Abstract
In the last two decades, there has been growing interest in mRNA-based technology for the development of prophylactic vaccines against infectious diseases. Technological advancements in RNA biology, chemistry, stability, and delivery systems have accelerated the development of fully synthetic mRNA vaccines. Potent, long-lasting, and safe immune responses observed in animal models, as well as encouraging data from early human clinical trials, make mRNA-based vaccination an attractive alternative to conventional vaccine approaches. Thanks to these data, together with the potential for generic, low-cost manufacturing processes and the completely synthetic nature, the prospects for mRNA vaccines are very promising. In addition, mRNA vaccines have the potential to streamline vaccine discovery and development, and facilitate a rapid response to emerging infectious diseases. In this review, we overview the unique attributes of mRNA vaccine approaches, review the data of mRNA vaccines against infectious diseases, discuss the current challenges, and highlight perspectives about the future of this promising technology.
Keywords: infectious diseases, RNA-based vaccine, self-amplifying mRNA, synthetic vaccine, vaccine on demand
Main Text
Vaccination is one of the most effective public health interventions to prevent and control infectious diseases. Since the first known clinical trial conducted with cowpox in 1796,1 vaccines have resulted in the eradication of many infectious diseases, and today ∼30 diseases worldwide can be prevented by vaccination.2, 3 Over the last two centuries, vaccinology has evolved from Pasteur’s principles of pathogen “isolation, inactivation, and injection” to a rational vaccine design based on genetic engineering, immunology, structural biology, and systems biology.4, 5
Despite the advancement in conventional vaccine approaches, challenges remain, and new vaccine technologies are necessary. The list of unmet medical needs includes vaccines against pathogens causing chronic infections that can evade adaptive immune responses or require cellular immune responses, and against emerging diseases, such as Zika, Ebola, Nipah, and pandemic influenza.6 Epidemic outbreaks caused by virus infections are emerging or reemerging almost every year and in all cases are characterized by unpredictability, high morbidity, exponential spread, and substantial social impact. A “vaccine on demand” approach that enables rapid development, large-scale production, and distribution of the vaccine would be desirable. Such an approach may not be compatible with conventional vaccine technology platforms that often require complex and lengthy research and development processes.
Nucleic acid-based vaccines, including viral vectors, plasmid DNA (pDNA), and mRNA, will be suitable for rapid response applications because of their ability to induce broadly protective immune responses and their potential of being produced by rapid and flexible manufacturing processes. Because the manufacturing processes of nucleic acid-based vaccines are independent of encoded antigens, different vaccines based on the same nucleic acid platform can utilize the same production and purification methods, as well as manufacturing facilities, requiring adaptations only in validation methods, therefore cutting both costs and time for vaccine production.7 Upon vaccination, nucleic acid-based vaccines mimic a viral infection to express vaccine antigens in situ, resulting in induction of both humoral and cytotoxic T cell responses.8 This advantage is critical for the elimination of intracellular pathogens or infections in which potent humoral and cellular immune responses are required to achieve protective efficacy. In addition, nucleic acid vaccines have intrinsic adjuvant properties because of their recognition by specific pattern recognition receptors (PRRs) and elicitation of innate immune responses, which are critical for maturation of dendritic cells (DCs) to enhance the induction of subsequent adaptive immune responses.9, 10 pDNA and viral vectors have been evaluated as vaccine platforms in human clinical trials and demonstrated to be safe and immunogenic. However, pDNA delivery into the nucleus of target cells is rather inefficient, and viral vectors can induce interfering vector-specific immune responses against viral structural proteins especially upon boosting. A growing interest in and research activities into mRNA-based vaccines have started to emerge in the past decade.11
mRNA-based vaccines have several advantages over pDNA and viral vector-based vaccines. mRNA vaccines do not generate infectious particles or integrate in the genome of the host cells. They can be delivered for antigen expression in situ without the need to cross the nuclear membrane barrier for protein expression, and can express complex antigens without packaging constraints.12 mRNA vaccines can be produced rapidly, possibly within days of obtaining gene sequence information, using completely synthetic manufacturing processes.13 The platform is versatile and amenable to multiple targets, and thereby ideal for rapid responses to newly emerging pathogens.14, 15, 16
The seminal work from Wolff et al.17 in 1990 provided the first successful example of in vitro-transcribed mRNA-expressing reporter proteins in the muscle after injection, which was followed by the studies of Martinon et al.,18 Conry et al.,19 and Hoerr et al.20 demonstrating that vaccinations with viral or cancer antigen-encoding mRNA elicited antigen-specific immune responses. Despite the early promising results, it was only in the last decade that technological advancements in RNA biology, chemistry, and delivery systems have enabled the efficient and stable manufacture of mRNA products.
The recent Ebola and Zika outbreaks have demonstrated how quickly emerging infectious diseases can spread, and underline the crucial need for having a rapid response vaccine on demand platform technology in place.6 mRNA vaccines have all of the attributes of a vaccine on demand, even though clinical validation remains to be confirmed.16
In this manuscript, we review the current state of mRNA vaccine approaches against infectious diseases, summarize the latest preclinical and clinical proof-of-concept data, and provide perspectives on the future of this promising new technology.
mRNA Vaccines: Types, Biology, and Latest Advancement
The concept for the development of an mRNA vaccine is rather straightforward. Once the antigen of choice from the pathogen target is identified, the gene is sequenced, synthesized, and cloned into the DNA template plasmid. mRNA is transcribed in vitro, and the vaccine is delivered into the subject. The mRNA vaccine utilizes the host cell machinery for in vivo translation of mRNA into the corresponding antigen, thereby mimicking a viral infection to elicit potent humoral and cellular immune responses. The final cellular location of the antigen is determined by the signal peptide and transmembrane domain. This can be intrinsic to the natural protein sequence or engineered to direct the protein to the desired cellular compartment.21 Therefore, the antigen can be expressed as intracellular, secreted, or membrane-bound protein. Importantly, given its fully synthetic nature, virtually any sequence could be designed in silico, synthesized, delivered as an mRNA vaccine, and tested rapidly in vivo in animal models. For example, tagging antigen sequences with targeting sequences to major histocompatibility complex (MHC) class II compartments, with MHC class I trafficking signals, or with immunodominant helper CD4 T cell epitopes could amplify antigen presentation efficiency and enhance cellular immune responses. Arrays of antigen sequences can also be designed and rapidly tested to generate vaccines with efficient leader sequences, optimal codon usage, enhanced neutralization capacity, or reduced undesired cross-reactivity, as recently shown by Zika mRNA vaccines developed by Richner et al.15
Due to the ability of the host’s innate system to sense and respond to RNA sequences of viral origin (reviewed in Chen et al.9 and Vabret et al.22), mRNA vaccines induce robust innate responses, including production of chemokines and cytokines such as interleukin-12 (IL-12) and tumor necrosis factor (TNF) at the injection site.23, 24, 25 These are factors crucial to successful induction of effective adaptive responses against the encoded antigen.26 Currently, two forms of mRNA vaccines have been developed: conventional mRNA encoding the antigen of interest flanked by 5′ and 3′ UTRs, and self-amplifying mRNA derived from the genome of positive-stranded RNA viruses. Self-amplifying mRNA encodes not only the antigen but also the viral replication machinery required for intracellular RNA amplification leading to high levels of antigen expression (Figure 1). Unique attributes of each mRNA technology, as well as the roadblocks that need to be overcome for advancement, are summarized in Table 1.
Figure 1.
Schematic Representation of mRNA Vaccines and Mechanism of Antigen Expression
Conventional mRNA carries the coding sequence of the antigen of interest (GOI) flanked by 5′ and 3′ UTRs, a terminal 5′ cap structure, and a 3′ poly(A) tail. Once delivered into the cell and released from the endosome into the cytoplasm, the mRNA is translated immediately. The self-amplifying mRNA is often derived from the genome of positive-sense single-stranded RNA viruses, such as alphaviruses. It encodes both the antigen of interest and viral nonstructural proteins (nsPs) required for intracellular RNA amplification and high levels of antigen expression. The self-amplifying mRNA can direct its self-amplification to generate RNA intermediates and many copies of antigen-encoding subgenomic mRNA, producing high levels of the encoded antigen. Both conventional mRNA and self-amplifying mRNA vaccines require a delivery system for cell uptake, usually by endocytosis, which is followed by unloading of mRNA cargo from the endosome into the cytosol, where translation and protein processing for MHC presentation occur. Once delivered in the cell, the mRNA is almost immediately sensed by pattern recognition receptors (PRRs) in the endosome and in the cytoplasm. PRRs such as Toll-like receptors TLR3, TLR7, and TLR8 are localized in the endosome, and cytosolic sensors such as RIG-I, MDA5, PKR, and OAS also recognize double-stranded and single-stranded RNAs in the cytoplasm. GOI, gene of interest; MHC, major histocompatibility complex; nsPs, nonstructural proteins.
Table 1.
Advantages and Disadvantages of Conventional or Self-Amplify mRNA Vaccines
| Vaccines | Advantages | Disadvantages |
|---|---|---|
| Conventional mRNA Self-amplifying mRNA |
synthetic production; egg and cell free | concerns with instability |
| rapid and scalable production compared with other vaccine platforms (e.g., subunit proteins, viral vectors) | limited immunogenicity data in humans | |
| noninfectious, non-integrating, and naturally degraded | potential toxic effect of free extracellular mRNA164, 165 | |
| expression in situ to produce antigens with structure unaltered by in vitro manufacturing process | inflammation due to enhanced type I IFN activation | |
| expression in situ to stimulate innate immune response, enhancing broad T and B cell immune responses | efficient delivery required to deliver and launch self-amplifying mRNA | |
| – | efficient delivery required to deliver and/or provide adjuvanting effect for conventional mRNA | |
| – | unproven toxicity profiles of delivery system components | |
| Conventional mRNA | shorter RNA length compared with self-amplifying mRNA | potential toxicity from modified nucleotides |
| applicable to nucleoside base modification | shorter duration of expression | |
| direct antigen expression from mRNA | higher effective RNA doses | |
| no risk of anti-vector immunity | – | |
| Self-amplifying mRNA | enhanced and prolonged antigen expression | potential elevated inflammation due to self-amplification |
| lower effective RNA doses, potentially resulting in better safety | longer RNA length, may lead to more challenging production of high-quality RNA compared with conventional mRNA | |
| intrinsic adjuvant effect | interaction between nsPs and host factors yet to be addressed | |
| potential apoptosis of vaccine-carrying cells due to vaccine self-amplification, leading to enhanced cross-presentation | – | |
| option for single-vector delivery of multiple or complex antigens | – |
IFN, interferon; nsPs, nonstructural proteins.
Cell-free Production and Purification of mRNA Vaccines
Both conventional mRNA and self-amplifying mRNA vaccine approaches share essential elements of an eukaryotic mRNA: a cap structure [m7Gp3N (N, any nucleotide)], a 5′ UTR, an open reading frame (ORF), a 3′ UTR, and a tail of 40–120 adenosine residues [poly(A) tail].27 Both types of mRNAs are produced in a cell-free system, using an enzymatic transcription reaction from a linearized pDNA template.12, 28 Manufacturing of mRNA vaccines against different disease targets requires the replacement of the sequence encoding the target antigen, without affecting the overall physicochemical characteristics of the RNA molecule (Figure 2). The first step in RNA production is the construction of a pDNA that contains a promoter sequence with high binding affinity to a DNA-dependent RNA polymerase (e.g., T7, SP6, or T3) and the sequence encoding the specific mRNA vaccine. The pDNA is linearized with a restriction enzyme and used as a template for an in vitro transcription (IVT) reaction using a DNA-dependent RNA polymerase. The enzyme moves along the template, elongating the RNA transcript until it runs off the end of the template. The template DNA is then degraded by incubation with DNase, and a cap [m7Gp3N] is enzymatically added to the 5′ end of the mRNA.29 Alternatively, a synthetic cap analog can be added during the IVT reaction in a single-step procedure.30, 31 The presence of a 5′ cap structure is crucial for efficient translation in vivo and protects mRNA from intracellular nuclease digestion.32, 33 Once synthesis is complete, mRNA is purified to remove reaction components, including enzymes, residual template DNA, truncated transcripts, or aberrant double-stranded transcripts. A highly purified RNA drug substance is critical for the potency of an mRNA vaccine, because contaminants can activate non-specific or undesirable innate sensors.34, 35, 36, 37 Karikó et al.,36, 38 and more recently Foster et al.,39 have demonstrated that purification of mRNA from double-stranded RNA (dsRNA) contaminants can enhance in vivo translation and reduce innate immune activation, and is critical for the success of RNA-based gene therapy and CAR T cell therapy applications. After purification, mRNA is exchanged into a final storage buffer or formulated with the delivery system for use.
