Abstract
With the presence of the blood-brain barrier (BBB), successful immunotherapeutic drug delivery to CNS malignancies remains a challenge. Immunomodulatory agents, such as cytokines, can reprogram the intratumoral microenvironment; however, systemic cytokine delivery has limited access to the CNS. To bypass the limitations of systemically administered cytokines, we investigated if RNA-modified T cells could deliver macromolecules directly to brain tumors. The abilities of T cells to cross the BBB and mediate direct cytotoxic killing of intracranial tumors make them an attractive tool as biological carriers. Using T cell mRNA electroporation, we demonstrated that activated T cells can be modified to secrete granulocyte macrophage colony-stimulating factor (GM-CSF) protein while retaining their inherent effector functions in vitro. GM-CSF RNA-modified T cells effectively delivered GM-CSF to intracranial tumors in vivo and significantly extended overall survival in an orthotopic treatment model. Importantly, GM-CSF RNA-modified T cells demonstrated superior anti-tumor efficacy as compared to unmodified T cells alone or in combination with systemic administration of recombinant GM-CSF. Anti-tumor effects were associated with increased IFN-γ secretion locally within the tumor microenvironment and systemic antigen-specific T cell expansion. These findings demonstrate that RNA-modified T cells may serve as a versatile platform for the effective delivery of biological agents to CNS tumors.
Keywords: RNA, T cells, brain tumor, immunotherapy, adoptive cellular therapy
Biological drug delivery to brain tumors remains a challenge due to the invasive nature of CNS malignancies and a restrictive blood-brain barrier. By genetically modifying T cells with RNA encoding for cytokines, Pohl-Guimarães et al. describe a novel and effective way to deliver immunomodulatory macromolecules locally within intracranial tumors.
Introduction
Cytokines such as granulocyte macrophage colony-stimulating factor (GM-CSF), an important hematopoietic growth factor and immunomodulator, have been applied as adjuvants in cancer immunotherapy, and they may be utilized to overcome the immunosuppressive tumor microenvironment.1 However, systemically delivered cytokines have limited access to brain tumors due to the selective blood-brain barrier (BBB), and they may require high doses to reach therapeutic concentrations, thus increasing the risk of side effects.2 Despite tremendous efforts on research investigating invasive and non-invasive drug delivery to CNS tumors (e.g., osmotic BBB disruption, receptor-mediated blood-brain-tumor barrier [BBTB] opening, convection-enhanced drug delivery, nanoparticles, and gene therapy), the BBB remains a major limiting factor for the development of novel and successful therapeutics for malignant brain tumors. Many studies have demonstrated the benefits of viral vector-based expression of cytokines using cellular therapy for solid tumors.3, 4, 5, 6 Recently, transgenic engineering of interleukin (IL)-15-expressing IL-13Rα-chimeric antigen receptor (CAR) T cells enhanced anti-tumor activity of IL-13Rα-CAR T cells in a glioma xenograft.7 Compared to unmodified vascular endothelial growth factor (VEGF)-CAR T, others have shown that IL-12-expressing VEGF-CAR T cells can eradicate multiple vascularized tumors in mice.3
While viral vector-based cell therapies can be utilized as a platform for delivery to intracranial tumors, due to the complexity and cost of clinical vector production, iterative clinical trials incorporating gene modification via vector-based strategies may be difficult to implement.8 Conversely, non-viral-based cell therapies using RNA transfection offer several advantages for gene expression over viral vectors. First, RNA is readily translated in the cytoplasm, yielding high transfection efficiency, thus bypassing the need for delivery to the nucleus for gene expression. Moreover, we have previously demonstrated that multiple RNAs can be combined for simultaneous gene expression in activated T cells,9 and the transient nature of gene expression from RNA templates allows for the pharmacological titration of both T cell dosing and RNA-based gene delivery.10 Lastly, circumventing long-lived expression of inflammatory molecules in adoptively transferred lymphocytes may bypass the potential safety concerns in the use of stably transfected T cells expressing immunopotentiating agents.
Adoptive T cell transfer (ACT), using autologous tumor-specific lymphocytes expanded ex vivo, has a major advantage over other cancer immunotherapies available, because T cells are known to cross the BBB11 and migrate to invasive intracranial tumors.12 Additionally, ex vivo expansion of T cells allows for the genetic T cell reprogramming. Transient gene expression using RNA-based modification of T cells is a relevant and contemporary approach under active investigation within the immune-oncology field. Consequently, successful electroporation (EP) of mRNA into primary T lymphocytes has now been developed for genetically modified T cells in preclinical studies,9, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 and it is already being evaluated in clinical trials in patients with advanced malignancies and recently completed clinical trials (ClinicalTrials.gov: NCT01355965, NCT01897415, NCT03060356, NCT02624258, and NCT01837602). To establish a proof of concept, we investigated T cells as biological carriers of RNA encoding for GM-CSF in an intracranial tumor mouse model. We demonstrated that murine activated T cells can be successfully modified to secrete GM-CSF protein while retaining effector T cell functions in vitro. GM-CSF RNA-modified T cells delivered enhanced protein levels of GM-CSF to intracranial tumors in vivo. By increasing local interferon gamma (IFN-γ) secretion at the tumor site and expanding systemic antigen-specific T cells, GM-CSF RNA-modified T cells prolonged overall survival outcomes in a murine intracranial tumor model.
Results
High Transfection Efficiency of GFP RNA-Modified T Cells by EP
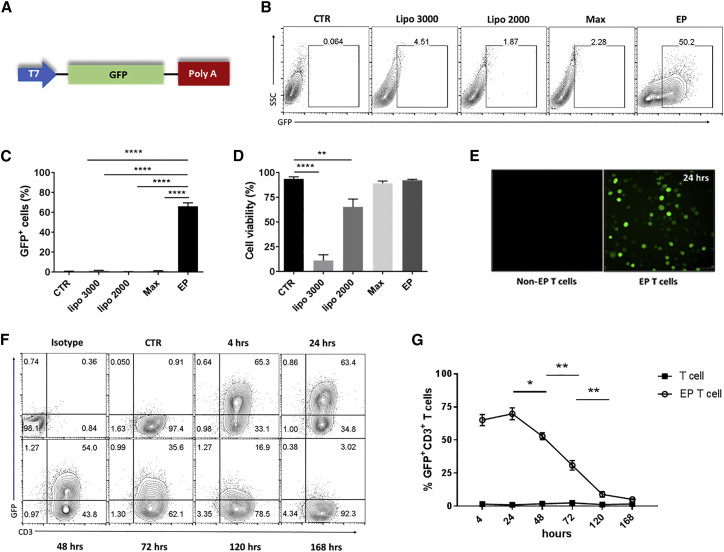
To identify the optimal non-viral method to modify T cells, we compared transfection efficiency and cell viability of concanavalin A (ConA)-activated T cells transfected with GFP RNA using four different techniques as follows: lipofectamine 2000 (lipo 2000), lipofectamine 3000 (lipo 3000), messenger max (Max), and EP. We demonstrated that EP is the most effective non-viral method screened. Compared to lipofectamine methods, EP yielded high percentages of GFP+ cells (>65% in repeated experiments) (****p < 0.0001, one-way ANOVA) (Figures 1B and 1C). Compared to unmodified T cells, a high percentage of cell viability is retained 24 hr post-EP (∼98%), while the percentage of cell viability after lipo 3000 transfection was dramatically reduced by 8-fold (11%) (****p < 0.0001, one-way ANOVA) (Figure 1D).
Figure 1.
High and Transient Transfection Efficiency of GFP RNA-Modified T Cells by Electroporation
Spleen was harvested from naive C57BL/6 mice and expanded with 1 μg/mL concanavalin A (ConA). At day 8, activated T cells were transfected with 10 μg GFP RNA using the indicated non-viral methods. Cells were harvested 24 hr after transfection, unless otherwise indicated. (A) Schematic representation of the expression vector construct encoding for GFP used for mRNA synthesis in vitro. (B and C) Representative contour plot (B) and the percentage of GFP+ cells detected by flow cytometry (C). (D and E) Percentage of cell viability (D) and fluorescence microscopy of GFP+ spleen cells (E) 24 hr post-transfection. (F and G) Representative contour plot (F) and the percentage of GFP+ CD3+ ConA-activated T cells detected by flow cytometry (G) at the indicated time points following GFP RNA electroporation. EP, electroporated; CTR, non-transfected cells were used as a control of GFP-electroporated cells for all experiments (*p < 0.05, **p < 0.01, and ****p < 0.0001, one-way ANOVA). Values indicated are the mean ± SEM.
Based on these data, we selected EP as a transfection method for subsequent experiments. RNA-modified T cells electroporated with an irrelevant RNA showed no significant differences of the FL-1 median fluorescence intensity (MFI) when compared to the mock and untransfected T cell groups (data not shown), excluding the possibility of autofluorescence from modified T cells potentially due to the transfection method. We next detected the kinetics of GFP expression after T cell EP with GFP RNA. While GFP expression in CD3+ T cells was highly detected (∼60%) at 4 and 24 hr, GFP expression decreased at 48 hr (*p < 0.05, two-way ANOVA) and 72 hr (**p < 0.01, two-way ANOVA), reaching basal levels by day 5 post-EP (**p < 0.01, two-way ANOVA) (Figures 1F and 1G).
