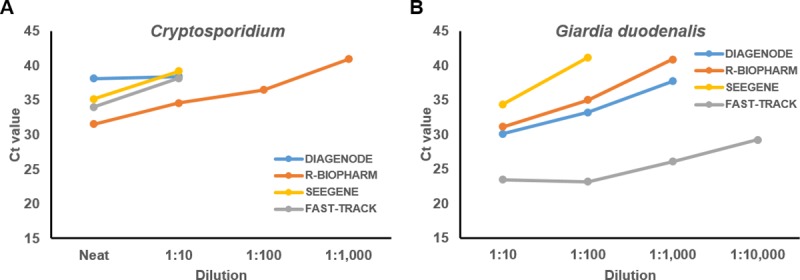
Relative detection limit analyses of the four commercial multiplex real-time PCR assays evaluated in this comparative study using serial dilutions of Cryptosporidium-positive (panel A) and Giardia-positive (panel B) individual DNA samples.
Discussion
In this comparative study we evaluated four commercial multiplex qPCR methods for the identification of the three most clinically relevant protozoan enteric parasites, namely Cryptosporidium spp., Giardia duodenalis, and Entamoeba histolytica. The diagnostic performance of both the Diagenode and the Fast Track assays have been individually assessed and compared to that obtained by microscopy in previous studies [3,13,15]. However, this is the first comprehensive survey reporting the diagnostic performance of the R-Biopharm and Seegene methods. Another significant methodological contribution of this work is that, in an attempt to normalize starting experimental conditions and strengthen the robustness of the obtained data, analyses were conducted against a reference panel of well-characterize DNA samples. Our results evidenced marked detection differences among tests depending on the targeted parasite species considered, with diagnostic sensitivities typically ranging from 53–88% for Cryptosporidium spp., and from 68–100% for G. duodenalis. The diagnostic sensitivity of these methods for the detection of E. histolytica was not fully assessed due to the insufficient number of positive samples to this pathogen available. Despite the highly variable sensitivity values observed, all four methods did not cross-react with any of the DNA samples from other protozoan or helminthic parasite species tested, suggesting a specificity near 100%. However, the exact extent of this statement should be confirmed in future studies including a larger panel of DNA samples from other potentially cross-reacting enteric pathogen and commensal species including Cyclospora cayetanensis, Entamoeba coli, Endolimax nana, Blastocystis sp., and Dientamoeba fragilis.
Regarding Cryptosporidium detection, the R-Biopharm method achieved the best performance in terms of diagnostic sensitivity (87.5%) and detection limit (two order of magnitude superior to that of the other tests). In contrast, the Diagenode (diagnostic sensitivity: 75.0%) and the Fast Track (diagnostic sensitivity: 53.1%) assays performed comparatively worse. This finding is hardly surprising when considering that both methods at best equalled the diagnostic performance of conventional microscopy when attempting to detect Cryptosporidium in clinical samples [3,13]. However, the Fast Track method was much better suited for the detection of G. duodenalis than the other three assays evaluated here, achieving a 100% sensitivity and a detection limit at least one order of magnitude superior to that of the other three methods. This is well in agreement with previous findings in patients with diarrhoea from community or hospital sources, where the Fast Track assay detected 83% more G. duodenalis-infected cases than microscopy examination [13]. In contrast, the Diagenode method achieved the lowest diagnostic sensitivity (68.1%) for the identification of G. duodenalis of all four assays compared here. This finding is also in line with previous data demonstrating that this technique performs only moderately better than conventional microscopy during routine examination of clinical stool samples [3,15]. Regarding E. histolytica, all methods tested, excepting Diagenode, performed similarly well for the detection of E. histolytica, with the Fast Track assay providing the highest diagnostic sensitivity values. However, these figures should be interpreted with caution due to the comparatively low number of E. histolytica DNA samples available for the analyses. Considering all the above, our data demonstrated large differences in the diagnostic performance of the compared methods. In addition to the test sensitivity/specificity evaluated here, other factors including cost, patient population surveyed, laboratory workflow, and diagnostic algorithm should be carefully considered when choosing the most appropriate multiplex PCR platform for the simultaneous detection of Cryptosporidium spp., G. duodenalis, and E. histolytica in human stool samples.
Our study has some limitations. First, the reference panel contained a restricted number of DNA samples, particularly those positive to E. histolytica and to other intestinal pathogen and commensal species. This fact may have hampered the accuracy of some of the obtained results. In order to minimize this issue, statistical analyses were based on the Shapiro-Wilk's test as non-parametric test, a method particularly suited for small to moderate samples sizes. Other potential confounding factors are the design of the primer sets used to amplify the selected targets and the use of different qPCR instruments depending on the method evaluated.