Figure 2.
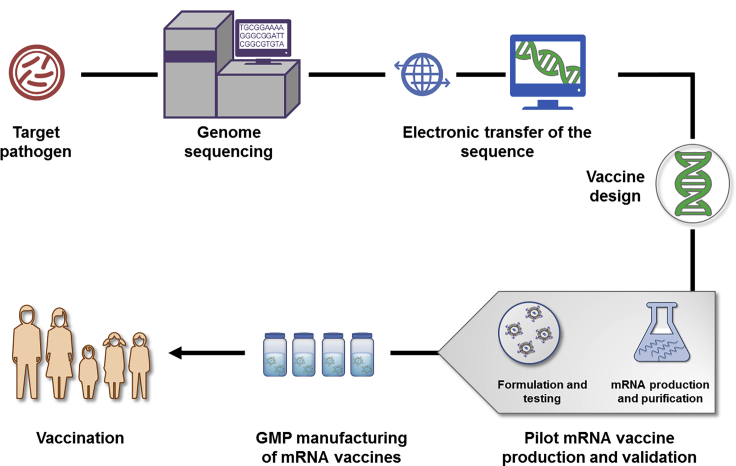
Schematic Illustration of mRNA Vaccine Production
Once a pathogen is identified or an outbreak declared, the genome of the pathogen and antigen(s) are determined, if not already available, by the combined sequencing, bioinformatics, and computational approach. Candidate vaccine antigen sequences are deposited electronically and available globally for in silico design of mRNA vaccines, followed by construction of plasmid DNA template by molecular cloning or synthesis. Pilot vaccine batches are generated in a cell-free system by in vitro transcription and capping of the mRNA, purification, and formulation with the delivery system. In-process analytic and potency tests are performed to assess the quality of pilot mRNA vaccine batches. If needed, pilot mRNA vaccine batches can be further tested in the immunogenicity and/or disease animal model. The final mRNA vaccine is scaled up and manufactured through a generic process with minimal modifications, rapidly tested, and dispatched for use. GMP, Good Manufacturing Practice.
These approaches are appropriate to produce almost any mRNA sequence, with low batch-to-batch variability, potentially saving time and reducing costs compared with other vaccine platforms. The mRNA drug substance and formulated drug product undergo testing and in-process analytics to assess identity, appearance, content, integrity, residual DNA, endotoxin contamination, and sterility.40 A potency test is used to verify the ability of the mRNA to be translated into a desired protein product after delivery into target cells. Depending on the specific mRNA construct, the above-described procedure may be slightly modified to accommodate modified nucleosides, capping strategies, or purification protocols.
All components needed for mRNA production are available at the Good Manufacturing Practice (GMP) grade, and industrial-scale production facilities designed to produce up to 30 million doses of RNA-based products per year are being established.41, 42
Conventional mRNA Vaccines
A conventional mRNA vaccine carries only the coding sequence of the antigen of interest flanked by regulatory regions (Figure 1). The major advantages of the conventional mRNA vaccine approach are the simplicity and relatively small size of the RNA molecule. In the simplest form, the stability and activity of the conventional mRNA in vivo is limited, because of propensity of cells to limit duration of expression. However, optimization of RNA structural elements and formulation can increase antigen expression and durability.43 The cap (often a cap 1 structure15, 44, 45, 46), UTRs, and the poly(A) tail are crucial for stability of the mRNA molecule, accessibility to ribosomes, and interaction with the translation machinery,47, 48, 49, 50 therefore representing targets for optimization (reviewed in Ross,43 Lundstrom,51 and Sahin et al.52). Additionally, codon usage can have a beneficial impact on protein translation, and replacing rare codons with frequently used synonymous codons is a common practice to enhance protein expression from DNA, RNA, and viral vector vaccines. However, this approach has been recently reviewed, suggesting that codon optimization may not necessarily increase protein production for mRNA therapeutics.53, 54
Unlike proteins, mRNA vaccines need to be expressed in situ to induce an antigen-specific immune response; therefore, understanding the magnitude and durability of antigen expression after mRNA injection is important for vaccine optimization. Results by Vogel et al.55 showed that naked, unmodified mRNA induced protein expression in vivo at 12–24 h after intramuscular (i.m.) injection and lasted for at least 6 days. Kinetics and magnitude of translation can be influenced by mRNA formulation, route of administration, nucleoside modifications, and sequence optimization.45, 56, 57
Nucleoside base modification has been used to produce so-called modified mRNA. In this review, the term modified mRNA or unmodified mRNA refers to conventional mRNA with or without chemically modified nucleotides, respectively. Nucleoside base modification, often coupled with chromatographic purification to remove dsRNA contaminants, is a fast-emerging approach to improve mRNA potency by reducing activation of PRRs. Several RNA structures, such as dsRNA produced by annealing of complementary RNAs or short RNA stem loops, as byproducts of IVT, or by viral RNA replication intermediates, are immunostimulatory.9, 58, 59, 60 Although activation of innate immunity is required for vaccine application, its improper or excessive activation can also interfere with antigen expression and adaptive immunity.61, 62 For example, Karikó et al.57 and others44, 63, 64, 65, 66, 67, 68, 69 have shown that pseudouridine-containing mRNAs reduce activation of Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I), protein kinase R (PKR), and 2′-5′-oligoadenylate synthetase (OAS), while increasing translational activity, resistance to RNase L-mediated degradation, and in vivo stability. The highest levels of protein production and immunogenicity are observed when nucleoside-modified mRNA is also purified by high-performance liquid chromatography (HPLC) or fast protein liquid chromatography (FPLC), where aberrant double-stranded transcripts are removed.38, 46 This enhanced protective immune response has been demonstrated in mice, ferrets, and non-human primates (NHPs) against influenza70, 71 and Zika viruses,14, 15 among other targets.
In addition to modified nucleosides, sequence optimization is also used to ensure robust protein expression and immunogenicity. Thess et al.45 have shown that an mRNA sequence enriched in G:C content and carrying optimized UTRs is superior to a nucleoside-modified counterpart in vitro and in vivo. Sequence-optimized mRNA delivered with lipid nanoparticles (LNPs) also elicited robust functional antibody titers against rabies and influenza antigens in NHPs.72 Conversely, Pardi et al.46 reported that 1-methylpseudouridine-modified mRNA delivered by LNPs produced 20 times more protein than its sequence-optimized but nucleoside-unmodified counterpart after intradermal (i.d.) injection in mice. Codon-optimized, FPLC-purified, unmodified mRNA encoding an influenza antigen induced weaker CD4+ T cell, T follicular helper (Tfh), and germinal center (GC) responses than the 1-methylpseudouridine-modified counterpart. The discrepancies among these findings are not yet clear and might be due to the difference in routes of administration, sequence optimization algorithms, modified nucleosides, and experimental conditions. Nonetheless, these data show that both approaches are superior to unmodified mRNA. Both sequence-optimized RNA and nucleoside-modified RNA delivered by LNPs induce a robust activation of innate immunity, rapid infiltration of neutrophils, monocytes, and DCs to the injection site and the draining lymph nodes (dLNs) in mice and NHPs, as well as upregulation of key co-stimulatory receptors (CD80 and CD86) and type I interferon (IFN)-inducible genes, including Mx1 and CXCL10.23, 72
In addition to mRNA modification, increasing adjuvant properties in formulation might also be advantageous for vaccination, as shown by the RNActive technology. This vaccine contains a naked, unmodified, sequence-optimized mRNA, whose potency depends on its carrier that consists of a non-coding RNA complexed with protamine, a cationic protein activating TLR7.25, 73, 74, 75, 76, 77, 78, 79 Loomis et al.80 also explored the effects of adjuvants and activation of local innate responses on mRNA vaccination by coupling naked 1-methylpseudouridine-modified mRNA to small-molecule TLR2 and TLR7 agonists. By using ovalbumin as the model vaccine antigen, they observed increased innate, cellular, and humoral immune responses in mice compared with naked mRNA.
In summary, the most advanced conventional mRNA vaccines under evaluation in preclinical and clinical trials consist of unmodified but sequence-optimized mRNA, nucleoside-modified mRNA, or mRNA adjuvanted with protamine-complexed non-coding RNA.
Self-Amplifying mRNA Vaccines
Self-amplifying mRNA vaccines are commonly based on the engineered RNA genome of positive-sense single-stranded RNA viruses, such as alphaviruses, flaviviruses, and picornaviruses.81, 82 In all cases, the mRNA mimics the replicative features of positive-sense single-stranded RNA viruses with the goal of increasing the duration and magnitude of the expression, as well as subsequent immunogenicity of the encoded antigen. The best-studied self-amplified mRNA molecules are derived from alphavirus genomes, such as those of the Sindbis virus (SINV), Semliki Forest virus (SFV), and Venezuelan equine encephalitis viruses (VEEVs) (reviewed in Ljungberg and Liljeström83 and Atkins et al.84). Negative-sense single-stranded RNA viruses, such as rhabdoviruses and measles viruses, can also be utilized for the development of RNA-based vaccines (reviewed in Mühlebach85 and Humphreys and Sebastian86). However, the negative-sense RNA genomes need to be rescued by reverse genetics where cell culture-based systems are required, and therefore are not the focus of this review.