Phenotypic Analysis of the GFP RNA-Modified T Cells
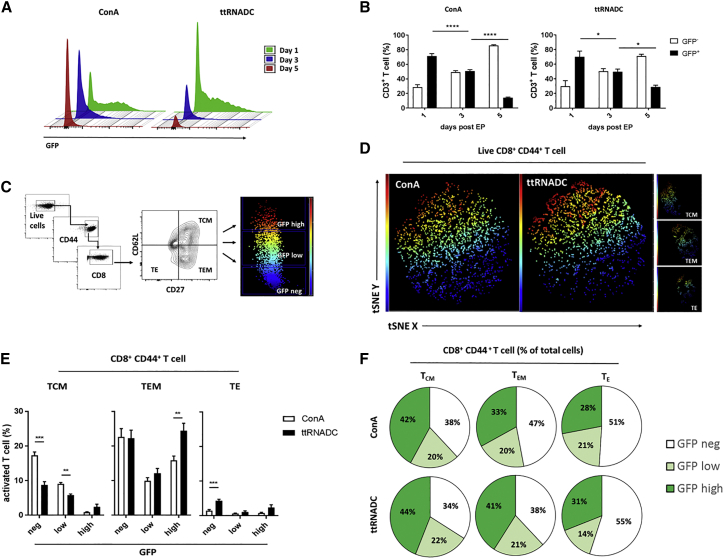
Different tools are available to expand T cells in vitro (e.g., anti-CD3 antibodies, mitogens, and CD3/CD28 beads). We next investigated if the method utilized to perform T cell activation would affect T cell transfection efficiency. We compared GFP transfection efficiency of murine T cells activated with IL-2 and ConA (a mitogen known to activate T cells in an antigen-presenting cell [APC]-independent manner) or total tumor RNA-pulsed dendritic cell (ttRNADC) platform plus IL-2 (a physiologically relevant APC-dependent antigen-specific T cell activation platform established in our laboratory25, 26). GFP expression in CD3+ T cells was detected by flow cytometry at 1, 3, and 5 days after GFP RNA T cell EP. At day 1, CD3+ T cells showed similar GFP expression (∼65%) between ConA and ttRNADC activation groups and decreased GFP expression by ∼1.5-fold at day 3 (****p < 0.0001, *p < 0.05, two-way ANOVA). While ConA-activated T cells decreased GFP expression by ∼3.5-fold at day 5 (****p < 0.0001, two-way ANOVA), ttRNADC-activated T cells retained a superior fraction of GFP+ T cells at day 5 (*p < 0.05, two-way ANOVA) (Figures 2A and 2B).
Figure 2.
Phenotypic Analysis of GFP RNA-Modified T Cells
Spleen was harvested from naive or ttRNADC-vaccinated C57BL/6 mice. Single-cell suspension was expanded with ttRNADC or ConA for 5 and 8 days, respectively. Activated T cells were electroporated with 10 μg GFP RNA. Cells were harvested for phenotypic analysis of GFP+ cells by flow cytometry at the indicated time points. (A and B) Representative histogram (A) and percentages of GFP+ CD3+ T cells expanded with ConA and ttRNADC (B). (C) Gating strategy to distinguish GFP expression within TCM (CD62L+, CD27+), TEM (CD62L−, CD27+), and TE (CD62L−, CD27−) compartments under the gate of CD8+ CD44+ T cells. (D and E) Representative t-SNE analysis (D) and percentages of TCM, TEM, and TE (E) negative for GFP or expressing low or high GFP, 24 hr after electroporation. (F) Percentages of CD8+ CD44+ T cell TCM, TEM, and TE negative for GFP or expressing low or high GFP of total cells, 24 hr after electroporation. TCM, central memory T cell; TEM, effector memory T cell; TE, effector T cell; t-SNE, t-distributed stochastic neighbor embedding; ttRNADC, total tumor RNA-pulsed dendritic cell (*p < 0.05; ***p < 0.001; ****p < 0.0001, two-way ANOVA; **p < 0.01, unpaired t test). Values indicated are the mean ± SEM.
To evaluate the relationship between T cell proliferation and transgene expression, we sorted GFP+ cells out of GFP-electroporated ConA- or ttRNADC-activated T cells at 4 hr post-EP (Figure S1A). GFP expression on CD8+CD44+ T cells was detected by flow cytometry every 24 hr for a period of 5 days (24, 48, 72, 96, and 120 hr). We demonstrated that the percentage and MFI of GFP from the GFP-transfected ConA and ttRNADC CD8+CD44+ T cell groups decreased over time (**p < 0.01, ***p < 0.001, and ****p < 0.0001, two-way ANOVA) (Figures S1B–S1D). These data indicate that activated T cells were losing the transgene as the T cells divided, likely due to both the division and degradation of the non-renewal resource of GFP RNA within the transfected T cells.
Since the T activation platforms used in this experiment skewed expansion toward CD8 T cells (****p < 0.0001, unpaired t test) (Figure S2A), we next evaluated the GFP transgene expression within CD8+ CD44+ central memory T cells (TCM cells; CD62L+, CD27+), effector memory T cells (TEM cells; CD62L−, CD27+), and effector T cells (TE cells; CD62L−, CD27−). Since naive T lymphocytes are mostly refractory to RNA EP, prior activation of T cells is necessary to achieve efficient transfection. Therefore, CD44+ was used as an activation marker to distinguish activated from naive T cells. The short-lived activation of T cells in vitro for 8 days that leads to significant upregulation of CD44 expression on T cells prompted us to use the designation as an activation marker in this context.
Under the live gate, CD8+ CD44+ T cells were gated on TCM, TEM, and TE cells based on their correspondent expression marker (mentioned above). We further characterized these cells as GFPneg, GFPlow, or GFPhigh (Figure 2C), based on their MFI, and we applied the t-distributed stochastic neighbor embedding (t-SNE) for visualization of the distinct T cell subsets expressing various amounts of GFP (Figure 2C; Figure S2B). The different T cell activation platforms resulted in differences in the GFP transgene expression across the defined T cell subsets. Compared to the ttRNADC-stimulated T cells, the percentage of TCM cells in the GFPlow CD44+ CD8+ T cell compartment was superior in the ConA-activated T cell group (**p < 0.01, unpaired t test). In contrast, the percentage of TEM cells in the GFPhigh CD44+ CD8+ T cell compartment was higher in the ttRNADC-activated T cell group (**p < 0.01, unpaired t test) (Figures 2D and 2E). Figure 2F shows the percentage of ConA- or ttRNADC-activated CD8+ CD44+ T cells (TCM, TEM, and TE cells) for each compartment (GFPneg, GFPlow, and GFPhigh) of the total amount of cells. While overall transfection efficiencies were similar across the T cell stimulation platforms, these results demonstrate that stimulation of T cells through antigen presentation by DCs results in prolonged RNA-based gene expression and slightly higher proportions of GFPhigh cells within TCM and TEM T cell subsets.
GM-CSF RNA-Modified T Cells Secrete Transgene In Vitro and Retain Effector Functions
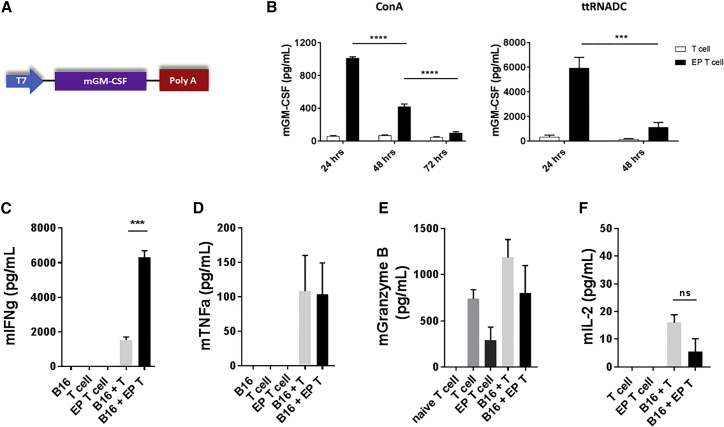
In solid tumors, previous studies have modified T cells with RNA to express membrane-bound proteins, such as T cell receptor (TCR) and CARs, to enhance T cell antigen specificity or T cell function.15, 22 Here, we evaluated if activated T cells could be modified with RNA to secrete a cytokine. After generating the murine GM-CSF in vitro-transcribed (IVT) RNA (Figure 3A), we tested the RNA in vitro by electroporating ConA- or ttRNADC-activated T cells with GM-CSF RNA. Compared to control unmodified T cells, mouse GM-CSF protein levels were significantly higher (Figure 3B) at 24 hr after GM-CSF RNA EP of ConA-activated T cells; similar results were found in the ttRNADC-expanded T cell group. GM-CSF secreted by ConA EP T cells decreased at 48 hr (****p < 0.0001, unpaired t test) (Figure 3B), and it reached basal levels by 72 hr (****p < 0.0001, unpaired t test) (Figure 3B). ttRNADC-stimulated T cells demonstrated similar kinetics of GM-CSF expression (***p < 0.001, unpaired t test) (Figure 3B). Compared to unmodified T cells, the percentage of CD3+ T cells and viability of GM-CSF-transfected T cells were similar at 96 hr post-EP (Figures S3A and S3B), confirming that GM-CSF RNA transfection had no deleterious effects on T cell survival.
