Abstract
First attempts to use exogenous mRNA for protein expression in vivo were made more than 25 years ago. However, widespread appreciation of in vitro transcribed mRNA as a powerful technology for supplying therapeutic proteins to the body has evolved only during the past few years. Various approaches to turning mRNA into a potent therapeutic have been developed. All of them share utilization of specifically designed, rather than endogenous, sequences and thorough purification protocols. Apart from this, there are two fundamental philosophies, one promoting the use of chemically modified nucleotides, the other advocating restriction to unmodified building blocks. Meanwhile, both strategies have received broad support by successful mRNA-based protein treatments in animal models. For such in vivo use, specifically optimized mRNA had to be combined with potent formulations to enable efficient in vivo delivery. The present review analyzes the applicability of mRNA technology to antibody therapy in all main fields: antitoxins, infectious diseases, and oncology.
Keywords: mRNA, antibody, passive immunization, lipid nanoparticle, LNP, antitoxin, infectious disease, oncology
Main Text
As a fundamental biological concept, the cellular machinery exploits mRNA as a transient carrier of information for synthesizing genetically encoded proteins. Hence, from a theoretical standpoint, mRNA should be capable of replacing DNA or recombinant proteins for therapeutic purposes. The first evidence supporting this hypothesis was provided in 1990 when direct injection of mRNA into mouse muscle elicited detectable protein expression.1 Shortly afterward, administration of vasopressin mRNA to the rat CNS suggested that even therapeutic effects could be obtained.2
Synthetic mRNA usually mimics natural transcripts with respect to design. mRNA elements and manufacturing by in vitro transcription (IVT) have been described elsewhere.3, 4 Although endogenous mRNAs typically have cap1 or cap2 structures, IVT mRNA can be produced with either cap0 or cap1, but not yet with cap2, by cotranscriptional capping using cap analogs or enzymatic capping (reviewed in Schlake et al.3, 4). However, capping may leave some mRNA molecules uncapped, incompletely capped, or with otherwise unnatural 5′ ends (reviewed in Schlake et al.3). Moreover, IVT can generate aberrant RNAs in addition to the desired mRNA, such as short transcripts caused by abortive cycling or double-stranded RNA (dsRNA) by self-complementary 3′ extension caused by primer extension.5, 6 Variant cap structures and dsRNA are among the pathogen-associated molecular patterns (PAMPs) recognized by various receptors in eukaryotic cells in the process of repelling pathogen invasion.3, 4 Receptor engagement or activation can stimulate immune responses that may suppress protein translation.7 Hence, IVT mRNA should be made free of such contaminants by means of either production or extensive purification.
Pathogen RNA can also differ from endogenous eukaryotic RNA in the pattern of base modifications. Notably, recent studies unveiled the existence of modified nucleosides, even in eukaryotic mRNA.8, 9, 10, 11, 12 The overall level of endogenous mRNA modification is rather low, however, compared with tRNA and rRNA. In contrast, modified IVT mRNA typically has 100% replacement of an unmodified nucleoside. Modifications can weaken RNA recognition by cellular receptors and may reduce the generation of dsRNA during IVT.13, 14, 15, 16 They can decrease immunostimulation and increase protein expression in mice, as has been demonstrated, for instance, for pseudouridine-modified IVT mRNA.17, 18 However, modification alone is usually insufficient to make IVT mRNA immunosilent and can even reduce expression under certain circumstances.19, 20, 21 Interestingly, unmodified mRNA can be on a par with modified mRNA in immunostimulation and protein expression.22 Stringent purification of IVT mRNA has been recognized as one of the critical parameters; chromatography, such as high-performance liquid chromatography (HPLC), can remove smaller and larger by-products, including abortive transcripts and dsRNA.19, 23 Enrichment of functional transcripts and depletion of contaminants that may cause unwanted immunostimulation and translational repression are probably the main drivers for enhanced protein expression.19, 24 Whereas chemical modification often increases protein expression compared to unmodified mRNA, HPLC reduces the difference between modified and unmodified transcripts in certain instances.19, 21 The combination of HPLC purification with specific mRNA sequence designs that support translation and silence immunostimulation can even make nucleotide modifications unnecessary for obtaining a very potent and largely immunosilent mRNA.19, 21 The critical role of the mRNA sequence was further corroborated by the recent finding that a U-depleted ORF (open reading frame) sequence gives rise to more efficient gene editing in primary human cells and less cytokine induction in whole human blood.25 Today, ample in vivo evidence for high protein expression and efficacious treatment supports either philosophy: the use of modified nucleotides or the combination of specific sequence design and stringent purification without modifications.18, 21, 26, 27, 28, 29, 30, 31, 32, 33 These studies suggest that mRNA may be used for passive immunization, as well, i.e., the delivery of antibody-encoding mRNA (Figure 1). However, such applications may be particularly dependent on potent mRNA molecules and their efficacious in vivo delivery.
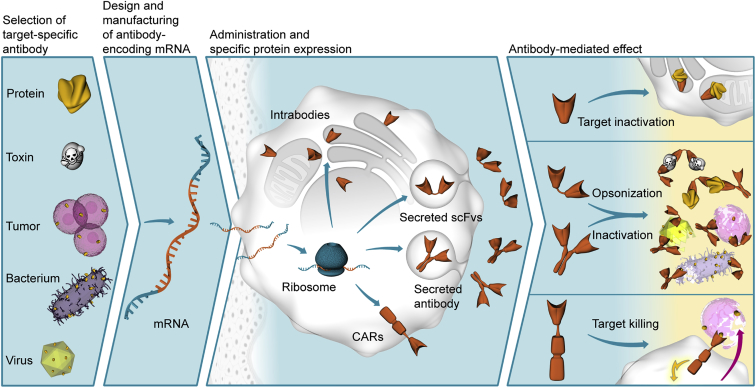
Figure 1.
Schematic Illustration of mRNA-Based Antibody Treatment
In the first step, effective antibodies have to be raised or selected for the target of interest. The respective amino acid sequence can then be encoded in an mRNA designed to produce high amounts of protein. Upon in vivo administration, typically using specific formulations for delivery and mRNA protection against degradation, transfected cells produce the encoded protein, which is not limited as to antibody format or localization. Depending on its mode of action, the mRNA-encoded protein finally triggers the desired therapeutic effect.
In vivo administration usually needs formulation of mRNA into nanoparticles to protect against degradation by ubiquitous RNases.34, 35 Moreover, formulation is required to direct the mRNA to the desired target cells and for their efficient transfection.36 Upon i.v. administration, nanoparticles are typically routed mainly to the liver.36 As a consequence, addressing tissues other than the liver still poses a major challenge. Initial in vivo studies on mRNA used well-known transfection reagents.18, 29, 37 Today, various polymer- and lipid-based nanoparticles are investigated for therapeutic mRNA delivery.38 Based on the experience in the small interfering RNA (siRNA) field,39 lipid nanoparticles (LNPs) appear to be the most advanced and thus are currently used in most instances.26, 31, 36 Although LNPs were initially developed for i.v. administration, alternative delivery routes are feasible, as well.36 LNPs usually contain four components: an ionizable or amino-lipid, cholesterol, a phospholipid (also termed helper lipid), and lipid-anchored polyethylene glycol (PEG).38, 40 While keeping this general composition, efforts to improve efficacy and tolerability of LNP formulations focused on relative proportions of components and the identity and features of the ionizable or amino-lipid.40 The latter is a major determinant of efficacy by controlling endosomal escape, and its potency and biodegradability can affect tolerability.32, 41, 42 Further developments address stability of LNP formulations and suggest that lyophilization may enable long-term storage at ambient temperature.43 Overall, the need for formulations adds to the complexity and challenges of mRNA approaches, including the chemistry, manufacturing, and control (CMC) aspects, which must cover additional parameters, such as particle size, lipid concentration, mRNA content, and encapsulation.
Today, recombinant antibodies are typically full-size immunoglobulins, mostly of the IgG type. Since they are heterotetramers requiring disulfide bridges and glycosylation can have a functional impact, most therapeutic antibodies are currently produced in mammalian cells.44, 45 To enable more cost-efficient expression in traditional hosts such as E. coli, antibody fragments, such as single-chain variable fragments (scFv), have been developed.46 In contrast to full-size antibodies, fragments usually show much shorter plasma half-lives.47 Other antibody fragments, heavy-chain-only VH (VHH) domains, or nanobodies, are derived from single-domain antibodies of camelids and sharks that lack light chains.48, 49 Antibody fragments are also the basis of bispecific antibodies that today form a huge family comprising a multitude of different formats.50, 51, 52, 53 Of note, antigen-specific scFvs are also part of chimeric antigen receptors (CARs), which are currently exploited for adoptive T cell transfer, a potent means of fighting cancer and infectious diseases.54, 55, 56, 57
mRNA is capable of encoding any of those antibody variants. Since exogenous mRNA instructs cells of the body to produce these proteins, they should be properly assembled and receive a natural glycosylation pattern. However, mRNA can deliver only antibodies with natural post-translational modifications. Hence, antibody conjugates and modifications, such as PEGylation for prolonged serum half-life of antibody fragments, cannot be recapitulated with mRNA.58 Partial compensation for the latter may come from the mRNA by expressing protein for a period of time instead of providing a protein pulse. Nevertheless, different areas of antibody therapy have their own challenges, and it has to be determined in detail whether antibody-encoding mRNA can meet the specific requirements. In the following sections, we introduce relevant fields of antibody treatment and review the respective mRNA applications.