Alphavirus RNA genomes, typically around 10–11 kb, encode four non-structural proteins, nsP1–4, which are translated directly from the genomic RNA and together form the RNA-dependent RNA polymerase (RDRP) complex.87, 88 RDRP interacts with host factors to form replication factories on the cytoplasmic side of modified membrane structures to synthesize the negative-sense RNA intermediates.89, 90, 91 This results in the generation of new viral genomes (up to 2 × 105 copies/cell)92 and, depending on temperature, of an even larger amount of capped subgenomic mRNA encoding structural proteins.93, 94
Self-amplifying mRNA replicons are created by replacing viral structural genes with the antigen gene of interest, which, upon delivery in the cytoplasm of target cells, are capable of RNA amplification to express high levels of the antigen of interest. Because these replicons lack endogenous viral structural genes, they are unlikely to produce infectious virions or virus-like vesicles at the injection site in subjects after vaccination, hence greatly reducing safety concerns associated with a live attenuated virus vaccine. The full-length self-amplifying mRNA can be readily produced by IVT from a pDNA template using the process previously described and delivered as either viral replicon particles (VRPs) when structural genes are provided in trans, or as synthetically formulated RNA. Alternatively, self-amplifying RNA can also be produced directly in target cells, for example, by delivering a pDNA expressing the alphavirus-derived RDRP complex and the antigen of interest into such target cells, which has been reviewed elsewhere.82 The efficacy of alphavirus VRPs as vaccine in preclinical models and humans has been previously described and has been largely attributed to the production of high levels of correctly processed heterologous proteins and to their ability to deliver antigen to a variety of cell types, including antigen-presenting cells.95, 96, 97, 98, 99, 100, 101, 102, 103 Activation of DCs upon VRPs infection results in a wave of cytokine secretion and subsequently a robust adjuvanting effect, which markedly amplifies the magnitude of vaccine-elicited adaptive immune responses.104
In the 1990s, Zhou et al.105 were the first to report that the self-amplifying SFV-derived IVT RNA could be used to elicit an immune response against a heterologous antigen. In 2001, the same group reported that as little as 10 μg of naked SFV-derived IVT RNA expressing respiratory syncytial virus (RSV) fusion protein (F), influenza virus hemagglutinin (HA), or Louping ill virus pre-membrane and envelope (prME) were sufficient to induce significant antibody responses and partial protection against the lethal challenge of RSV, influenza, or Louping ill virus in vaccinated mice, respectively.106 An important step for self-amplifying mRNA vaccine development was the use of complexing agents to protect and formulate IVT RNA. Geall et al.107 reported that encapsulation of a 9-kb self-amplifying mRNA with LNP considerably improved the durability and magnitude of in vivo antigen production compared with naked RNA, and as little as 0.1 μg of LNP-formulated self-amplifying mRNA elicited potent T and B cell responses in mice. Similar results were later reported using a cationic nanoemulsion (CNE) to formulate RNA.108
Self-amplifying mRNA vaccines enable high levels of antigen production from low doses of vaccine for an extended duration, likely because of their self-amplification in cells. By using a secreted alkaline phosphatase (SEAP) as reporter gene, Brito et al.108 showed that antigen expression from self-amplifying mRNA peaked at 3 days and remained measurable at more than 14 days postadministration in NHPs. In a recent study, Vogel et al.55 compared antigen expression kinetics, immune responses, and protective efficacy after injection of self-amplifying mRNA or unmodified conventional mRNA in mice. They showed that the self-amplifying mRNA vaccine expressed much higher levels of the protein for a longer period of time and induced more potent immune responses than the conventional mRNA vaccine. Similar superior kinetics and potency have been previously described for CNE-formulated self-amplifying mRNA.108 Leyman et al.109 recently compared luciferase expression of self-amplifying mRNA, nucleoside-modified conventional mRNA, and unmodified conventional mRNA after i.d. electroporation in pigs. Both conventional mRNAs reached their peak expression 1 day after electroporation, followed by a 2.5- to 5-fold decrease at day 6. Self-amplifying mRNA-driven expression in the skin of pigs was similar to that of conventional mRNA at day 1, then slightly increased at day 6, and persisted until day 12. The high levels of antigen expressed from the self-amplifying mRNA vaccine may play an important role in its potency, as reported for other nucleic acid vaccines. Using VRP-delivered alphavirus self-amplifying mRNA, Kamrud et al.110 demonstrated that increased antigen expression from VRP-delivered self-amplifying mRNA vaccines correlated with increased protection in immunized mice after challenge.
When used in an influenza challenge model, both naked self-amplifying mRNA and conventional mRNA protected against infection with the homologous virus.55 However, 64-fold less self-amplifying mRNA (1.25 μg of self-amplifying mRNA versus 80 μg of mRNA) was required to generate a similar level of protection. Considering that self-amplifying mRNA is much larger than mRNA (9.3 versus 2.2 kb), this suggested that 270-fold fewer self-amplifying mRNA molecules are required to achieve the similar vaccine efficacy. The dose-sparing quality of self-amplifying mRNA vaccines may facilitate scale-up and manufacturing large numbers of vaccine doses. On the other hand, the production process and stability of the longer self-amplifying mRNA molecule could be more challenging than conventional mRNA products and require process improvement.
Another factor, in addition to the magnitude and duration of antigen expression, which may also contribute to the increased immune response elicited by self-amplifying mRNA, is the dsRNA amplification intermediates that trigger host sensing machinery, activate innate immunity, and confer an adjuvant effect. Indeed, co-administration of LNP-formulated self-amplifying mRNA, regardless of the encoded antigen (GFP, matrix protein 1, or nucleoprotein), could act as an adjuvant for the subunit monovalent inactivated influenza vaccine to elicit H1-specific effector CD8+ T cells, increase the magnitude of H1-specific CD4+ T cells, and shift their polarization toward a Th1 phenotype.111
However, the role of innate immunity, such as type I IFN induction, in self-amplifying mRNA vaccination is complex. Delivery of self-amplifying mRNA by LNPs has strong local pro-inflammatory effects with activation of innate immune, anti-viral, and inflammatory signaling pathways.24 Early and robust induction of type I IFN and IFN-stimulated responses was detected at the site of injection in mice within a few hours. However, although balanced IFN activation is beneficial for generating potent immune responses, elevated type I IFN early responses impair self-amplifying mRNA expression and potency.112, 113 Consistently, antigen expression and immunogenicity were both enhanced in the absence of type I IFN signaling, as previously shown with conventional mRNA.24, 114, 115 The negative impact from excessive IFN activation could also derive at the level of T cells. Although type I IFN can determine the differentiation of antigen-primed CD8+ T cells into cytolytic effectors, they may also promote T cell exhaustion.62 Whether type I IFNs inhibit or stimulate the CD8+ T cell response to mRNA vaccines might depend on the timing and intensity of type I IFNs induced.116
The usage of modified nucleosides to silence self-amplifying mRNA recognition by innate immune sensors is not an option, because it could impair RDRP-mediated mRNA amplification in target cells, and the effect of modified nucleoside would also be lost after the first round of amplification. Beissert et al.117 have demonstrated that antigen expression from self-amplifying mRNA can be significantly improved in vitro and in vivo by co-transferring mRNAs encoding IFN and PKR inhibitory proteins, such as viral protein B18R from vaccinia virus. These data are consistent with previous results showing an increase in translation from an alphavirus self-amplifying mRNA co-expressing a dominant-negative PKR118 or co-delivered with a recombinant vaccinia B18R protein.119 Other potential strategies to enhance the potency of self-amplifying mRNA vaccines include sequence modification to generate IFN-insensitive RNA, novel formulations to limit IFN induction upon vaccine delivery, and inclusion of small-molecule modulators to target various components of the IFN signaling cascade.61 Finally, potential interactions between the encoded nsPs and host factors, as well as their immunogenicity, merit additional investigation. So far, no evidence has been generated in preclinical models that immune responses against the nsPs interfere with the immune response induced by subsequent booster doses,120 but this needs to be confirmed with synthetic self-amplifying mRNA vaccines.
Another attribute of the self-amplifying mRNA vaccine platform is its ability to encode multiple antigens in the same replicon. This could allow the development of a vaccine expressing both a target antigen and an immuno-modulatory biological molecule (e.g., biological adjuvant) for enhanced potency, vaccine encoding both B and T cell antigens, single combo vaccine simultaneously targeting multiple pathogens, or vaccine targeting multi-subunit complex antigen. For certain pathogens, neutralizing antibody responses are directed toward conformational epitopes resulting from protein complex formation, rather than the individual components. Using viral delivery of alphavirus replicon, Wen et al.121 showed that a self-amplifying mRNA replicon can express five full-length subunits (gH, gL, UL128, UL130, and UL131A) of the human cytomegalovirus (HCMV) pentameric complex, form the properly folded pentameric complex, and elicit potent and broadly neutralizing antibodies in mice. Given no packaging constraints in a synthetic delivery system, the coding capacity of self-amplifying mRNA might be pushed even further. Proof-of-concept studies of multi-protein antigen delivery with self-amplifying mRNA formulated with LNPs were reported by Brito et al.12 using the HCMV gH/gL protein complex and Magini et al.111 using influenza virus nucleoprotein (NP) and M1 proteins. The self-amplifying mRNA vaccine encoding gH/gL induced antibodies and CD4+ T cells at levels similar to those induced by an MF59-adjuvanted gH/gL protein complex. Immunization of mice with very low doses of self-amplifying mRNA vaccine (0.1–0.2 μg) encoding influenza M1 and/or NP elicited potent antigen-specific T cell responses and protection from viral challenge. Such an approach might be particularly useful for the generation of a cross-protective influenza single-vector vaccine able to induce both T cell- and B cell-mediated immunity. Indeed, a swine fever virus-derived self-amplifying mRNA vaccine encoding influenza HA and NP was shown to induce antigen-specific T and B cell responses in mice.122, 123
In summary, the self-amplifying mRNA vaccine platform is a versatile technology that can be used to develop single- or multi-antigen vaccines, with the benefit of low effective dosage thanks to its high antigen expression and strong intrinsic adjuvant effect. Insights into the mechanisms controlling the interaction of self-amplifying mRNA replicons with host innate immunity and subsequent modulation of antigen-specific immune responses will enable the rational design of improved self-amplifying mRNA vaccines.
Synthetic Delivery Vehicles for mRNA Vaccines
mRNA vaccines need a delivery system to reach their full potential, because naked RNA not only is prone to nuclease degradation, it is also too large and negatively charged to passively cross the cell membrane.124 It has been speculated that the cellular uptake rate of naked mRNA is less than 1 in 10,000 molecules.52 As such, the mRNA delivery field has focused on the discovery and development of methods and materials that can transport RNA into cells, benefiting from extensive prior research into non-viral delivery of small interfering RNA (siRNA), recently reviewed elsewhere.125, 126 There are several major delivery vehicles currently under investigation, some encapsulating mRNA molecules into particles and others using positively charged polymers to bind to RNA through charge interactions. Several groups are also attempting to create particles that behave like viruses able to shield mRNA from endo-, 5′ exo-, and 3′ exo-nucleases, deliver mRNA efficiently into cells, and achieve a sufficient level of the encoded protein.127, 128, 129 In all cases, the vehicle must pass through the target cell membrane, and after cell uptake, usually by endocytosis, it must escape the endosome and unload its mRNA cargo into the cytosol, where translation occurs. For some mRNAs, the delivery system may also influence the quantity and quality of local gene expression patterns, innate immune stimulation, and can provide a synergistic adjuvant effect.24, 130
One of the most promising and commonly used systems for non-viral delivery of RNA is LNPs. LNPs are formulated using precise molar ratios of phospholipids that enhance fusogenicity and endosomal escape, cationic-ionizable amino lipids that condense with nucleic acid at low pH, poly(ethylene) glycol (PEG) lipids that provide steric stabilization of the formulation before use, and cholesterols that enable vesicle stability both in vivo and in vials.131 Several amino lipids have been developed for siRNA delivery over the past two decades, and the first LNP-delivered therapeutic siRNA was approved by the US Food and Drug Administration (FDA) in 2018.132 Leveraging the knowledge from siRNA delivery, many laboratories have used LNP delivery of self-amplifying mRNA and conventional mRNAs against various pathogens, such as rabies, Zika, and influenza viruses, and demonstrated rapid and robust immune responses in mice, ferrets, and NHPs.13, 14, 15, 70, 71, 72
LNPs enter cells by exploiting membrane-derived endocytic pathways. Endosomal processing and release of the encapsulated genetic material into the cytosol is the rate-limiting step of delivery and a major area of research for formulation improvement. Gilleron et al.133 have estimated that only less than 2% of LNP-delivered siRNAs taken up by cells are released into the cytosol, and endosomal escape occurs only during a brief phase of endolysosomal maturation. Using CRISPR-based genetic disruption of different stages of the lysosomal pathway, Patel et al.135 demonstrated that, in addition to fusogenicity, late endosome-lysosome formation in the endocytic process is essential during LNP trafficking for mRNA release in the cytosol.134 LNP-mediated mRNA delivery can be modulated through incorporation of bioactive lipids modulating cellular signaling or enriching in endo-lysosomal compartments. The link between endosomal escape and LNP potency has been further explored by Sabnis et al.,136 who demonstrated that the improved delivery efficiency of their novel amino lipid, designed by a rational chemistry approach, was due to improved fusogenicity and endosomal escape. The use of high-throughput screening of nanocarriers in vitro toward the development of mRNA vaccine formulation with enhanced endosomal escape is hindered by the discrepancies observed between in vivo and in vitro data. Recent encouraging results using skeletal muscle cells grown on an in vitro hydrogel may provide a better prediction of mRNA uptake, endosomal entry, release, and protein expression in vivo upon i.m. injection.137
Although not as clinically advanced as lipid-based systems for mRNA delivery, polymers have shown considerable potential in mRNA vaccine development. One example is the dendrimer-based nanoparticle system developed by Chahal et al.,138, 139 which comprises molecules of high amine density with organizing branching structures condensing around mRNA to form monodisperse spheres. After a single immunization, the dendrimer-encapsulated self-amplifying mRNA vaccine elicited CD8+ T cell and antibody responses that fully protected against lethal challenge by Ebola virus, H1N1 influenza, and Zika viruses in mice, with 40 μg as the optimal RNA dosage. This dendrimer technology was used to generate a multiplexed vaccine consisting of six self-amplifying mRNA replicons simultaneously formulated in the same nanoparticle, which protected mice against lethal Toxoplasma gondii challenge. Although promising, this technology is still at the early stage and needs further optimization, given the high RNA amount required.