Figure 3.
GM-CSF RNA-Modified T Cells Secrete Transgene In Vitro and Retain Effector T Cell Function
Spleen was harvested from ttRNADC-vaccinated C57BL/6 mice or OT-1 transgenic mice. Single-cell suspension was expanded with ttRNADC or ConA for 5 and 8 days, respectively. Activated T cells were electroporated with 10 μg murine GM-CSF RNA. GM-CSF-expressing T cells were co-cultured with B16F10-OVA (ratio of 10:1). Supernatant was harvested and cytokine levels were detected by ELISA or CBA. (A) Schematic representation of the GM-CSF expression vector construct used for mRNA synthesis in vitro. (B) GM-CSF levels detected in the supernatant at the indicated time points post-electroporation. (C–F) Levels of (C) IFN-γ, (D) TNF-α, (E) Granzyme B, and (F) IL-2 detected in the supernatant 24 hr after co-culture. B16, B16F10-OVA; CBA; cytometric bead array; EP, electroporated; ttRNADC, total tumor RNA-pulsed dendritic cell (***p < 0.001, unpaired t test; ****p < 0.0001, two-way ANOVA; ns, not significant). Values indicated are the mean ± SEM.
We next determined the transfection efficiency of ConA-activated T cells using increasing concentrations of GM-CSF RNA (2.5, 5, 10, 15, and 20 μg). Compared to mock EP T cells, the increasing concentration of GM-CSF RNA enhanced transfection efficiency in an RNA dose-dependent manner (Figure S3C). We previously demonstrated the capacity of human T cells to express multiple genes through an RNA-based EP platform. To investigate the feasibility of co-transfection in murine lymphocytes, ConA-activated T cells were electroporated with GFP RNA and RNA encoding for Gaussia luciferase (Luc, a secreted version of firefly luciferase). No difference in GFP+ T cell expression (Figures S3D and S3E) or luciferase activity (Figure S3F) was observed after EP with GFP or Luc RNA alone, or in combination. These data suggest that murine lymphocytes can be efficiently co-transfected with multiple RNAs, allowing for the reprogramming of the tumor microenvironment.
We then assessed if effector T cell function was being disrupted following T cell EP. We evaluated the secretion of well-known immunostimulating cytokines involved with anti-tumor cytotoxic T cell responses, such as IFN-γ, tumor necrosis factor alpha (TNF-α), IL-2, and Granzyme B.27 Using a functional assay, B16F10-ovalbumin (OVA) tumor cells were co-cultured with GM-CSF-expressing ConA-activated OT-1 T cells, and immunostimulating cytokines were measured in the supernatant at 24 hr after co-culture. Interestingly, compared to unmodified OT-1 T cells (cultured with tumor cells), GM-CSF-expressing OT-1 T cells displayed a dramatic enhancement of the IFN-γ secretion (∼4-fold) (***p < 0.001, unpaired t test) (Figure 3C). However, IL-2, TNF-α, and Granzyme B levels remained similar between groups after co-culture with target tumor cells (Figures 3D–3F).
Enhanced GM-CSF Levels within the Brain Tumor Microenvironment following Intravenous Delivery of GM-CSF RNA-Modified T Cells
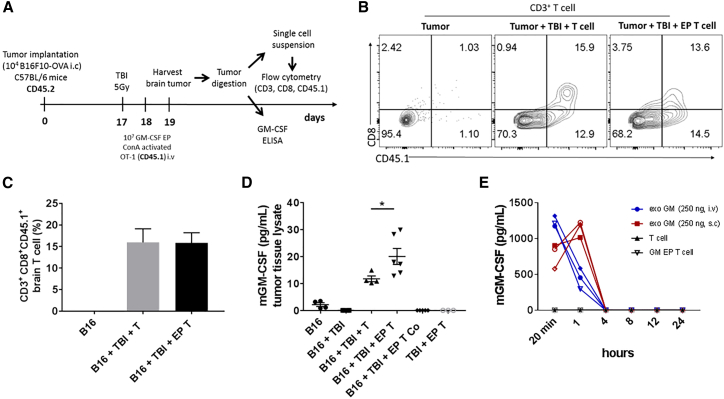
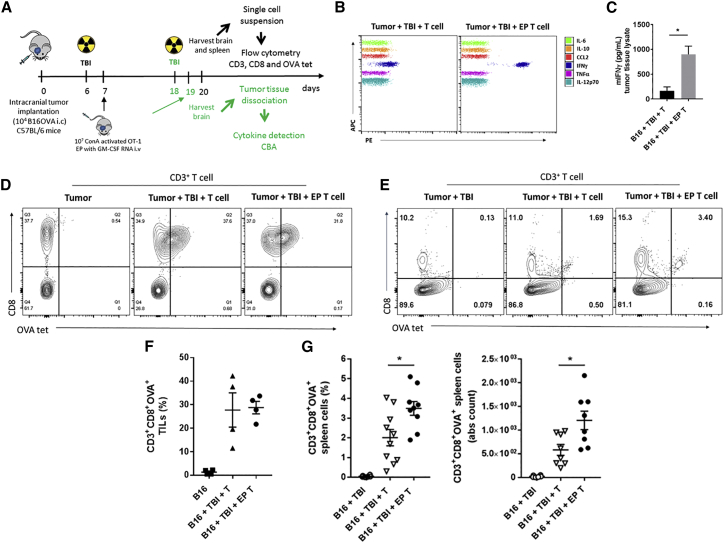
To investigate if genetically modified T cells with RNA could migrate locally to brain tumors, we injected GM-CSF-expressing OT-1 T cells (CD45.1 congenic background) intravenously (i.v.) into B16F10-OVA tumor-bearing C57BL/6 mice (CD45.2 congenic background). The brain tumor was excised at 24 hr after the T cell injection to detect adoptively transferred OT-1 T cells (identified by CD3, CD8, and CD45.1 markers). The percentage of CD3+ CD8+ CD45.1+ T cells was similar in both groups injected with unmodified or GM-CSF-expressing T cells, indicating successful localization of the injected T cells into intracranial tumors (Figures 4B and 4C).
Figure 4.
Enhanced GM-CSF Levels within the Brain Tumor following Systemic Injection of GM-CSF RNA-Modified T Cells
Spleen was harvested from OT-1 transgenic mice. Single cells suspension was expanded with ConA for 8 days and EP with 10 μg of murine GM-CSF RNA. 10 × 106 of GM-CSF-expressing OT-1 (CD45.1) T cells were injected intravenously (i.v.), shortly after electroporation, into lymphodepleted (5 Gy) tumor-bearing naive C57BL/6 mice (CD45.2) at 18 days post intracranial (i.c.) B16F10-OVA implantation (1 × 104 cells/mouse). 24 hr later, brain was harvested and tumor tissue was dissociated. Antigen-specific OT-1 T cells were detected by flow cytometry using anti-CD3, anti-CD8 and anti-CD45.1 antibodies. (A) Schematic figure depicting experimental design. (B and C) Representative contour plot (B) and percentages of antigen-specific T cells (C) detected within the brain tumor at 24 hr following a single injection of GM-CSF-expressing T cells. (D) GM-CSF levels detected within the tumor tissue lysate at 24 hr after GM-CSF-expressing T cell injection. (E) Serum GM-CSF levels detected in non-tumor-bearing C57BL/6 mice receiving systemic (i.v. or s.c.) injection of exogenous GM-CSF (exoGM-CSF) or i.v. injection of GM-CSF-expressing T cells. B16, B16F10-OVA; Co, contralateral; EP, electroporated; exoGM-CSF, exogenous recombinant GM-CSF protein; Gy, gray; i.v., intravenous; i.c., intracranial; TBI, total body irradiation; s.c., subcutaneous (*p < 0.05, one-way ANOVA). Values indicated are the mean ± SEM.
To measure whether cytokine secretion by RNA-modified T cells could be detected locally at the tumor site, we injected GM-CSF-expressing T cells i.v. into established B16F10-OVA tumor-bearing C57BL/6 mice (18 days after tumor implantation), and we quantified GM-CSF levels within the extracted brain tumor tissue lysate 24 hr following T cell injection. Compared to the unmodified T cell group, GM-CSF protein levels were increased nearly 2-fold in the GM-CSF-expressing T cell group (*p < 0.05, one-way ANOVA) (Figure 4D). Notably, unmodified T cells showed moderate levels of GM-CSF after co-culture with target tumor cells in vitro (Figure S4), indicating that unmodified activated OT-1 T cells secrete their own GM-CSF upon tumor recognition in vivo.