Antibodies against Toxins
In 1890, the toxin-protective properties of serum of animals immunized with diphtheria or tetanus toxin were discovered.59 Four years later, serum therapy was successfully administered for the first time to children suffering from diphtheria. At the same time, two independent reports demonstrated that serum of immunized animals can protect against snake venoms.60, 61 After antibodies had been demonstrated to be the active component of toxin-neutralizing serum, the serum was replaced by immunoglobulins or F(ab′)2 antibody fragments purified from serum and, later, plasma.62, 63 Removal of the Fc fragment that shortens serum half-life was primarily introduced to reduce the probability of adverse reactions such as complement activation. Nevertheless, non-human immunoglobulin preparations are prone to inducing an immune response that hampers efficacy.64 If human sources are used, availability of appropriate donors could be challenging. Despite these and other issues, immunoglobulin preparations against toxins are still widely used. To date, antitoxins that neutralize venoms from various spiders, scorpions, and snakes, to name but a few, have been developed. As a further example, the standard treatment for botulism is a trivalent equine antitoxin.65, 66 The use of recombinant mAbs as antitoxins could overcome the limitations of immunoglobulin preparations. However, Bezlotoxumab, binding to Clostridium difficile toxin, is the only recombinant antitoxin that has been approved.67
Current antitoxins are often suboptimal and restricted in availability. Hence, the World Health Organization (WHO) put snakebites on its list of neglected tropical diseases in 2017. Moreover, for instance, botulinum toxin is not only a natural threat, but has a history of use as a bioweapon and is thus classified as a category A bioterror agent. These aspects may explain the extensive research on novel and better antitoxins. Numerous mAbs have been developed for botulinum neurotoxin, various scorpion and snake venoms, and other toxins, such as ricin, staphylococcal enterotoxin B, and C. difficile.68, 69, 70, 71, 72, 73 Several studies have indicated that combining different mAbs targeting the same toxin improve neutralization.71, 72, 73, 74 Compared to IgG mAbs, VHH domains of single-domain antibodies from camelids or shark provide various advantages, including better thermal and pH stability, and facilitate tissue penetration, which can be of particular benefit for antitoxins.75, 76 Thus, a plethora of studies investigated VHH-based antibodies for a variety of different toxins, including anthrax toxin, botulinum neurotoxin, Shiga toxin, ricin, C. difficile toxin, staphylococcal enterotoxin B, and scorpion and snake venoms.77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88 As for mAbs, combining different binding domains that can be easily assembled into bi- or oligovalent VHH-based neutralizing agents (VNAs) provides particularly potent reagents.80, 83, 85
mRNA as Antitoxin
Beyond efficacy, time to neutralization is a critical parameter of any antitoxin in an emergency, i.e., in the case of post-exposure treatment. As the immediate protein precursor, antibody-encoding mRNA may represent a suitable alternative to protein preparations. In a recent study, Thran et al.,89 for the first time, investigated whether mRNA could indeed meet the requirements of an antitoxin. To this end, they encoded a VNA that had been shown previously to potently neutralize botulinum neurotoxin.80 It consists of two linked VHHs binding distinct epitopes and an albumin-binding peptide that prolongs serum half-life to 1–2 days (Figure 2).80 The same design was used for a second VNA targeting Shiga toxin. For their investigations, Thran et al.89 built on sequence-optimized, chemically unmodified mRNA. The VNA-ORF was maximized for GC content and flanked with 5′ and 3′ UTRs for human hydroxysteroid (17-β) dehydrogenase-4 and human albumin, respectively, which had been successfully used in a previous study.21 Enzymatic steps introduced a cap1 structure at the 5′ end and a poly(A) tail at the 3′ end. Together with a template-encoded poly(A) sequence and linker, the latter formed a bipartite poly(A) element, structurally similar to the 3′ end of mRNA used for the expression of bi-specific T cell engagers (BiTEs).90 mRNA was purified by HPLC and formulated in LNPs for i.v. administration. Thus, the mRNA was primarily delivered to the liver, representing a bioreactor for antibody expression.91
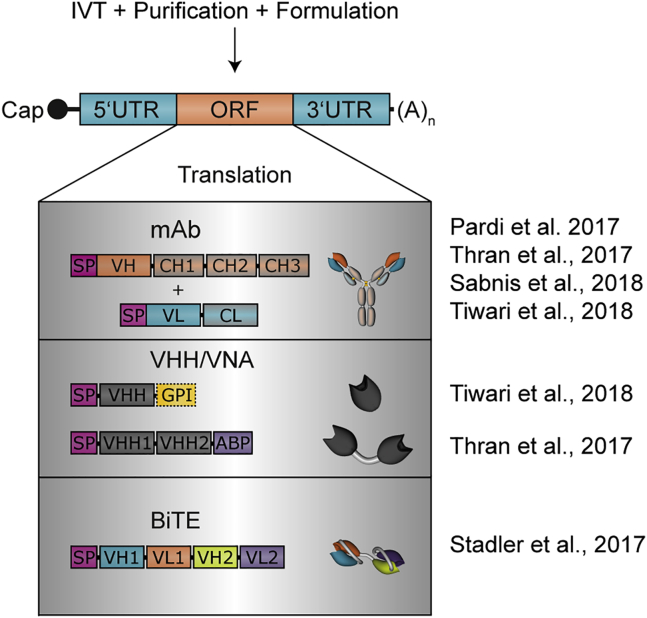
Figure 2.
Schematic Representation of mRNA and Antibody Designs Used in Recent Studies
All studies on mRNA-based antibody expression conducted so far have in common the use of thoroughly purified mRNA from in vitro transcription (IVT). Whereas most studies formulated the mRNA to enable i.v. injection, intratracheal administration by Tiwari et al.106 was mostly done with uncomplexed mRNA. Although differing in details and particularly regulatory sequence elements, the basic structure of mRNAs was identical: the open reading frame (ORF) was flanked by specific UTR sequences, and the 5′ and 3′ ends were formed by a cap structure and a poly(A) tail, respectively. In the various studies, different antibody formats were used; the principle domain and protein structure are depicted here. SP, signal peptide; VH, variable (domain) heavy; VL, variable (domain) light; CH, constant (domain) heavy; CL, constant (domain) light; VHH, VH domain, heavy-chain-only; VNA, VHH-based neutralizing agents; ABP, albumin-binding peptide.
Comparison of mRNA-derived VNAs produced in eukaryotic cells and recombinant proteins prepared from E. coli revealed equivalent potencies, both for the anti-botulinum neurotoxin serotype A (BoNTA) and the anti-Shiga toxin VNA. For subsequent in vivo studies, outbred CD1 mice were used. Upon i.v. administration of 40 μg VNA-mRNA in LNPs, serum titers 24 h later were remarkably different. Whereas the anti-BoNTA VNA reached serum titers of 200–400 μg/mL, those of the other antibody were approximately 10-fold lower. This supports an earlier statement that unlocking the full potential of mRNA requires sophisticated target-specific optimization.21 In vivo titration showed a more-than-linear increase in serum titers with elevated mRNA-LNP doses, a phenomenon that was observed for other antibody formats as well, but the reason for it is not yet understood.89, 92 Onset of in vivo expression was very fast; substantial protein levels (about one-tenth of maximum serum titers) were already detectable after 2 h, the earliest time analyzed. This finding is in line with observations that mRNA accumulates in hepatocytes within minutes after i.v. administration of LNPs and gives rise to considerable protein levels within a few hours.26, 32 Serum titers peaked at approximately 24 h after administration for BoNTA-VNA and apparently slightly faster, at 6–24 h, for Stx2-VNA. During the first 3 days after mRNA administration, serum VNA levels appeared to decline more slowly than suggested by the reported half-life of the recombinant proteins and compared with later serum titer kinetics, indicating that early kinetics benefitted from enduring mRNA expression. Protective capacity of BoNTA-VNA mRNA was assessed in an intoxication model. To this end, the authors chose a post-exposure setting to challenge mRNA as an emergency therapeutic.89 Regardless of whether mRNA-LNP was administered 2, 4, or 6 h after intravenous (i.v.) intoxication, all animals expressing BoNTA-VNA survived, whereas control mice receiving mRNA for an irrelevant VNA succumbed. Even in the most challenging setting (6 h after exposure) in which animals developed mild clinical symptoms before they fully recovered, mRNA was on par with recombinant BoNTA-VNA. Notably, the dose of recombinant VNA was lower than the applied mRNA amount. According to serum titers measured in preceding experiments, a much lower dose of mRNA-LNP should have been sufficient to obtain protection. However, it is also possible that a higher dose of mRNA-LNP compared with recombinant protein is required, to get almost immediate toxin neutralization, since mRNA inevitably slightly lags behind recombinant approaches regarding kinetics of protein availability. In summary, this first and, as yet, only report suggests that mRNA may be an appropriate antitoxin platform. (For currently known or announced applications, see Table 1.)