The capability to generate viable, potent single-dose vaccines against multiple antigens or even pathogens may have broad utility and reduce the number and frequency of vaccinations. Future research should also evaluate the lack or presence of potential interference in activation of target-specific immune responses among co-delivered mRNA vaccines encoding antigens from multiple pathogens.
As an alternative to a single-vial formulation, such as LNP, a two-vial approach where the delivery formulation can be manufactured and stockpiled separate from the target mRNA has been developed by Brito et al.108 In this context, an emulsion-based delivery vehicle was used to bind self-amplifying mRNA to the nanoparticle surface, prior to administration. The CNE-delivered self-amplifying mRNA was shown to elicit potent immune responses after two or three immunizations in mice, rats, rabbits, and NHPs with a variety of viral, bacterial, and recently, parasite antigens.21, 108, 140, 141, 142 This two-vial approach allows for the delivery vehicle to be stockpiled and mixed with any target RNA, designed, and produced whenever an outbreak occurs.
There are additional aspects of delivery to consider for the efficacy of mRNA vaccines. The route of administration is critical for RNA uptake and expression. Vaccines are typically administered via i.d., i.m., or subcutaneous (s.c.) injections, in order to target tissues, such as the skin and skeletal muscle, which are densely populated by DCs. For LNP-formulated nucleoside-modified mRNA vaccine, the greatest magnitude and duration of antigen expression were observed after i.m. or i.d. injection in mice.56 However, although both i.d. and i.m. administration of LNP-formulated nucleoside-modified mRNA influenza H10 vaccine induced protective titers in NHPs, the responses occurred more rapidly after i.d. vaccination.143 DC targeting is another active area of research for mRNA delivery. In an effort to improve targeting of DCs via s.c. injection, Englezou et al.144 explored and optimized cationic lipids and lipoplex formation to increase the uptake, release in the cytoplasm, and translation of a self-amplifying mRNA molecule in DCs. The lipoplex, optimized for increased in vitro translation in DCs, elicited pro-inflammatory cytokines, humoral responses, and T cellular responses against the self-amplifying mRNA-encoded influenza NP antigen after s.c. vaccination in mice and in an adoptive transfer model. Although extensive studies have been performed for ex vivo mRNA loading of DCs to generate cell-mediated immunity against cancer or for therapeutic vaccination (reviewed in Benteyn et al.145 and Gornati et al.146), in vivo targeting to DCs of synthetic mRNA vaccines against infectious diseases is still in the early phase. Formulation design, optimization, and combination with adjuvants or surface molecules to target DCs after immunization might increase the potency of synthetic mRNA vaccines, given the central role of DCs in processing antigens and initiating antigen-specific adaptive immune responses.
Thermal stability of vaccines can pose a major logistical problem for stockpiling and distribution, particularly in countries that lack infrastructure to maintain the cold chain. Jones et al.147 reported that a Kunjin virus-derived self-amplifying mRNA freeze-dried in trehalose was stable for 10 months when stored at 4°C–6°C, indicating the feasibility of using lyophilization to stabilize RNA during storage. In addition, a lyophilized protamine-complexed mRNA vaccine was shown to retain its full biological activity even after exposure to thermal stress conditions for several weeks and up to 36 months at 5°C–25°C.73, 75 Similarly, oscillating temperatures between +4°C and +56°C for 20 cycles, to simulate interruptions of the cold chain during transport, did not impact the protective efficacy of a lyophilized mRNA vaccine in a lethal challenge mouse model.148
The development of a formulation enabling potent single-dose mRNA vaccines that are thermostable, can be stockpiled, and can simultaneously target multiple antigens and pathogens will have broad utility for a range of diseases, reduce the number and frequency of vaccinations, and alleviate healthcare worker burden.
mRNA Vaccines in the Prevention of Infectious Diseases: From Preclinical Proof of Concept to Initial Clinical Data
Early clinical evaluation of mRNA vaccines mostly addressed cancer immunotherapy, often with no validated benchmarks to compare.149 These studies have been pivotal to prove that large-scale GMP production of mRNA is feasible, and mRNA vaccines have a favorable safety profile. However, the use of mRNA as a therapeutic drug or in cancer vaccination is not the focus of this review and has been extensively reviewed elsewhere.52, 150, 151, 152
In contrast with cancer, where the lack of conventional vaccines makes it difficult to weigh the effectiveness of RNA as a platform, infectious diseases offer a portfolio of traditional vaccines to benchmark. To this end, considerable preclinical research (Tables 2 and 3) and early clinical trials (Table 4) on infectious diseases have been conducted.
Table 2.
Summary of Representative Preclinical Studies of Conventional mRNA Vaccines for Prevention of Infectious Diseases
| Antigen (Pathogen) | mRNA and Delivery | Route | Elicited Responses | Models Tested | References |
|---|---|---|---|---|---|
| HA (influenza virus) | sequence-optimized protamine-RNA | i.d. | humoral, cellular, protection | mice, ferrets, pigs | 73 |
| GP (rabies virus) | sequence-optimized protamine | i.d. | humoral, cellular, protection | mice, pigs | 76, 148 |
| HA (influenza virus) | sequence-optimized LNP | i.m. | humoral, cellular, innate | mice, NHPs | 72 |
| HA (influenza virus) | nucleoside-modified LNP | i.d., i.m. | humoral, cellular, innate, protection | mice, ferrets, NHPs | 23, 46, 70, 71, 143 |
| prM-E (Zika virus) | nucleoside-modified LNP | i.d., i.m. | humoral, protection | mice, NHPs | 14, 161, 166 |
| PC, gB, pp65 (HCMV) | nucleoside-modified LNP | i.m. | humoral, cellular | mice, NHPs | 163 |
| GP (Ebola virus) | nucleoside-modified LNP | i.m. | humoral, protection | guinea pigs | 167 |
| Env (HIV) | nucleoside-modified LNP | i.d. | humoral, cellular | mice, NHPs | 46 |
| NP (influenza virus) | unmodified liposome | s.c. | cellular | mice | 18 |
| Gag (HIV) | unmodified cationic nanomicelles | s.c. | humoral | mice | 168 |
Env, envelope; gB, glycoprotein B; GP, glycoprotein; HA, hemagglutinin; HCMV, human cytomegalovirus; i.d., intradermal; i.m., intramuscular; i.v., intravenous; LNP, lipid nanoparticle; NHP, nonhuman primate; PC, pentameric complex; prM-E, pre-membrane and envelope; s.c., subcutaneous.
Table 3.
Summary of Representative Preclinical Studies of Self-Amplifying mRNA Vaccines for Prevention of Infectious Diseases
| Antigen (Pathogen) | Delivery | Route | Elicited Responses | Models Tested | References |
|---|---|---|---|---|---|
| HA (influenza virus) | CNE, LNP, MDNP | i.m. | humoral, cellular, protection | mice, ferrets | 13, 138, 142 |
| gp140 (HIV) | CNE | i.m. | humoral, cellular | mice, rabbits, NHPs | 108, 140 |
| gB, pp65-IE1 (HCMV) | CNE | i.m. | humoral, cellular | NHPs | 108 |
| SLOdm, BP-2a (streptococci) | CNE | i.m. | humoral, protection | mice | 21 |
| PMIF (malaria) | CNE | i.m. | humoral, cellular, protection | mice | 141 |
| F (RSV) | CNE, LNP | i.m. | humoral, cellular, protection | mice, cotton rats | 107, 108 |
| GP (rabies), gH/gL (HCMV) | LNP | i.m. | humoral | mice | 12 |
| NP, M1 (influenza virus) | LNP | i.m. | humoral, cellular, protection | mice | 111 |
| prM-E (Louping ill virus), F (RSV), HA (influenza virus) | naked | i.m. | humoral, cellular, partial protection | mice | 106 |
| HA, NP (influenza virus) | PEI, chitosan | s.c. | humoral, cellular | mice, rabbits | 122, 123 |
| prM-E (Zika virus) | MDNP, NLC | i.m. | humoral | mice, guinea pigs | 139, 162 |
| GP (Ebola virus) | MDNP | i.m. | humoral, cellular, protection | mice | 138 |
| Six antigensa (Toxoplasma gondii) | MDNP | i.m. | protection | mice | 138 |
BP-2a, group B Streptococcus pilus 2a backbone protein; CNE, cationic nanoemulsion; F, fusion protein; gB, glycoprotein B; GP, glycoprotein; HA, hemagglutinin; HCMV, human cytomegalovirus; i.d., intradermal; i.m., intramuscular; i.v., intravenous; LNP, lipid nanoparticle; M1, matrix protein 1; MDNP, modified dendrimer nanoparticle; NHP, nonhuman primate; NLC, nanostructured lipid carrier; NP, nucleoprotein; PC, pentameric complex; PEI, polyethylenimine; PMIF, plasmodium macrophage migration inhibitory factor; prM-E, pre-membrane and envelope; Ref. References; RSV, respiratory syncytial virus; s.c., subcutaneous; SLOdm, double-mutated group A Streptococcus Streptolysin-O.
AMA1, GRA6, ROP2A, ROP18, SAG1, and SAG2A from Toxoplasma gondii.
Table 4.
mRNA Vaccines for Infectious Diseases in Clinical Trials
| Sponsor | Indication | mRNA Vaccine: Delivery | Trial No. (ClinicalTrials.gov) | Stage | Status and References |
|---|---|---|---|---|---|
| CureVac | rabies | CV7201 (sequence-optimized mRNA: protamine-RNA) | NCT02241135 | phase I | completed75 PCD: February 2018 |
| CureVac | rabies | CV7202 (sequence-optimized mRNA: LNPs) | NCT03713086 | phase I | recruiting160 PCD: December 2019 |
| Moderna | influenza H10N8 | mRNA-1440 (nucleoside-modified mRNA: LNPs) | NCT03076385 | phase I | completed71 PCD: October 2018 |
| Moderna | influenza H7N9 | mRNA-1851 (nucleoside-modified mRNA: LNPs) | NCT03345043 | phase I | active PCD: September 2018 |
| Moderna | hMPV/HPIV3 | mRNA-1653 (nucleoside-modified mRNA: LNPs) | NCT03392389 | phase I | active PCD: July 2019 |
| Moderna | Zika | mRNA-1325 (nucleoside-modified mRNA: LNPs) | NCT03014089 | phase I/II | active PCD: September 2018 |
| Moderna | HCMV | mRNA-1647 and mRNA-1443 (nucleoside-modified mRNA: LNPs) | NCT03382405 | phase I | recruiting PCD: February 2020 |
| Moderna | chikungunya | mRNA-1388 (nucleoside-modified mRNA: LNPs) | NCT03325075 | phase I | active PCD: March 2019 |
The table summarizes clinical trials that evaluate vaccination by direct injection of mRNA vaccines registered at ClinicalTrials.gov as of January 8, 2018. Clinical evaluation of vaccination with dendritic cells loaded with antigen-expressing mRNAs is reviewed elsewhere.169 HCMV, human cytomegalovirus; hMPV, human metapneumovirus; HPIV3, human parainfluenza virus type 3; LNP, lipid nanoparticle; Moderna, Moderna Therapeutics; PCD, estimated primary completion date; Ref., References.
mRNA Vaccines Targeting Influenza Virus Infection
mRNA vaccines against influenza virus are among the most extensively studied because of the availability of tools to measure T and B cell responses, the ease of testing efficacy in small-animal models, and the benefits that an mRNA-based influenza vaccine could bring. Production of conventional, FDA-approved vaccines against new influenza pandemic viruses could take at least 6 months, leaving the population unprotected during this period.153 By comparison, Hekele et al.13 have demonstrated that once the genetic sequence of the target influenza HA antigen is known, a self-amplifying mRNA vaccine can be generated within a very short period of time. In 2013, during an outbreak of a deadly strain of H7N9 influenza in China, the HA gene was cloned in a self-amplifying mRNA vaccine pDNA template, and the self-amplifying mRNA vaccine was produced within 8 days after publication of the HA gene sequence.
