Abstract
It has previously been shown that engineered zinc finger nucleases (ZFNs) can be packaged into adeno-associated viruses (AAVs) and delivered intravenously into mice, non-human primates, and most recently, humans to induce highly efficient therapeutic genome editing in the liver. Lipid nanoparticles (LNPs) are synthetic delivery vehicles that enable repeat administration and are not limited by the presence of preexisting neutralizing antibodies in patients. Here, we show that mRNA encoding ZFNs formulated into LNP can enable >90% knockout of gene expression in mice by targeting the TTR or PCSK9 gene, at mRNA doses 10-fold lower than has ever been reported. Additionally, co-delivering mRNA-LNP containing ZFNs targeted to intron 1 of the ALB locus with AAV packaged with a promoterless human IDS or FIX therapeutic transgene can result in high levels of targeted integration and subsequent therapeutically relevant levels of protein expression in mice. Finally, we show repeat administration of ZFN mRNA-LNP after a single AAV donor dose results in significantly increased levels of genome editing and transgene expression compared to a single dose. These results demonstrate LNP-mediated ZFN mRNA delivery can drive highly efficient levels of in vivo genome editing and can potentially offer a new treatment modality for a variety of diseases.
Conway and colleagues demonstrate for the first time non-viral in vivo genome editing using zinc finger nucleases, achieving therapeutically relevant levels of targeted gene knockout as well as targeted integration of therapeutic transgenes within murine liver at mRNA doses 10-fold lower than has ever been reported.
Introduction
Genome editing holds great potential for addressing numerous medical conditions. Programmable nucleases, such as zinc finger nucleases (ZFNs),1 transcription-activator like effector nucleases (TALENs),2 and the clustered regularly interspersed palindromic repeats (CRISPR)/CRISPR-associated (Cas) enzyme system3 can be engineered to direct a DNA double-stranded break (DSB) at essentially any site in the genome. When programmable nucleases are expressed in a cell in the presence of a donor DNA template that contains homology to the region flanking the target DSB, homology-directed repair (HDR) mechanisms can correct a deleterious mutation4 or insert an expression cassette for a therapeutic protein.5, 6 In the absence of a donor DNA, repair by the non-homologous end joining (NHEJ) pathway can lead to random insertions and deletions (indels) at the DSB site that can disrupt the target gene.7 Although programmable nucleases hold promise for addressing some diseases, safe and effective delivery of these agents to patients remains a challenge.
In vivo genome editing was first reported several years ago for editing of the FIX and ALB loci in mice using adeno-associated virus (AAV) vectors to deliver site-specific nucleases and donor DNA.5, 6 Viral vectors based on AAV have been commonly used as delivery agents for transgenes, as they are not generally pathogenic in humans and can provide prolonged expression of the introduced gene product.8 However, use of these vectors is hampered by the high incidence of preexisting and induced immune response,1 and persistent expression of programmable nucleases can lead to excessive off-target cleavage.9
Recently, lipid nanoparticles (LNPs) have become an attractive option for delivering in vitro transcribed mRNAs encoding programmable nucleases to target cells. Much progress in the use of LNPs as a delivery vehicle for RNA has been made with small interfering RNAs used to inhibit expression of disease-associated proteins. These studies showed that LNP made with lipids containing an ionizable amino head group that maintains a neutral charge in circulation associate with plasma apolipoprotein E and are effectively taken up by hepatocytes via receptor-mediated endocytosis.10, 11, 12, 13 In the acidic environment of the endosome, the ionizable head group acquires a positive charge promoting release of the RNA cargo into the cytoplasm. After intravenous administration, mRNAs formulated into LNP are primarily taken up by the liver where they direct a transient burst of translation that peaks within a few hours and diminishes after a few days.14 LNP-formulated mRNAs have been used to direct transient expression of reporter genes,14 Cas9 nuclease,15, 16, 17, 18, 19, 20 erythropoietin,21, 22 and human factor IX protein21 in mice, small pigs, and non-human primates. mRNA offers several advantages for delivering programmable nucleases, including ease of production and the desirable rapid and transient protein expression profile. Exogenously administered mRNA can activate innate immune anti-viral responses23 that induce counterproductive pathways, such as RNA degradation and inhibition of protein synthesis. But, effective engineering of the in vitro transcribed mRNA by ensuring efficient capping,24, 25 including optimal 5′ and 3′ UTRs in the mRNA,26, 27 and incorporating a poly(A) tail28 enhances stability and activity of the message. Synthesizing the transcript with modified nucleosides29 and high-pressure liquid chromatography (HPLC) purification of the final mRNA30 have also been shown to reduce degradation and innate immune activation.
In this study, we demonstrate that ZFN mRNAs formulated in LNP made with a novel ionizable lipid are effectively delivered to the liver after intravenous injection into mice and result in highly efficient genome editing. ZFN administration via this route was well tolerated, even after repeated dosing, resulting in accumulative increases in genome editing.
Results
Novel Ionizable Lipid Enables Effective Delivery of ZFN mRNA to Hepatocytes In Vivo
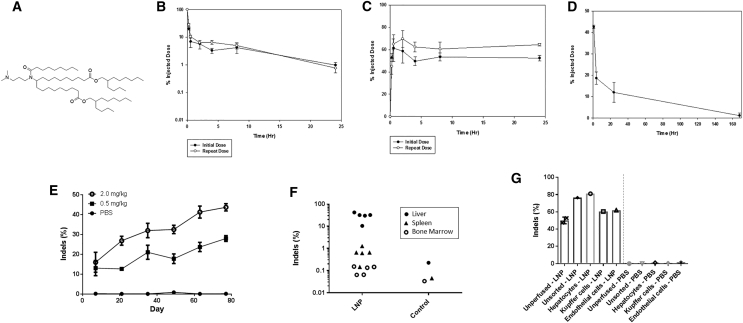
Desirable characteristics for ionizable lipids used in lipid delivery vehicles for mRNA in vivo include a conical shape and ionizable head group that support effective cellular delivery and subsequent endosomal release of the RNA cargo12 and biocompatible chemistry that promotes rapid clearance of the lipid as nontoxic metabolites13. From a screen of ionizable lipids synthesized to address these parameters, one lead was selected for further characterization (Figure 1A). To explore its utility in vivo, the lipid was formulated into LNPs with a pair of well characterized, in vitro transcribed ZFN mRNAs (31523/48641) targeting the first intron of the murine ALB (mALB) gene.6 The LNPs were rapidly cleared from circulation after injection into the tail vein, with less than 5% of injected dose remaining after 4 h (Figure 1B) and subsequent accumulation of approximately 60%–70% in the liver (Figure 1C). Importantly, analysis of the ionizable lipid showed rapid elimination from the liver (Figure 1D) with about 10% of the injected material remaining after 24 h, which diminished to undetectable levels by 7 days.
Figure 1.