Table 1.
Applications of mRNA-Encoded Antibodies
| Medical Area | Antibody Format | Indication | Developmental State | Company/Research Group |
|---|---|---|---|---|
| Oncology | bispecific | solid tumor | research | BioNTech AG90 |
| undisclosed | solid tumors | pre-clinical | BioNTech AG | |
| IgG | CD20 | research | CureVac AG89 | |
| undisclosed | superficial tumors | pre-clinical | CureVac AG | |
| Infectious diseases | IgG | HIV | research | University of Pennsylvania92 |
| IgG | rabies | research | CureVac AG89 | |
| IgG | influenza B | research | CureVac AG89 | |
| IgG | influenza A | research | Moderna Therapeutics42 | |
| various | RSV | research | Georgia Institute of Technology and Emory University106 | |
| undisclosed (mRNA-1944) | Chikungunya virus | phase 1 | Moderna Therapeutics/DARPA | |
| Toxins | VNA | Botulinum toxin | research | CureVac AG89 |
| VNA | Shiga toxin | research | CureVac AG89 | |
| Undisclosed | undisclosed | molecular therapies | pre-clinical | CureVac AG |
DARPA, Defense Advanced Research Projects Agency.
Antibodies against Infectious Diseases
Sera for passive immunization to treat infectious diseases were initially generated by vaccinating horses, but, in a large number of patients, the equine sera induced “serum sickness” caused by hypersensitivity reactions, a problem that could be avoided by using pooled human sera prepared from individuals with high antibody titers (hyperimmune sera). This approach using animal or human sera has also been applied to treat or prevent viral infections. In 1996, RespiGam (respiratory syncytial virus [RSV] immune globulin) was approved to prevent RSV infection in children under 2 years of age with additional risk factors.93, 94, 95 Hyperimmune human sera have also been used to treat or prevent other viral diseases, including cytomegalovirus (CMV), hepatitis A and B, and measles.96, 97 The most common passive immunization still applied today is the use of rabies immunoglobulin and, more recently, purified equine rabies immunoglobulin fragments, F(ab′)2, after a proven or potential exposure to infectious rabies virus during post-exposure prophylaxis (PEP).98
The field of passive immunization took a further leap forward when mAbs were used instead of human sera, based on the groundbreaking work published in 1975 by Köhler and Milstein,99 who were also awarded a Nobel Prize in 1984. The first licensed product against an infectious disease based on mAbs was Palivizumab, used to prevent RSV infection in at-risk infants.100, 101 It is a humanized mAb of the IgG1 isotype. It neutralizes RSV by interacting with the RSV fusion protein F and is effective against both RSV serotypes. The efficacy of Palivizumab was demonstrated in premature babies (born at ≤35 weeks) or infants with bronchopulmonary dysplasia (BPD) when it was applied in five intramuscular injections of 15 mg/kg each. With respect to the primary endpoint, hospitalization with confirmed RSV infection, Palivizumab prophylaxis provided a 55% reduction over placebo treatment.100 Because other prophylaxis or treatment options such as a licensed RSV vaccine are lacking, Palivizumab is still the standard of care. More than 30 mAbs have subsequently been licensed for human use, but only 2 of them are for treatment of infectious diseases (anthrax and rabies). The latest mAb, Rabishield, produced by the Serum Institute of India, was approved in India in 2016 for the prevention of rabies.102 Again, the mode of action of this antibody is virus neutralization, and the mAb can be used for post-exposure prophylaxis of rabies in the event of an exposure to infectious rabies virus. Many more therapeutic or preventive mAbs are in development for infectious diseases including, but not limited to, HIV, Ebola virus, and influenza.103
mRNA Antibody Therapy
A drawback of these passive immunization approaches against infectious diseases is the need for large amounts of purified immunoglobulins or mAbs to provide adequate protection. Hence, the next iteration in the ongoing developments in this field is to improve the efficiency by generation of protective mAbs in the host by administering the respective genetic material.
Two recent publications investigated i.v. administration of mRNA-encoded antibodies as a means of protecting against infections with HIV or rabies virus.89, 92 Both studies used humanized IgG antibodies with proven virus neutralizing capacity. For mRNA-based expression, the ORF sequences for heavy and light chains of the mAbs were encoded on individual mRNAs and mixed in either an equimolar92 or experimentally determined optimal ratios,89 prior to formulation in LNPs. Although the LNPs were obtained from the same company, it is not known whether the composition was identical in both studies. With respect to mRNA design, major differences beyond applying distinct antibodies and UTR elements include the use of 100% N1-methyl-pseudouridine, a cap1 structure and a monopartite poly(A) tail of approximately 100 nucleotides, for HIV,92 compared with exclusively unmodified nucleotides, a cap0 structure and a long, bipartite poly(A) tail for the rabies virus.89
Given their heterotetrameric nature, mAb functionality requires correct assembly of heavy and light chains before secretion. Starting the initial characterization with three different humanized mAbs against rabies, influenza, and HIV, Thran et al.89 demonstrated that, upon in vitro cell transfection, the mRNA produced functional antibodies in all instances. Moreover, direct comparison to recombinant protein for one of the mAbs, VRC01 against HIV, revealed equivalence with respect to in vitro virus neutralization. Both groups observed a disproportionately high increase in antibody serum titers in mice in response to increasing mRNA-LNP doses given i.v.89, 92 Whereas serum titers in BALB/c mice generated by VRC01-mRNA in Pardi et al.92 showed an accelerated decrease after 4–6 days and became virtually undetectable after 11 days, Thran et al.89 measured a steady serum half-life of about 1 week for their anti-influenza IgG during the observation period of more than 3 weeks in outbred NIH Swiss albino mice. In contrast, their anti-rabies mAb showed the same kinetics in only half of the animals. Clearance from the serum started to accelerate in the remaining mice after 10–14 days, for which the authors could demonstrate a correlation to the development of anti-drug antibodies (ADAs). Obviously, an ADA is strongly dependent on the specific antibody and is not an intrinsic consequence of the use of mRNA-LNP. However, ADA cannot be responsible for the VRC01 pharmacokinetics in the study by Pardi et al.,92 since immunocompromised NSG mice that cannot develop ADA reveal antibody kinetics similar to BALB/c animals. In an independent expression study conducted in non-human primates, which are expected to tolerate human IgGs much better than mice, mAb serum titers after i.v. injection of mRNA-LNP lasted for several weeks, suggesting the absence of ADA.42 However, Pardi et al.92 did not investigate whether ADA responses developed later or after repeated treatment, as has been observed, for instance, in AAV-based approaches.104, 105
The studies of Pardi et al.92 and Thran et al.89 both successfully demonstrated that mRNA-mediated antibody expression can protect mice from virus infection. To this end, Pardi et al.92 conducted HIV-1 challenge experiments in NSG mice with a reconstituted human immune system, generated by injection of human CD34+ hematopoietic stem cells. Remarkably, i.v. administration of 30 μg of mRNA (approximately 1.4 mg/kg) generated 2-fold higher antibody serum titers than 600 μg of the corresponding recombinant protein, but an mRNA dose of 0.7 mg/kg had already been found to be sufficient to fully protect mice when given 24 h prior to exposure to the virus. Thran et al.89 used a very similar maximum dose of 40 μg of mRNA and focused on the anti-rabies mAb for in vivo efficacy studies in NIH Swiss mice. Consistent with a fast onset of antibody expression, peaking as early as about 4 h, mice were fully protected, not only in a pre- but also in a post-exposure scenario. In addition to efficacy, good tolerability is an important parameter for mRNA-based antibody expression to become a viable approach for passive immunization, not least because it is a prerequisite for repeated treatment if needed. Pardi et al.92 demonstrated in C57BL/6 mice that mRNA-LNP administration is possible without inducing pro-inflammatory cytokines, such as interleukin-6 (IL-6), interferon-α (IFN-α), and tumor necrosis factor-α (TNF-α). In line with previous reports, incorporation of modified nucleotides was insufficient; thorough purification using chromatography was required in addition. Moreover, the authors were able to maintain elevated mAb serum levels by weekly injections into NSG mice, without loss of efficacy. However, since these analyses of tolerability used immunocompromised mice or different mRNAs (not coding for VRC01 mAb), formulations, or administration routes compared to the challenge studies, the properties of i.v. administered mRNA-LNPs expressing the VRC01 mAb can be deduced indirectly at best. Although the experimental setting of Thran et al.89 appeared to elicit some weak cytokine responses in outbred NIH Swiss mice, they were obviously well below levels considered critical. The authors observed neither suppression of antibody expression nor the occurrence of adverse effects, such as abnormal liver histopathology.