The potential risks of systemic cytokine administration have been demonstrated in many patients. Additionally, achieving biologically meaningful concentrations of cytokines within the CNS through systemic circulation may be limiting due to the short half-lives of cytokines and the BBB. To evaluate systemic concentrations of GM-CSF delivered by RNA-modified T cells compared to systemic drug delivery of recombinant GM-CSF, we administered GM-CSF-expressing T cells i.v. into non-tumor-bearing C57BL/6 mice, and we measured GM-CSF levels in the serum over a period of 24 hr. 250 ng/mouse of the recombinant GM-CSF (exoGM-CSF) was also injected systemically (i.v. or subcutaneously [s.c.]) into non-tumor-bearing animals as a comparator group. GM-CSF protein levels were detected 20 min after exoGM-CSF i.v. and s.c. injections. As expected, due to differences in the route of administration, GM-CSF levels dramatically decreased at 1 hr after exoGM-CSF i.v. injection, whereas GM-CSF levels increased transiently following s.c. injection, presumably as GM-CSF was absorbed into the blood compartment from s.c. depot. In both cases, GM-CSF reached basal levels by 4 hr post-injection. In contrast, following i.v. injection of GM-CSF-expressing T cells, we found undetectable levels of GM-CSF in the serum at all the indicated time points (Figure 4E), despite high GM-CSF protein expression from T cells in vitro (Figure S5). Taken together, these data demonstrate that RNA-modified T cells deliver increased concentrations of a cytokine within the brain tumor microenvironment while not modulating systemic concentrations.
GM-CSF RNA-Modified T Cells Enhance Overall Survival in a Murine Brain Tumor Model
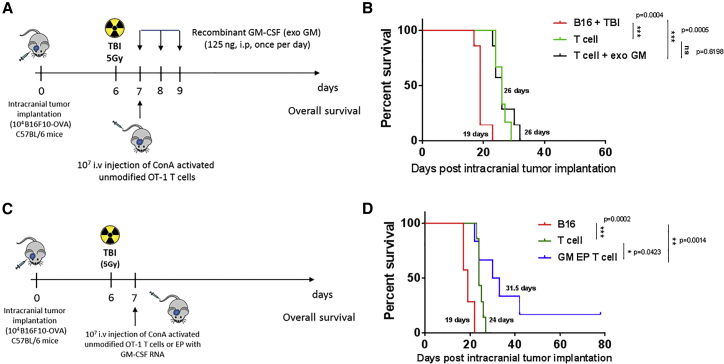
To justify the rationale of using RNA-modified T cell as a platform to deliver biological agents to brain tumors, we first evaluated the anti-tumor efficacy of systemic injection of recombinant GM-CSF (exoGM-CSF) along with adoptive transfer of activated tumor-specific lymphocytes in vivo. At day 7 after tumor implantation, a single i.v. injection of unmodified ConA-activated OT-1 T cell alone or in combination with 125 ng/mouse exoGM-CSF (3 equal doses, every 24 hr) was administered into B16F10-OVA tumor-bearing C57BL/6 mice (Figure 5A). Compared to the unmodified T cell-injected group, the exoGM-CSF plus OT-1 T cell-injected group did not show any additional survival benefit (Figure 5B). We then injected GM-CSF RNA-modified OT-1 T cells into B16F10-OVA intracranial tumor-bearing C57BL/6 mice at day 7 after tumor implantation (Figure 5C). Compared to the unmodified OT-1 T cell group, the GM-CSF-expressing OT-1 T cells prolonged overall survival (Figure 5D) (*p < 0.05, Gehan-Breslow-Wilcoxon test), with approximately 14% (1 of 7) of long-term survivals.
Figure 5.
GM-CSF RNA-Modified T Cells Prolong Overall Survival in a Murine Brain Tumor Model
Spleen was harvested from OT-1 transgenic mice. Cells were expanded with ConA. At day 8, activated T cells were electroporated with 10 μg murine GM-CSF RNA. 10 × 106 GM-CSF-expressing T cells were injected i.v. (shortly after electroporation) into intracranial tumor-bearing C57BL/6 mice at day 7 after tumor B16F10-OVA implantation (1 × 104 cells/mouse). Mice received TBI (5 Gy) 24 hr prior to ACT. Mice were monitored until they reached endpoint. (A and C) Schematic figure illustrating the experimental design of the murine brain tumor model. (B) Overall survival of intracranial B16F10-OVA tumor-bearing mice receiving unmodified ConA-activated OT-1 T cells alone, in combination with exogenous GM-CSF (125 ng, i.p., once a day/3 times), (D) or GM-CSF EP OT-1 intravenously. ACT, adoptive cell transfer; B16, B16F10-OVA; EP, electroporated; exoGM-CSF, exogenous recombinant GM-CSF protein; Gy, gray; i.v., intravenous; i.c., intracranial; TBI, total body irradiation (*p < 0.05, **p < 0.01, and ***p < 0.001, Gehan-Breslow-Wilcoxon test).
While we demonstrated that RNA-modified T cells produced over 600–1,000 pg/mL GM-CSF in vitro within 24 hr after EP (Figure 3B; Figure S5), in vivo detection capabilities are shown to be reliable at as little as 10 pg/mL (Figure 4D). Given the relatively small total blood volume of a mouse (1.5–2.0 mL), we find it particularly relevant that RNA-modified T cells deliver increased levels of GM-CSF to the tumor microenvironment without increasing systemic levels of GM-CSF detectable in the blood, while systemic GM-CSF administration (which is not effective in enhancing anti-tumor immunity during adoptive T cell therapy) leads to high levels of circulating GM-CSF. These experiments highlight the pharmacologic, biologic, and adjuvant effect differences in the delivery of GM-CSF through RNA-modified T cells versus systemic administration. While we did not observe any toxicity in mice treated with RNA-modified T cells, extensive toxicity evaluation of this approach and detection of local concentrations of GM-CSF in other organ sites (e.g., lung, spleen, and lymph node [LN], where lymphocytes may accumulate) must be considered in further clinical development.
To determine the local effects by which GM-CSF RNA-modified T cells potentiated the overall survival, we injected GM-CSF-expressing OT-1 T cells i.v. into mice with established tumors (18 days after tumor implantation), and we analyzed the IFN-γ levels within the brain tumor microenvironment 24 hr after T cell injection (Figure 6A). Compared to the unmodified T cell group, GM-CSF RNA-modified T cells increased IFN-γ secretion at the tumor site nearly 2.5-fold (*p < 0.05, unpaired t test) (Figures 6B and 6C). By using OT-1- (CD3, CD8, and OVA tetramer) (Figure 6A) and DC- (major histocompatibility complex [MHC] class II and CD11c) (Figure S6A) specific markers, we then determined the local and systemic effects of GM-CSF-expressing T cell on antigen-specific T cell and APCs within the brain and/or spleen (harvested 13 days after T cell injection). Compared to the unmodified OT-1 T cell group, while GM-CSF-expressing OT-1 T cell injection increased the percentage (∼2-fold) and absolute count (∼2-fold) of CD3+ CD8+ OVA tetramer+ T cells in the spleen (*p < 0.05, unpaired t test) (Figures 6E and 6G), the percentage of antigen-specific T cells remained similar within the brain tumor-infiltrating lymphocyte compartment (Figures 6D and 6F). The overall percentage, absolute count, and MFI of MHC class II and CD11c remained unchanged (Figures S6B–S6D).
Figure 6.
GM-CSF RNA-Modified T Cells Increase Local IFN-γ Secretion and Potentiate Systemic T Cell Expansion
Spleen was harvested from OT-1 transgenic mice. Single-cell suspension was expanded with ConA for 8 days and EP with 10 μg murine GM-CSF RNA. 10 × 106 GM-CSF-expressing OT-1 T cells were injected i.v. (shortly after RNA electroporation) into tumor-bearing C57BL/6 mice at 7 or 18 days after i.c. B16F10-OVA tumor implantation (1 × 104 cells/mouse). Mice received TBI (5 Gy) 24 hr prior to ACT. Mice were sacrificed at 20 days after tumor implantation. Brain and spleen were harvested 24 hr or 13 days after T cell injection, respectively. Antigen-specific OT-1 T cells were detected by flow cytometry using OVAtet, anti-CD3, and anti-CD8 antibodies. Inflammatory cytokine secretion was detected in the tumor tissue lysate using cytokine bead array. (A) Schematic figure illustrating two different experimental designs (green color indicates the design specifically for (B) and (C). (B and C) Representative plot (B) and IFN-γ levels (C) detected in the tumor tissue lysate 24 hr after GM-CSF-expressing T cell injection. (D and F) Representative contour plot (D) and percentage of OVA-specific T cells (F) within the brain TILs 13 days after T cell injection. (E and G) Representative contour plot (E) and percentage and absolute count (G) of OVA-specific T cells detected in the spleen 13 days after T cell injection. ACT, adoptive cell transfer; B16, B16F10-OVA; EP, electroporated; Gy, gray; i.c., intracranial; i.v., intravenous; MFI, median fluorescence intensity; TBI, total body irradiation; T, T cell; TILs, tumor-infiltrating lymphocytes (*p < 0.05, unpaired t test). Values indicated are the mean ± SEM.
