Abstract
Whooping cough’s primary etiological agent is Bordetella pertussis. The closely related Bordetella parapertussis rarely causes severe disease. Here we report an unusual case of bacteremia caused by B. parapertussis, review the literature, and characterize the genomic sequence of the bacterial isolate in comparison with B. parapertussis isolates from respiratory infections.
Keywords: bacteremia, Bordetella parapertussis, children, genomics
Bordetella pertussis and Bordetella parapertussis are the main causative agents of whooping cough, a disease that is still endemic worldwide despite high pertussis vaccine coverage of young children. B. parapertussis is much less often involved in the disease than B. pertussis, as it is responsible for only 2%–20% of cases and generally causes a less severe respiratory illness [1–3].
Invasive illnesses with Bordetella spp. are generally caused by B. holmesii and B. bronchiseptica in immunocompromised hosts, such as patients with asplenia, chronic obstructive pulmonary disease, HIV infections, hematological disorders, or transplantation [4–6]. Reports of systemic infection with B. parapertussis are scarce, with only 3 cases reported in the literature [2, 7]; 2 of them had a known underlying disease. This systemic manifestation of B. parapertussis infection might thus be at least in part determined by host susceptibility.
Phylogenetic analyses of B. pertussis and B. parapertussis indicated that these 2 closely related human-adapted species have evolved independently from B. bronchiseptica–like ancestors [8]. Further, the population of B. parapertussis isolates from humans is highly homogeneous genetically [9, 10]. Unlike B. pertussis, B. parapertussis does not express the pertussis toxin because of many mutations, particularly within the toxin gene promoter region [11]. In addition, B. parapertussis has an O-antigen that confers protection from complement-mediated immunity, and can therefore be considered a virulent factor [12]. The possibility that specific virulence determinants of B. parapertussis isolates are implicated in invasive infections remains to be investigated, as genomic sequences from B. parapertussis causing systemic disease have not been reported so far.
In this study, we report a clinical case of B. parapertussis bacteremia. Further, we determined the genomic sequence of the isolate and compared it with all publicly available genomic sequences of B. parapertussis from respiratory infections.
CASE REPORT
The patient was a 3-year-old female without notable medical history. She had received 3 injections for primary vaccination against pertussis (acellular pertussis vaccine) before 1 year of age. She was living in Algeria when an acute leukemia was suspected by the presence of fever, lethargy, and hepatosplenomegaly at physical examination. The diagnosis of acute lymphoblastic leukemia was confirmed in France in April 2017 by hemogram, with an abnormal white blood cell count 42×109/L with 33×109/L blasts, anemia, and thrombocytopenia, and by the bone marrow diffuse infiltration of undifferentiated blasts (88%) characterized on immunophenotyping analysis. A central venous catheter was implanted, and chemotherapy was started according to the French Acute Lymphoblastic Leukemia Study Group guidelines. One day before chemotherapy started, the patient presented with fever (38.1°C) associated with a moderate cough, which resolved within the next 24 hours. Inflammatory parameters were slightly elevated, with plasma levels of C-reactive protein at 33 mg/L, then 53 mg/L at day 2. Intravenous piperacillin–tazobactam treatment was started 5 days later, when gram-negative bacilli were observed on the first blood culture, and was switched to oral azithromycin when Bordetella parapertussis was identified by Matrix-Assisted Laser Desorption Ionization Time of Flight (MALDI-TOF) mass spectrometry. Real-time polymerase chain reaction targeting the IS1001 insertion sequence was performed on a nasopharyngeal aspirate [13]. Its positivity suggested the presence of B. parapertussis. The patient did not present any complications, and the antileukemia chemotherapy was continued. The patient was discharged on day 35 of hospitalization and followed the French guidelines for lymphoblastic acute leukemia chemotherapy without any other infectious complication.