Characterization of In Vivo LNP Delivery
(A) Structure of ionizable lipid used in LNP formulation. Pharmacokinetics of lipid in plasma (B) and biodistribution to liver (C) after single and repeat intravenous administration of LNPs in mice (n = 5). (D) Elimination of lead ionizable lipid from liver following intravenous injection in mice (n = 5). (E) Genome-editing results from mouse bulk liver tissue following six repeat intravenous administrations of ionizable lipid-containing LNPs comprising mRNAs encoding ZFNs 48641 and 31523 (n = 4–6 per data point) (p < 0.0001, R2 = 0.5557, F = 32.52, degrees of freedom = 26 for 0.5 mg/kg from first to sixth dose; p < 0.0001, R2 = 0.6996, F = 55.9, degrees of freedom = 24 for 2.0 mg/kg from first to sixth dose). (F) Biodistribution of genome modification following a single 2-mg/kg administration of lead ionizable lipid-containing LNP comprising mRNAs encoding ZFNs 48641 and 31523 (n = 5 per LNP group organ). Note background detection level of indels of 0.1%–0.2% (p = 0.00054 comparing liver versus spleen; p = 0.00048 comparing liver versus bone marrow). (G) Genome modification of mice from (F), which were either sacrificed and unmanipulated prior to liver harvest (unperfused) or perfused through the hepatic portal vein with buffered saline prior to liver harvest to remove blood cells within the liver (unsorted) as well as to prepare the liver for perfusion with a collagenase solution. A fraction of the perfused livers was also FACS-sorted into individual liver cell populations (n = 1–2 per group).
When the double-stranded DNA break directed by ZFNs is repaired by the NHEJ pathway, small insertions or deletions of nucleotides (indels) can occur at the cut site, which are an indicator of genome-editing activity. Analysis of liver-derived genomic DNA at the target cleavage site indicated that LNPs could be administered repeatedly yielding accumulative increases in genome-editing activity with subsequent injections at 14-day intervals for up to six doses in a dose-dependent manner (Figure 1E). Genome-editing activity after the sixth dose was much greater in the liver than in the spleen or bone marrow (Figure 1F), indicating that LNPs are predominantly taken up by the liver when administered intravenously, consistent with previous studies.10, 14 The measured level of genome-editing activity increased if the mouse liver was perfused via the hepatic portal vein with buffered saline, which potentially may be due to removal of contaminating blood cells prior to harvesting DNA for analysis or by some other unknown mechanism (Figure 1G). Finally, analysis of DNA prepared from different liver cell types isolated by fluorescence-activated cell sorting (FACS)-sorting after collagenase digestion demonstrated that genome editing occurred in 80% of hepatocytes and to a lesser extent in Kupffer cells and endothelial cells (Figure 1G).
Optimization of Factors to Enable Robust In Vivo Genome Editing
Using LNP-delivered ZFN mRNAs targeting the murine ALB gene, we explored a number of parameters that have been reported to affect mRNA activity, stability, and immunogenicity.
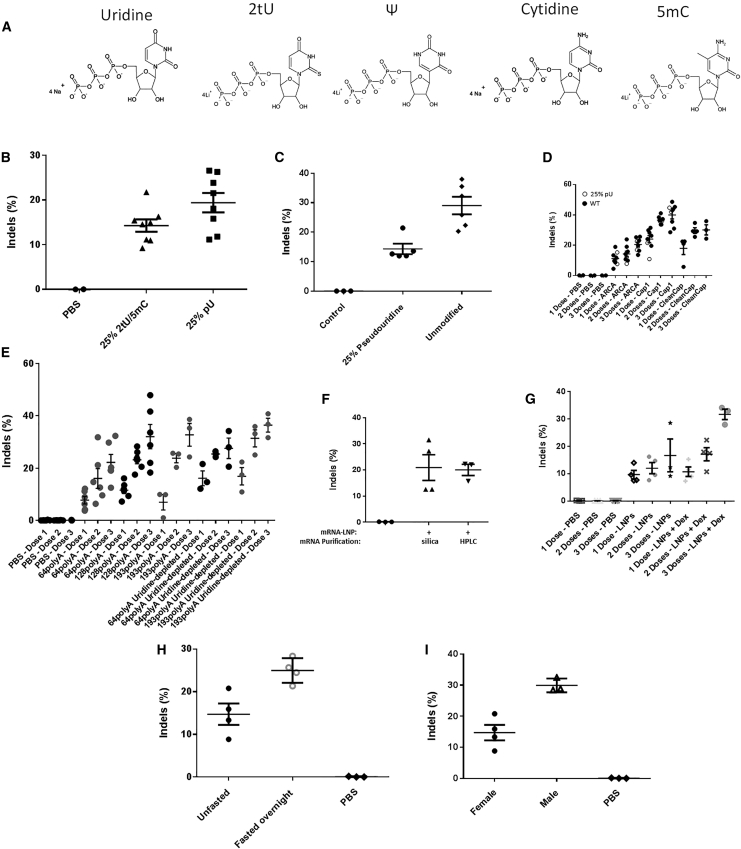
Activation of the innate anti-viral response has been commonly observed after administration of in vitro transcribed mRNAs, likely due to activation of pattern recognition receptors31 and incorporation of modified nucleosides, particularly uridine analogs (Figure 2A) improved the activity of mRNAs both in cultured cells29, 32, 33, 34 and in mice.32, 33 However, ZFN mRNAs where 25% of the cytosine and uridine nucleosides were replaced with 5-methyl-cytosine (5mC) and 2-thiouridine (2tU), produced less genomic editing activity in DNA prepared from liver than mRNAs produced with 25% pseudouridine (pU) (Figure 2B), and mRNAs containing 25% pU were less active (Figure 2C) or no more active than mRNAs containing unmodified nucleosides (Figure 2D). Because of the redundancy of the genetic code, uridine content of mRNA can also be reduced by selecting codons that lack uridine in the “wobble” position.35 Such “uridine-depleted” mRNAs generated higher genome-editing activity after each of three repeated doses than their original sequence counterparts (Figure 2E).
Figure 2.
Optimization of mRNA Construct and Animal Pretreatment
(A) Natural and modified nucleosides used during the in vitro transcription reactions to create unmodified and modified mRNA, respectively. (B) Liver genome-editing results from mice injected with a single 2 mg/kg dose of LNP containing 48641/31523 ZFN mRNAs (n = 8 per group) (p = 0.067). (C) Genome modification results following a single administration of 0.5 mg/kg LNP comprising 25% pU substituted or unmodified mRNAs encoding ZFNs 48641 and 31523 (n = 5–6 per group) (p = 0.0029). (D) Genome-editing results from repeat dosing (14-day intervals) of 2 mg/kg LNP containing 48641/31523 ZFN mRNAs with 25% pU nucleoside substitution (open circles) or no nucleoside substitution (closed circles) (n = 8 per group) (p = 0.000016 comparing three doses of ARCA versus Cap1). (E) Repeat dosing (14-day intervals, 0.5 mg/kg) of LNP containing 48641/31523 ZFN mRNAs containing different poly(A) lengths and composition of uridines in the protein coding sequence as indicated. Livers were harvested 7 days post-dosing. “64polyA,” “128polyA,” and “193polyA” refer, respectively, to 64, 128, or 193 long poly(A) regions while “uridine-depleted” refers to polynucleotides with at a percentage of uridines deleted from the wobble positions in the codons (n = 3 per group) (p = 0.089 comparing 1 dose of original versus uridine-depleted 193poly(A)); (n = 3–6 per group) (p = 0.021 comparing one dose of original versus uridine-depleted 64poly(A)); (n = 3–6 per group) (p = 0.085 comparing three doses of original 64poly(A) versus 193poly(A)). (F) Single dose of LNP containing 48641/31523 ZFN mRNAs, purified either via silica column or HPLC and injected into mice at 1 mg/kg and livers harvested 28 days post-dosing. (G) Genome-editing results from repeat dosing (14-day intervals, 2 mg/kg) of LNP containing 48641/31523 ZFN mRNAs (n = 3 per group) (p = 0.075 comparing 3 doses with and without Dex). (H) Genome-modification results following a single administration 0.5 mg/kg LNP containing 25% pU substituted 48641/31523 ZFN mRNAs into mice which were either fed ad libitum or mice that were fasted overnight prior to LNP dosing (n = 4 per group) (p = 0.012). (I) Genome-modification results following a single administration of 0.5 mg/kg LNP containing 25% pU substituted 48641/31523 ZFN mRNAs into either male or female C57BL6 wild-type mice (n = 4 females and 3 males) (p = 0.0047).