Discussion
Effective drug delivery to brain tumors requires therapeutic concentrations locally at the tumor. However, successful drug delivery to the CNS remains a challenge, and it is often hampered by the BBB and factors characteristic of intracranial tumors, such as the tumor islands disseminated throughout brain parenchyma, intratumoral pressure, and abnormal vasculature within the tumors.28, 29, 30 Strategies to deliver therapeutic macromolecules to the CNS tumor microenvironment, at levels that mediate biological and clinical responses, have included local delivery with implantable diffusion devices (e.g., carmucistine wafer Gliadel), convection-enhanced delivery employing one or more implanted catheters, nanoparticle delivery strategies, BBB disruption approaches with systemic delivery, and the use of gene-modified cellular vehicles to deliver therapeutic agents.31, 32, 33
While immunomodulatory agents can be utilized to reprogram the intratumoral microenvironment, repeated direct intratumoral injection poses many practical challenges, and systemic administration is hindered not only by barrier restrictions but also by dose-limiting toxicities. To circumvent the challenges of drug delivery to brain tumors, we hypothesized that T cells could be modified with RNA to deliver a soluble molecule directly to the brain tumor microenvironment. Although the BBB is an impediment in the context of systemic drug delivery, T cells are an attractive biological carrier due their inherent capability to cross the BBB11, 34 and migrate to areas of invasive neoplastic growth.12
While previous studies used genetic modification of T cells with RNA as a strategy to improve T cell function or redirect tumor-antigen specificity, our study demonstrated the capacity for GM-CSF RNA-modified T cells to effectively deliver therapeutic macromolecules locally to intracranial tumors and mediate enhanced anti-tumor immunity. GM-CSF, a pleiotropic cytokine, is a promising adjuvant utilized to potentiate cancer vaccines in preclinical studies and clinical trials.1, 35 The anti-tumor effects of GM-CSF are likely due to indirect effects on T cells through resident or recruited APCs, such as DCs, and direct effects on DC differentiation and maturation by increasing co-stimulatory molecules, such as CD80, CD86, and CD1a.36, 37, 38 Based on these data, we evaluated if local delivery of GM-CSF by T cells within brain tumors would increase anti-tumor immunity and enhance survival outcomes in tumor-bearing mice. We show that systemic injection of GM-CSF RNA-modified T cells significantly prolongs survival in mice bearing intracranial tumors. Importantly, exogenous GM-CSF combined with unmodified T cells did not enhance overall survival in the same orthotopic brain tumor model. Flow cytometric analyses reveal that such responses are associated with locally increased secretion of IFN-γ in the tumor microenvironment and systemic antigen-specific T cell expansion. Recent studies using CAR T cell therapy in clinical settings demonstrated the occurrence of inflammatory toxicity, in particular cytokine storm and neurotoxicity. To date, elevated IL-6 levels have been a hallmark of cytokine release storm and previously reported in viral-based T cell therapy.39 Unlike viral vector approaches, our RNA-modified T cell platform led to undetectable levels of IL-6 locally at the tumor site (Figure 6B), suggesting a low risk of neurotoxicity. While GM-CSF RNA-modified T cell-treated animals used in this study did not exhibit any symptoms of neurotoxicity or cytokine storm, an extensive toxicity evaluation of such an approach before translation into human clinical trials would be warranted.
Cytokines are tightly regulated and capable of causing severe toxic adverse effects.2 Therefore, it is highly relevant to understand transfection efficiency in different T cell populations for further application in ACT. Previous reports have shown the characterization of RNA-transfected CD4+ T and CD8+ T cells.22, 23 We extended these findings and revealed a detailed phenotypic analysis of RNA-modified CD8+ T cell subtypes, such as TCM, TEM, and TE. As a proof of principle, we used GFP RNA-modified T cells to address this question, and we found a variation of GFP T cell transfection efficiency in different T cell subsets. These results can potentially aid in choosing the appropriate T cell subset for genetic modification and ACT, based on transgene expression levels of each T cell population.
We show that murine T cells activated with different T cell activation platforms can be modified to secrete GM-CSF protein in vitro. Gene expression is differentially regulated during T cell activation using antigen-specific or antigen-independent processes.40 Compared to ConA T cell activation, we demonstrate that ttRNADC T cells mediate superior cytokine level secretion by GM-CSF-expressing T cells. The variations observed on protein levels may be a consequence of distinct T cell activation machinery utilized by T cells during expansion via APCs (ttRNADC) or mitogens (ConA). Various mechanisms could be taken into account for the observed increases in transgene production from DC-stimulated T cells: for instance, an increase in RNA uptake, superior stability of the RNA in DC-stimulated cells compared to ConA stimulation, slower metabolic turnover of the RNA and/or protein in the T cell populations, and/or differences in the proliferative rate of T cells in the two platforms that results in dilution among progeny at a different rate. Our data demonstrated that, although the percentages of the GFP+ CD3+ T cells are similar between the ConA and ttRNADC groups (Figure 2B; Figure S2C) at 24 hr after T cell EP, when we compared the GFP MFI between these two groups, we found increased MFI in the ttRNADC-activated T cell group (Figure S2D). Another potential explanation may be accounted for by a slower rate of GFP decay by the GFP-transfected ttRNADC-activated T cell group when compared to the ConA-activated T cell group (Figure S2E). These results with GFP expression demonstrate that ttRNADC-stimulated T cells express higher and more sustained levels of transgene after EP compared to ConA-stimulated T cells. The mechanistic underpinnings of such a phenomenon are unclear, but they are consistent with the higher levels of cytokine expression observed between the two stimulation platforms. Nonetheless, this observation is noteworthy and potentially relevant to the further development of RNA-modified T cells for potential clinical use.
Interestingly, not only do GM-CSF-expressing T cells maintain effector function but also GM-CSF RNA-modified T cells secreted significantly higher levels of IFN-γ following co-culture with cognate antigen-expressing tumor cells in vitro. This finding corroborates with the increased levels of IFN-γ secretion found at the tumor site following i.v. injection of GM-CSF RNA-modified T cells into tumor-bearing mice. The mechanisms of how GM-CSF potentiates IFN-γ release from T cells are under active investigation.
The therapeutic benefits of transgenic expression of cytokines using a cellular therapy approach in solid tumors have been previously demonstrated.3, 4, 5, 6, 7 However, previous studies did not address whether cytokine production by genetically modified T cells occurs locally within the tumor microenvironment. Compared to the unmodified T cell group, we demonstrated enhanced GM-CSF protein levels within the brain tumor lysate following systemic injection of GM-CSF-expressing T cells into tumor-bearing mice. Importantly, lympho-depleted control animals and the contralateral side of the brain from animals receiving GM-CSF RNA-modified T cells showed undetectable levels of GM-CSF, demonstrating effective local secretion of the transgene within the tumor microenvironment by RNA-modified T cells. While GM-CSF is known to be produced in vitro and in vivo by a wide range of cell types, including tumor cells,41, 42, 43, 44 our observed increases of intratumoral cytokine occurred only with GM-CSF RNA-modified T cells when compared to unmodified T cells. Furthermore, the enhanced GM-CSF levels within the tumor microenvironment and undetectable serum GM-CSF levels found in mice receiving systemic RNA-modified T cell injection rule out the possibility of other sources of enhanced GM-CSF at the tumor site.
While the majority of published RNA-modified T cell therapy studies have utilized xenograft models, which do not recapitulate the immunosuppressive tumor microenvironment found in most cancers types,45 our syngeneic murine brain tumor model allowed us to study cytokine delivery in situ by antigen-specific T cells modified with GM-CSF RNA in an immunocompetent brain tumor model. GM-CSF is mostly known for its pro-inflammatory effects; however, it can also have anti-inflammatory responses by modulating myeloid cells to a tolerigenic state,46 further leading to a regulatory immune response. Such a phenomenon appears to be dose dependent, and it can be affected by the interaction of GM-CSF with other relevant cytokines (e.g., IL-12). Although this potential immunosuppressive effect may have implications for use in immune therapy, our study demonstrates the feasibility and applicability of the RNA-modified T cell platform as a tool to deliver biological agents to brain tumors in a preclinical setting, and it can be potentially applied to other activating cytokines (e.g., IL-2, IL-7, and IL-15). Moreover, such therapy may bypass the need for potentially high and toxic concentrations to be achieved through systemic delivery in order to modulate local concentrations within the brain. The capacity to titrate cytokine production using different amounts of RNA during EP along with the capacity to combine several RNAs lends the potential for combinatorial approaches that may further increase efficacy without additive systemic toxicity.