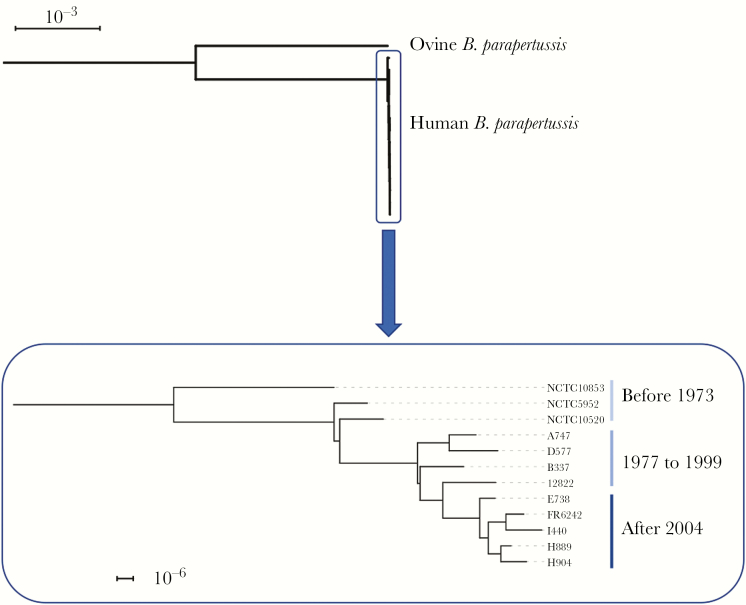
The organism collected from blood culture was sent to the National Reference Center for further investigations (Supplementary Data). The isolate (named FR6242) displayed a brown pigment and a visible hemolysis on Bordet-Gengou agar medium, as expected for B. parapertussis. Western blots revealed that it was positive for filamentous hemagglutinin (FHA) production and negative for pertussis toxin, but also negative for pertactin (PRN) production, as observed for most human B. parapertussis isolates since 2007 in France [14]. There are no clinical breakpoints for determination of susceptibility or resistance of Bordetellae from European Committee on Antimicrobial Susceptibility Testing (EUCAST). No resistance to erythromycin and azithromycin has been reported for B. parapertussis, but previous studies have found that B. pertussis isolates resistant to erythromycin and azithromycin have, in most cases, a minimum inhibitory concentration (MIC) >256 mg/L [15, 16]. The isolate was considered susceptible to azithromycin, clarithromycin, and erythromycin (MIC, 0.12, 0.19, and 0.125 mg/L, respectively) and trimethoprim-sulfamethoxazole (MIC of trimethoprim, 0.012 mg/L). The genomic sequence was assembled into 78 contigs, with a total size of 4 720 641 base pairs, and was compared with 11 human B. parapertussis genomes (mainly collected in the United States) available in public sequence repositories. According to the 7-gene multilocus sequence typing (MLST) scheme [10], FR6242 belonged to ST19, as did the reference B. parapertussis strain 12822 (isolated in 1993 from an infant with cough illness) [17] and the 10 other isolates. Genotyping of virulence-related genes revealed the sequence identity between isolate FR6242 and the other isolates for the main targets including pertussis toxin subunit-encoding genes (Supplementary Table 1). Only the gene brkB, involved in Bordetella serum resistance, showed variation, with allele 2 in reference strain 12822 and allele 6 in all isolates, including FR6242. This change corresponds to a nonsynonymous SNP in position 742, leading to the modification of AA248 of the BrkB protein (threonine vs alanine). Analysis of the prn gene coding for pertactin revealed that FR6242 had 2 deletions within this gene. A total of 54 whole-genome single nucleotide polymorphisms (SNP) were identified between FR6242 and reference strain 12822. Among them, 23 nonsynonymous SNPs were shared with the other 10 isolates (Supplementary Table 2). Of these, FR6242 shared 19 SNPs with the most recent isolates (collected after 2004), and more particularly with strain I440 collected in 2012 (unspecified clinical origin). Phylogenetic relationships among the 12 B. parapertussis genomes (Figure 1) showed that strain FR6242 was related to I440 and more broadly to a clade comprising the recent isolates, which were clearly grouped into a clade separated from more ancient isolates. Only 2 nonsynonymous substitutions and 1 intergenic nucleotide substitution were specific to the invasive strain FR6242: (1) within gene BPP_RS12865/BPP2546, encoding a putative short chain dehydrogenase; (2) within gene BPP_RS18460/BPP3661, encoding a putative oxidoreductase; and (3) at an intergenic position located between 2 conserved hypothetical proteins (Supplementary Table 2).
Figure 1.
Single nucleotide polymorphism–based phylogenetic tree of Bordetella parapertussis isolates rooted on ovine Bpp5. The analysis focused on 12 human B. parapertussis isolates from the period 1935–2017, including FR6242 (bottom box). The analysis was performed by a mapping approach using the genome sequence of B. parapertussis strain 12822 (NC_002928.3) as the reference. The scale bar indicates the number of nucleotide substitutions per site.