Natural mRNAs contain a cap structure at the 5′ end consisting of an N7-methylguanosine linked by a reverse 5′ to 5′ triphosphate bond to the first nucleotide of the transcript (Cap 0). The first nucleotide or first two nucleotides are subsequently methylated at the 2′ position to form the mature cap (Cap1 or Cap2, respectively). The cap is important for translation and to mask the mRNA from the innate immune anti-viral machinery.36 Several different cap variants were tested for their effect on mALB ZFN activity, including (1) an anti-reverse capping analog (ARCA) that is added to the transcript during synthesis in vitro;37 (2) a Cap1 structure that was enzymatically added to the mRNA after in vitro transcription (Cap1); and (3) a Cap1 structure added to the mRNA during synthesis using CleanCap technology (CleanCap). The enzymatically added Cap1 produced higher levels of genomic editing after each of three repeated doses (Figure 2D).
A poly(A) tail is added to most mRNAs during transcription and enhances translational activity and stability.38 To explore how poly(A) tail length would affect genome editing activity, templates were prepared for the mALB ZFN mRNAs that would add 64, 128, or 193 adenosine residues at the end of mRNA during in vitro transcription. In general, greater genome-editing activity was detected in liver DNA by ZFN mRNAs with longer poly(A) tails (Figure 2E).
Previous studies have indicated that purification of mRNAs by HPLC produced improved translational activity,30 although we did not consistently observe greater genome-editing activity when HPLC-purified mRNAs were compared with mRNAs purified by silica columns in mice (Figure 2F). Pretreatment of mice with dexamethasone, a synthetic glucocorticoid, has been shown to inhibit inflammation induced by administration of siRNA nanoparticles.39 Dexamethasone pretreatment trended toward increased genome-editing activity of mALB ZFNs, particularly after the first dose in mice receiving multiple injections of the LNP (Figure 2G). Interestingly, fasting the mice the night before LNP injection improved genome-editing activity (Figure 2H). The reduced concentration of chylomicrons in the bloodstream, which would increase the levels of LDL receptors on the surface of hepatocytes, is the hypothesized mechanism.40 Finally, since it has been previously reported that sex significantly influences transduction of AAV in the murine liver,41 we explored if there was differential transduction of LNPs in male mice compared to female mice. We observed that genome-editing activity was indeed higher in male mice than female mice for a given dose of LNPs (Figure 2I), though the associated mechanism is still not fully understood.
Complete Knockout of Therapeutically Relevant Gene Targets in the Liver Using Very Low mRNA Doses
Due to the error-prone nature of the NHEJ pathway following nuclease-mediated DSB, a properly positioned cleavage site within a protein-coding region of a gene can disrupt expression of the target gene. To explore how effectively LNPs could deliver ZFN mRNAs targeted to a therapeutically relevant gene, ZFNs utilizing a recently developed novel architecture42 were generated to target the murine TTR gene (mTTR). Transthyretin is predominantly expressed by hepatocytes and normally serves to transport thyroid hormones and vitamin A in circulation, but in pathological situations the protein aggregates into deposits that damage the heart and nervous system.43 Reducing expression levels has been shown to correlate with clinically significant benefits in patients with hereditary TTR amyloidosis.44
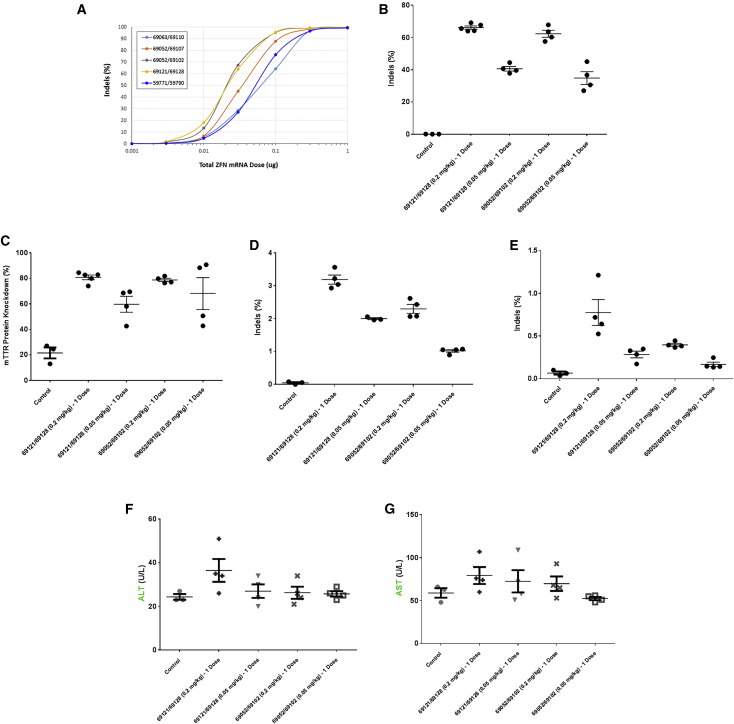
A panel of in vitro transcribed ZFN mRNA pairs targeting mTTR were prepared, formulated into LNPs, and added onto Hepa1-6 cells in culture. A dose response of genome-editing activity was determined for the target site (Figure 3A). Two pairs of ZFN mRNAs with the highest on-target genome-editing activity were selected for further analysis in mice (69121/69128 and 69052/69102). When formulated with the ionizable lipid into LNPs, both ZFN pairs showed dose-dependent genome-editing activity in liver DNA after a single tail-vein injection (Figure 3B), generating more than 60% of gene disruption in bulk liver tissue at the 0.2 mg/kg dose. This level of gene editing resulted in an 80% decrease in the level of transthyretin protein in the plasma 35 days after LNP dosing (Figure 3C). mTTR genome-editing levels were much lower in the spleen (Figure 3D) and the kidney (Figure 3E). Delivery of ZFN mRNAs by LNPs seemed to be well tolerated, as no significant increase was observed in plasma levels of the liver enzymes alanine aminotransferase (ALT; Figure 3F) or aspartate aminotransferase (AST; Figure 3G) measured the day after injection for either dose.
Figure 3.
Knockout of Therapeutically Relevant Liver Gene Target TTR
(A) Genome-editing results from a panel of optimized ZFNs targeted to the murine TTR gene electroporated as mRNA in dose curves in murine hepatocarcinoma cell line Hepa1-6. (B) Genome-editing activity in mouse bulk liver tissue 35 days following a single 0.05 or 0.2 mg/kg dose of LNP containing 69121/69128 or 69052/69102 ZFN mRNAs into mice (n = 4–5 per group) (p = 0.0000013 comparing 69121/69128 0.05 versus 0.2 mg/kg doses). (C) Murine TTR ELISA assay in plasma collected from the mice described in (B) (n = 4–5 per group) (p = 0.0088 comparing 69121/69128 0.05 versus 0.2 mg/kg doses). (D) Genome editing in off-target organ (spleen) collected at the same time as livers from (B). (E) Genome editing in off-target organ (kidney) collected at the same time as livers from (B). (F and G) Results of liver function test (LFT) in serum collected from the mice described in (B) 1 day post-dosing: (F) alanine transaminase (ALT) and (G) aspartate transaminase (AST).