In summary, we present RNA-modified T cells as a novel modality that can overcome the limitations of biologic drug delivery presented by the BBB and be used as an effective cellular vehicle to deliver therapeutic macromolecules to invasive brain tumors. Such an approach not only suggests a novel therapeutic delivery strategy to control and direct cellular immune responses against CNS tumors but also highlights the potential to reprogram the complex tumor microenvironment. Although we recognize the limitations of our study that may not recapitulate invasive and heterogeneous tumors seen in human patients, as a proof of concept, our demonstration that T cells can be modified, using an RNA-based gene delivery strategy to deliver local concentrations of cytokines to the tumor microenvironment and mediate enhanced anti-tumor immunity, is innovative, and it opens the door to the utilization of T cells for local delivery of novel combinations of RNA transgenes for maximizing potential clinical utility. The ease and reproducibility of transient RNA-based gene expression allows for the potential of multiple sequential infusions of RNA-modified T cells for bio-response modifier delivery, without concerns of prolonged expression of potentially toxic macromolecules from stably transfected T cells. Furthermore, our demonstration of the capacity to express more than one protein product in T cells, through simple co-EP of multiple mRNAs, offers significant versatility in exploring combinatorial therapeutics. Lastly, the simplicity and safety of such an approach reduce manufacturing complexity, and they mediate the capacity for rapid screening of therapeutic molecules of potential utility in a preclinical setting. Of interest would be exploration of the use of RNA-modified T cells for drug delivery in neuroinflammatory or neurodegenerative disorders as well.
Materials and Methods
Animals and Cell Lines
C57BL/6 wild-type and C57BL/6-Tg (TcraTcrb) 1100Mjb/J (OT-1) transgenic mice were obtained from Jackson Laboratory. OT-1 transgenic mice (previously described) express a transgenic TCR that recognizes the SIINFEKL sequence peptide derived from residues 257–264 of OVA.47 The OVA-transfected B16F10 (B16F10-OVA) cell line was a kind gift from Dr. Richard G. Vile, PhD, Mayo Clinic.48, 49 The luciferase-transfected KR158B (Kluc) astrocytoma cell line (originally isolated from a spontaneously arising astrocytoma in an NF1;Trp53 mutant mouse with a C57BL/6 background) was kindly given by Dr. Tyler Jacks (Massachusetts Institute of Technology, Boston, MA).50 B16F10-OVA and Kluc cell lines were cultured in DMEM containing 10% fetal bovine serum (FBS) without sodium pyruvate or with sodium pyruvate, respectively. All animals were housed in specific pathogen-free facilities. Experiments were performed according to University of Florida Institutional Animal Care and Use Committee (IACUC)-approved protocols.
In Vitro Transcription of mRNA and ttRNA Extraction
The plasmids encoding for mouse GM-CSF, and GFP were digested with the restriction enzyme SpeI to linearize the DNA, whereas Gaussia luciferase (Luc) plasmid was digested with SmaI restriction enzyme. Linear DNA was used as a template for the IVT RNA using mMessage mMachine T7 (Ambion, Waltham, MA), according to the manufacturer’s protocol. IVT RNA was purified, and ttRNA was extracted from the dissociated Kluc tumor cell line using RNAeasy kit (QIAGEN, Hilden, Germany). RNA was quantified by spectrophotometry (Nanodrop, Waltham, MA).
DC Generation and Vaccination
Bone marrow-derived DCs were generated from 5-week-old wild-type C57BL/6 mice as previously described.25 Briefly, bone marrow from both rear legs (tibia and femur) and sternum was harvested. Single-cell suspension was obtained and filtered through a 70-μm cell strainer. Cells were centrifuged and re-suspended in ammonium-chloride-potassium (ACK) lysis buffer (Gibco, Waltham, MA). Following lysis, cells were washed and re-suspended in RPMI containing 5% FBS, L-glutamine, non-essential amino acids, 1% HEPES, GM-CSF (20 ng/mL), and IL-4 (20 ng/mL). Immature DCs were electroporated at day 8. At day 9, naive C57BL/6 mice were vaccinated intradermally (i.d.) with ttRNA-pulsed DCs (1.25 × 105 cells per ear).
T Cell Isolation and Activation
Spleen was harvested from 4- to 8-week-old naive, ttRNADC-vaccinated C57BL/6 mice or OT-1 transgenic mice. Red blood cells (RBCs) were lysed using ACK lysis buffer. Cells were cultured in RPMI 1640 (Gibco, Waltham, MA) medium supplemented with 10% FBS and recombinant human (20 U/mL) or mouse IL-2 (50 U/mL). Single-cell suspension of cells was activated with 1 μg/mL ConA (Sigma, St. Louis, MO) at days 1 and 4 or ttRNA-pulsed DCs for 5–7 days.
T Cell and DC Transfection
At 8 days after ConA T cell activation, cells were harvested; washed; and transfected with lipofectamine 2000, 3000, or messenger Max (Invitrogen, Thermo Fisher Scientific), according to the manufacturer’s protocol, or resuspended in 200 μL Opti-MEM. 5 × 106 cells were electroporated in 4-mm cuvettes with 10 μg GFP, Luc (Gaussia Luciferase), or GM-CSF RNA. At 9 days post-BM-derived DC generation, immature DCs were harvested, washed, and resuspended in Opti-MEM. 5 × 106 cells in 200 μL were electroporated in 2-mm cuvettes with 25 μg ttRNA from Kluc. EP was performed by using an Electro Square Porator (ECM 830, BTX, Holliston, MA), as previously described.25 Following transfection, cells were incubated in RPMI medium at 37°C in a humidified atmosphere containing 5% CO2.
Functional T Cell Assay
ConA-activated OT-1 T cells were co-cultured with B16F10-OVA (ratio of 10:1) in a round-bottom 96-well plate containing RPMI 1640; 24 hr later, the plate was centrifuged for 2 min at 200 × g. Supernatant was collected and stored at −80°C for analysis.
Tumor Implantation
B16F10-OVA cells were cultured and harvested with 0.05% trypsin (Gibco, Waltham, MA). Tumor cells were resuspended in 1× PBS and mixed with methylcellulose (1:1 ratio) (R&D Systems). 8- to 10-week-old naive C57BL/6 mice were anesthetized with isoflurane and placed at a stereotactic frame. Intracranial (i.c.) implantation of tumor cells (1 × 104 B16F10-OVA in 2.5 μL/mouse) was performed 2 mm to the right of the bregma and 4 mm below the skull using a 25G needle attached to a 250-μL syringe (Hamilton, Reno, NV). Mice were monitored and sacrificed before reaching endpoint.
Adoptive T Cell Transfer
Randomization of mice was performed after tumor implantation prior to ACT. Unmodified or GM-CSF RNA ConA-activated OT-1 T cells (10 × 106) were washed, re-suspended in 100 μL PBS, and then injected i.v., shortly after RNA EP, into mice at day 7 or 18 after tumor implantation. To engraft T cells more efficiently, all experimental group animals were lympho-depleted by total body irradiation (TBI) using 500 rad (5 Gy) the day prior to T cell injection.
Tumor Tissue Dissociation
Brain tumor tissue was harvested 24 hr after T cell injection (day 19 post-tumor implantation). Tissue was chopped up, transferred to an Eppendorf tube, weighted, and digested with papain (56 U/mL/mg tumor tissue) (Worthington, Lakewood, NJ). Samples were placed in the 37°C shaker for 40 min at 225 rpm. Following incubation, tubes were centrifuged at maximum speed for 10 min. Supernatant was stored at −80°C for analysis.
Cytokine Analysis
For cytokine kinetic experiments, blood was collected in a microtainer tube. Samples were centrifuged at 1,000 × g for 10 min, and serum was stored at −80°C for further analysis. For murine GM-CSF measurement in the supernatant, samples were harvested at 24, 48, and 72 hr after T cell transfection. For murine IFN-γ detection in the supernatant, samples were collected 24 hr after co-culture of T cells with tumor cells. Murine GM-CSF, IFN-γ, and IL-2 protein levels were detected in the supernatant, serum, or tissue tumor lysate by ELISA according to the manufacturer’s protocol (eBioscience, Waltham, MA). The plate was read using a microplate absorbance reader (Cytation 3, Biotek, Winooski, VT) and analyzed by Gene 5 Software. TFN-α protein levels were detected in the supernatant by cytometric bead array (CBA) according to the manufacturer’s protocol (BD Biosciences, San Jose, CA).
Flow Cytometry
Cells were washed with PBS containing 2% FBS prior to the addition of antibodies. Cells were stained with fluorophore-conjugated antibodies specific for mouse CD3-AF700 (BD Biosciences, San Jose, CA), CD4-Perc P5.5 (eBioscience, Waltham, MA), CD8-APC (eBioscience, Waltham, MA), CD8a-PECy7 (BD Pharmingen, San Jose, CA), CD27-PE (BD Biosciences, San Jose, CA), CD44-BV421 (BD Biosciences, San Jose, CA), CD62L-PECy7 (BD Biosciences, San Jose, CA), CD45.1 fluorescein isothiocyanate (FITC) (BD Biosciences, San Jose, CA), and OVA tetramer-PE (OVAtet) (MBL International, Woburn, MA) for 15 min at room temperature. Cells were washed twice with 1× PBS. Flow cytometry was performed on an LSRII flow cytometry system (BD Biosciences, San Jose, CA) or fluorescence-activated cell sorting (FACS) calibur instrument (Becton Dickinson, Franklin Lakes, NJ). Data were acquired with CELLQuest Software (BD Biosciences, San Jose, CA) and analyzed with the aid of FlowJo Software (Tree Star, Ashland, OR) and Cytobank Software.