These two studies addressed systemic delivery of antibody-encoding mRNA, which used the liver as a bioreactor to provide the systemic antibody. However, indications such as infection with RSV need protective antibodies only at distinct locations—the lung in the case of RSV. In such instances, local mRNA delivery and expression may be more effective than establishing systemic titers. Actually, intramuscular administration of the recombinant anti-RSV mAb Palivizumab merely delivers a small fraction of antibody to the relevant tissue. Hence, Tiwari et al.106 investigated whether mRNA-based antibody expression may cope with the challenges of RSV infection in another study, using BALB/c mice as an animal model. They applied mRNA harboring a cap1 structure, a poly(A) tail, and N1-methyl-pseudouridine. mRNA was purified with standard spin columns instead of by chromatographic means and was administered as intratracheal aerosol. Most in vivo experiments were conducted with naked mRNA, since it turned out to be at least as efficient as formulations with one of two polyethyleneimine (PEI) derivatives, confirming earlier reports that, surprisingly, unprotected mRNA somehow escapes degradation in the lung, at least partially.37, 107 As antibodies, the authors applied Palivizumab and an anti-RSV VHH, in both secreted and membrane-anchored form. Heavy and light chains of Palivizumab were encoded on individual mRNAs that were mixed in a 4:1 molar ratio, substantially differing from the work reviewed above. mRNA-derived antibodies were functional in vitro, demonstrated by suppression of cell infection. Upon intratracheal aerosol delivery, mRNA was distributed throughout the lung, leading to up to 45% of cells with detectable antibody expression. This gave rise to strongly reduced RSV F copy numbers (by about 90%), 4 days after infection, which took place after administration of virus preparations to each nostril 1 day after mRNA treatment. The dose response was somewhat inconclusive, since 100 and 40 μg of mRNA appeared to be similarly effective, whereas 20 μg did not reduce RSV F copy numbers at all. When VHH was anchored to the membrane, RSV was substantially inhibited even at 7 days after mRNA administration, and the protein was still detectable 28 days after transfection. Intratracheal aerosol delivery of naked mRNA did not induce significantly elevated cytokine levels in lungs 24 h after treatment.
In sum, current data strongly suggest that mRNA-mediated antibody expression has the potential to be included in the armamentarium against infectious diseases. (For currently known or announced applications, see Table 1.)
Antibodies in Oncology
Since the first approval of an antibody for cancer treatment, the pace of development of antibody-based therapies in oncology has continuously accelerated, leading to more than 30 approved antibodies today and further candidates currently under clinical evaluation. The majority of approved therapies are based on IgG mAbs, but antibody-drug conjugates and bispecific antibodies have gained in importance.
The fundamental idea of antibody-based cancer therapy dates back to observations in the 1960s that human cancer cells can be identified by specific antigens, which are overexpressed, mutated, or selectively expressed compared to healthy tissue and therefore are targetable by antibodies.108 Antibody-based therapies in cancer can be subdivided into three major therapeutic approaches.109
-
(1)
Antibodies that are able to mediate direct tumor cell killing after binding of the target receptor through withdrawal of essential growth signals or delivery of apoptosis signals. For instance, Trastuzumab blocks HER2/ERBB2 receptor dimerization, kinase activation, and downstream signaling, leading to starvation of cancer cells and, ultimately, to cancer cell death.110 Agonistic antibodies binding to apoptosis-inducing receptors on cancer cells like TNF-related apoptosis-inducing ligand (TRAIL) receptors and antibody-drug conjugates (ADCs) delivering toxic payloads specifically to cancer cells are also able to induce direct tumor cell killing.111, 112
-
(2)Antibodies that induce immune-mediated cell killing.
-
(2.1)IgG antibodies activate immune-mediated cell killing by binding to a cancer cell-specific antigen resulting in complement-dependent cytotoxicity (CDC) or antibody-dependent cellular cytotoxicity (ADCC).113
-
(2.2)Antibodies blocking CTLA4 or PD1, inhibitory co-receptors on immune cells that are involved in immune evasion, induce immune-mediated cell killing through reinvigoration of antigen-specific T cell responses directed against the cancer cells.114
-
(2.3)Antibodies with dual specificity like blinatumomab redirect T cells to cancer cells and induce immune-mediated cell killing by direct engagement.115
-
(2.1)
-
(3)
The last approach is the ablation of tumor vasculature or tumor stroma through targeting of vascular growth factors or stromal factors that deprive the tumor tissue of essential nutrients and oxygen and lead ultimately to tumor cell death.116
The success of antibody-based therapies for cancer over the past years, especially immune checkpoint blockade, has invigorated immuno-oncology research and discovery of additional co-inhibitory receptors, such as LAG-3, TIM-3, and TIGIT, as well as co-stimulatory receptors, including CD40, OX40, 4-1BB, ICOS, and GITR. Currently, several mAb-based therapies have been developed that target these immunomodulators.117 Moreover, major efforts are invested in the development of, e.g., smaller mAb fragments or bispecific antibodies. Compared to mAbs, mAb fragments and engineered variants such as diabodies, triabodies, minibodies, and single-domain antibodies provide unique properties, including higher tissue penetration and the opportunity to combine several targeting moieties.118 Bispecific antibodies can be divided into three main categories based on their biological target and mode of action.115
-
(a)
Cytotoxic effector cell redirectors that target T cells or natural killer (NK) cells directly to tumor cells expressing a specific antigen.119, 120
-
(b)
Tumor-targeted immunomodulators that enable tumor-infiltrating immune cells and tumor cells to co-stimulate immune cells via CD40 or 4-1BB activation.115
-
(c)
Dual immunomodulators that simultaneously bind to two immunomodulating targets expressed on immune cells. These immunomodulators can block inhibitory immune pathways, deplete suppressive immune cells or activate effector cells.115, 121
mRNA-Based Antibody Therapy for Cancer
Recombinant antibody-based therapies in cancer face some challenges related to manufacturing issues, including poor stability during long-term storage, tendency to aggregate over time, and the presence of various impurities intrinsic to the production process, as well as properties intrinsic to the drug product.118 Especially bispecific antibodies have a low serum half-life of a few hours, necessitating an infusion pump for continuous delivery in patients.122 The use of mRNA resulting in the generation of therapeutic antibodies directly in patients represents a promising approach to overcoming these limitations.
In a recent study using immune-mediated killing via bispecific antibodies (category “a” bispecifics with mode of action 2.3 above), Stadler et al.90 demonstrated that IVT mRNA encoding a bispecific T cell engager targeting the tumor-associated antigens claudin 6 (CLDN6), claudin 18.2 (CLDN18.2), or epithelial cell adhesion molecule (EpCAM) can be translated into proteins in cells named RiboMABs. The obtained RiboMABs were able to induce specific lysis of target cells at picomolar concentration with potency comparable to the corresponding recombinant protein. The authors used engineered mRNA with specific 5′ and 3′ UTRs, a cap1 structure, and a bipartite poly(A) tail to improve intracellular stability and translational efficiency. Modified nucleotides were incorporated into the mRNA during IVT, and mRNA was purified by HPLC to ensure translation and avoid systemic proinflammatory cytokine release. In vivo administration of unpurified conventional mRNA without modified nucleosides resulted in systemic cytokine release and minimal RiboMAB protein in human peripheral blood mononuclear cell (PBMC)-engrafted NSG mice. According to luciferase expression data in BALB/c mice, the i.v. injection of CD3 x CLDN6 mRNA formulated with a polymer/lipid-based transfection reagent (TransIT) into NSG mice resulted in uptake and abundant translation in the liver. The encoded bispecific antibody was detectable in the plasma starting 15–30 min after administration, peaking after 6 h, and remaining measurable, even after 72 h. In contrast, after single administration of a comparable dose of recombinant CD3 x CLDN6 protein, the plasma level dropped sharply after 6 h and was barely detectable after 24 h, because of the short serum half-life of the protein. An ex vivo cytotoxicity assay demonstrated the ability of plasma from mRNA-treated mice to induce specific lysis of CLDN6-expressing target cells. The maximum lysis of 90% was reached in plasma 6 h after treatment, in line with peak expression at the same time, and remained above half-maximal level for up to 6 days after mRNA injection. In contrast, the cytotoxic activity of plasma derived from mice receiving recombinant protein dropped to background levels quickly (after 24 h). An extended dose-response analysis demonstrated strong and sustained ex vivo cytotoxic activity after a single injection of 0.05 μg of CD3 x CLDN6 BiTE-encoding mRNA. Safety evaluation of RiboMAB mRNA in human PBMC-engrafted NSG mice revealed no evidence of liver toxicity or systemic cytokine release caused by non-specific T cell activation.90
The therapeutic efficacy of mRNA was investigated for two bispecific antibodies: CD3 x CLDN6 and EpCAM x CD3. To this end, NSG mice were subcutaneously inoculated with human OV-90 ovarian carcinoma cells expressing the tumor-associated antigens CLDN6 and EpCAM. After tumor establishment, mice were engrafted with human PBMCs 1 week prior to the start of mRNA treatment. Mice received 3 μg of CD3 x CLDN6 mRNA once a week for a consecutive 3 weeks or 200 μg/kg body weight of the corresponding recombinant antibody, administered three times a week for a total of 10 doses. Complete tumor rejection was reached in all mice treated with CD3 x CLDN6 mRNA or protein, whereas all control tumor-bearing mice treated with an unspecific mRNA progressed. A significant increase in tumor-infiltrating T cells was detectable only in tumors derived from mice treated with CD3 x CLDN6 mRNA but not in mice treated with a non-specific mRNA. The results observed with CD3 x CLDN6 mRNA could be reproduced with EpCAM x CD3 mRNA in the same model. Notably, weekly treatment with mRNA resulted in anti-tumoral efficacy comparable to that obtained with administration three times weekly of an equivalent recombinant protein dose.90
Thran et al.89 also demonstrated anti-tumoral therapeutic efficacy of an mRNA-encoded antibody, but in a human B cell lymphoma model. The heavy and light chain sequence of rituximab, a clinically approved IgG1 antibody against the human tumor-associated antigen CD20 (mode of action 2.1 above), was encoded by two separate mRNAs. In contrast to the investigations performed by Stadler et al.,90 optimized, HPLC-purified IVT mRNA with natural, unmodified nucleotides was used in the experiments. Further major differences regarding the mRNA were: (1) cap0 instead of cap1 5′ structure, (2) different 5′ and 3′ UTRs, and (3) formulation in LNP instead of TransIT. Similar to the first study, the principal translation of IVT mRNA into a functional antibody was demonstrated in cells. For in vivo efficacy, NOD/SCID mice were challenged i.v. with Raji B cells, a human B cell lymphoma cell line expressing CD20, stably transfected with luciferase to enable in vivo monitoring of tumor progression. Twice weekly treatment with different doses of rituximab mRNA-LNP harboring the two mRNAs in an equimolar ratio started 4 days after tumor cell inoculation. The formulated mRNA was administered i.v. to enable uptake and abundant translation in the liver.21, 36 Complete tumor rejection was reached in the majority of mice treated with a 50 μg dose of rituximab mRNA-LNP, and mice treated with a 10 μg dose demonstrated significantly decreased tumor growth. Notably, a control group treated with 200 μg recombinant antibody progressed faster in comparison to the high-dose rituximab mRNA-LNP-treated group.89
Taken together, both studies clearly show that mRNA-mediated antibody expression is applicable to oncology and may provide advantages compared to recombinant biologics in certain instances such as short-lived antibody formats. (For currently known or announced applications, see Table 1.)