ZFN mRNAs were designed and constructed for the murine equivalent of a second therapeutically relevant gene, proprotein convertase subtilisin/kexin type 9 (PCSK9). PCSK9 is a plasma protein produced predominantly by hepatocytes. It binds to the LDL receptor on hepatocytes reducing LDL cholesterol clearance. Rare variants with increased activity are associated with high-plasma LDL cholesterol levels and increased rates of cardiovascular disease.45 PCSK9-targeted ZFN mRNAs were also effectively delivered to the liver by these LNPs, inducing efficient genome editing (Figure S1A) and reducing plasma levels of the PCSK9 protein more than 90% after a single dose via tail-vein injection (Figure S1B).
In Vivo Therapeutic Protein Replacement via Co-delivery of Viral and Non-viral Genome-Editing Components
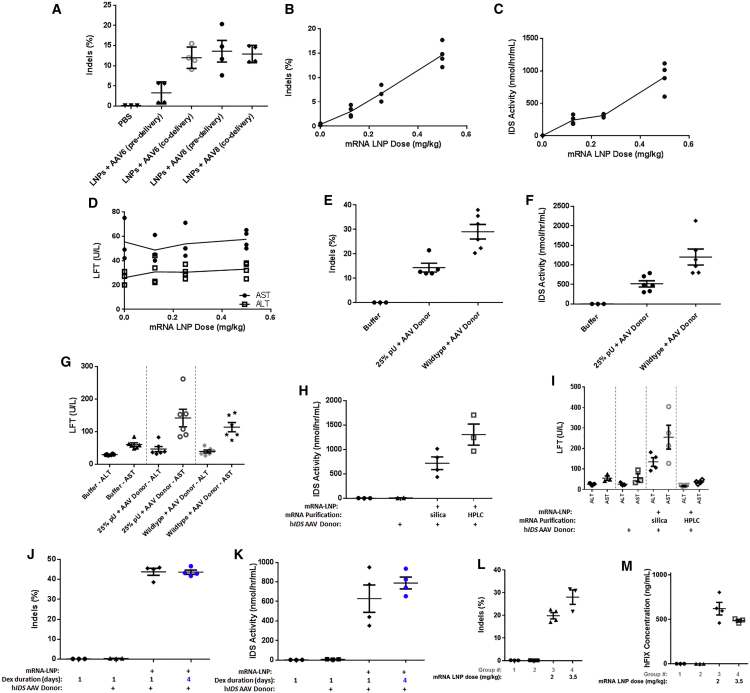
For gene-knockout applications, delivery of ZFN mRNA to hepatocytes is sufficient as a potential therapeutic strategy. However, many diseases involve loss-of-function mutations in genes that require either gene correction of the aberrant mutation or gene insertion of the wild-type form of the entire protein coding sequence of the gene. To perform either of these strategies, delivery of a DNA donor template to allow for HDR and targeted integration of the desired sequence within the genome is required. Currently, viral vectors are the most efficient vehicles for delivering DNA to the nucleus of cells in vivo, as they have evolved over millennia to do so. We harnessed the highly efficient nuclear delivery capability of AAV to deliver a single-stranded DNA transgene containing (1) the coding sequence for the therapeutic human enzyme IDS (hIDS), which is mutated in MPS II patients, (2) a splice acceptor sequence to allow for splicing into an upstream exon, and (3) homology arms that flank the ZFN cut site within murine ALB intron 1. To facilitate integration into the genome and subsequent splicing and expression of the transcript, mALB ZFNs targeting intron 1 were produced into mRNA, formulated into LNPs, and either co-delivered or delivered 24 h following delivery of either AAV6 or AAV8 containing a human IDS transgene donor. Interestingly, while co-delivery of the LNPs and AAV6 seemed to enhance genome modification, there did not seem to be any difference in efficacy with AAV8 (Figure 4A). Presumably, the difference in AAV serotype-specific co-receptors on hepatocytes may be able to explain this difference. Since LNPs and AAV co-delivery yielded the best liver transduction results for both serotypes, this dosing regimen was maintained for all subsequent experiments.
Figure 4.
Optimization of ZFN mRNA-LNP and AAV Transgene Donor Co-delivery for Targeted Integration of Therapeutic Transgene In Vivo
(A) Liver genome-editing results from mice injected with a single 2 mg/kg dose of LNP containing 48641/31523 ZFN mRNAs (ARCA-capped) either 1 day after (predelivery) or at the same time as (co-delivery) 1.5e12 vector genomes (vg) AAV6 or AAV8 encoding a human IDS transgene donor with homology arms flanking the ZFN cut site and a splice acceptor just upstream of the transgene coding region (n = 4 per group) (p = 0.0037 comparing AAV6 pre- and co-delivery). (B) Liver genome-editing results from mice injected with a single dose of LNP containing 48641/31523 ZFN mRNAs over the indicated dose range along with 1.5e12 vector genomes (vg) AAV8 encoding a human IDS transgene donor with homology arms flanking the ZFN cut site and a splice acceptor just upstream of the transgene coding region (p = 0.003, R2 = 0.994, F = 331.7, degrees of freedom = 2). (C) IDS activity assay in plasma collected from the mice described in (B) (p = 0.0181, R2 = 0.9641, F = 53.75, degrees of freedom = 2). (D) Results of liver function test (LFT) in serum collected from the mice described in (B) 1 day post-dosing. LFT, liver function test; ALT, alanine transaminase; AST, aspartate transaminase. (E) Liver genome-modification results following a single 0.5 mg/kg administration of LNP containing 25% pU substituted or unmodified 48641/31523 ZFN mRNAs co-injected with 1.5e12 vg/mouse AAV8 hIDS donor (n = 5–6 per group) (p = 0.0029). (F) IDS activity in plasma was collected from the mice described in (E) (n = 6 per group) (p = 0.011). (G) Results of liver function test (LFT) in serum collected from the mice described in (E) 1 day post-dosing. (H) Plasma IDS enzymatic activity measured from mice 28 days after co-injection with a single 1 mg/kg dose of LNP containing 48641/31523 ZFN mRNAs, purified either via silica column or HPLC and 1.5e12 vg/mouse AAV8 hIDS donor (n = 3–4 per group) (p = 0.055). (I) Results of liver function test (LFT) in serum collected from the mice described in (H) 1 day post-dosing (n = 4 per group) (p = 0.0015 comparing silica versus HPLC ALT; p = 0.0095 comparing AST). (J) Liver genome-editing activity results following single doses of LNP containing 48641/31523 ZFN mRNAs (enzymatic Cap1) injected into mice at 2 mg/kg at the same time as 1.5e12 vector genomes (vg) AAV8 encoding a human IDS transgene donor. Animals were either pretreated with dexamethasone just prior to LNP dosing or just prior to and for an additional 3 days after dosing. (K) IDS activity in plasma was collected from the mice described in (J). (L) Liver genome-editing results following single doses of LNP containing 48641/31523 ZFN mRNAs injected into mice at 2 or 3.5 mg/kg at the same time as 1.5e12 vector genomes (vg) AAV8 encoding a human FIX transgene donor. The FIX donor comprised homology arms flanking the ZFN cut site and a splice acceptor just upstream of the transgene coding region (n = 3–4 per group) (p = 0.047). (M) FIX protein expression results in plasma collected from the mice described in (L).