Luciferase Assay
Supernatant was collected 24 hr after T cell EP to detect luciferase secretion. Luciferase assay was performed according to the manufacturer’s protocol (Promega, Madison, WI). Relative light units were measured with a microplate absorbance reader (Cytation 3, Biotek, Winooski, VT) and analyzed by Gene 5 Software.
Statistical Analysis
One-way ANOVA and unpaired t test were used to determine statistical significance for in vitro and ex vivo experiments. Survival data from the animal studies were analyzed by the Gehan-Breslow-Wilcoxon test. Statistical significance was set for p values less than 0.05. GraphPad Prism (La Jolla, CA) and Microsoft Excel (Redmond, WA) were used to conduct all analyses. Values indicated are the mean ± SEM.
Author Contributions
Concept and Design, F.P.-G. and D.A.M.; Development and Methodology, F.P.-G., C.F., and D.A.M.; Data Acquisition, F.P.-G. and C.Y.; Data Analysis and Interpretation, F.P.-G., C.F., C.Y., D.A.M., E.J.S., J.H., K.A.D., and T.J.W.; Technical and Material Support, C.Y., J.D., K.A.D., T.J.W., and D.A.M.; Writing and Manuscript Review, F.P.-G., C.Y., E.J.S., and D.A.M.
Conflicts of Interest
D.A.M. holds ownership interest (including patents) in iOncologi, Inc., and D.A.M. has patented immunotherapy-related technology that has been licensed by Annias Immunotherapeutics, Inc.; Celldex Therapeutics, Inc.; and Immunomic Therapeutics, Inc. The other authors declare no conflicts of interest.
Acknowledgments
The authors would like to thank Howard R. Seay and Andrew R. Schultz, from the University of Florida Diabetes Institute, for their critical input on t-SNE analysis. This research was supported by the National Cancer Institute (1R01CA195563) and the Preston A. Wells, Jr. Endowment at the University of Florida. This study was also supported in part by the University of Florida Health Cancer Center and University of Florida Clinical and Translational Sciences Award (5UL1TR001427-03).
Footnotes
Supplemental Information includes six figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.10.007.
Supplemental Information
References
- 1.Arellano M., K Waller E. Granulocyte-macrophage-colony-stimulating factor and other cytokines: as adjuncts to cancer immunotherapy, stem cell transplantation, and vaccines. Curr. Hematol. Rep. 2004;3:424–431. [PubMed] [Google Scholar]
- 2.Panelli M.C., White R., Foster M., Martin B., Wang E., Smith K., Marincola F.M. Forecasting the cytokine storm following systemic interleukin (IL)-2 administration. J. Transl. Med. 2004;2:17. doi: 10.1186/1479-5876-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinnasamy D., Yu Z., Kerkar S.P., Zhang L., Morgan R.A., Restifo N.P., Rosenberg S.A. Local delivery of interleukin-12 using T cells targeting VEGF receptor-2 eradicates multiple vascularized tumors in mice. Clin. Cancer Res. 2012;18:1672–1683. doi: 10.1158/1078-0432.CCR-11-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L., Morgan R.A., Beane J.D., Zheng Z., Dudley M.E., Kassim S.H., Nahvi A.V., Ngo L.T., Sherry R.M., Phan G.Q. Tumor-infiltrating lymphocytes genetically engineered with an inducible gene encoding interleukin-12 for the immunotherapy of metastatic melanoma. Clin. Cancer Res. 2015;21:2278–2288. doi: 10.1158/1078-0432.CCR-14-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu D., Song L., Wei J., Courtney A.N., Gao X., Marinova E., Guo L., Heczey A., Asgharzadeh S., Kim E. IL-15 protects NKT cells from inhibition by tumor-associated macrophages and enhances antimetastatic activity. J. Clin. Invest. 2012;122:2221–2233. doi: 10.1172/JCI59535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koneru M., Purdon T.J., Spriggs D., Koneru S., Brentjens R.J. IL-12 secreting tumor-targeted chimeric antigen receptor T cells eradicate ovarian tumors in vivo. OncoImmunology. 2015;4:e994446. doi: 10.4161/2162402X.2014.994446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krenciute G., Prinzing B.L., Yi Z., Wu M.F., Liu H., Dotti G., Balyasnikova I.V., Gottschalk S. Transgenic expression of IL15 improves antiglioma activity of IL13Rα2-CAR T cells but results in antigen loss variants. Cancer Immunol. Res. 2017;5:571–581. doi: 10.1158/2326-6066.CIR-16-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin D.Y., Huang Y., Li D., Wang Y.S., Wang W., Wei Y.Q. Paralleled comparison of vectors for the generation of CAR-T cells. Anticancer Drugs. 2016;27:711–722. doi: 10.1097/CAD.0000000000000387. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell D.A., Karikari I., Cui X., Xie W., Schmittling R., Sampson J.H. Selective modification of antigen-specific T cells by RNA electroporation. Hum. Gene Ther. 2008;19:511–521. doi: 10.1089/hum.2007.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett D.M., Liu X., Jiang S., June C.H., Grupp S.A., Zhao Y. Regimen-specific effects of RNA-modified chimeric antigen receptor T cells in mice with advanced leukemia. Hum. Gene Ther. 2013;24:717–727. doi: 10.1089/hum.2013.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sallusto F., Impellizzieri D., Basso C., Laroni A., Uccelli A., Lanzavecchia A., Engelhardt B. T-cell trafficking in the central nervous system. Immunol. Rev. 2012;248:216–227. doi: 10.1111/j.1600-065X.2012.01140.x. [DOI] [PubMed] [Google Scholar]
- 12.Miao H., Choi B.D., Suryadevara C.M., Sanchez-Perez L., Yang S., De Leon G., Sayour E.J., McLendon R., Herndon J.E., 2nd, Healy P. EGFRvIII-specific chimeric antigen receptor T cells migrate to and kill tumor deposits infiltrating the brain parenchyma in an invasive xenograft model of glioblastoma. PLoS ONE. 2014;9:e94281. doi: 10.1371/journal.pone.0094281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ang W.X., Li Z., Chi Z., Du S.H., Chen C., Tay J.C., Toh H.C., Connolly J.E., Xu X.H., Wang S. Intraperitoneal immunotherapy with T cells stably and transiently expressing anti-EpCAM CAR in xenograft models of peritoneal carcinomatosis. Oncotarget. 2017;8:13545–13559. doi: 10.18632/oncotarget.14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uslu U., Schuler G., Dörrie J., Schaft N. Combining a chimeric antigen receptor and a conventional T-cell receptor to generate T cells expressing two additional receptors (TETARs) for a multi-hit immunotherapy of melanoma. Exp. Dermatol. 2016;25:872–879. doi: 10.1111/exd.13095. [DOI] [PubMed] [Google Scholar]
- 15.Caruso H.G., Torikai H., Zhang L., Maiti S., Dai J., Do K.A., Singh H., Huls H., Lee D.A., Champlin R.E. Redirecting T-Cell Specificity to EGFR Using mRNA to Self-limit Expression of Chimeric Antigen Receptor. J. Immunother. 2016;39:205–217. doi: 10.1097/CJI.0000000000000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schutsky K., Song D.G., Lynn R., Smith J.B., Poussin M., Figini M., Zhao Y., Powell D.J., Jr. Rigorous optimization and validation of potent RNA CAR T cell therapy for the treatment of common epithelial cancers expressing folate receptor. Oncotarget. 2015;6:28911–28928. doi: 10.18632/oncotarget.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pato A., Eisenberg G., Machlenkin A., Margalit A., Cafri G., Frankenburg S., Merims S., Peretz T., Lotem M., Gross G. Messenger RNA encoding constitutively active Toll-like receptor 4 enhances effector functions of human T cells. Clin. Exp. Immunol. 2015;182:220–229. doi: 10.1111/cei.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Höfflin S., Prommersberger S., Uslu U., Schuler G., Schmidt C.W., Lennerz V., Dörrie J., Schaft N. Generation of CD8(+) T cells expressing two additional T-cell receptors (TETARs) for personalised melanoma therapy. Cancer Biol. Ther. 2015;16:1323–1331. doi: 10.1080/15384047.2015.1070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh N., Liu X., Hulitt J., Jiang S., June C.H., Grupp S.A., Barrett D.M., Zhao Y. Nature of tumor control by permanently and transiently modified GD2 chimeric antigen receptor T cells in xenograft models of neuroblastoma. Cancer Immunol. Res. 2014;2:1059–1070. doi: 10.1158/2326-6066.CIR-14-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y., Moon E., Carpenito C., Paulos C.M., Liu X., Brennan A.L., Chew A., Carroll R.G., Scholler J., Levine B.L. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer Res. 2010;70:9053–9061. doi: 10.1158/0008-5472.CAN-10-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon S.H., Lee J.M., Cho H.I., Kim E.K., Kim H.S., Park M.Y., Kim T.G. Adoptive immunotherapy using human peripheral blood lymphocytes transferred with RNA encoding Her-2/neu-specific chimeric immune receptor in ovarian cancer xenograft model. Cancer Gene Ther. 2009;16:489–497. doi: 10.1038/cgt.2008.98. [DOI] [PubMed] [Google Scholar]