Outlook
Although studies on mRNA-based antibody expression are still very limited in number, data support the notion that it may develop into a competitive therapeutic approach. However, results on expression and efficacy have been mostly restricted to small rodent models. Hence, translation to larger animals and finally humans has yet to be demonstrated. Unrelated work on mRNA-mediated protein expression provides the first evidence that application in much larger animals, including non-human primates (NHPs), is possible.21, 33 Moreover, a recent publication on the development of LNP formulations indicated that substantial antibody expression can be obtained in NHPs by improving endosomal escape.42 However, efficacy of LNPs appeared to drop from mice to NHPs. Thus, establishing mRNA-mediated antibody expression as a viable therapeutic option in humans may require further progress with respect to mRNA and formulation technology to reach effective protein titers, especially in those instances that need particularly high serum levels.
Besides efficacy, passive immunization demands an excellent safety profile. Different strategies have been developed to avoid inappropriate stimulation of cellular RNA sensors and, thus, adverse events such as immune responses or strong induction of cytokine secretion. Accordingly, the mRNA-based antibody treatments described here did not reveal critical tolerability issues, in line with various mRNA studies on protein (replacement) therapy. However, repeated doses of nanoparticles can induce complement activation-related pseudoallergy (CARPA)123, to which mice are rather insensitive. Hence, tolerability has to be analyzed in more detail, and it may turn out that mRNA-based antibody therapy requires further advancement of formulations. Biocompatibility of LNPs, for instance, can be improved by facilitating degradability.32, 42, 124 The trend toward subcutaneous injection of antibodies for improved convenience and reduced therapy costs poses an additional challenge. Although administration via routes other than i.v. has been shown for mRNA,36 it has still to be demonstrated that mRNA-based antibody therapy remains efficacious with the use of such routes. Finally, it has to be effective at a competitive cost.
Importantly, mRNA-based antibody expression may offer benefits compared to recombinant proteins (Box 1). First, mRNA gives rise to protein expression for typically a few days instead of providing a single protein pulse upon administration. Indeed, extension of therapeutic levels of short-lived proteins was demonstrated for two VNAs with serum half-lives of 1–2 days and even more impressively for a BiTE antibody with a serum half-life of just a few hours.89, 90 Thus, mRNA could reduce the frequency of treatments if repeated doses are required. Second, mRNA offers the advantage that individual sequences are much more similar regarding physicochemical characteristics than different antibodies, thereby enabling a generic process that reduces costs. This may also facilitate manufacturing of cocktails, but requires a solution that avoids unwanted antibody chimeras. Whereas in the case of long-lived IgGs, sequential administration is at least an option due to the transience of mRNA expression, a more convenient and true cocktail approach would need more sophisticated solutions such as the knob-into-hole concept.125 Third, mRNA-mediated antibody expression can address intracellular targets, since nucleic acids can be readily transfected into cells. In contrast, delivery of proteins through the cell membrane is challenging.126, 127 However, such intrabody approaches depend on appropriate mRNA delivery systems. Whereas targeting to the liver appears to be solved by LNPs, formulations delivering to other tissues are largely lacking. However, with the emergence of mRNA as a much-appreciated therapeutic platform, it is very likely that there is more to come in the near future, including passive immunization.
Box 1. The Pros and Cons of mRNA Compared with Recombinant Protein Regarding Passive Immunization.
Pros
Cell-free production
Uniform physiochemistry of drug substances
Same technology for all antibody (Ab) formats
Potential for storage at ambient temperature
mRNA half-life confers improved pharmacokinetics (PK; in case of short-lived Ab)
Accessibility of intracellular targets
Potentially improved tolerability due to patient-specific Ab modification
Opportunity to reduce systemic exposure by in situ synthesis
Potential of high Ab titers with low doses of drug substance
Cons
Two-component drug product (mRNA and formulation)
Lag phase to reach Ab peak level
No Ab conjugates
No PEGylation for improved PK
Potentially variable Ab modification (e.g., among patients)
Potential activation of cellular immune sensors
Administration limited to i.v. route as yet (subcutaneous still unproven)
Technology at early developmental stage
Acknowledgments
We thank our colleagues Andreas Thess and Michael Stolz for critical reading of the manuscript. We are grateful to Bettina Danker for excellent graphical illustrations. We would like to note that there are many more important publications in the fields of passive immunization and the use of mRNA that could have been cited. Because of space limitations, we had to restrict the citations, and thus we apologize to all authors affected.