The first study demonstrating immunogenicity of an mRNA-encoding influenza NP in mice was published in 1993.18 However, only in 2012 was the first complete protection from influenza virus after mRNA vaccination demonstrated.73 Petsch et al.73 showed that i.d. injection with unmodified conventional mRNA encoding various influenza virus antigens combined with a protamine-complexed RNA adjuvant was immunogenic in mice, ferrets, and domestic pigs, matching the immune responses conferred from a licensed inactivated virus vaccine. This pivotal work was followed by several publications (reviewed in Scorza and Pardi154) showing that mRNA-based influenza vaccines, either as conventional mRNA or self-amplifying mRNA formulated in LNP or CNE, induce broadly protective T and B cell immune responses. Chitosan, polyethylenimine (PEI), and dendrimer-based formulations of self-amplifying mRNA HA have also been shown to be effective.55, 123, 138
Lutz et al.72 demonstrated for the first time that a single i.m. injection with 10 μg of LNP-formulated HA-encoding mRNA induced protective hemagglutination inhibition (HAI) titers in NHPs comparable with or even higher than vaccination with a full human dose of licensed vaccines based on inactivated virus. Responses could be boosted by a second dose and remained stable for over a year. Only limited reactogenicity at the injection site and minor changes in systemic cytokine and chemokine concentrations were reported. Lipid-complexed mRNA of influenza HA gene segments was also tested by intravenous immunization in mice and showed T cell activation following a single dose.155
Lindgren et al.143 have characterized the quality of B cell immune responses to an LNP-formulated N1-methyl-psudouridine-modified conventional mRNA encoding HA of H10N8 pandemic influenza strain after both i.d. and i.m. vaccination of NHPs. They showed that circulating H10-specific memory B cells expanded after each immunization, along with a transient appearance of plasmablasts. They showed for the first time the formation of a GC in dLNs after mRNA vaccination, along with an increase in circulating H10-specific Tfh cells, previously shown to correlate with high-avidity antibody responses after seasonal influenza vaccination in humans and to be predictive of seroconversion.156, 157, 158
More recently, LNP-formulated mRNA has been used for the development of a “universal” influenza vaccine capable of inducing potent immune responses against viral epitopes conserved among virus strains. One current target for this approach is the immune subdominant HA stalk that is less tolerant to escape mutations.159 Using a nucleoside-modified, FPLC-purified mRNA-LNP vaccine expressing influenza full-length HA, Pardi et al.70 demonstrated that a single immunization raised durable antibodies against the stalk of HA in mice, ferrets, and rabbits, and was protective against both homologous and heterologous influenza challenges in mice.
The first human trial of an mRNA-based influenza vaccine, using an LNP-formulated, nucleoside-modified conventional mRNA encoding an H10N8 HA antigen, has been recently reported (ClinicalTrials.gov: NCT03076385).71 Interim findings from 23 individuals vaccinated with 100 μg of the RNA vaccine i.m. showed that the vaccine was immunogenic in all subjects at 43 days after vaccination, with 100% and 87% of vaccinated subjects achieving HAI titers ≥40 and micro-neutralization titers ≥20, respectively. These results are encouraging, although antibody titers were lower compared with those reported in animal models. Most subjects reported an acceptable mild-to-moderate reactogenicity profile (injection-site pain, headache, fatigue, and chills) similar to adjuvanted vaccines.
mRNA Vaccines Targeting Rabies Virus Infection
Data from a first time in humans study of protamine-formulated, sequence-optimized conventional mRNA vaccine encoding rabies virus glycoprotein (RABV-G) have recently been published.75 This vaccine had been previously shown to induce protective immunity against a lethal intracerebral virus challenge in mice and a potent neutralizing antibody response in pigs.76 In the human study, subjects received 80–640 μg of mRNA vaccine three times by needle-syringe or needle-free devices, either i.d. or i.m. Within a 7-day window after vaccination, nearly all subjects reported mild-to-moderate injection-site reactions, and 78% reported solicited systemic adverse events, including fever, fatigue, and pain. The vaccine was generally safe, with a reasonable tolerability profile and with only one case of adverse events (Bell’s palsy) in a subject vaccinated at a 640-μg mRNA dose. Unexpectedly, the vaccine failed to induce an adequate antibody response by needle injection, independent of dose or route of administration. Needle-free delivery provided superior results to needle-syringe injection, consistent with observations in animal models. However, immune responses declined after 1 year in subjects who were followed up long term. Subsequent preclinical work has demonstrated that the longevity of immune responses generated by this rabies mRNA vaccine can be improved by optimizing the delivery system.72 LNP formulation of the above-described rabies mRNA showed induction in NHPs of virus neutralization titers above the World Health Organization (WHO) reference titer of 0.5 IU/mL, which could be boosted by a second dose and remained stable above this threshold for at least 5 months. A phase I clinical trial has been recently initiated to evaluate this LNP-formulated sequence-optimized mRNA vaccine (ClinicalTrials.gov: NCT03713086).160
mRNA Vaccines Targeting Zika Virus Infection
An LNP-formulated nucleoside-modified conventional mRNA vaccine targeting Zika virus has been recently moved into clinical evaluation (ClinicalTrials.gov: NCT03014089).
The preclinical assessment of this ZIKA mRNA vaccine had been described by Richner et al.15, 161 In parallel, Pardi et al.14 independently described their own LNP-formulated, nucleoside-modified conventional mRNA against Zika infection. Both groups demonstrated impressive levels of neutralizing titers and protection against lethal challenge in mice after two 10-μg i.m. or one 30-μg i.d. vaccination, respectively, and in NHPs after a single 50-μg i.d. vaccination. Richner et al.15, 161 also tested an mRNA vaccine encoding a modified Zika prM-E antigen that carried mutations destroying the conserved fusion-loop epitope in domain II of the E protein, potentially further improving the safety of the vaccine. They demonstrated that this vaccine remained protective against Zika challenge while diminishing production of antibodies enhancing Dengue virus infection in cells or mice, which is a major concern for Dengue and Zika virus vaccines. The same vaccine was evaluated in a mouse model of maternal vaccination and fetal infection, where two immunizations protected against maternal, placental, and fetal infection from Zika virus and completely rescued a defect in fetal viability. Finally, Erasmus et al.162 have also tested a Zika self-amplifying mRNA vaccine formulated with nanostructured lipid carriers (hybrid formulation between oil-in-water nanoemulsion and LNPs), and showed that a single i.m. vaccination at doses as low as 0.01 μg resulted in durable immune responses and completely protected mice against a lethal virus challenge.
mRNA Vaccines Targeting Multivalent Antigens
Immunization against more than one antigen concurrently is an interesting concept, with the potential of generating immunity against multiple pathogens, different antigens of a same pathogen, or complex polyproteins. This is particularly important when it is necessary to target diverse antigens to stimulate different arms of immunity or to target antigen expressed in multiple life cycles of a pathogen, as in the case of Toxoplasma gondii. The work from Chahal et al.138 demonstrated that six self-amplifying mRNA replicons can be co-formulated, express each of the encoded antigens, and induce protective immunity. A similar approach has been recently reported with a conventional mRNA vaccine. John et al.163 used the HCMV pentameric complex as the model antigen to study multi-valency and interference of conventional mRNA vaccines. LNP encapsulation of all five nucleoside-modified mRNAs, each one encoding one subunit of the pentameric protein complex, was shown to allow complex formation in vitro, efficient delivery in vivo, and generation of robust immune responses in mice and NHPs. The inclusion of an additional mRNA encoding HCMV glycoprotein B (gB) did not affect the levels of antibodies against either the pentameric complex or gB, suggesting a lack of interference by combining the different antigens. The combination of the six mRNAs encoding the HCMV pentameric complex and gB is currently under evaluation in a phase I clinical trial (ClinicalTrials.gov: NCT03382405). Although preclinical data are promising, efficacy, safety, and manufacturability will need to be carefully evaluated to understand whether this multivalent conventional mRNA vaccine approach is feasible and effective, or if a single multivalent self-amplifying mRNA vector vaccine should be pursued.
mRNA Vaccines Targeting Bacterial and Parasitical Infections
In addition to viral targets, a self-amplifying mRNA vaccine has been shown to elicit immune responses against bacterial pathogens, such as group A and group B streptococci, and provided partial protection through passive and active immunization in the mouse challenge model.21 After vaccination, protection was also transferred to newborns, suggesting that the self-amplifying mRNA vaccine may also be suitable for maternal immunization approaches.
Recently, the utility of self-amplifying mRNA against parasitic diseases has been evaluated by using malaria as target disease.141 A self-amplifying mRNA vaccine encoding plasmodium macrophage migration inhibitory factor (PMIF), which suppresses memory T cells and allows the parasites to evade the immune system, protected against reinfection. Self-amplifying mRNA vaccination elicited both cellular and humoral immune responses against PMIF, and anti-PMIF immunoglobulin G (IgG) blocked the proinflammatory action of PMIF. It delayed blood-stage latency after sporozoite infection, augmented Tfh cell and GC response, and enhanced the differentiation of antigen-experienced memory CD4+ T cells and liver-resident CD8+ T cells. Moreover, mice that were cured from infection but received a second sporozoite challenge were fully protected against reinfection. Protection from reinfection was recapitulated by the adoptive transfer of CD8+ or CD4+ T cells. This work demonstrated the utility of self-amplifying mRNA vaccination to block an immune-evasion mechanism employed by the parasite.
Other Clinical Studies of mRNA Vaccines
Additional LNP-formulated nucleoside-modified conventional mRNA vaccines currently under clinical evaluation include vaccines targeting influenza H7N9 (ClinicalTrials.gov: NCT03345043), chikungunya (ClinicalTrials.gov: NCT03325075), and human metapneumovirus and human parainfluenza virus type 3 (ClinicalTrials.gov: NCT03392389) viruses, but details of these studies are currently unavailable.
Conclusions and Perspectives
mRNA-based vaccines are a promising novel platform with the potential to be highly versatile, potent, streamlined, scalable, inexpensive, and cold-chain free. Importantly, mRNA-based vaccines may fill the gap between emerging pandemic infectious diseases and a rapid, abundant supply of effective vaccines. The mRNA vaccine technology holds great potential over conventional vaccines. This versatile RNA vaccine platform offers benefits in the speed and cost of discovery and development, the probability of success for many targets, and rapid production of an effective vaccine against novel threats (Figure 2). However, it is still too early to fully understand its safety and effectiveness in humans. Recently published results from two clinical trials of conventional mRNA vaccines against infectious diseases showed an overall good tolerability profile and promising immunogenicity, but with more modest responses than expected based on results from animals.71, 75 Further insights into the mechanism of action are needed to understand the impact of innate immune responses generated both by the mRNA and the delivery system, and to determine how learning from animal species will translate to humans.
Several questions remain unanswered, including the relative utility of nucleoside-modified versus unmodified mRNAs, self-amplifying mRNAs versus conventional mRNAs, the most efficient and safest delivery system, and the best route of administration. In addition, the safety and tolerability profile of these emerging technologies needs to be fully elucidated, including the relative roles of the RNA and delivery systems in stimulating proinflammatory innate immune responses, and how the doses of these components contribute. A further understanding of the sequence of events leading to antigen expression, innate activation, and adaptive responses could guide the development of a portfolio of mRNA molecules and delivery systems with differential attributes. This will create a toolbox to tackle different applications, such as prophylactic versus therapeutic vaccines, infectious disease versus host disease (e.g., cancer) targets, or delivery of vaccine antigen versus therapeutic molecules (e.g., molecular antibodies).