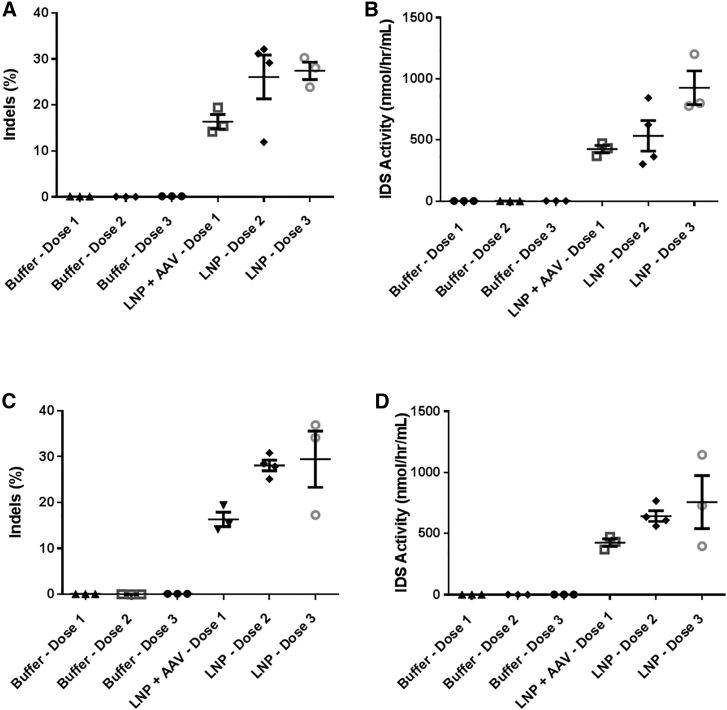
To determine a dose-response of the ZFN mRNA-LNP, a fixed dose of hIDS AAV donor was co-delivered with ascending doses of LNPs. Seven days post-dosing, a ZFN mRNA-LNP dose-dependent increase in the levels of liver genome modification (Figure 4B) and concomitant IDS enzymatic activity within mouse plasma (Figure 4C) was observed with no associated liver toxicity, as assessed by transient serum liver function test (LFT) elevation (Figure 4D). In addition to observing lower levels of genome editing from ZFN mRNA incorporating nucleoside modification (Figures 2C and 4E), we also observed lower levels of plasma IDS enzymatic activity (Figure 4F) with no significant difference in associated liver toxicity (Figure 4G). However, while no difference in genome editing was observed with HPLC-purified ZFN mRNA compared to silica column-purified mRNA (Figure 2F), there was an increase in the level of IDS enzymatic activity using HPLC-purified ZFN mRNA (Figure 4H) which was also associated with a lower level of LFT elevation (Figure 4I). Furthermore, using silica-purified ZFN mRNA, we observed similar levels of genome editing using standard 1-day dexamethasone pretreatment compared to mice that were dosed with dexamethasone for an additional 3 days following LNP dosing (Figure 4J). However, we did observe a trend toward higher levels of IDS enzymatic activity with the longer dexamethasone treatment regimen (Figure 4K). These data indicate that lowering innate immune response using HPLC-purified mRNA or extended corticosteroid treatment can potentially increase productive expression of an integrated transgene in mice, particularly at higher mRNA-LNP doses.
To determine whether the non-viral ZFN co-delivery with AAV transgene strategy was translatable to additional integrated therapeutic transgenes, we designed AAV donors containing a human FIX coding sequence. We observed high levels of liver genome editing and hFIX protein expression within plasma at 2 and 3.5 mg/kg ZFN mRNA-LNP doses (Figures 4L and 4M), indicating our strategy was indeed translatable to additional therapeutic programs being pursued for in vivo genome editing in the clinic.
Repeat Administration of ZFNs via LNPs Enables Increased Levels of Therapeutic Protein Production from the Liver
One major advantage of LNP delivery of site-specific nucleases compared to viral delivery is the lack of adaptive immune response formed against the LNPs. This opens up the potential for repeat administration of the nuclease to allow for increased genome editing and targeted integration, since the AAV-delivered DNA will remain episomally within the mostly non-dividing hepatocytes for quite some time46 and be able to continue to act as a DNA donor template for targeted integration via either HDR or NHEJ-mediated end capture. For clinical applications, the option to potentially re-dose patients if therapeutic levels of transgene expression are not achieved after a first dose would be quite attractive.
To determine if AAV-delivered hIDS transgene DNA persisted long enough in mouse hepatocytes to act as an effective DNA donor, a set of experiments were performed investigating repeat administration of ZFN mRNA-LNP employing differential dosing intervals. Continuing with the standard LNP re-dosing interval of 14 days, AAV donor was co-delivered with ZFN mRNA-LNP into mice, and just the LNP was re-dosed 14 days apart for two or three total doses. After three total doses, there was an approximately 2-fold increase in the level of liver genome editing (Figure 5A) as well as IDS enzymatic activity within mouse plasma (Figure 5B) compared to a single dose, indicating increased levels of targeted integration had indeed occurred over this time course. Since a fraction of hepatocytes are slowly dividing and dilution of the AAV episome can occur over time (especially within pediatric populations), an LNP re-dosing interval of 7 days was investigated. The level of genome modification (Figure 5C) and plasma IDS enzymatic activity (Figure 5D) also increased approximately 2-fold after multiple doses of ZFN mRNA-LNP compared to a single dose using this shortened dosing interval.
Figure 5.
Repeat Administration of ZFN mRNA-LNP with Single AAV Donor Delivery for Increasing Targeted Integration of Therapeutic Transgene In Vivo
(A) Liver genome-editing results of repeat dosing (14-day intervals, 2 mg/kg) of LNP containing 48641/31523 ZFN mRNAs into mice. The first dosing also included co-delivery of 1.5e12 vector genomes (vg) AAV8 encoding a human IDS transgene donor with homology arms flanking the ZFN cut site in the ALB gene and a splice acceptor just upstream of the transgene coding region. (n = 3 per group) (p = 0.011 comparing doses 1 and 3). (B) IDS activity in plasma that was collected from the mice described in (A) (n = 3 per group) (p = 0.023 comparing doses 1 and 3). (C) Liver genome-editing results of repeat dosing (7-day intervals, 2 mg/kg) of LNP containing 48641/31523 ZFN mRNAs into mice. The first dosing also included co-delivery of 1.5e12 vector genomes (vg) AAV8 encoding the same human IDS transgene donor described in (A) (n = 3–4 per group) (p = 0.0017 comparing doses 1 and 2). (D) IDS activity in plasma that was collected from the mice described in (C) (n = 3-4 per group) (p = 0.013 comparing doses 1 and 2).
Discussion
In this work, we show for the first time that an engineered site-specific nuclease delivered intravenously in a non-viral carrier can mediate targeted integration of a large therapeutic transgene in mice. Sangamo Therapeutics is currently conducting three in vivo genome-editing clinical trials (NCT03041324, NCT02702115, and NCT02695160) using AAV vectors to deliver site-specific nucleases to the liver of patients. Due to the adaptive immune response against the viral capsids, re-dosing patients with nuclease to increase the levels of therapeutic genome modification is currently not an option due to neutralizing antibodies that are generated against the viral delivery vehicle. This work demonstrates for the first time that repeat in vivo administration of site-specific nucleases delivered via a non-viral carrier can increase the levels of therapeutic protein expressed from hepatocytes. This treatment modality may have broad impact in allowing for improved therapeutic gene modification in strategies that are currently being employed in clinical trials.