DISCUSSION
B. parapertussis is circulating worldwide [1] and can cause outbreaks [18]. In France, this species represents 5%–6% of whooping cough cases [19]. It is associated with mild forms of respiratory disease. Invasive infections by Bordetella species are typically observed in immunocompromised hosts, and in most described cases they have been associated with initial respiratory diseases. Most reported bacteremia cases have involved B. holmesii and B. bronchiseptica. Before this study, only 3 B. parapertussis bacteremia cases had been reported, to our knowledge [2, 7]. These cases occurred (1) in a patient with severe asthma needing steroid treatment, which is known to cause immunological dysfunction; (2) in a patient with T-cell acute lymphoblastic leukemia; and (3) in an infant without known medical history who died from respiratory arrest. All 3 cases had concomitant lower respiratory tract infection. Here, we report a fourth case, who had acute leukemia and could be considered immunocompromised with associated susceptibility to infection, even if the patient had not started chemotherapy yet and was not neutropenic at the time of infection. As serology for HIV was negative, no immune functional tests were performed. However, the patient did not display severe symptoms of infection, and blood culture had been performed systematically due to fever occurring in an immunocompromised background.
The B. parapertussis isolate FR6242 from the present case was not distinguishable from respiratory B. parapertussis isolates based on its antigens and virulence factor gene sequences. Genome-wide analysis revealed 2 amino acid changes and 1 intergenic nucleotide substitution unique to this isolate, as well as a number of other changes shared with subsets of B. parapertussis isolates. However, it is not immediately clear how the genomic features disclosed here might be associated with invasive disease, and they may as well represent natural variation with no effect of the capacity of the bacterium to cause invasive infection. Knowing whether these changes are associated with B. parapertussis bacteremia will require the availability of genomic sequences of multiple other isolates and experimental demonstration of possible effects on invasiveness. In B. holmesii, no differences were found between isolates of respiratory and bacteremia origins, using either genomic analysis or cellular/animal models [20, 21]. Therefore, the bacterial determinants of Bordetella bacteremia remain elusive.
Clearly, B. parapertussis bacteremia might also be related to the host immune response, rather than primarily to bacterial determinants, as found for B. holmesii and B. bronchiseptica [4–6]. In addition, B. parapertussis might induce a different immune response from B. pertussis, as it differs in the structure of lipopolysaccharide (LPS) [22]. In vitro, purified B. parapertussis LPS is a stronger activator of the innate immune response than purified B. pertussis lipooligosaccharide in terms of maturation of human dendritic cells and cytokine production [23], but the TLR4 response induced by B. parapertussis LPS seems less efficient, allowing the organism to escape from a robust inflammatory response, efficient bacterial clearance [24], and subsequent bacterial dissemination in an immune-compromised host.
Possible cross-immunity against B. parapertussis, induced by acellular vaccines that target B. pertussis virulence, was underlined in several studies [25]. Such a cross-protection could be due to greater anti-FHA antibody response induced by acellular FHA-containing vaccines, as compared with whole-cell vaccines. In our study, the patient was immunized against B. pertussis; however, vaccine-induced immunity was not sufficient to protect against B. parapertussis.
In conclusion, we report the fourth case of B. parapertussis bacteremia in the literature, and the first for which the genomic sequence of the infectious isolate was determined. This new case underlines that B. parapertussis can cause invasive infection, mostly in immunocompromised hosts. Investigation of bacterial genomic determinants potentially associated with invasive infection will require the genomic study of additional isolates.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Author contributions. J.T. and S.M. wrote the report. A.S. managed the patient. S.Bo. carried out laboratory testing and identified B. parapertussis. A.L. and S.G. carried out the culture of the strain. V.B. and S.Br. carried out genome sequencing and interpreted the data. All authors were involved in the revision of the manuscript. Written consent to publication was obtained.
Consent for publication. Written consent from the patient’s parents was obtained before writing of the manuscript. All potentially identifying information was removed from the text.
Sequence availability. Raw sequence data (fastq files) were deposited in the European Nucleotide Archive and are available under study accession number PRJEB29316.
Financial support. This work was supported by Santé Publique France (Saint-Maurice, France).