The next 5 years will be very important for the field of mRNA vaccines, with results from several human clinical trials providing a clearer understanding of the true prospects of the technology and insights into the strengths and weaknesses of the various mRNA technologies and delivery systems under development.
Author Contributions
G.M. and D.Y. planned the manuscript. G.M. wrote and coordinated the draft. C.Z. wrote a part of the draft. J.B.U., J.L., and D.Y. reviewed the manuscript.
Conflicts of Interest
G.M., J.B.U., and D.Y. are employees of the GSK group of companies and report ownership of GSK shares and/or restricted GSK shares.
Acknowledgments
We thank Robert van den Berg and Nicolas Delahaye for their critical reading of the manuscript.
Contributor Information
Giulietta Maruggi, Email: giulietta.x.maruggi@gsk.com.
Dong Yu, Email: dong.x.yu@gsk.com.
References
- 1.Jenner E. Sampson Low; 1978. An Inquiry into the Causes and Effects of the Variolae Vaccinae, a Disease Discovered in Some of the Western Counties of England, Particularly Gloucestershire, and Known by the Name of the Cow Pox. [Google Scholar]
- 2.Standaert B., Rappuoli R. Towards a more comprehensive approach for a total economic assessment of vaccines?: 1. The building blocks for a health economic assessment of vaccination. J. Mark. Access Health Policy. 2017;5:1335162. doi: 10.1080/20016689.2017.1335162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Younger D.S., Younger A.P., Guttmacher S. Childhood vaccination: implications for global and domestic public health. Neurol. Clin. 2016;34:1035–1047. doi: 10.1016/j.ncl.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Plotkin S.A., Plotkin S.L. The development of vaccines: how the past led to the future. Nat. Rev. Microbiol. 2011;9:889–893. doi: 10.1038/nrmicro2668. [DOI] [PubMed] [Google Scholar]
- 5.Kazmin D., Nakaya H.I., Lee E.K., Johnson M.J., van der Most R., van den Berg R.A., Ballou W.R., Jongert E., Wille-Reece U., Ockenhouse C. Systems analysis of protective immune responses to RTS,S malaria vaccination in humans. Proc. Natl. Acad. Sci. USA. 2017;114:2425–2430. doi: 10.1073/pnas.1621489114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom D.E., Black S., Rappuoli R. Emerging infectious diseases: a proactive approach. Proc. Natl. Acad. Sci. USA. 2017;114:4055–4059. doi: 10.1073/pnas.1701410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu M.A. Immunologic basis of vaccine vectors. Immunity. 2010;33:504–515. doi: 10.1016/j.immuni.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Chen N., Xia P., Li S., Zhang T., Wang T.T., Zhu J. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life. 2017;69:297–304. doi: 10.1002/iub.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 11.DeFrancesco L. The ‘anti-hype’ vaccine. Nat. Biotechnol. 2017;35:193–197. doi: 10.1038/nbt.3812. [DOI] [PubMed] [Google Scholar]
- 12.Brito L.A., Kommareddy S., Maione D., Uematsu Y., Giovani C., Berlanda Scorza F., Otten G.R., Yu D., Mandl C.W., Mason P.W. Self-amplifying mRNA vaccines. Adv. Genet. 2015;89:179–233. doi: 10.1016/bs.adgen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Hekele A., Bertholet S., Archer J., Gibson D.G., Palladino G., Brito L.A., Otten G.R., Brazzoli M., Buccato S., Bonci A. Rapidly produced SAM(®) vaccine against H7N9 influenza is immunogenic in mice. Emerg. Microbes Infect. 2013;2:e52. doi: 10.1038/emi.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;169:176. doi: 10.1016/j.cell.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 16.Ulmer J.B., Mansoura M.K., Geall A.J. Vaccines ‘on demand’: science fiction or a future reality. Expert Opin. Drug Discov. 2015;10:101–106. doi: 10.1517/17460441.2015.996128. [DOI] [PubMed] [Google Scholar]
- 17.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 18.Martinon F., Krishnan S., Lenzen G., Magné R., Gomard E., Guillet J.G., Lévy J.P., Meulien P. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur. J. Immunol. 1993;23:1719–1722. doi: 10.1002/eji.1830230749. [DOI] [PubMed] [Google Scholar]
- 19.Conry R.M., LoBuglio A.F., Wright M., Sumerel L., Pike M.J., Johanning F., Benjamin R., Lu D., Curiel D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995;55:1397–1400. [PubMed] [Google Scholar]
- 20.Hoerr I., Obst R., Rammensee H.G., Jung G. In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur. J. Immunol. 2000;30:1–7. doi: 10.1002/1521-4141(200001)30:1<1::AID-IMMU1>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 21.Maruggi G., Chiarot E., Giovani C., Buccato S., Bonacci S., Frigimelica E., Margarit I., Geall A., Bensi G., Maione D. Immunogenicity and protective efficacy induced by self-amplifying mRNA vaccines encoding bacterial antigens. Vaccine. 2017;35:361–368. doi: 10.1016/j.vaccine.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 22.Vabret N., Bhardwaj N., Greenbaum B.D. Sequence-specific sensing of nucleic acids. Trends Immunol. 2017;38:53–65. doi: 10.1016/j.it.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Röhss J., John S., Hassett K., Yuzhakov O., Bahl K. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther. 2017;25:2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pepini T., Pulichino A.M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., Debasitis J.C., Maruggi G., Otten G.R., Geall A.J. Induction of an IFN-mediated antiviral response by a self-amplifying RNA vaccine: implications for vaccine design. J. Immunol. 2017;198:4012–4024. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards D.K., Jasny E., Yoon H., Horscroft N., Schanen B., Geter T., Fotin-Mleczek M., Petsch B., Wittman V. Adjuvant effects of a sequence-engineered mRNA vaccine: translational profiling demonstrates similar human and murine innate response. J. Transl. Med. 2017;15:1. doi: 10.1186/s12967-016-1111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwasaki A., Medzhitov R. Control of adaptive immunity by the innate immune system. Nat. Immunol. 2015;16:343–353. doi: 10.1038/ni.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geall A.J., Mandl C.W., Ulmer J.B. RNA: the new revolution in nucleic acid vaccines. Semin. Immunol. 2013;25:152–159. doi: 10.1016/j.smim.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Pardi N., Muramatsu H., Weissman D., Karikó K. In vitro transcription of long RNA containing modified nucleosides. Methods Mol. Biol. 2013;969:29–42. doi: 10.1007/978-1-62703-260-5_2. [DOI] [PubMed] [Google Scholar]
- 29.Martin S.A., Moss B. Modification of RNA by mRNA guanylyltransferase and mRNA (guanine-7-)methyltransferase from vaccinia virions. J. Biol. Chem. 1975;250:9330–9335. [PubMed] [Google Scholar]
- 30.Malone R.W., Felgner P.L., Verma I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stepinski J., Waddell C., Stolarski R., Darzynkiewicz E., Rhoads R.E. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA. 2001;7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip. Rev. RNA. 2010;1:253–265. doi: 10.1002/wrna.15. [DOI] [PubMed] [Google Scholar]
- 33.Marcotrigiano J., Gingras A.C., Sonenberg N., Burley S.K. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 34.Pascolo S. Messenger RNA-based vaccines. Expert Opin. Biol. Ther. 2004;4:1285–1294. doi: 10.1517/14712598.4.8.1285. [DOI] [PubMed] [Google Scholar]
- 35.Pascolo S. Vaccination with messenger RNA (mRNA) Handb. Exp. Pharmacol. 2008;183:221–235. doi: 10.1007/978-3-540-72167-3_11. [DOI] [PubMed] [Google Scholar]
- 36.Weissman D., Pardi N., Muramatsu H., Karikó K. HPLC purification of in vitro transcribed long RNA. Methods Mol. Biol. 2013;969:43–54. doi: 10.1007/978-1-62703-260-5_3. [DOI] [PubMed] [Google Scholar]
- 37.Mu X., Greenwald E., Ahmad S., Hur S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018;46:5239–5249. doi: 10.1093/nar/gky177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foster J.B., Choudhari N., Perazzelli J., Storm J., Hofmann T.J., Jain P., Storm P.B., Pardi N., Weissman D., Waanders A.J. Purification of mRNA encoding chimeric antigen receptor is critical for generation of a robust T-cell response. Hum. Gene Ther. 2018 doi: 10.1089/hum.2018.145. Published online October 2, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muralidhara B.K., Baid R., Bishop S.M., Huang M., Wang W., Nema S. Critical considerations for developing nucleic acid macromolecule based drug products. Drug Discov. Today. 2016;21:430–444. doi: 10.1016/j.drudis.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 41.CureVac. (2017). CureVac announces groundbreaking of industrial-scale GMP production facility for RNA therapeutics. https://www.curevac.com/news/curevac-announces-groundbreaking-of-industrial-scale-gmp-production-facility-for-rna-therapeutics/
- 42.Moderna. (2018). Moderna opens new manufacturing site for Norwood, MA. https://investors.modernatx.com/news-releases/news-release-details/moderna-opens-new-manufacturing-site-norwood-ma.