Notably, this work also demonstrates complete non-viral carrier-delivered site-specific nuclease-mediated liver gene knockout at the lowest mRNA dose ever reported. Additionally, data presented here indicates rapid elimination of LNPs, minimizing likelihood of accumulation of carrier components in the target tissue. For clinical translation, minimizing the total exposure of nucleic acid and lipid drug products to patients is beneficial from a potential accumulated toxicity standpoint47 as well as from a manufacturing standpoint. The cost of goods to manufacture an in vitro transcribed mRNA product for in vivo administration becomes prohibitive for clinical-scale use past the doses used in this study, especially to be able to treat clinical indications with large patient populations. The results reported in this study demonstrate promising results potentially scalable to non-human primate and clinical applications.
To date, the only clinical programs employing genome-editing strategies that have shown signs of safety and efficacy have utilized either ZFNs48 or TALENs.49 All previously reported in vivo non-viral delivery studies have focused on the CRISPR/Cas9 system, which requires the added complexity of delivering both a genomic DNA (gRNA) and Cas9 mRNA. In addition, CRISPR has yet to demonstrate safety or efficacy in a clinical setting for any ex vivo or in vivo genome-editing application using viral or non-viral delivery. This work is the first reported example of ZFN-mediated non-viral in vivo genome editing, which, given the track record of clinical safety and efficacy, may present a clearer path toward clinical translation than previously reported CRISPR/Cas9 studies, requiring clinical scale manufacturing of both gRNA and Cas9 mRNA.
While this and other works have shown highly efficient delivery of site-specific nucleases to the liver, significant efforts still need to be invested in efficiently targeting other therapeutically relevant organs. A handful of investigators have begun to show signs of efficacy in non-viral RNA delivery to the lung,50, 51 endothelium,52 and spleen;53 however, much work is still needed to optimize these systems before they are ready for clinical—particularly for mRNA delivery of genome editing reagents.
In conclusion, this work demonstrates that therapeutically relevant levels of gene knockout as well as therapeutic transgene expression in hepatocytes is possible utilizing non-viral delivery of site-specific ZFN mRNAs. In vivo delivery of mRNA has broad applications for addressing numerous diseases with high unmet medical need, which the findings in this study will hopefully enable new and current investigators to more effectively pursue. The insights we present here may allow for the acceleration of the deployment of in vivo mRNA delivery into more challenging applications such as in disease mouse models, non-human primates, and ultimately for human therapeutic development.
Materials and Methods
ZFN mRNA Design and Production
ZFNs targeting intron 1 of the murine ALB gene, exon 2 of the murine PCSK9 gene, and exon 2 of the murine TTR gene were subcloned into individual vectors (pVAX-GEM) containing a T7 RNA polymerase promoter, a 5′ UTR containing a sequence derived from the Xenopus beta-globin gene, a 3′ UTR containing the woodchuck hepatitis virus response element (WPRE) sequence, and a poly(A) tract.
mRNA was produced from the linearized ZFN construct plasmids using in vitro transcription (IVT) at Trilink Biotechnologies with either unmodified residues or a fraction of modified nucleosides. mRNA was capped either co-transcriptionally with an anti-reverse cap analog (ARCA) cap, enzymatically post-IVT using the vaccinia virus capping enzyme along with the mRNA Cap 2′-O-methyltransferase enzyme to produce “Cap1” mRNA or chemically to produce “Cap1” (CleanCap, Trilink Biotechnologies). mRNA was purified through a silica bead column (silica-purified) or subsequently run through an HPLC column and fractionated to remove double-stranded RNA species (HPLC-purified).
LNP Formulation
LNPs were formulated as previously described.14 In brief, a 1:1 mass ratio of the two individual ZFN mRNAs in aqueous solution at pH 4.0 was mixed with a combination of four lipids (cholesterol; 1,2-distearoyl-sn-glycero-3-phosphocholine [DSPC]; a PEG2000 lipid; with a C14 anchor; and a proprietary ionizable lipid—chemical structure shown in Figure 1A, NMR and MS spectra shown in Figure S2) in an ethanol solution at a 0.035 (w/w) mRNA:lipid ratio by in-line mixing at Acuitas Therapeutics, allowing the combination of the lipids and mRNAs to self-assemble. Resultant LNPs were subsequently dialyzed overnight in aqueous solution to remove residual ethanol. LNP batches used in these studies had a size (intensity weighted average) of 76 ± 4 nm with a polydispersity index of 0.04 ± 0.02 as determined by dynamic light scattering (Malvern NanoZS Zetasizer, Saint-Laurent, QC, Canada) and an encapsulation efficiency of 96% ± 1%. For pharmacokinetic and biodistribution (PK/BD) studies, LNPs were radiolabeled with 0.002% tritiated cholesterylhexadecylether (3H-CHE, 48,000 Ci/mol, PerkinElmer, Waltham, MA, USA).
AAV Donor Design and Production
The hIDS donor construct has been previously described.54 The hIDS donor construct contains a codon-optimized hIDS cDNA lacking the endogenous IDS signal peptide sequence, a splice acceptor sequence derived from human FIX, and arms of homology to the mouse ALB target site of approximately 600 bp in length in total. Recombinant AAV2/6 or AAV2/8 vectors (comprised of AAV2 ITRs and the AAV6 or AAV8 capsid, respectively) were produced by triple transfection of HEK293 cells in 10-chamber CELLSTACK culture chambers (Corning), purified by cesium chloride density gradient centrifugation followed by dialysis, and titered as previously described.54
In Vitro ZFN mRNA Transfection
To assess the activity of ZFN mRNA, mRNA were electroporated into Hepa1-6 cells using a BTX 830 Square Wave Electroporation System (Harvard Biosciences) then cells were allowed to grow for 3 days. Transfected cells were then harvested for gDNA using QIAGEN spin columns.
In Vivo LNP Transduction
8- to 10-week-old male or female C57BL/6 purchased from Charles River were injected intravenously through the tail vein with 200 μL of an aqueous solution containing diluted LNPs or a mixture of diluted LNPs and AAV donor encoding a human IDS or human FIX transgene containing homology arms flanking the ZFN cut site and a splice acceptor just upstream of the coding sequence of the transgene. Some animals were injected with 5 mg/kg dexamethasone intraperitoneally 30 min prior to LNP dosing. Animals were then sacrificed 7 days post-LNP-dosing or re-dosed with LNPs at either a 7- or 14-day interval before subsequent sacrifice and harvesting of liver tissue. Livers were snap frozen, and a small portion of both the left and right lobes were dissected and harvested for gDNA using a FastPrep-24 Homogenizer (MP Biomedicals), Lysis Matrix D solution (MP Biomedicals), and a MasterPure DNA Purification kit (Epicenter).
Genome-Editing Quantification
Primers were designed to amplify approximately 200 bp of total genomic DNA sequence containing the ZFN cut site. Amplicons were then run on either a Miseq or Nextseq (Illumina) and indels from the wild-type genomic sequence were quantified.