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Watanabe M, Nagai M. Whooping cough due to Bordetella parapertussis: an unresolved problem. Expert Rev Anti Infect Ther 2004; 2:447–54. [DOI] [PubMed] [Google Scholar]
- 2. Wallihan R, Selvarangan R, Marcon M, et al. Bordetella parapertussis bacteremia: two case reports. Pediatr Infect Dis J 2013; 32:796–8. [DOI] [PubMed] [Google Scholar]
- 3. Cherry JD, Seaton BL. Patterns of Bordetella parapertussis respiratory illnesses: 2008-2010. Clin Infect Dis 2012; 54:534–7. [DOI] [PubMed] [Google Scholar]
- 4. Matic NA, Bunce PE. Isolation of Bordetella bronchiseptica from blood and a pancreatic abscess. J Clin Microbiol 2015; 53:1778–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pittet LF, Emonet S, Schrenzel J, et al. Bordetella holmesii: an under-recognised Bordetella species. Lancet Infect Dis 2014; 14:510–9. [DOI] [PubMed] [Google Scholar]
- 6. Powers HR, Shah K. Bordetella bronchiseptica bloodstream infection in a renal transplant patient. Transpl Infect Dis 2017; 19(6). [DOI] [PubMed] [Google Scholar]
- 7. Correa-Londono A, Ellner PD. Case report. Clin Microbiol Newsletter 1980; 2:4. [Google Scholar]
- 8. Linz B, Ivanov YV, Preston A, et al. Acquisition and loss of virulence-associated factors during genome evolution and speciation in three clades of Bordetella species. BMC Genomics 2016; 17:767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bouchez V, Brun D, Dore G, et al. Bordetella parapertussis isolates not expressing pertactin circulating in France. Clin Microbiol Infect 2011; 17:675–82. [DOI] [PubMed] [Google Scholar]
- 10. Diavatopoulos DA, Cummings CA, Schouls LM, et al. Bordetella pertussis, the causative agent of whooping cough, evolved from a distinct, human-associated lineage of B. bronchiseptica. PLoS Pathog 2005; 1:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aricò B, Rappuoli R. Bordetella parapertussis and Bordetella bronchiseptica contain transcriptionally silent pertussis toxin genes. J Bacteriol 1987; 169:2847–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang X, Rodríguez ME, Harvill ET. O antigen allows B. parapertussis to evade B. pertussis vaccine-induced immunity by blocking binding and functions of cross-reactive antibodies. PLoS One 2009; 4:e6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roorda L, Buitenwerf J, Ossewaarde JM, van der Zee A. A real-time PCR assay with improved specificity for detection and discrimination of all clinically relevant Bordetella species by the presence and distribution of three insertion sequence elements. BMC Res Notes 2011; 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bouchez V, Guiso N. Bordetella pertussis, B. parapertussis, vaccines and cycles of whooping cough. Pathog Dis 2015; 73. [DOI] [PubMed] [Google Scholar]
- 15. Guillot S, Descours G, Gillet Y, et al. Macrolide-resistant Bordetella pertussis infection in newborn girl, France. Emerg Infect Dis 2012; 18:966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Z, Cui Z, Li Y, et al. High prevalence of erythromycin-resistant Bordetella pertussis in Xi’an, China. Clin Microbiol Infect 2014; 20:O825–30. [DOI] [PubMed] [Google Scholar]
- 17. Parkhill J, Sebaihia M, Preston A, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet 2003; 35:32–40. [DOI] [PubMed] [Google Scholar]
- 18. Koepke R, Bartholomew ML, Eickhoff JC, et al. Widespread Bordetella parapertussis infections—Wisconsin, 2011-2012: clinical and epidemiologic features and antibiotic use for treatment and prevention. Clin Infect Dis 2015; 61:1421–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tubiana S, Belchior E, Guillot S, et al. ; Renacoq Participants Monitoring the impact of vaccination on pertussis in infants using an active hospital-based pediatric surveillance network: results from 17 years’ experience, 1996-2012, France. Pediatr Infect Dis J 2015; 34:814–20. [DOI] [PubMed] [Google Scholar]
- 20. Bouchez V, AlBitar-Nehme S, Novikov A, Guiso N, Caroff M. Bordetella holmesii: lipid A structures and corresponding genomic sequences comparison in three clinical isolates and the reference strain ATCC 51541. Int J Mol Sci 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harvill ET, Goodfield LL, Ivanov Y, et al. Genome sequences of nine Bordetella holmesii strains isolated in the United States. Genome Announc 2014; 2(3):e00438–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Caroff M, Aussel L, Zarrouk H, et al. Structural variability and originality of the Bordetella endotoxins. J Endotoxin Res 2001; 7:63–8. [PubMed] [Google Scholar]
- 23. Fedele G, Nasso M, Spensieri F, et al. Lipopolysaccharides from Bordetella pertussis and Bordetella parapertussis differently modulate human dendritic cell functions resulting in divergent prevalence of Th17-polarized responses. J Immunol 2008; 181:208–16. [DOI] [PubMed] [Google Scholar]
- 24. Wolfe DN, Buboltz AM, Harvill ET. Inefficient Toll-like receptor-4 stimulation enables Bordetella parapertussis to avoid host immunity. PLoS One 2009; 4:e4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liko J, Robison SG, Cieslak PR. Do pertussis vaccines protect against Bordetella parapertussis? Clin Infect Dis 2017; 64:1795–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.