- 43.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 45.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatterjee S., Pal J.K. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 48.Jansen R.P. mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- 49.Bashirullah A., Cooperstock R.L., Lipshitz H.D. Spatial and temporal control of RNA stability. Proc. Natl. Acad. Sci. USA. 2001;98:7025–7028. doi: 10.1073/pnas.111145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 51.Lundstrom K. Latest development on RNA-based drugs and vaccines. Future Sci. OA. 2018;4:FSO300. doi: 10.4155/fsoa-2017-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics—developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 53.Mauro V.P., Chappell S.A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2014;20:604–613. doi: 10.1016/j.molmed.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fåhraeus R., Marin M., Olivares-Illana V. Whisper mutations: cryptic messages within the genetic code. Oncogene. 2016;35:3753–3759. doi: 10.1038/onc.2015.454. [DOI] [PubMed] [Google Scholar]
- 55.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H. Self-amplifying RNA vaccines give equivalent protection against influenza to mRNA vaccines but at much lower doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 58.Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 59.Pichlmair A., Schulz O., Tan C.P., Rehwinkel J., Kato H., Takeuchi O., Akira S., Way M., Schiavo G., Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nallagatla S.R., Hwang J., Toroney R., Zheng X., Cameron C.E., Bevilacqua P.C. 5′-Triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 61.Iavarone C., O’hagan D.T., Yu D., Delahaye N.F., Ulmer J.B. Mechanism of action of mRNA-based vaccines. Expert Rev. Vaccines. 2017;16:871–881. doi: 10.1080/14760584.2017.1355245. [DOI] [PubMed] [Google Scholar]
- 62.De Beuckelaer A., Pollard C., Van Lint S., Roose K., Van Hoecke L., Naessens T., Udhayakumar V.K., Smet M., Sanders N., Lienenklaus S. Type I interferons interfere with the capacity of mRNA lipoplex vaccines to elicit cytolytic T cell responses. Mol. Ther. 2016;24:2012–2020. doi: 10.1038/mt.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 64.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nallagatla S.R., Bevilacqua P.C. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karikó K., Muramatsu H., Keller J.M., Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012;20:948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Svitkin Y.V., Cheng Y.M., Chakraborty T., Presnyak V., John M., Sonenberg N. N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res. 2017;45:6023–6036. doi: 10.1093/nar/gkx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L., Tam Y.K., Karikó K., Barbosa C.J., Madden T.D. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lutz J., Lazzaro S., Habbeddine M., Schmidt K.E., Baumhof P., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Heidenreich R., Fotin-Mleczek M. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ Vaccines. 2017;2:29. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petsch B., Schnee M., Vogel A.B., Lange E., Hoffmann B., Voss D., Schlake T., Thess A., Kallen K.J., Stitz L., Kramps T. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat. Biotechnol. 2012;30:1210–1216. doi: 10.1038/nbt.2436. [DOI] [PubMed] [Google Scholar]
- 74.Kallen K.J., Heidenreich R., Schnee M., Petsch B., Schlake T., Thess A., Baumhof P., Scheel B., Koch S.D., Fotin-Mleczek M. A novel, disruptive vaccination technology: self-adjuvanted RNActive(®) vaccines. Hum. Vaccin. Immunother. 2013;9:2263–2276. doi: 10.4161/hv.25181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 76.Schnee M., Vogel A.B., Voss D., Petsch B., Baumhof P., Kramps T., Stitz L. An mRNA vaccine encoding rabies virus glycoprotein induces protection against lethal infection in mice and correlates of protection in adult and newborn pigs. PLoS Negl. Trop. Dis. 2016;10:e0004746. doi: 10.1371/journal.pntd.0004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rauch S., Lutz J., Kowalczyk A., Schlake T., Heidenreich R. RNActive® technology: generation and testing of stable and immunogenic mRNA vaccines. Methods Mol. Biol. 2017;1499:89–107. doi: 10.1007/978-1-4939-6481-9_5. [DOI] [PubMed] [Google Scholar]
- 78.Fotin-Mleczek M., Duchardt K.M., Lorenz C., Pfeiffer R., Ojkić-Zrna S., Probst J., Kallen K.J. Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J. Immunother. 2011;34:1–15. doi: 10.1097/CJI.0b013e3181f7dbe8. [DOI] [PubMed] [Google Scholar]
- 79.Kowalczyk A., Doener F., Zanzinger K., Noth J., Baumhof P., Fotin-Mleczek M., Heidenreich R. Self-adjuvanted mRNA vaccines induce local innate immune responses that lead to a potent and boostable adaptive immunity. Vaccine. 2016;34:3882–3893. doi: 10.1016/j.vaccine.2016.05.046. [DOI] [PubMed] [Google Scholar]
- 80.Loomis K.H., Lindsay K.E., Zurla C., Bhosle S.M., Vanover D.A., Blanchard E.L., Kirschman J.L., Bellamkonda R.V., Santangelo P.J. In vitro transcribed mRNA vaccines with programmable stimulation of innate immunity. Bioconjug. Chem. 2018;29:3072–3083. doi: 10.1021/acs.bioconjchem.8b00443. [DOI] [PubMed] [Google Scholar]
- 81.Tews B.A., Meyers G. Self-replicating RNA. Methods Mol. Biol. 2017;1499:15–35. doi: 10.1007/978-1-4939-6481-9_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lundstrom K. Replicon RNA viral vectors as vaccines. Vaccines (Basel) 2016;4:E39. doi: 10.3390/vaccines4040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ljungberg K., Liljeström P. Self-replicating alphavirus RNA vaccines. Expert Rev. Vaccines. 2015;14:177–194. doi: 10.1586/14760584.2015.965690. [DOI] [PubMed] [Google Scholar]
- 84.Atkins G.J., Fleeton M.N., Sheahan B.J. Therapeutic and prophylactic applications of alphavirus vectors. Expert Rev. Mol. Med. 2008;10:e33. doi: 10.1017/S1462399408000859. [DOI] [PubMed] [Google Scholar]
- 85.Mühlebach M.D. Vaccine platform recombinant measles virus. Virus Genes. 2017;53:733–740. doi: 10.1007/s11262-017-1486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Humphreys I.R., Sebastian S. Novel viral vectors in infectious diseases. Immunology. 2018;153:1–9. doi: 10.1111/imm.12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Strauss J.H., Strauss E.G. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rupp J.C., Sokoloski K.J., Gebhart N.N., Hardy R.W. Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 2015;96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lemm J.A., Rümenapf T., Strauss E.G., Strauss J.H., Rice C.M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.LaPointe A.T., Gebhart N.N., Meller M.E., Hardy R.W., Sokoloski K.J. The identification and characterization of Sindbis virus RNA:host protein interactions. J. Virol. 2018;92 doi: 10.1128/JVI.02171-17. e02171-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Frolova E.I., Gorchakov R., Pereboeva L., Atasheva S., Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J. Virol. 2010;84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lundstrom K. Biology and application of alphaviruses in gene therapy. Gene Ther. 2005;12(Suppl 1):S92–S97. doi: 10.1038/sj.gt.3302620. [DOI] [PubMed] [Google Scholar]
- 93.Sawicki D.L., Kaariainen L., Lambek C., Gomatos P.J. Mechanism for control of synthesis of Semliki Forest virus 26S and 42s RNA. J. Virol. 1978;25:19–27. doi: 10.1128/jvi.25.1.19-27.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Keränen S., Kääriäinen L. Functional defects of RNA-negative temperature-sensitive mutants of Sindbis and Semliki Forest viruses. J. Virol. 1979;32:19–29. doi: 10.1128/jvi.32.1.19-29.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Perri S., Greer C.E., Thudium K., Doe B., Legg H., Liu H., Romero R.E., Tang Z., Bin Q., Dubensky T.W., Jr. An alphavirus replicon particle chimera derived from venezuelan equine encephalitis and sindbis viruses is a potent gene-based vaccine delivery vector. J. Virol. 2003;77:10394–10403. doi: 10.1128/JVI.77.19.10394-10403.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lundstrom K. Alphavirus-based vaccines. Methods Mol. Biol. 2016;1404:313–328. doi: 10.1007/978-1-4939-3389-1_22. [DOI] [PubMed] [Google Scholar]
- 97.Bernstein D.I., Reap E.A., Katen K., Watson A., Smith K., Norberg P., Olmsted R.A., Hoeper A., Morris J., Negri S. Randomized, double-blind, Phase 1 trial of an alphavirus replicon vaccine for cytomegalovirus in CMV seronegative adult volunteers. Vaccine. 2009;28:484–493. doi: 10.1016/j.vaccine.2009.09.135. [DOI] [PubMed] [Google Scholar]
- 98.Wecker M., Gilbert P., Russell N., Hural J., Allen M., Pensiero M., Chulay J., Chiu Y.L., Abdool Karim S.S., Burke D.S., HVTN 040/059 Protocol Team. NIAID HIV Vaccine Trials Network Phase I safety and immunogenicity evaluations of an alphavirus replicon HIV-1 subtype C gag vaccine in healthy HIV-1-uninfected adults. Clin. Vaccine Immunol. 2012;19:1651–1660. doi: 10.1128/CVI.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mogler M.A., Kamrud K.I. RNA-based viral vectors. Expert Rev. Vaccines. 2015;14:283–312. doi: 10.1586/14760584.2015.979798. [DOI] [PubMed] [Google Scholar]
- 100.Liljeström P., Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (N. Y.) 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 101.Gardner J.P., Frolov I., Perri S., Ji Y., MacKichan M.L., zur Megede J., Chen M., Belli B.A., Driver D.A., Sherrill S. Infection of human dendritic cells by a sindbis virus replicon vector is determined by a single amino acid substitution in the E2 glycoprotein. J. Virol. 2000;74:11849–11857. doi: 10.1128/jvi.74.24.11849-11857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.MacDonald G.H., Johnston R.E. Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol. 2000;74:914–922. doi: 10.1128/jvi.74.2.914-922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nishimoto K.P., Laust A.K., Wang K., Kamrud K.I., Hubby B., Smith J.F., Nelson E.L. Restricted and selective tropism of a Venezuelan equine encephalitis virus-derived replicon vector for human dendritic cells. Viral Immunol. 2007;20:88–104. doi: 10.1089/vim.2006.0090. [DOI] [PubMed] [Google Scholar]
- 104.Tonkin D.R., Whitmore A., Johnston R.E., Barro M. Infected dendritic cells are sufficient to mediate the adjuvant activity generated by Venezuelan equine encephalitis virus replicon particles. Vaccine. 2012;30:4532–4542. doi: 10.1016/j.vaccine.2012.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou X., Berglund P., Rhodes G., Parker S.E., Jondal M., Liljeström P. Self-replicating Semliki Forest virus RNA as recombinant vaccine. Vaccine. 1994;12:1510–1514. doi: 10.1016/0264-410x(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 106.Fleeton M.N., Chen M., Berglund P., Rhodes G., Parker S.E., Murphy M., Atkins G.J., Liljeström P. Self-replicative RNA vaccines elicit protection against influenza A virus, respiratory syncytial virus, and a tickborne encephalitis virus. J. Infect. Dis. 2001;183:1395–1398. doi: 10.1086/319857. [DOI] [PubMed] [Google Scholar]
- 107.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brito L.A., Chan M., Shaw C.A., Hekele A., Carsillo T., Schaefer M., Archer J., Seubert A., Otten G.R., Beard C.W. A cationic nanoemulsion for the delivery of next-generation RNA vaccines. Mol. Ther. 2014;22:2118–2129. doi: 10.1038/mt.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leyman B., Huysmans H., Mc Cafferty S., Combes F., Cox E., Devriendt B., Sanders N.N. Comparison of the expression kinetics and immunostimulatory activity of replicating mRNA, nonreplicating mRNA, and pDNA after intradermal electroporation in pigs. Mol. Pharm. 2018;15:377–384. doi: 10.1021/acs.molpharmaceut.7b00722. [DOI] [PubMed] [Google Scholar]
- 110.Kamrud K.I., Custer M., Dudek J.M., Owens G., Alterson K.D., Lee J.S., Groebner J.L., Smith J.F. Alphavirus replicon approach to promoterless analysis of IRES elements. Virology. 2007;360:376–387. doi: 10.1016/j.virol.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Magini D., Giovani C., Mangiavacchi S., Maccari S., Cecchi R., Ulmer J.B., De Gregorio E., Geall A.J., Brazzoli M., Bertholet S. Self-amplifying mRNA vaccines expressing multiple conserved influenza antigens confer protection against homologous and heterosubtypic viral challenge. PLoS ONE. 2016;11:e0161193. doi: 10.1371/journal.pone.0161193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hua Z., Hou B. TLR signaling in B-cell development and activation. Cell. Mol. Immunol. 2013;10:103–106. doi: 10.1038/cmi.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maruggi G., Shaw C.A., Otten G.R., Mason P.W., Beard C.W. Engineered alphavirus replicon vaccines based on known attenuated viral mutants show limited effects on immunogenicity. Virology. 2013;447:254–264. doi: 10.1016/j.virol.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 114.Pollard C., Rejman J., De Haes W., Verrier B., Van Gulck E., Naessens T., De Smedt S., Bogaert P., Grooten J., Vanham G., De Koker S. Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol. Ther. 2013;21:251–259. doi: 10.1038/mt.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sheehan K.C., Lai K.S., Dunn G.P., Bruce A.T., Diamond M.S., Heutel J.D., Dungo-Arthur C., Carrero J.A., White J.M., Hertzog P.J., Schreiber R.D. Blocking monoclonal antibodies specific for mouse IFN-alpha/beta receptor subunit 1 (IFNAR-1) from mice immunized by in vivo hydrodynamic transfection. J. Interferon Cytokine Res. 2006;26:804–819. doi: 10.1089/jir.2006.26.804. [DOI] [PubMed] [Google Scholar]
- 116.De Beuckelaer A., Grooten J., De Koker S. Type I interferons modulate CD8+ T cell immunity to mRNA vaccines. Trends Mol. Med. 2017;23:216–226. doi: 10.1016/j.molmed.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 117.Beissert T., Koste L., Perkovic M., Walzer K.C., Erbar S., Selmi A., Diken M., Kreiter S., Türeci Ö., Sahin U. Improvement of in vivo expression of genes delivered by self-amplifying RNA using vaccinia virus immune evasion proteins. Hum. Gene Ther. 2017;28:1138–1146. doi: 10.1089/hum.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gorchakov R., Frolova E., Williams B.R., Rice C.M., Frolov I. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 2004;78:8455–8467. doi: 10.1128/JVI.78.16.8455-8467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim Y.G., Baltabekova A.Z., Zhiyenbay E.E., Aksambayeva A.S., Shagyrova Z.S., Khannanov R., Ramanculov E.M., Shustov A.V. Recombinant Vaccinia virus-coded interferon inhibitor B18R: expression, refolding and a use in a mammalian expression system with a RNA-vector. PLoS ONE. 2017;12:e0189308. doi: 10.1371/journal.pone.0189308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Uematsu Y., Vajdy M., Lian Y., Perri S., Greer C.E., Legg H.S., Galli G., Saletti G., Otten G.R., Rappuoli R. Lack of interference with immunogenicity of a chimeric alphavirus replicon particle-based influenza vaccine by preexisting antivector immunity. Clin. Vaccine Immunol. 2012;19:991–998. doi: 10.1128/CVI.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wen Y., Monroe J., Linton C., Archer J., Beard C.W., Barnett S.W., Palladino G., Mason P.W., Carfi A., Lilja A.E. Human cytomegalovirus gH/gL/UL128/UL130/UL131A complex elicits potently neutralizing antibodies in mice. Vaccine. 2014;32:3796–3804. doi: 10.1016/j.vaccine.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 122.McCullough K.C., Bassi I., Milona P., Suter R., Thomann-Harwood L., Englezou P., Démoulins T., Ruggli N. Self-replicating replicon-RNA delivery to dendritic cells by chitosan-nanoparticles for translation in vitro and in vivo. Mol. Ther. Nucleic Acids. 2014;3:e173. doi: 10.1038/mtna.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Démoulins T., Milona P., Englezou P.C., Ebensen T., Schulze K., Suter R., Pichon C., Midoux P., Guzmán C.A., Ruggli N., McCullough K.C. Polyethylenimine-based polyplex delivery of self-replicating RNA vaccines. Nanomedicine (Lond.) 2016;12:711–722. doi: 10.1016/j.nano.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 124.Houseley J., Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 125.Tatiparti K., Sau S., Kashaw S.K., Iyer A.K. siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials (Basel) 2017;7:E77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Johannes L., Lucchino M. Current challenges in delivery and cytosolic translocation of therapeutic RNAs. Nucleic Acid Ther. 2018;28:178–193. doi: 10.1089/nat.2017.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stanton M.G. Current status of messenger RNA delivery systems. Nucleic Acid Ther. 2018;28:158–165. doi: 10.1089/nat.2018.0726. [DOI] [PubMed] [Google Scholar]
- 128.Hajj K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:17056. [Google Scholar]
- 129.Li B., Zhang X., Dong Y. Nanoscale platforms for messenger RNA delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2018;1530:e1530. doi: 10.1002/wnan.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kanasty R.L., Whitehead K.A., Vegas A.J., Anderson D.G. Action and reaction: the biological response to siRNA and its delivery vehicles. Mol. Ther. 2012;20:513–524. doi: 10.1038/mt.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cullis P.R., Hope M.J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alnylam Pharmaceuticals. (2018). Alnylam announces first-ever FDA approval of an RNAi therapeutic, ONPATTRO™ (patisiran) for the treatment of the polyneuropathy of hereditary transthyretin-mediated amyloidosis in adults. http://investors.alnylam.com/news-releases/news-release-details/alnylam-announces-first-ever-fda-approval-rnai-therapeutic.