For quantification of targeted integration (Figure S3), a forward primer that binds upstream of the left hIDS donor homology arm was used in combination with a reverse primer that binds either within the splice acceptor sequence of the hIDS donor or within exon 2 of the ALB locus to amplify HDR or wild-type alleles, respectively. Both reverse primers were designed to have similar base composition as well as yield similarly sized amplicons to minimize PCR amplification bias. The relative fraction of each allele within either control or LNP + AAV edited bulk mouse liver tissue was then quantified via densitometry following running the PCR products on an agarose gel.
For long amplicon analyses (Figure S4), primers were designed to amplify approximately 10 kb of total genomic DNA sequence equally flanking the murine ALB ZFN cut site. Amplicons were barcoded using native barcoding kit (EXP-NBD103) and then ran on an Oxford Nanopore MinION. Barcoded sequences were demultiplexed, and then the percentage of aligned sequences containing long deletions (>100 bp) were subsequently quantified.
Serum Protein Analysis
Mouse plasma was diluted 1:100, 1:100, or 1:20,000 in PBS and ran for human FIX, murine PCSK9, or murine TTR protein levels, respectively, on a sandwich ELISA kit (Affinity Biologicals, Boster Biological Technology, or Cusabio, respectively). Absorbance was read at 450 nm using a microplate reader.
IDS Enzymatic Activity Assay
At animal sacrifice, mouse blood was collected into tubes containing sodium citrate, and the cell fraction was removed to yield mouse plasma. Plasma was then diluted 1:100 in water and incubated with iduronate-2-sulfatase (IDS) substrate (4-methylumbelliferyl-α-iduronate 2-sulfate) for 4 h at 37°C. A 0.4 M sodium phosphate solution was then added to halt the reaction. Recombinant human iduronidase (IDUA) was then added, and the solution was then incubated for 24 h at 37°C. IDS substrate that has been successfully cleaved will yield a fluorescent product, which is then measured on a fluorescent plate reader at 365 nm excitation/450 nm emission.
Liver Transaminase Quantification
24 h following LNP dosing, blood was collected and processed to serum, and serum preparations were sent to IDEXX laboratories (3 Centennial Dr., North Grafton, MA, USA). A liver specific chemistry panel was measured by chemistry analyzer as described by IDEXX.
In Vivo LNP PK, BD, and Elimination
8- to 10-week-old female CD-1 mice were injected intravenously via the tail vein with 200 μL of an aqueous LNP solution at a dose of 2 mg/kg. For PK/BD, tissues were collected at various time points post-administration of radiolabeled LNP, processed (for blood, centrifuged to plasma; for liver, homogenized using a FastPrep-24 Homogenizer, MP Biomedicals), and the amount of radioactivity determined measured using a liquid scintillation counter (expressed as % injected dose). For liver elimination, liver samples were collected at various times after administration and flash frozen. Tissue was homogenized, and lipid components recovered using a two-phase liquid extraction with 2:1 chloroform:methanol. The organic phase was evaporated to dryness and then reconstituted in ethanol for analysis by ultra performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS; Water Acuity with Xevo TQD, Mississauga, ON, Canada).
Liver Cell Subpopulation Isolation
Mice were perfused with 20 mL of PBS followed by perfusion with a digestive enzyme solution with collagenase type I (Sigma-Aldrich), collagenase XI (Sigma-Aldrich), and hyaluronidase (Sigma-Aldrich) through the hepatic portal vein. The liver was isolated immediately following perfusion. Tissues were digested for 45 min at 37°C and 550 rpm. Digested tissues were passed through a 70-μm filter. Red blood cells (RBCs) were lysed using lysis buffer. Cells were resuspended in FACS buffer (2% FBS in 1 × PBS).
Cells were stained to identify specific cell populations and sorted using the BD FACS Aria cell sorters in the University of British Columbia cellular analysis core. Antibodies used for staining were CD31 (clone 390, Thermo Fisher), F4/80 (clone BM8, Thermo Fisher), and CD45 (clone 104, Thermo Fisher). We defined cell populations in the following manner: liver endothelial cells (CD45−CD31+F4/80−), Kupffer cells (CD45+CD31−F4/80+), and hepatocytes (CD45−CD31−F4/80−).
Statistical Analysis
All values are presented as mean ± SEM. GraphPad Prism and Excel software were used to perform statistical analyses. Differences among treated groups were evaluated using one-way ANOVA. Linear regression analysis was used to determine statistical significance of increased editing activity following repeat LNP dosing or with ascending dose.
Author Contributions
A.C. and G.L. designed the studies; M.M., K.K., and A.B. performed the molecular biology, biochemistry, and cell culture work; K.M. performed the long amplicon bioinformatics analysis; L.Z., J.C.M., and D.E.P. designed the ZFNs; R.C.D. developed the IDS activity assay and hIDS donor; B.L.M., Y.K.T, P.J.C.L., and C.B. designed and formulated the LNP; P.J.C.L. and Y.K.T. performed many of the animal studies; B.L.M., P.J.C.L., Y.K.T., C.B., and T.R. provided critical guidance on LNP and study design; A.C. wrote the manuscript; and M.C.H. and G.L. edited and provided feedback on the manuscript.
Conflicts of Interest
All authors are current or former employees of Sangamo Therapeutics or Acuitas Therapeutics.
Acknowledgments
The authors would like to thank Lisa King, Carolyn Gaspar, and Kathleen Meyer for assistance with animal study coordination and animal study design as well as Pacific BioLabs (Hercules, CA, USA) for performing many of the animal studies in this manuscript. Many thanks to Sarah Hinkley and her team for assembling and providing all of the ZFNs for screening, Patrick Li and his team for providing much of the bioinformatics and deep sequencing support, and Richard Surosky and his team for providing high quality AAV for the animal studies. This work was supported by Sangamo Therapeutics.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.03.003.