Accurate and fast detection of clinically relevant enteric protozoa is highly desirable for prompt, adequate, and effective treatment [10]. In this context, multiplex qPCR platforms have been reliably shown to increase the positivity rates of protozoan pathogens compared to conventional methods [13,22–24]. Even more importantly, multiplex molecular panels including viral, bacterial, and parasitic pathogens enable the syndromic testing of patients with gastrointestinal symptoms in a cost-effective manner [25]. Taken together, these facts explain why these methodologies are being increasingly used in diagnostic laboratories and reference centres. However, the choice of the most suitable assay should be based on a careful evaluation of variable including optimal laboratory workflow and diagnostic algorithm, test performance, and patient population to be tested.
Supporting information
(XLSX)
Acknowledgments
The Authors thank Dr Ismael Thuissard for assisting with statistical analyses. The preliminary results of this survey have been previously presented at the 6th International Giardia & Cryptosporidium Conference held at La Havana (Cuba) on April 26–28, 2017.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was funded by the European University of Madrid (Spain), under project 2015/UEM21 (MM). Additional funding was also provided by the Health Institute Carlos III (ISCIII), Ministry of Economy and Competitiveness (Spain), under projects CP12/03081 (DC) and PI13/01106 (IF).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kotloff KL. The burden and etiology of diarrheal illness in developing countries. Pediatr Clin North Am. 2017; 64: 799–814. 10.1016/j.pcl.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 2.Fletcher SM, Stark D, Harkness J, Ellis J. Enteric protozoa in the developed world: a public health perspective. Clin Microbiol Rev. 2012; 25: 420–449. 10.1128/CMR.05038-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laude A, Valot S, Desoubeaux G, Argy N, Nourrisson C, Pomares C, et al. Is real-time PCR-based diagnosis similar in performance to routine parasitological examination for the identification of Giardia intestinalis, Cryptosporidium parvum/Cryptosporidium hominis and Entamoeba histolytica from stool samples? Evaluation of a new commercial multiplex PCR assay and literature review. Clin Microbiol Infect. 2016; 22: 190.e1-190–e8.. [DOI] [PubMed] [Google Scholar]
- 4.Food and Agriculture Organization of the United Nations/World Health Organization. Multicriteria-based ranking for risk management of food-borne parasites. Microbiological Risk Assessment Series, Rome 2014; 23: 302.
- 5.Efstratiou A, Ongerth JE, Karanis P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2011–2016. Water Res. 2017; 114: 14–22. 10.1016/j.watres.2017.01.036 [DOI] [PubMed] [Google Scholar]
- 6.Rosado-García FM, Guerrero-Flórez M, Karanis G, Hinojosa MDC, Karanis P. Water-borne protozoa parasites: The Latin American perspective. Int J Hyg Environ Health. 2017; 220: 783–798. 10.1016/j.ijheh.2017.03.008 [DOI] [PubMed] [Google Scholar]
- 7.Verweij JJ. Application of PCR-based methods for diagnosis of intestinal parasitic infections in the clinical laboratory. Parasitology 2014; 141: 1863–1872. 10.1017/S0031182014000419 [DOI] [PubMed] [Google Scholar]
- 8.van Lieshout L, Roestenberg M. Clinical consequences of new diagnostic tools for intestinal parasites. Clin Microbiol Infect. 2015; 21: 520–528. 10.1016/j.cmi.2015.03.015 [DOI] [PubMed] [Google Scholar]
- 9.Ryan U, Paparini A, Oskam C. New technologies for detection of enteric parasites. Trends Parasitol. 2017; 33: 532–546. 10.1016/j.pt.2017.03.005 [DOI] [PubMed] [Google Scholar]
- 10.van Lieshout L, Verweij JJ. Newer diagnostic approaches to intestinal protozoa. Curr Opin Infect Dis. 2010; 23: 488–493. 10.1097/QCO.0b013e32833de0eb [DOI] [PubMed] [Google Scholar]
- 11.Verweij JJ, Stensvold CR. Molecular testing for clinical diagnosis and epidemiological investigations of intestinal parasitic infections. Clin Microbiol Rev. 2014; 27: 371–418. 10.1128/CMR.00122-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binnicker MJ. Multiplex molecular panels for diagnosis of gastrointestinal infection: performance, result interpretation, and cost-effectiveness. J Clin Microbiol. 2015; 53: 3723–3728. 10.1128/JCM.02103-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McAuliffe GN, Anderson TP, Stevens M, Adams J, Coleman R, Mahagamasekera P, et al. Systematic application of multiplex PCR enhances the detection of bacteria, parasites, and viruses in stool samples. J Infect. 2013; 67: 122–129. 10.1016/j.jinf.2013.04.009 [DOI] [PubMed] [Google Scholar]