References
- 1.Wolff J.A., Malone R.W., Williams P., Chong W., Acsadi G., Jani A., Felgner P.L. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 2.Jirikowski G.F., Sanna P.P., Maciejewski-Lenoir D., Bloom F.E. Reversal of diabetes insipidus in Brattleboro rats: intrahypothalamic injection of vasopressin mRNA. Science. 1992;255:996–998. doi: 10.1126/science.1546298. [DOI] [PubMed] [Google Scholar]
- 3.I T., Thess A., Thran M., Jordan I. mRNA as novel technology for passive immunotherapy. Cell. Mol. Life Sci. 2019;76:301–328. doi: 10.1007/s00018-018-2935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlake T., Thess A., Fotin-Mleczek M., Kallen K.J. Developing mRNA-vaccine technologies. RNA Biol. 2012;9:1319–1330. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Triana-Alonso F.J., Dabrowski M., Wadzack J., Nierhaus K.H. Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J. Biol. Chem. 1995;270:6298–6307. doi: 10.1074/jbc.270.11.6298. [DOI] [PubMed] [Google Scholar]
- 6.Gholamalipour Y., Karunanayake Mudiyanselage A., Martin C.T. 3′ end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character-RNA-Seq analyses. Nucleic Acids Res. 2018;46:9253–9263. doi: 10.1093/nar/gky796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nallagatla S.R., Hwang J., Toroney R., Zheng X., Cameron C.E., Bevilacqua P.C. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 8.Harcourt E.M., Kietrys A.M., Kool E.T. Chemical and structural effects of base modifications in messenger RNA. Nature. 2017;541:339–346. doi: 10.1038/nature21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X., Zhu P., Ma S., Song J., Bai J., Sun F., Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 10.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X., Xiong X., Wang K., Wang L., Shu X., Ma S., Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 2016;12:311–316. doi: 10.1038/nchembio.2040. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert W.V., Bell T.A., Schaening C. Messenger RNA modifications: Form, distribution, and function. Science. 2016;352:1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Mu X., Greenwald E., Ahmad S., Hur S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018;46:5239–5249. doi: 10.1093/nar/gky177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kariko K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kariko K., Muramatsu H., Keller J.M., Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012;20:948–953. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchida S., Kataoka K., Itaka K. Screening of mRNA Chemical Modification to Maximize Protein Expression with Reduced Immunogenicity. Pharmaceutics. 2015;7:137–151. doi: 10.3390/pharmaceutics7030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlack T. Sequence-engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kauffman K.J., Mir F.F., Jhunjhunwala S., Kaczmarek J.C., Hurtado J.E., Yang J.H., Webber M.J., Kowalski P.S., Heartlein M.W., DeRosa F., Anderson D.G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pascolo S. Messenger RNA-based vaccines. Expert Opin. Biol. Ther. 2004;4:1285–1294. doi: 10.1517/14712598.4.8.1285. [DOI] [PubMed] [Google Scholar]
- 24.Probst J., Weide B., Scheel B., Pichler B.J., Hoerr I., Rammensee H.G., Pascolo S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007;14:1175–1180. doi: 10.1038/sj.gt.3302964. [DOI] [PubMed] [Google Scholar]
- 25.Vaidyanathan S., Azizian K.T., Haque A.K.M.A., Henderson J.M., Hendel A., Shore S., Antony J.S., Hogrefe R.I., Kormann M.S.D., Porteus M.H., McCaffrey A.P. Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther. Nucleic Acids. 2018;12:530–542. doi: 10.1016/j.omtn.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeRosa F., Guild B., Karve S., Smith L., Love K., Dorkin J.R., Kauffman K.J., Zhang J., Yahalom B., Anderson D.G., Heartlein M.W. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 2016;23:699–707. doi: 10.1038/gt.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nabhan J.F., Wood K.M., Rao V.P., Morin J., Bhamidipaty S., LaBranche T.P., Gooch R.L., Bozal F., Bulawa C.E., Guild B.C. Intrathecal delivery of frataxin mRNA encapsulated in lipid nanoparticles to dorsal root ganglia as a potential therapeutic for Friedreich’s ataxia. Sci. Rep. 2016;6:20019. doi: 10.1038/srep20019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin C.Y., Perche F., Ikegami M., Uchida S., Kataoka K., Itaka K. Messenger RNA-based therapeutics for brain diseases: An animal study for augmenting clearance of beta-amyloid by intracerebral administration of neprilysin mRNA loaded in polyplex nanomicelles. J. Control. Release. 2016;235:268–275. doi: 10.1016/j.jconrel.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Später D., Xu H., Tabebordbar M., Gorbatov R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Su H.H., Yang Y., Hu Y., Zhang L., Blancafort P., Huang L. Systemic delivery of modified mRNA encoding herpes simplex virus 1 thymidine kinase for targeted cancer gene therapy. Mol. Ther. 2013;21:358–367. doi: 10.1038/mt.2012.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. USA. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.An D., Schneller J.L., Frassetto A., Liang S., Zhu X., Park J.S., Theisen M., Hong S.J., Zhou J., Rajendran R. Systemic Messenger RNA Therapy as a Treatment for Methylmalonic Acidemia. Cell Rep. 2017;21:3548–3558. doi: 10.1016/j.celrep.2017.11.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carlsson L., Clarke J.C., Yen C., Gregoire F., Albery T., Billger M., Egnell A.C., Gan L.M., Jennbacken K., Johansson E. Biocompatible, Purified VEGF-A mRNA Improves Cardiac Function after Intracardiac Injection 1 Week Post-myocardial Infarction in Swine. Mol. Ther. Methods Clin. Dev. 2018;9:330–346. doi: 10.1016/j.omtm.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Probst J., Brechtel S., Scheel B., Hoerr I., Jung G., Rammensee H.G., Pascolo S. Characterization of the ribonuclease activity on the skin surface. Genet. Vaccines Ther. 2006;4:4. doi: 10.1186/1479-0556-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 38.Haji K.A., Whitehead K.A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:17056. [Google Scholar]
- 39.Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 40.Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 41.Gilleron J., Querbes W., Zeigerer A., Borodovsky A., Marsico G., Schubert U., Manygoats K., Seifert S., Andree C., Stöter M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 42.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ball R.L., Bajaj P., Whitehead K.A. Achieving long-term stability of lipid nanoparticles: examining the effect of pH, temperature, and lyophilization. Int. J. Nanomedicine. 2016;12:305–315. doi: 10.2147/IJN.S123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jäger V., Büssow K., Wagner A., Weber S., Hust M., Frenzel A., Schirrmann T. High level transient production of recombinant antibodies and antibody fusion proteins in HEK293 cells. BMC Biotechnol. 2013;13:52. doi: 10.1186/1472-6750-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walsh G. Biopharmaceutical benchmarks 2014. Nat. Biotechnol. 2014;32:992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- 46.Holliger P., Hudson P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 47.Tabrizi M.A., Tseng C.M., Roskos L.K. Elimination mechanisms of therapeutic monoclonal antibodies. Drug Discov. Today. 2006;11:81–88. doi: 10.1016/S1359-6446(05)03638-X. [DOI] [PubMed] [Google Scholar]
- 48.Könning D., Zielonka S., Grzeschik J., Empting M., Valldorf B., Krah S., Schröter C., Sellmann C., Hock B., Kolmar H. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr. Opin. Struct. Biol. 2017;45:10–16. doi: 10.1016/j.sbi.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 49.Hultberg A., Temperton N.J., Rosseels V., Koenders M., Gonzalez-Pajuelo M., Schepens B., Ibañez L.I., Vanlandschoot P., Schillemans J., Saunders M. Llama-derived single domain antibodies to build multivalent, superpotent and broadened neutralizing anti-viral molecules. PLoS ONE. 2011;6:e17665. doi: 10.1371/journal.pone.0017665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi B.D., Kuan C.T., Cai M., Archer G.E., Mitchell D.A., Gedeon P.C., Sanchez-Perez L., Pastan I., Bigner D.D., Sampson J.H. Systemic administration of a bispecific antibody targeting EGFRvIII successfully treats intracerebral glioma. Proc. Natl. Acad. Sci. USA. 2013;110:270–275. doi: 10.1073/pnas.1219817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fournier P., Schirrmacher V. Bispecific antibodies and trispecific immunocytokines for targeting the immune system against cancer: preparing for the future. BioDrugs. 2013;27:35–53. doi: 10.1007/s40259-012-0008-z. [DOI] [PubMed] [Google Scholar]