- 133.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 134.Heyes J., Palmer L., Bremner K., MacLachlan I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release. 2005;107:276–287. doi: 10.1016/j.jconrel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 135.Patel S., Ashwanikumar N., Robinson E., DuRoss A., Sun C., Murphy-Benenato K.E., Mihai C., Almarsson Ö., Sahay G. Boosting intracellular delivery of lipid nanoparticle-encapsulated mRNA. Nano Lett. 2017;17:5711–5718. doi: 10.1021/acs.nanolett.7b02664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bhosle S.M., Loomis K.H., Kirschman J.L., Blanchard E.L., Vanover D.A., Zurla C., Habrant D., Edwards D., Baumhof P., Pitard B., Santangelo P.J. Unifying in vitro and in vivo IVT mRNA expression discrepancies in skeletal muscle via mechanotransduction. Biomaterials. 2018;159:189–203. doi: 10.1016/j.biomaterials.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 138.Chahal J.S., Khan O.F., Cooper C.L., McPartlan J.S., Tsosie J.K., Tilley L.D., Sidik S.M., Lourido S., Langer R., Bavari S. Dendrimer-RNA nanoparticles generate protective immunity against lethal Ebola, H1N1 influenza, and Toxoplasma gondii challenges with a single dose. Proc. Natl. Acad. Sci. USA. 2016;113:E4133–E4142. doi: 10.1073/pnas.1600299113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chahal J.S., Fang T., Woodham A.W., Khan O.F., Ling J., Anderson D.G., Ploegh H.L. An RNA nanoparticle vaccine against Zika virus elicits antibody and CD8+ T cell responses in a mouse model. Sci. Rep. 2017;7:252. doi: 10.1038/s41598-017-00193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bogers W.M., Oostermeijer H., Mooij P., Koopman G., Verschoor E.J., Davis D., Ulmer J.B., Brito L.A., Cu Y., Banerjee K. Potent immune responses in rhesus macaques induced by nonviral delivery of a self-amplifying RNA vaccine expressing HIV type 1 envelope with a cationic nanoemulsion. J. Infect. Dis. 2015;211:947–955. doi: 10.1093/infdis/jiu522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Baeza Garcia A., Siu E., Sun T., Exler V., Brito L., Hekele A., Otten G., Augustijn K., Janse C.J., Ulmer J.B. Neutralization of the Plasmodium-encoded MIF ortholog confers protective immunity against malaria infection. Nat. Commun. 2018;9:2714. doi: 10.1038/s41467-018-05041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Brazzoli M., Magini D., Bonci A., Buccato S., Giovani C., Kratzer R., Zurli V., Mangiavacchi S., Casini D., Brito L.M. Induction of broad-based immunity and protective efficacy by self-amplifying mRNA vaccines encoding influenza virus hemagglutinin. J. Virol. 2015;90:332–344. doi: 10.1128/JVI.01786-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lindgren G., Ols S., Liang F., Thompson E.A., Lin A., Hellgren F., Bahl K., John S., Yuzhakov O., Hassett K.J. Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+ PD-1+ CXCR3+ T follicular helper cells. Front. Immunol. 2017;8:1539. doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Englezou P.C., Sapet C., Démoulins T., Milona P., Ebensen T., Schulze K., Guzman C.A., Poulhes F., Zelphati O., Ruggli N., McCullough K.C. Self-amplifying replicon RNA delivery to dendritic cells by cationic lipids. Mol. Ther. Nucleic Acids. 2018;12:118–134. doi: 10.1016/j.omtn.2018.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Benteyn D., Heirman C., Bonehill A., Thielemans K., Breckpot K. mRNA-based dendritic cell vaccines. Expert Rev. Vaccines. 2015;14:161–176. doi: 10.1586/14760584.2014.957684. [DOI] [PubMed] [Google Scholar]
- 146.Gornati L., Zanoni I., Granucci F. Dendritic cells in the cross hair for the generation of tailored vaccines. Front. Immunol. 2018;9:1484. doi: 10.3389/fimmu.2018.01484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jones K.L., Drane D., Gowans E.J. Long-term storage of DNA-free RNA for use in vaccine studies. Biotechniques. 2007;43:675–681. doi: 10.2144/000112593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stitz L., Vogel A., Schnee M., Voss D., Rauch S., Mutzke T., Ketterer T., Kramps T., Petsch B. A thermostable messenger RNA based vaccine against rabies. PLoS Negl. Trop. Dis. 2017;11:e0006108. doi: 10.1371/journal.pntd.0006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Grunwitz C., Kranz L.M. mRNA cancer vaccines—messages that prevail. Curr. Top. Microbiol. Immunol. 2017;405:145–164. doi: 10.1007/82_2017_509. [DOI] [PubMed] [Google Scholar]
- 150.Kaczmarek J.C., Kowalski P.S., Anderson D.G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9:60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fiedler K., Lazzaro S., Lutz J., Rauch S., Heidenreich R. mRNA cancer vaccines. Recent Results Cancer Res. 2016;209:61–85. doi: 10.1007/978-3-319-42934-2_5. [DOI] [PubMed] [Google Scholar]
- 152.Pastor F., Berraondo P., Etxeberria I., Frederick J., Sahin U., Gilboa E., Melero I. An RNA toolbox for cancer immunotherapy. Nat. Rev. Drug Discov. 2018;17:751–767. doi: 10.1038/nrd.2018.132. [DOI] [PubMed] [Google Scholar]
- 153.Krammer F., Palese P. Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 2015;14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 154.Scorza F.B., Pardi N. New kids on the block: RNA-based influenza virus vaccines. Vaccines (Basel) 2018;6:E20. doi: 10.3390/vaccines6020020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 156.Crotty S. Follicular helper CD4 T cells (TFH) Annu. Rev. Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 157.Suan D., Sundling C., Brink R. Plasma cell and memory B cell differentiation from the germinal center. Curr. Opin. Immunol. 2017;45:97–102. doi: 10.1016/j.coi.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 158.Pilkinton M.A., Nicholas K.J., Warren C.M., Smith R.M., Yoder S.M., Talbot H.K., Kalams S.A. Greater activation of peripheral T follicular helper cells following high dose influenza vaccine in older adults forecasts seroconversion. Vaccine. 2017;35:329–336. doi: 10.1016/j.vaccine.2016.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Krammer F., Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr. Opin. Virol. 2013;3:521–530. doi: 10.1016/j.coviro.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.CureVac. (2018). CureVac announces first study participant enrolled in phase I clinical trial testing prophylactic mRNA. https://www.curevac.com/newsroom/news/curevac-announces-first-study-participant-enrolled-in-phase-i-clinical-trial-testing-prophylactic-mr/.
- 161.Richner J.M., Jagger B.W., Shan C., Fontes C.R., Dowd K.A., Cao B., Himansu S., Caine E.A., Nunes B.T.D., Medeiros D.B.A. Vaccine mediated protection against Zika virus-induced congenital disease. Cell. 2017;170:273–283.e12. doi: 10.1016/j.cell.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Erasmus J.H., Khandhar A.P., Guderian J., Granger B., Archer J., Archer M., Gage E., Fuerte-Stone J., Larson E., Lin S. A nanostructured lipid carrier for delivery of a replicating viral RNA provides single, low-dose protection against Zika. Mol. Ther. 2018;26:2507–2522. doi: 10.1016/j.ymthe.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.John S., Yuzhakov O., Woods A., Deterling J., Hassett K., Shaw C.A., Ciaramella G. Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine. 2018;36:1689–1699. doi: 10.1016/j.vaccine.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 164.Fischer S., Gerriets T., Wessels C., Walberer M., Kostin S., Stolz E., Zheleva K., Hocke A., Hippenstiel S., Preissner K.T. Extracellular RNA mediates endothelial-cell permeability via vascular endothelial growth factor. Blood. 2007;110:2457–2465. doi: 10.1182/blood-2006-08-040691. [DOI] [PubMed] [Google Scholar]
- 165.Kannemeier C., Shibamiya A., Nakazawa F., Trusheim H., Ruppert C., Markart P., Song Y., Tzima E., Kennerknecht E., Niepmann M. Extracellular RNA constitutes a natural procoagulant cofactor in blood coagulation. Proc. Natl. Acad. Sci. USA. 2007;104:6388–6393. doi: 10.1073/pnas.0608647104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhu Z., Gorman M.J., McKenzie L.D., Chai J.N., Hubert C.G., Prager B.C., Fernandez E., Richner J.M., Zhang R., Shan C. Correction: Zika virus has oncolytic activity against glioblastoma stem cells. J. Exp. Med. 2017;214:3145. doi: 10.1084/jem.2017109309122017c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Meyer M., Huang E., Yuzhakov O., Ramanathan P., Ciaramella G., Bukreyev A. Modified mRNA-based vaccines elicit robust immune responses and protect guinea pigs from Ebola virus disease. J. Infect. Dis. 2018;217:451–455. doi: 10.1093/infdis/jix592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhao M., Li M., Zhang Z., Gong T., Sun X. Induction of HIV-1 gag specific immune responses by cationic micelles mediated delivery of gag mRNA. Drug Deliv. 2016;23:2596–2607. doi: 10.3109/10717544.2015.1038856. [DOI] [PubMed] [Google Scholar]
- 169.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines—a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]