Supplemental Information
References
- 1.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller J.C., Tan S., Qiao G., Barlow K.A., Wang J., Xia D.F., Meng X., Paschon D.E., Leung E., Hinkley S.J. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 3.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Y., Liang D., Wang Y., Bai M., Tang W., Bao S., Yan Z., Li D., Li J. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. doi: 10.1016/j.stem.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 5.Li H., Haurigot V., Doyon Y., Li T., Wong S.Y., Bhagwat A.S., Malani N., Anguela X.M., Sharma R., Ivanciu L. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475:217–221. doi: 10.1038/nature10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma R., Anguela X.M., Doyon Y., Wechsler T., DeKelver R.C., Sproul S., Paschon D.E., Miller J.C., Davidson R.J., Shivak D. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126:1777–1784. doi: 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi G., Choi J.G., Bharaj P., Abraham S., Dang Y., Kafri T., Alozie O., Manjunath M.N., Shankar P. CCR5 Gene Editing of Resting CD4(+) T Cells by Transient ZFN Expression From HIV Envelope Pseudotyped Nonintegrating Lentivirus Confers HIV-1 Resistance in Humanized Mice. Mol. Ther. Nucleic Acids. 2014;3:e198. doi: 10.1038/mtna.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 9.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W., Stebbing D., Crosley E.J., Yaworski E. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 13.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn J.D., Smith A.R., Patel M.C., Shaw L., Youniss M.R., van Heteren J., Dirstine T., Ciullo C., Lescarbeau R., Seitzer J. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Yin H., Song C.Q., Dorkin J.R., Zhu L.J., Li Y., Wu Q., Park A., Yang J., Suresh S., Bizhanova A. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin H., Song C.Q., Suresh S., Wu Q., Walsh S., Rhym L.H., Mintzer E., Bolukbasi M.F., Zhu L.J., Kauffman K. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017;35:1179–1187. doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M., Zuris J.A., Meng F., Rees H., Sun S., Deng P., Han Y., Gao X., Pouli D., Wu Q. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. USA. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J.B., Zhang S., Kos P., Xiong H., Zhou K., Perelman S.S., Zhu H., Siegwart D.J. Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. Engl. 2017;56:1059–1063. doi: 10.1002/anie.201610209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang C., Mei M., Li B., Zhu X., Zu W., Tian Y., Wang Q., Guo Y., Dong Y., Tan X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017;27:440–443. doi: 10.1038/cr.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeRosa F., Guild B., Karve S., Smith L., Love K., Dorkin J.R., Kauffman K.J., Zhang J., Yahalom B., Anderson D.G., Heartlein M.W. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 2016;23:699–707. doi: 10.1038/gt.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA Without Chemical Nucleoside Modifications Enables an Effective Protein Therapy in Large Animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 24.Grudzien E., Kalek M., Jemielity J., Darzynkiewicz E., Rhoads R.E. Differential inhibition of mRNA degradation pathways by novel cap analogs. J. Biol. Chem. 2006;281:1857–1867. doi: 10.1074/jbc.M509121200. [DOI] [PubMed] [Google Scholar]
- 25.Krieg P.A., Melton D.A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donello J.E., Loeb J.E., Hope T.J. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J. Virol. 1998;72:5085–5092. doi: 10.1128/jvi.72.6.5085-5092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malone R.W., Felgner P.L., Verma I.M. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA. 1989;86:6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munroe D., Jacobson A. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 1990;10:3441–3455. doi: 10.1128/mcb.10.7.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 30.Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 32.Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kormann M.S., Hasenpusch G., Aneja M.K., Nica G., Flemmer A.W., Herber-Jonat S., Huppmann M., Mays L.E., Illenyi M., Schams A. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 2011;29:154–157. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 34.Warren L., Manos P.D., Ahfeldt T., Loh Y.H., Li H., Lau F., Ebina W., Mandal P.K., Smith Z.D., Meissner A. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mauro V.P., Chappell S.A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2014;20:604–613. doi: 10.1016/j.molmed.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramanathan A., Robb G.B., Chan S.H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stepinski J., Waddell C., Stolarski R., Darzynkiewicz E., Rhoads R.E. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogs 7-methyl(3′-O-methyl)GpppG and 7-methyl (3′-deoxy)GpppG. RNA. 2001;7:1486–1495. [PMC free article] [PubMed] [Google Scholar]
- 38.Jalkanen A.L., Coleman S.J., Wilusz J. Determinants and implications of mRNA poly(A) tail size—does this protein make my tail look big? Semin. Cell Dev. Biol. 2014;34:24–32. doi: 10.1016/j.semcdb.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abrams M.T., Koser M.L., Seitzer J., Williams S.C., DiPietro M.A., Wang W., Shaw A.W., Mao X., Jadhav V., Davide J.P. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol. Ther. 2010;18:171–180. doi: 10.1038/mt.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper A.D. Hepatic uptake of chylomicron remnants. J. Lipid Res. 1997;38:2173–2192. [PubMed] [Google Scholar]
- 41.Davidoff A.M., Ng C.Y., Zhou J., Spence Y., Nathwani A.C. Sex significantly influences transduction of murine liver by recombinant adeno-associated viral vectors through an androgen-dependent pathway. Blood. 2003;102:480–488. doi: 10.1182/blood-2002-09-2889. [DOI] [PubMed] [Google Scholar]
- 42.Paschon D.E., Lussier S., Wangzor T., Xia D.F., Li P.W., Hinkley S.J., Scarlott N.A., Lam S.C., Waite A.J., Truong L.N. Diversifying the structure of zinc finger nucleases for high-precision genome editing. Nat Commun. 2019 doi: 10.1038/s41467-019-08867-x. Published online March 8, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gertz M.A., Benson M.D., Dyck P.J., Grogan M., Coelho T., Cruz M., Berk J.L., Plante-Bordeneuve V., Schmidt H.H.J., Merlini G. Diagnosis, Prognosis, and Therapy of Transthyretin Amyloidosis. J. Am. Coll. Cardiol. 2015;66:2451–2466. doi: 10.1016/j.jacc.2015.09.075. [DOI] [PubMed] [Google Scholar]
- 44.Adams D., Gonzalez-Duarte A., O’Riordan W.D., Yang C.C., Ueda M., Kristen A.V., Tournev I., Schmidt H.H., Coelho T., Berk J.L. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018;379:11–21. doi: 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- 45.Seidah N.G., Awan Z., Chrétien M., Mbikay M. PCSK9: a key modulator of cardiovascular health. Circ. Res. 2014;114:1022–1036. doi: 10.1161/CIRCRESAHA.114.301621. [DOI] [PubMed] [Google Scholar]
- 46.Nakai H., Yant S.R., Storm T.A., Fuess S., Meuse L., Kay M.A. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khvorova A. Oligonucleotide Therapeutics - A New Class of Cholesterol-Lowering Drugs. N. Engl. J. Med. 2017;376:4–7. doi: 10.1056/NEJMp1614154. [DOI] [PubMed] [Google Scholar]
- 48.Tebas P., Stein D., Tang W.W., Frank I., Wang S.Q., Lee G., Spratt S.K., Surosky R.T., Giedlin M.A., Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qasim W., Zhan H., Samarasinghe S., Adams S., Amrolia P., Stafford S., Butler K., Rivat C., Wright G., Somana K. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl. Med. 2017;9:eaaj2013. doi: 10.1126/scitranslmed.aaj2013. [DOI] [PubMed] [Google Scholar]
- 50.Kaczmarek J.C., Patel A.K., Kauffman K.J., Fenton O.S., Webber M.J., Heartlein M.W., DeRosa F., Anderson D.G. Polymer-Lipid Nanoparticles for Systemic Delivery of mRNA to the Lungs. Angew. Chem. Int. Ed. Engl. 2016;55:13808–13812. doi: 10.1002/anie.201608450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Y., Xiong H., Zhang X., Cheng Q., Siegwart D.J. Systemic mRNA Delivery to the Lungs by Functional Polyester-based Carriers. Biomacromolecules. 2017;18:4307–4315. doi: 10.1021/acs.biomac.7b01356. [DOI] [PubMed] [Google Scholar]
- 52.Dahlman J.E., Barnes C., Khan O., Thiriot A., Jhunjunwala S., Shaw T.E., Xing Y., Sager H.B., Sahay G., Speciner L. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 2014;9:648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fenton O.S., Kauffman K.J., Kaczmarek J.C., McClellan R.L., Jhunjhunwala S., Tibbitt M.W., Zeng M.D., Appel E.A., Dorkin J.R., Mir F.F. Synthesis and Biological Evaluation of Ionizable Lipid Materials for the In Vivo Delivery of Messenger RNA to B Lymphocytes. Adv. Mater. 2017;29 doi: 10.1002/adma.201606944. 1606944. [DOI] [PubMed] [Google Scholar]
- 54.Judge L.M., Haraguchiln M., Chamberlain J.S. Dissecting the signaling and mechanical functions of the dystrophin-glycoprotein complex. J. Cell Sci. 2006;119:1537–1546. doi: 10.1242/jcs.02857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.