- 22.Parkhurst M.R., Joo J., Riley J.P., Yu Z., Li Y., Robbins P.F., Rosenberg S.A. Characterization of genetically modified T-cell receptors that recognize the CEA:691-699 peptide in the context of HLA-A2.1 on human colorectal cancer cells. Clin. Cancer Res. 2009;15:169–180. doi: 10.1158/1078-0432.CCR-08-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birkholz K., Hombach A., Krug C., Reuter S., Kershaw M., Kämpgen E., Schuler G., Abken H., Schaft N., Dörrie J. Transfer of mRNA encoding recombinant immunoreceptors reprograms CD4+ and CD8+ T cells for use in the adoptive immunotherapy of cancer. Gene Ther. 2009;16:596–604. doi: 10.1038/gt.2008.189. [DOI] [PubMed] [Google Scholar]
- 24.Schaft N., Dörrie J., Müller I., Beck V., Baumann S., Schunder T., Kämpgen E., Schuler G. A new way to generate cytolytic tumor-specific T cells: electroporation of RNA coding for a T cell receptor into T lymphocytes. Cancer Immunol. Immunother. 2006;55:1132–1141. doi: 10.1007/s00262-005-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flores C., Pham C., Snyder D., Yang S., Sanchez-Perez L., Sayour E., Cui X., Kemeny H., Friedman H., Bigner D.D. Novel role of hematopoietic stem cells in immunologic rejection of malignant gliomas. OncoImmunology. 2015;4:e994374. doi: 10.4161/2162402X.2014.994374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell D.A., Batich K.A., Gunn M.D., Huang M.N., Sanchez-Perez L., Nair S.K., Congdon K.L., Reap E.A., Archer G.E., Desjardins A. Tetanus toxoid and CCL3 improve dendritic cell vaccines in mice and glioblastoma patients. Nature. 2015;519:366–369. doi: 10.1038/nature14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen D.S., Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 28.Leten C., Struys T., Dresselaers T., Himmelreich U. In vivo and ex vivo assessment of the blood brain barrier integrity in different glioblastoma animal models. J. Neurooncol. 2014;119:297–306. doi: 10.1007/s11060-014-1514-2. [DOI] [PubMed] [Google Scholar]
- 29.Rascher G., Fischmann A., Kröger S., Duffner F., Grote E.H., Wolburg H. Extracellular matrix and the blood-brain barrier in glioblastoma multiforme: spatial segregation of tenascin and agrin. Acta Neuropathol. 2002;104:85–91. doi: 10.1007/s00401-002-0524-x. [DOI] [PubMed] [Google Scholar]
- 30.Charles N.A., Holland E.C., Gilbertson R., Glass R., Kettenmann H. The brain tumor microenvironment. Glia. 2011;59:1169–1180. doi: 10.1002/glia.21136. [DOI] [PubMed] [Google Scholar]
- 31.Soni V., Jain A., Khare P., Gulbake A., Jain S.K. Potential approaches for drug delivery to the brain: past, present, and future. Crit. Rev. Ther. Drug Carrier Syst. 2010;27:187–236. doi: 10.1615/critrevtherdrugcarriersyst.v27.i3.10. [DOI] [PubMed] [Google Scholar]
- 32.Chacko A.M., Li C., Pryma D.A., Brem S., Coukos G., Muzykantov V. Targeted delivery of antibody-based therapeutic and imaging agents to CNS tumors: crossing the blood-brain barrier divide. Expert Opin. Drug Deliv. 2013;10:907–926. doi: 10.1517/17425247.2013.808184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Tellingen O., Yetkin-Arik B., de Gooijer M.C., Wesseling P., Wurdinger T., de Vries H.E. Overcoming the blood-brain tumor barrier for effective glioblastoma treatment. Drug Resist. Updat. 2015;19:1–12. doi: 10.1016/j.drup.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 34.Ransohoff R.M., Kivisäkk P., Kidd G. Three or more routes for leukocyte migration into the central nervous system. Nat. Rev. Immunol. 2003;3:569–581. doi: 10.1038/nri1130. [DOI] [PubMed] [Google Scholar]
- 35.Wen Q., Xiong W., Liu S., Zhou C., Ma L. [Effect of bifunctional IL2-GMCSF in promoting dendritic cell activation in vitro in simulated tumor-induced immune suppression] Nan Fang Yi Ke Da Xue Xue Bao. 2015;35:1239–1244. [PubMed] [Google Scholar]
- 36.Wada H., Noguchi Y., Marino M.W., Dunn A.R., Old L.J. T cell functions in granulocyte/macrophage colony-stimulating factor deficient mice. Proc. Natl. Acad. Sci. USA. 1997;94:12557–12561. doi: 10.1073/pnas.94.23.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dranoff G. GM-CSF-based cancer vaccines. Immunol. Rev. 2002;188:147–154. doi: 10.1034/j.1600-065x.2002.18813.x. [DOI] [PubMed] [Google Scholar]
- 38.Dranoff G., Jaffee E., Lazenby A., Golumbek P., Levitsky H., Brose K., Jackson V., Hamada H., Pardoll D., Mulligan R.C. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc. Natl. Acad. Sci. USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davila M.L., Riviere I., Wang X., Bartido S., Park J., Curran K., Chung S.S., Stefanski J., Borquez-Ojeda O., Olszewska M. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci. Transl. Med. 2014;6:224ra25. doi: 10.1126/scitranslmed.3008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tumeh P.C., Koya R.C., Chodon T., Graham N.A., Graeber T.G., Comin-Anduix B., Ribas A. The impact of ex vivo clinical grade activation protocols on human T-cell phenotype and function for the generation of genetically modified cells for adoptive cell transfer therapy. J. Immunother. 2010;33:759–768. doi: 10.1097/CJI.0b013e3181f1d644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cousins D.J., Staynov D.Z., Lee T.H. Regulation of interleukin-5 and granulocyte-macrophage colony-stimulating factor expression. Am. J. Respir. Crit. Care Med. 1994;150:S50–S53. doi: 10.1164/ajrccm/150.5_Pt_2.S50. [DOI] [PubMed] [Google Scholar]
- 42.Nimer S.D., Uchida H. Regulation of granulocyte-macrophage colony-stimulating factor and interleukin 3 expression. Stem Cells. 1995;13:324–335. doi: 10.1002/stem.5530130402. [DOI] [PubMed] [Google Scholar]
- 43.Metcalf D. The colony-stimulating factors and cancer. Nat. Rev. Cancer. 2010;10:425–434. doi: 10.1038/nrc2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi Y., Liu C.H., Roberts A.I., Das J., Xu G., Ren G., Zhang Y., Zhang L., Yuan Z.R., Tan H.S. Granulocyte-macrophage colony-stimulating factor (GM-CSF) and T-cell responses: what we do and don’t know. Cell Res. 2006;16:126–133. doi: 10.1038/sj.cr.7310017. [DOI] [PubMed] [Google Scholar]
- 45.Rabinovich G.A., Gabrilovich D., Sotomayor E.M. Immunosuppressive strategies that are mediated by tumor cells. Annu. Rev. Immunol. 2007;25:267–296. doi: 10.1146/annurev.immunol.25.022106.141609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong I.S. Stimulatory versus suppressive effects of GM-CSF on tumor progression in multiple cancer types. Exp. Mol. Med. 2016;48:e242. doi: 10.1038/emm.2016.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright K.O., Murray D.A., Crispe N.I., Pierce R.H. Quantitative PCR for detection of the OT-1 transgene. BMC Immunol. 2005;6:20. doi: 10.1186/1471-2172-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez-Perez L., Kottke T., Diaz R.M., Ahmed A., Thompson J., Chong H., Melcher A., Holmen S., Daniels G., Vile R.G. Potent selection of antigen loss variants of B16 melanoma following inflammatory killing of melanocytes in vivo. Cancer Res. 2005;65:2009–2017. doi: 10.1158/0008-5472.CAN-04-3216. [DOI] [PubMed] [Google Scholar]
- 49.Daniels G.A., Sanchez-Perez L., Diaz R.M., Kottke T., Thompson J., Lai M., Gough M., Karim M., Bushell A., Chong H. A simple method to cure established tumors by inflammatory killing of normal cells. Nat. Biotechnol. 2004;22:1125–1132. doi: 10.1038/nbt1007. [DOI] [PubMed] [Google Scholar]
- 50.Reilly K.M., Loisel D.A., Bronson R.T., McLaughlin M.E., Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat. Genet. 2000;26:109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.