- 52.Zitron I.M., Thakur A., Norkina O., Barger G.R., Lum L.G., Mittal S. Targeting and killing of glioblastoma with activated T cells armed with bispecific antibodies. BMC Cancer. 2013;13:83. doi: 10.1186/1471-2407-13-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kontermann R.E. Dual targeting strategies with bispecific antibodies. MAbs. 2012;4:182–197. doi: 10.4161/mabs.4.2.19000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barrett D.M., Grupp S.A., June C.H. Chimeric Antigen Receptor- and TCR-Modified T Cells Enter Main Street and Wall Street. J. Immunol. 2015;195:755–761. doi: 10.4049/jimmunol.1500751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gottschalk S., Rooney C.M. Adoptive T-Cell Immunotherapy. Curr. Top. Microbiol. Immunol. 2015;391:427–454. doi: 10.1007/978-3-319-22834-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parida S.K., Poiret T., Zhenjiang L., Meng Q., Heyckendorf J., Lange C., Ambati A.S., Rao M.V., Valentini D., Ferrara G. T-Cell Therapy: Options for Infectious Diseases. Clin. Infect. Dis. 2015;61(Suppl 3):S217–S224. doi: 10.1093/cid/civ615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morgan R.A., Dudley M.E., Wunderlich J.R., Hughes M.S., Yang J.C., Sherry R.M., Royal R.E., Topalian S.L., Kammula U.S., Restifo N.P. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang K., Basu A., Wang M., Chintala R., Hsieh M.C., Liu S., Hua J., Zhang Z., Zhou J., Li M. Tailoring structure-function and pharmacokinetic properties of single-chain Fv proteins by site-specific PEGylation. Protein Eng. 2003;16:761–770. doi: 10.1093/protein/gzg093. [DOI] [PubMed] [Google Scholar]
- 59.Behring E., Kitasato S. Ueber das Zustandekommen der Diphtherie-Immunitat und der Tetanus-Immunitat bei thieren. Dtsch. Med. Wochenschr. 1890;16:1113–1114. [PubMed] [Google Scholar]
- 60.Phisalix C., Bertrand G. Sur la propriété antitoxique du sang des aeutraliaccinés contre le venin de vipère. Comptes Rendus de la Société de Biologie. 1894;46:111–113. [Google Scholar]
- 61.Calmette A. Contribution à l’étude du venin des serpents. Immunisation des animaux et traitement de l’envenimation. Ann. Inst. Pasteur (Paris) 1894;8:275–291. [Google Scholar]
- 62.Pope C.G. The action of proteolytic enzymes on the antitoxins and proteins in immune sera. I. True digestion of the proteins. Br. J. Exp. Pathol. 1939;20:132–149. [PMC free article] [PubMed] [Google Scholar]
- 63.Pope C.G. The action of proteolytic enzymes on the antitoxins and proteins in immune sera. II. Heat denaturation after partial enzyme action. Br. J. Exp. Pathol. 1939;20:201–212. [PMC free article] [PubMed] [Google Scholar]
- 64.Wilde H., Thipkong P., Sitprija V., Chaiyabutr N. Heterologous antisera and antivenins are essential biologicals: perspectives on a worldwide crisis. Ann. Intern. Med. 1996;125:233–236. doi: 10.7326/0003-4819-125-3-199608010-00012. [DOI] [PubMed] [Google Scholar]
- 65.American Academy of Pediatrics . Clostridial infections. In: Peter G., editor. 1997 Red Book: Report of the Committee on Infectious Diseases. Twenty-Fourth Edition. American Academy of Pediatrics; 1997. pp. 174–178. [Google Scholar]
- 66.Shapiro R.L., Hatheway C., Swerdlow D.L. Botulism in the United States: a clinical and epidemiologic review. Ann. Intern. Med. 1998;129:221–228. doi: 10.7326/0003-4819-129-3-199808010-00011. [DOI] [PubMed] [Google Scholar]
- 67.Wilcox M.H., Gerding D.N., Poxton I.R., Kelly C., Nathan R., Birch T., Cornely O.A., Rahav G., Bouza E., Lee C., MODIFY I and MODIFY II Investigators Bezlotoxumab for Prevention of Recurrent Clostridium difficile Infection. N. Engl. J. Med. 2017;376:305–317. doi: 10.1056/NEJMoa1602615. [DOI] [PubMed] [Google Scholar]
- 68.Fan Y., Barash J.R., Lou J., Conrad F., Marks J.D., Arnon S.S. Immunological Characterization and Neutralizing Ability of Monoclonal Antibodies Directed Against Botulinum Neurotoxin Type H. J. Infect. Dis. 2016;213:1606–1614. doi: 10.1093/infdis/jiv770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clot-Faybesse O., Juin M., Rochat H., Devaux C. Monoclonal antibodies against the Androctonus australis hector scorpion neurotoeutralizationnisation and use foreutralizationisation. FEBS Lett. 1999;458:313–318. doi: 10.1016/s0014-5793(99)01179-5. [DOI] [PubMed] [Google Scholar]
- 70.Trémeau O., Boulain J.C., Couderc J., Fromageot P., Ménez A. A monoclonal antibody which recognized the functional site of snake neurotoxins and which neutralizes all short-chain variants. FEBS Lett. 1986;208:236–240. doi: 10.1016/0014-5793(86)81024-9. [DOI] [PubMed] [Google Scholar]
- 71.Noy-Porat T., Alcalay R., Epstein E., Sabo T., Kronman C., Mazor O. Extended therapeutic window for post-exposure treatment of ricin intoxication conferred by the use of high-affinity antibodies. Toxicon. 2017;127:100–105. doi: 10.1016/j.toxicon.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 72.Dutta K., Varshney A.K., Franklin M.C., Goger M., Wang X., Fries B.C. Mechanisms mediating enhanced neutralization efficacy of staphylococcal enterotoxin B by combinations of monoclonal antibodies. J. Biol. Chem. 2015;290:6715–6730. doi: 10.1074/jbc.M114.630715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lowy I., Molrine D.C., Leav B.A., Blair B.M., Baxter R., Gerding D.N., Nichol G., Thomas W.D., Jr., Leney M., Sloan S. Treatment with monoclonal antibodies against Clostridium difficile toxins. N. Engl. J. Med. 2010;362:197–205. doi: 10.1056/NEJMoa0907635. [DOI] [PubMed] [Google Scholar]
- 74.Nowakowski A., Wang C., Powers D.B., Amersdorfer P., Smith T.J., Montgomery V.A., Sheridan R., Blake R., Smith L.A., Marks J.D. Potent neutralization of botulinum neurotoxin by recombinant oligoclonal antibody. Proc. Natl. Acad. Sci. USA. 2002;99:11346–11350. doi: 10.1073/pnas.172229899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E.B., Bendahman N., Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 76.van der Linden R.H., Frenken L.G., de Geus B., Harmsen M.M., Ruuls R.C., Stok W., de Ron L., Wilson S., Davis P., Verrips C.T. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta. 1999;1431:37–46. doi: 10.1016/s0167-4838(99)00030-8. [DOI] [PubMed] [Google Scholar]
- 77.Moayeri M., Leysath C.E., Tremblay J.M., Vrentas C., Crown D., Leppla S.H., Shoemaker C.B. A heterodimer of a VHH (variable domains of camelid heavy chain-only) antibody that inhibits anthrax toxin cell binding linked to a VHH antibody that blocks oligomer formation is highly protective in an anthrax spore challenge model. J. Biol. Chem. 2015;290:6584–6595. doi: 10.1074/jbc.M114.627943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vrentas C.E., Moayeri M., Keefer A.B., Greaney A.J., Tremblay J., O’Mard D., Leppla S.H., Shoemaker C.B. A Diverse Set of Single-domain Antibodies (VHHs) against the Anthrax Toxin Lethal and Edema Factors Provides a Basis for Construction of a Bispecific Agent That Protects against Anthrax Infection. J. Biol. Chem. 2016;291:21596–21606. doi: 10.1074/jbc.M116.749184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mukherjee J., Tremblay J.M., Leysath C.E., Ofori K., Baldwin K., Feng X., Bedenice D., Webb R.P., Wright P.M., Smith L.A. A novel strategy for development of recombinant antitoxin therapeutics tested in a mouse botulism model. PLoS ONE. 2012;7:e29941. doi: 10.1371/journal.pone.0029941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mukherjee J., Dmitriev I., Debatis M., Tremblay J.M., Beamer G., Kashentseva E.A., Curiel D.T., Shoemaker C.B. Prolonged prophylactic protection from botulism with a single adenovirus treatment promoting serum expression of a VHH-based antitoxin protein. PLoS ONE. 2014;9:e106422. doi: 10.1371/journal.pone.0106422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tremblay J.M., Mukherjee J., Leysath C.E., Debatis M., Ofori K., Baldwin K., Boucher C., Peters R., Beamer G., Sheoran A. A single VHH-based toxin-neutralizing agent and an effector antibody protect mice against challenge with Shiga toxins 1 and 2. Infect. Immun. 2013;81:4592–4603. doi: 10.1128/IAI.01033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herrera C., Tremblay J.M., Shoemaker C.B., Mantis N.J. Mechanisms of Ricin Toxin Neutralization Revealed through Engineered Homodimeric and Heterodimeric Camelid Antibodies. J. Biol. Chem. 2015;290:27880–27889. doi: 10.1074/jbc.M115.658070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vance D.J., Tremblay J.M., Mantis N.J., Shoemaker C.B. Stepwise engineering of heterodimeric single domain camelid VHH antibodies that passively protect mice from ricin toxin. J. Biol. Chem. 2013;288:36538–36547. doi: 10.1074/jbc.M113.519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Z., Schmidt D., Liu W., Li S., Shi L., Sheng J., Chen K., Yu H., Tremblay J.M., Chen X. A novel multivalent, single-domain antibody targeting TcdA and TcdB prevents fulminant Clostridium difficile infection in mice. J. Infect. Dis. 2014;210:964–972. doi: 10.1093/infdis/jiu196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schmidt D.J., Beamer G., Tremblay J.M., Steele J.A., Kim H.B., Wang Y., Debatis M., Sun X., Kashentseva E.A., Dmitriev I.P. A Tetraspecific VHH-Based Neutralizing Antibody Modifies Disease Outcome in Three Animal Models of Clostridium difficile Infection. Clin. Vaccine Immunol. 2016;23:774–784. doi: 10.1128/CVI.00730-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zanganeh S., Rouhani Nejad H., Mehrabadi J.F., Hosseini R., Shahi B., Tavassoli Z., Aramvash A. Rapid and Sensitive Detection of Staphylococcal Enterotoxin B by Recombinant Nanobody Using Phage Display Technology. Appl. Biochem. Biotechnol. 2019;187:493–505. doi: 10.1007/s12010-018-2762-y. [DOI] [PubMed] [Google Scholar]
