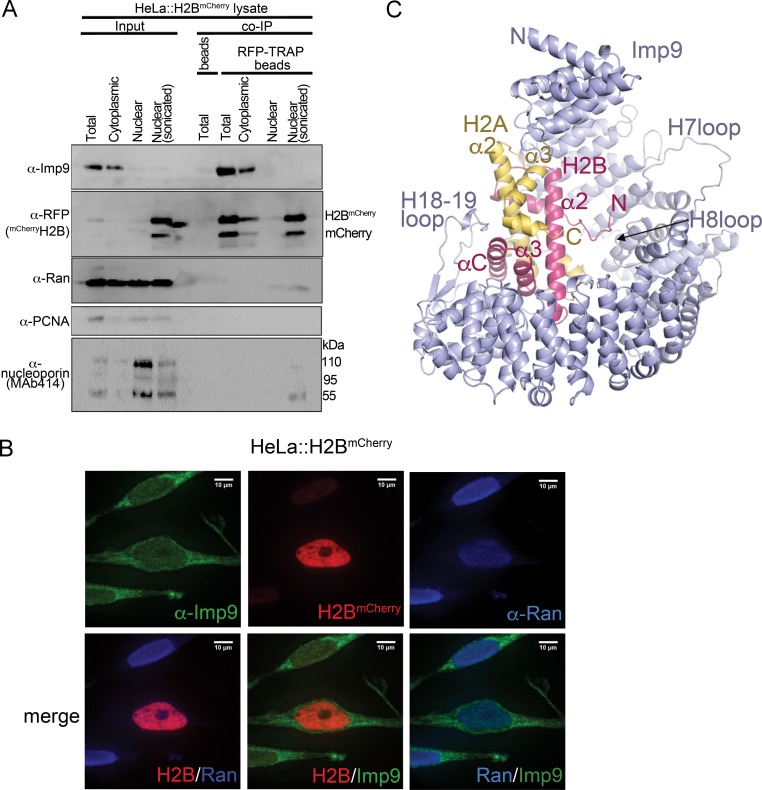
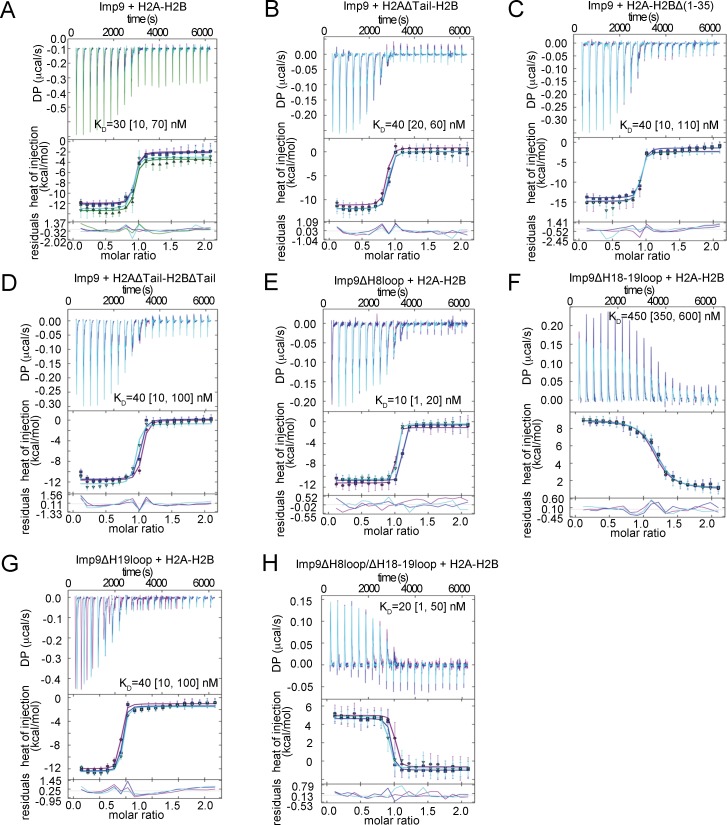
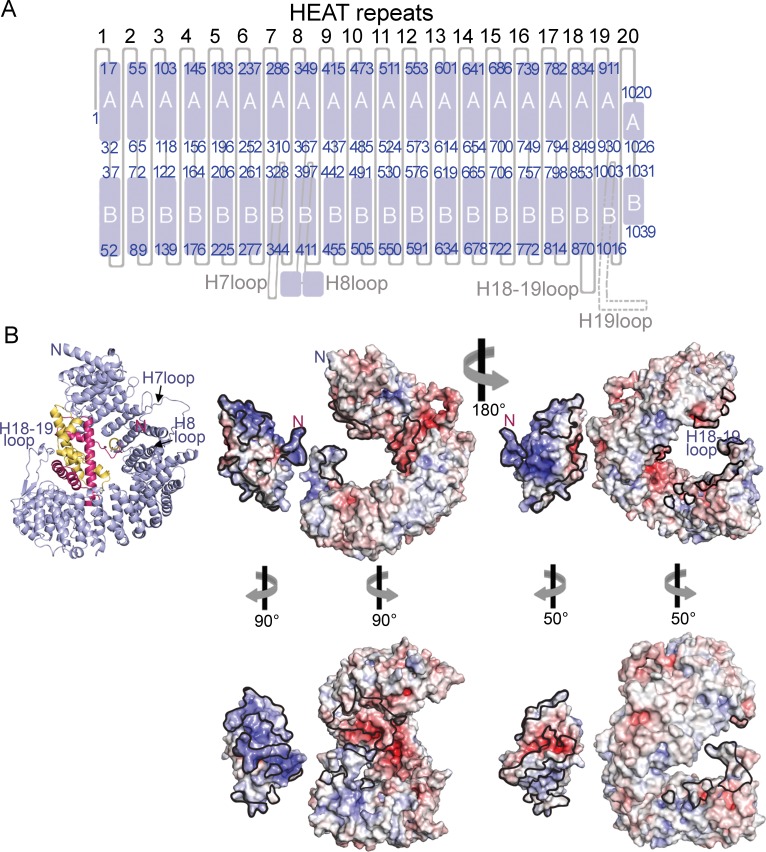
Figure 1. Interactions between Imp9 and H2A-H2B in the cell and crystal structure of the Imp9 •H2A-H2B complex.
(A) Coimmunoprecipitation (CoIP) studies of H2BmCherry from whole cell, cytoplasmic and nuclear fractions of the lysates from HeLa cells stably expressing H2BmCherry, followed by immunoblots with Imp9, Ran, RFP antibodies. PCNA and MAb414 antibodies are used as loading control antibodies. 10 µg of 1.5 mg lysates are analyzed as CoIP input. Blots are representative of three identical experiments. (B) Subcellular localization of Imp9 and Ran in Hela::H2BmCherry cells. HeLa cells were fixed, permeabilized, incubated with affinity-purified rabbit polyclonal Imp9 antibody and mouse monoclonal anti–Ran antibody, and visualized by confocal microscopy. The secondary antibodies were Alexa 488 conjugated anti–rabbit and Alexa 405 conjugated anti-mouse, respectively. The column on the right contains two-color merge images. (C). The crystal structure of human Imp9 (blue) in complex with X. laevis H2A (yellow)-H2B (red).