- 14.Mengelle C, Mansuy JM, Prere MF, Grouteau E, Claudet I, Kamar N, et al. Simultaneous detection of gastrointestinal pathogens with a multiplex Luminex-based molecular assay in stool samples from diarrhoeic patients. Clin Microbiol Infect. 2013; 19: E458–465. 10.1111/1469-0691.12255 [DOI] [PubMed] [Google Scholar]
- 15.Van Lint P, Rossen JW, Vermeiren S, Ver Elst K, Weekx S, Van Schaeren J, et al. Detection of Giardia lamblia, Cryptosporidium spp. and Entamoeba histolytica in clinical stool samples by using multiplex real-time PCR after automated DNA isolation. Acta Clin Belg. 2013; 68: 188–192. 10.2143/ACB.3170 [DOI] [PubMed] [Google Scholar]
- 16.Stark D, Roberts T, Ellis JT, Marriott D, Harkness J. Evaluation of the EasyScreen enteric parasite detection kit for the detection of Blastocystis spp., Cryptosporidium spp., Dientamoeba fragilis, Entamoeba complex, and Giardia intestinalis from clinical stool samples. Diagn Microbiol Infect Dis. 2014; 78: 149–152. 10.1016/j.diagmicrobio.2013.10.013 [DOI] [PubMed] [Google Scholar]
- 17.Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, et al. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol. 2015; 53: 915–925. 10.1128/JCM.02674-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ken Dror S, Pavlotzky E, Barak M. Evaluation of the NanoCHIP Gastrointestinal Panel (GIP) test for simultaneous detection of parasitic and bacterial enteric pathogens in fecal specimens. PLoS One. 2016; 11: e0159440 10.1371/journal.pone.0159440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madison-Antenucci S, Relich RF, Doyle L, Espina N, Fuller D, Karchmer T, et al. Multicenter evaluation of BD Max enteric parasite real-time PCR assay for detection of Giardia duodenalis, Cryptosporidium hominis, Cryptosporidium parvum, and Entamoeba histolytica. J Clin Microbiol. 2016; 54: 2681–2688. 10.1128/JCM.00765-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuurs TA, Koelewijn R, Brienen EAT, Kortbeek T, Mank TG, Mulder B, et al. , Harmonization of PCR-based detection of intestinal pathogens: experiences from the Dutch external quality assessment scheme on molecular diagnosis of protozoa in stool samples. Clin Chem Lab Med. 2018; 56(10): 1722–1727. 10.1515/cclm-2017-1057 [DOI] [PubMed] [Google Scholar]
- 21.Paulos S, Mateo M, de Lucio A, Hernández-de Mingo M, Bailo B, Saugar JM, et al. Evaluation of five commercial methods for the extraction and purification of DNA from human faecal samples for downstream molecular detection of the enteric protozoan parasites Cryptosporidium spp., Giardia duodenalis, and Entamoeba spp. J Microbiol Methods. 2016; 127: 68–73. 10.1016/j.mimet.2016.05.020 [DOI] [PubMed] [Google Scholar]
- 22.ten Hove R, Schuurman T, Kooistra M, Möller L, van Lieshout L, Verweij JJ. Detection of diarrhoea-causing protozoa in general practice patients in The Netherlands by multiplex real-time PCR. Clin Microbiol Infect. 2007; 13: 1001–1007. 10.1111/j.1469-0691.2007.01788.x [DOI] [PubMed] [Google Scholar]
- 23.Friesen J, Fuhrmann J, Kietzmann H, Tannich E, Müller M, Ignatius R. Evaluation of the Roche LightMix Gastro parasites multiplex PCR assay detecting Giardia duodenalis, Entamoeba histolytica, cryptosporidia, Dientamoeba fragilis, and Blastocystis hominis. Clin Microbiol Infect. In press. 10.1016/j.cmi.2018.03.025 [DOI] [PubMed] [Google Scholar]
- 24.Morio F, Valot S, Laude A, Desoubeaux G, Argy N, Nourrisson C, et al. Evaluation of a new multiplex PCR assay (ParaGENIE G-Amoeba Real-Time PCR kit) targeting Giardia intestinalis, Entamoeba histolytica and Entamoeba dispar/Entamoeba moshkovskii from stool specimens: evidence for the limited performances of microscopy-based approach for amoeba species identification. Clin Microbiol Infect. 2018; 24: 1205–1209. 10.1016/j.cmi.2018.02.007 [DOI] [PubMed] [Google Scholar]
- 25.Goldenberg SD, Bacelar M, Brazier P, Bisnauthsing K, Edgeworth JD. A cost benefit analysis of the Luminex xTAG Gastrointestinal Pathogen Panel for detection of infectious gastroenteritis in hospitalised patients. J Infect. 2015; 70: 504–511. 10.1016/j.jinf.2014.11.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.