- 87.Meddeb-Mouelhi F., Bouhaouala-Zahar B., Benlasfar Z., Hammadi M., Mejri T., Moslah M., Karoui H., Khorchani T., El Ayeb M. Immunized camel sera and derived immunoglobulin subclasses neutralizing Androctonus australis hector scorpion toxins. Toxicon. 2003;42:785–791. doi: 10.1016/j.toxicon.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 88.Richard G., Meyers A.J., McLean M.D., Arbabi-Ghahroudi M., MacKenzie R., Hall J.C. In vivo neutralization of α-cobratoxin with high-affinity llama single-domain antibodies (VHHs) and a VHH-Fc antibody. PLoS ONE. 2013;8:e69495. doi: 10.1371/journal.pone.0069495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thran M., Mukherjee J., Pönisch M., Fiedler K., Thess A., Mui B.L., Hope M.J., Tam Y.K., Horscroft N., Heidenreich R. mRNA mediates passive vaccination against infectious agents, toxins, and tumors. EMBO Mol. Med. 2017;9:1434–1447. doi: 10.15252/emmm.201707678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stadler C.R., Bähr-Mahmud H., Celik L., Hebich B., Roth A.S., Roth R.P., Karikó K., Türeci Ö., Sahin U. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 91.Shi B., Keough E., Matter A., Leander K., Young S., Carlini E., Sachs A.B., Tao W., Abrams M., Howell B., Sepp-Lorenzino L. Biodistribution of small interfering RNA at the organ and cellular levels after lipid nanoparticle-mediated delivery. J. Histochem. Cytochem. 2011;59:727–740. doi: 10.1369/0022155411410885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.L., Tam Y.K. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Groothuis J.R., Levin M.J., Rodriguez W., Hall C.B., Long C.E., Kim H.W., Lauer B.A., Hemming V.G., The RSVIG Study Group Use of intravenous gamma globulin to passively immunize high-risk children against respiratory syncytial virus: safety and pharmacokinetics. Antimicrob. Agents Chemother. 1991;35:1469–1473. doi: 10.1128/aac.35.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meissner H.C., Fulton D.R., Groothuis J.R., Geggel R.L., Marx G.R., Hemming V.G., Hougen T., Snydman D.R. Controlled trial to evaluate protection of high-risk infants against respiratory syncytial virus disease by using standard intravenous immune globulin. Antimicrob. Agents Chemother. 1993;37:1655–1658. doi: 10.1128/aac.37.8.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wandstrat T.L. Respiratory syncytial virus immune globulin intravenous. Ann. Pharmacother. 1997;31:83–88. doi: 10.1177/106002809703100114. [DOI] [PubMed] [Google Scholar]
- 96.Quiambao B.P., Dytioco H.Z., Dizon R.M., Crisostomo M.E., Laot T.M., Teuwen D.E. Rabies post-exposure prophylaxis in the Philippines: health status of patients having received purified equine F(ab’)(2) fragment rabies immunoglobulin (Favirab) PLoS Negl. Trop. Dis. 2008;2:e243. doi: 10.1371/journal.pntd.0000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Casadevall A., Dadachova E., Pirofski L.A. Passive antibody therapy for infectious diseases. Nat. Rev. Microbiol. 2004;2:695–703. doi: 10.1038/nrmicro974. [DOI] [PubMed] [Google Scholar]
- 98.Keller M.A., Stiehm E.R. Passive immunity in prevention and treatment of infectious diseases. Clin. Microbiol. Rev. 2000;13:602–614. doi: 10.1128/cmr.13.4.602-614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 100.The IMpact-RSV Study Group Palivizumab, a Humanized Respiratory Syncytial Virus Monoclonal Antibody, Reduces Hospitalization From Respiratory Syncytial Virus Infection in High-risk Infants. Pediatrics. 1998;102:531–537. [PubMed] [Google Scholar]
- 101.Meissner H.C., Welliver R.C., Chartrand S.A., Law B.J., Weisman L.E., Dorkin H.L., Rodriguez W.J. Immunoprophylaxis with palivizumab, a humanized respiratory syncytial virus monoclonal antibody, for prevention of respiratory syncytial virus infection in high risk infants: a consensus opinion. Pediatr. Infect. Dis. J. 1999;18:223–231. doi: 10.1097/00006454-199903000-00004. [DOI] [PubMed] [Google Scholar]
- 102.Nagarajan T., Marissen W.E., Rupprecht C.E. Monoclonal antibodies for the prevention of rabies: theory and clinical practice. Antibody Technology Journal. 2014;4:1–12. [Google Scholar]
- 103.Walker L.M., Burton D.R. Passive immunotherapy of viral infections: ‘super-antibodies’ enter the fray. Nat. Rev. Immunol. 2018;18:297–308. doi: 10.1038/nri.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Johnson P.R., Schnepp B.C., Zhang J., Connell M.J., Greene S.M., Yuste E., Desrosiers R.C., Clark K.R. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 2009;15:901–906. doi: 10.1038/nm.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Saunders K.O., Wang L., Joyce M.G., Yang Z.Y., Balazs A.B., Cheng C., Ko S.Y., Kong W.P., Rudicell R.S., Georgiev I.S. Broadly Neutralizing Human Immunodeficiency Virus Type 1 Antibody Gene Transfer Protects Nonhuman Primates from Mucosal Simian-Human Immunodeficiency Virus Infection. J. Virol. 2015;89:8334–8345. doi: 10.1128/JVI.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tiwari P.M., Vanover D., Lindsay K.E., Bawage S.S., Kirschman J.L., Bhosle S., Lifland A.W., Zurla C., Santangelo P.J. Engineered mRNA-expressed antibodies prevent respiratory syncytial virus infection. Nat. Commun. 2018;9:3999. doi: 10.1038/s41467-018-06508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mays L.E., Ammon-Treiber S., Mothes B., Alkhaled M., Rottenberger J., Müller-Hermelink E.S., Grimm M., Mezger M., Beer-Hammer S., von Stebut E. Modified Foxp3 mRNA protects against asthma through an IL-10-dependent mechanism. J. Clin. Invest. 2013;123:1216–1228. doi: 10.1172/JCI65351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rettig W.J., Old L.J. Immunogenetics of human cell surface differentiation. Annu. Rev. Immunol. 1989;7:481–511. doi: 10.1146/annurev.iy.07.040189.002405. [DOI] [PubMed] [Google Scholar]
- 109.Scott A.M., Wolchok J.D., Old L.J. Antibody therapy of cancer. Nat. Rev. Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 110.Hudis C.A. Trastuzumab: mechanism of action and use in clinical practice. N. Engl. J. Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 111.de Miguel D., Lemke J., Anel A., Walczak H., Martinez-Lostao L. Onto better TRAILs for cancer treatment. Cell Death Differ. 2016;23:733–747. doi: 10.1038/cdd.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thomas A., Teicher B.A., Hassan R. Antibody-drug conjugates for cancer therapy. Lancet Oncol. 2016;17:e254–e262. doi: 10.1016/S1470-2045(16)30030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Diamantis N., Banerji U. Antibody-drug conjugates: an emerging class of cancer treatment. Br. J. Cancer. 2016;114:362–367. doi: 10.1038/bjc.2015.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Page D.B., Postow M.A., Callahan M.K., Allison J.P., Wolchok J.D. Immune modulation in cancer with antibodies. Annu. Rev. Med. 2014;65:185–202. doi: 10.1146/annurev-med-092012-112807. [DOI] [PubMed] [Google Scholar]
- 115.Dahlén E., Veitonmäki N., Norlén P. Bispecific antibodies in cancer immunotherapy. Ther. Adv. Vaccines Immunother. 2018;6:3–17. doi: 10.1177/2515135518763280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang W.H., Xu J., Mu J.B., Xie J. Revision of the concept of anti-angiogenesis and its applications in tumor treatment. Chronic Dis Transl Med. 2017;3:33–40. doi: 10.1016/j.cdtm.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mayes P.A., Hance K.W., Hoos A. The promise and challenges of immune agonist antibody development in cancer. Nat. Rev. Drug Discov. 2018;17:509–527. doi: 10.1038/nrd.2018.75. [DOI] [PubMed] [Google Scholar]
- 118.Goswami S., Wang W., Arakawa T., Ohtake S. Developments and challenges for mAb-based therapeutics. Antibodies (Basel) 2013;2:452–500. [Google Scholar]
- 119.Klinger M., Benjamin J., Kischel R., Stienen S., Zugmaier G. Harnessing T cells to fight cancer with BiTE® antibody constructs: past developments and future directions. Immunol. Rev. 2016;270:193–208. doi: 10.1111/imr.12393. [DOI] [PubMed] [Google Scholar]
- 120.Wu J., Fu J., Zhang M., Liu D. AFM13: a first-in-class tetravalent bispecific anti-CD30/CD16A antibody for NK cell-mediated immunotherapy. J. Hematol. Oncol. 2015;8:96. doi: 10.1186/s13045-015-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lan Y., Zhang D., Xu C., Hance K.W., Marelli B., Qi J., Yu H., Qin G., Sircar A., Hernández V.M. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aan5488. eaan5488. [DOI] [PubMed] [Google Scholar]
- 122.Garber K. Bispecific antibodies rise again. Nat. Rev. Drug Discov. 2014;13:799–801. doi: 10.1038/nrd4478. [DOI] [PubMed] [Google Scholar]
- 123.Szebeni J. Complement activation-related pseudoallergy: a stress reaction in blood triggered by nanomedicines and biologicals. Mol. Immunol. 2014;61:163–173. doi: 10.1016/j.molimm.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 124.Prieve M.G., Harvie P., Monahan S.D., Roy D., Li A.G., Blevins T.L., Paschal A.E., Waldheim M., Bell E.C., Galperin A. Targeted mRNA Therapy for Ornithine Transcarbamylase Deficiency. Mol. Ther. 2018;26:801–813. doi: 10.1016/j.ymthe.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ridgway J.B., Presta L.G., Carter P. ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996;9:617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- 126.Marschall A.L., Zhang C., Frenzel A., Schirrmann T., Hust M., Perez F., Dübel S. Delivery of antibodies to the cytosol: debunking the myths. MAbs. 2014;6:943–956. doi: 10.4161/mabs.29268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.El-Sayed A., Futaki S., Harashima H. Delivery of macromolecules using arginine-rich cell-penetrating peptides: ways to overcome endosomal entrapment. AAPS J. 2009;11:13–22. doi: 10.1208/s12248-008-9071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]